Introduction
Signal Transduction ATPases with Numerous Domains (STAND), a large class of P-loop NTPases 1, belong to AAA+ ATPases 2–4. They include AP(apoptotic)-ATPases 5 (e.g. animal apoptosis regulators CED4/Apaf-1, plant disease resistance proteins, and bacterial AfsR-like transcription regulators), NACHT NTPases 6 (e.g. CARD4, NAIP, Het-E-1, TLP1) and several other less well-characterized families 1. STAND differ from other P-loop NTPases by their unique sequence motifs, which include an hhGRExE (h, hydrophobic; x, any residue) motif at the N-terminal region, a GxP/GxxP motif at the C-terminal region of the NTPase domain, in addition to a C-terminal helical domain and additional domains such as WD40, TPR, LRR or catalytic modules 1. Despite significant biological interests, structural coverage of STAND proteins is very limited and only two other structures are currently known: the cell death regulators Apaf-1 and CED-4 7,8. Here we report the crystal structure of SSO1545 from Sulfolobus solfataricus, which was determined using the semi-automated, high-throughput pipeline of the Joint Center for Structural Genomics (JCSG) 9, as part of the National Institute of General Medical Sciences' Protein Structure Initiative (PSI). SSO1545 (NP_342973.1), a representative of the archaeal STANDs 1, is a member of Pfam PF01637 10 and encodes a protein of 356 residues with calculated molecular weight and isoelectric point of 41.7 kD and 8.2 respectively.
Materials and Methods
Protein production and crystallization
The gene encoding SSO1545 (GenBank: AAK41763, GI:13814777; Swiss-Prot: Q97Y08) was amplified by polymerase chain reaction (PCR) from Sulfolobus solfataricus (DSM 1617; P2) genomic DNA using PfuTurbo DNA polymerase (Stratagene) and primers corresponding to the predicted 5' and 3' ends. The PCR product was cloned into plasmid pSpeedET, which encodes an expression and purification tag followed by a tobacco etch virus (TEV) protease cleavage site (MGSDKIHHHHHHENLYFQG) at the amino terminus of the full-length protein. The cloning junctions were confirmed by DNA sequencing. Expression was performed in a selenomethionine-containing medium using the Escherichia coli strain GeneHogs (Invitrogen). At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 µg/ml, and the cells were harvested and frozen. After one freeze/thaw cycle the cells were sonicated in lysis buffer [50 mM HEPES pH 8.0, 50 mM NaCl, 10 mM imidazole, 1 mM Tris (2-carboxyethyl)phosphine-HCl (TCEP)] and the lysate was clarified by centrifugation at 32,500 × g for 30 min. The soluble fraction was run over nickel-chelating resin (GE Healthcare) pre-equilibrated with lysis buffer, the resin washed with wash buffer [50 mM HEPES pH 8.0, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP], and the protein eluted with elution buffer [20 mM HEPES pH 8.0, 300 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP]. The eluate was buffer exchanged with HEPES crystallization buffer [20 mM HEPES pH 8.0, 200 mM NaCl, 40 mM imidazole, 1 mM TCEP] using a PD-10 column (GE Healthcare), and incubated with 1 mg of TEV protease per 15 mg of eluted protein. The protease-treated eluate was run over nickel-chelating resin (GE Healthcare) pre-equilibrated with HEPES crystallization buffer and the resin was washed with the same buffer. The flow-through and wash fractions were combined and concentrated for crystallization trials to 15 mg/ml by centrifugal ultrafiltration (Millipore). SSO1545 was crystallized using the nanodroplet vapor diffusion method 11 with standard JCSG crystallization protocols 9. The crystallization reagent that produced the SSO1545 crystal used for structure solution contained 20% PEG-8000 and 0.1M CHES pH 9.5. Ethylene glycol was added to the crystal as a cryoprotectant to a final concentration of 10% (v/v). Initial screening for diffraction was carried out using the Stanford Automated Mounting (SAM) system 12 and an X-ray microsource 13 installed on a Stanford Synchrotron Radiation Laboratory beamline (SSRL, Menlo Park, CA). The crystal was indexed in monoclinic space group P21. The molecular weight and oligomeric state of SSO1545 were determined using a 0.8 × 30cm Shodex Protein KW-803 column (Thomson Instruments) pre-calibrated with gel filtration standards (Bio-Rad).
Data collection, structure solution, and refinement
Multi-wavelength anomalous diffraction (MAD) data were collected at the Advanced Photon Source (APS) GM/CA CAT Beamline 23-ID-D at the Argonne National Laboratory. The data were collected at wavelengths corresponding to the high energy remote (λ1) and inflection (λ2) of a selenium MAD experiment. The data sets were collected at 100 K using a MarCCD 300 detector (MarResearch, USA). The data processing and structure solution were carried out using an automatic structure solution pipeline developed at JCSG. The MAD data were integrated and reduced using XDS and then scaled with the program XSCALE 14. Selenium sites were located with SHELXD 15. Phase refinement and automatic model building were performed using autoSHARP 16 and wARP 17. Model completion and refinement were performed with COOT 18 and REFMAC 19. CCP4 programs were used to for data conversion and other calculations 20. Data reduction and refinement statistics are summarized in Table I 20,21.
Table I.
Summary of crystal parameters, data collection, and refinement statistics for SSO154 (PDB: 2fna).
| Space group | P21 | |||
| Unit cell parameters | a=55.36Å, b=108.35Å, c=70.57Å, β=100.3° | |||
| Data collection | λ1 MADSe | λ2 MADSe | ||
| Wavelength (Å) | 0.9537 | 0.9800 | ||
| Resolution range (Å) | 29.20-2.00 | 29.20-2.00 | ||
| Number of observations | 176,708 | 172,434 | ||
| Number of unique reflections | 53,921 | 53,545 | ||
| Completeness (%) | 97.7 (96.0)a | 97.0 (92.0)a | ||
| Mean I/σ (I) | 12.6 (2.7)a | 12.0 (2.4)a | ||
| Rsym on I (%) | 0.075 (0.363)a | 0.083 (0.379)a | ||
| Highest resolution shell (Å) | 2.11-2.00 | 2.11-2.00 | ||
| Model and refinement statistics | ||||
| Resolution range (Å) | 29.20-2.00 | Data set used in refinement | λ1 MADSe | |
| No. of reflections (total) | 53,894b | Cutoff criteria | |F|>0 | |
| No. of reflections (test) | 2,733 | Rcryst | 0.174 | |
| Completeness (% total) | 97.5 | Rfree | 0.226 | |
| Stereochemical parameters | ||||
| Restraints (RMS observed) | ||||
| Bond angle (°) | 1.43 | |||
| Bond length (Å) | 0.014 | |||
| Average isotropic B-value (Å2) | 42.5 | |||
| ESU based on Rfree (Å) | 0.16 | |||
| Protein residues/atoms | 704 / 5743 | |||
| Water molecules / ions / ligands / cryoprotectant | 347 / 2 / 2 / 7 | |||
Highest resolution shell.
Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded due to systematic absences, negative intensities, and rounding errors in the resolution limits and cell parameters.
Rsym = ∑|Ii-<Ii>| / ∑|Ii| where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Rcryst = ∑| |Fobs|-|Fcalc| | / ∑|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.1% of the total reflections chosen at random and omitted from refinement
Validation and deposition
Analysis of the stereochemical quality of the model was accomplished using AutoDepInputTool 22 , MolProbity 23, SFcheck 4.0 20, and WHATIF 5.0 24. All molecular graphics were prepared with PyMOL (DeLano Scientific, http://pymol.sf.net). Figure 1B was generated with ESPRIPT 25. The electrostatic potentials for figure 4B were calculated with Delphi (default setting) 26. Atomic coordinates and experimental structure factors for SSO1545 at 2.0 Å resolution have been deposited in the Protein Data Bank (PDB, http://www.wwpdb.org/) under access code 2fna.
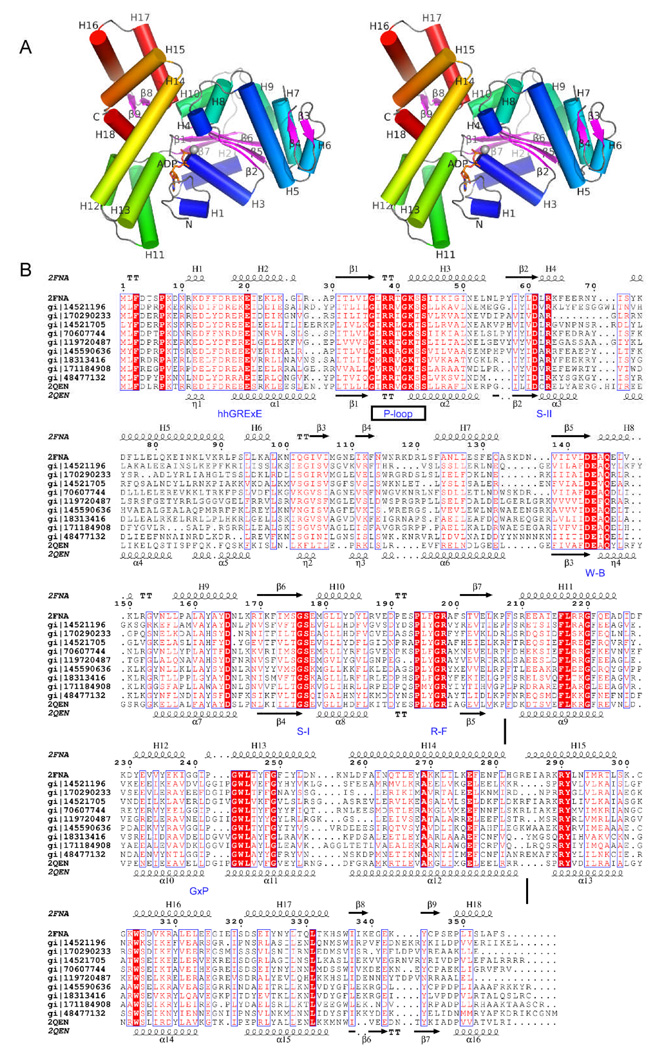
Figure 1.
Crystal structure of the archaeal ATPase SSO1545 from Sulfolobus solfataricus complexed with ADP and magnesium ion. (A). Stereo ribbon diagram of an SSO1545 monomer is where the helices are color-coded from N-terminus (blue) to C-terminus (red) and the β-strands are in purple. Helices H1–H18 and β-strands (β1–β9) are labeled. The ADP is shown in sticks (orange) and the magnesium is shown as a sphere (silver). (B) Sequence alignment between SSO1545 and several representative homologous proteins from archaea. The secondary structure elements and sequence numbering of SSO1545 are shown at the top. The secondary structure elements of Paby2304 (PDB 2qen) are shown at the bottom. The conserved sequence motifs, Walker A (P-loop), Walker B (W-B), sensor I (S-I), sensor II (S-II), arginine finger (R-F), as well as STAND specific hhGRExE and GxP motifs, are annotated at the bottom in blue. The domain boundaries are shown as vertical bars.
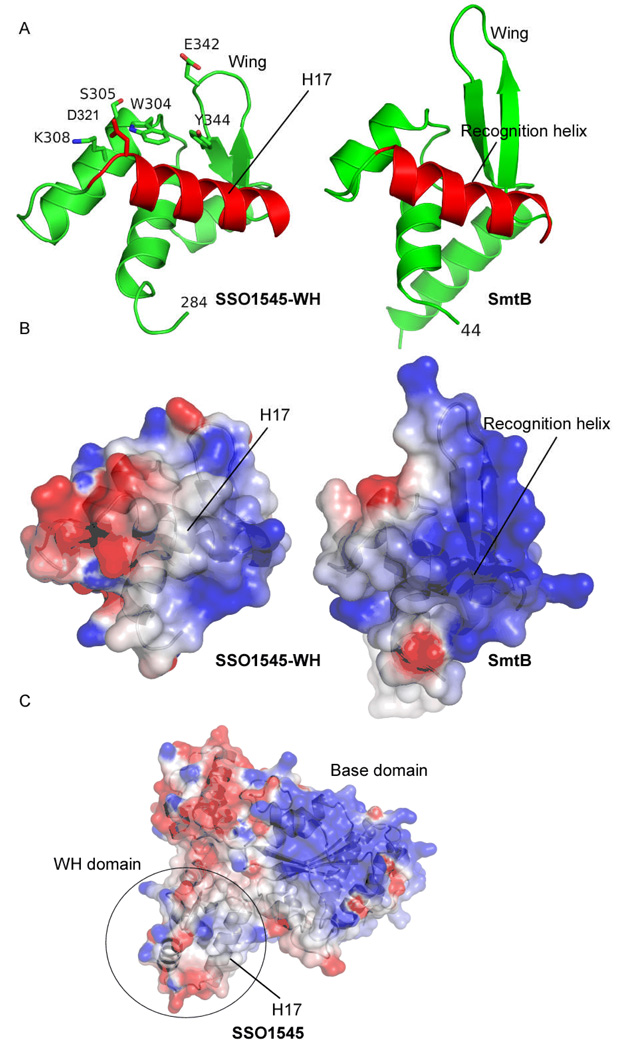
Figure 4.
Electrostatic potentials of SSO1545 and its WH domain, compared to the transcription repressor SmtB. (A). Ribbon representation of the WH domains of SSO1545 and SmtB. Both molecules are shown in superimposed orientations. The recognition helix of SmtB and the equivalent helix H17 of SSO1545 are highlighted in red. The highly conserved residues of SSO1545 are shown as sticks. (B). Electrostatic surfaces of the WH domains of SSO1545 (residues 284–356) and SmtB (residues 44–121). The orientations of both domains are the same as in (A). The same electrostatic potential scale was used to render both domains. The color was scaled from −3 to 3 kT (blue, positive; red, negative electrostatic potential). (C). Electrostatic surface of the full length SSO1545. The orientation of the SSO1545 is the same as in the right panel of Fig. 2A.
Results and Discussion
The selenomethionine derivative of the full length SSO1545 was expressed in Escherichia coli with an N-terminal TEV cleavable His-tag and purified by metal affinity chromatography. The crystal structure of SSO1545 was determined in space group P21 at 2.0 Å resolution using the MAD method (PDB 2fna). The final model of SSO1545 contains two monomers (residues 1 to 356 and the residual expression tag Gly0), a bound ADP and magnesium ion associated with the active site of each monomer, 447 water molecules and seven 1,2-ethanediol molecules. Residues 115–119 of both monomers were omitted due to the lack of interpretable electron density. The Matthews coefficient (Vm) 27 for SSO1545 is 2.5 Å3/Da, and the estimated solvent content is 50.7%. The structure was refined to Rcryst and Rfree of 17.4 and 22.6 %. The model displays good geometry with a 6.34 all-atom clash score, 97.7% residues in favorable regions of the Ramachandran plot (no outliers) and 98% favorable side chain rotamers according to MOLPROBITY 23. The crystal packing suggests that the ADP-bound SSO1545 is likely to exist as a monomer in solution in agreement with the size exclusion chromatography which indicated 80% monomer and additional high molecular weight aggregates. The two monomers in the asymmetric unit are related by a pseudo-translation and are essentially identical with a Cα RMSD (Root-Mean-Square Deviation) of 0.47 Å. Data collection, model, and refinement statistics are summarized in Table I.
SSO1545 contains 9 β-strands (β1–β9) and 18 α-helices (H1–H18), including three short 310 helices (H1, H4 and H8) [Fig. 1A] 25. SSO1545 contains three domains, an N-terminal NTPase domain (residues 1–283) and a C-terminal winged helix domain (WH domain, residues 284–356). The NTPase domain can be divided into two subdomains: a three-layered α/β RecA-like base domain (residues 1–207) and a helical lid domain (residues 208–282), that is a common feature of AAA+ ATPases. A search for proteins structurally similar to SSO1545 using the DALI server 28 identified many AAA+ ATPases. The most similar structures include archaeal Cdc6/Orc2 (PDB 1w5t, Z=19.6) 29 and Cdc6/Orc1 (PDB 2qby, Z=14.6) 30, apoptosis regulators CED-4 (PDB 2a5y, Z=17.2) 8 and Apaf-1 (PDB 1z6t, Z=12.0) 7, bacterial DNA replication initiator DnaA (PDB 2z4s, Z=11.9) 31. The structurally conserved regions encompass the entire NTPase domain. This, combined with the conserved AAA+ sequence motifs (Walker A, Walker B, senor I and “arginine finger”), identifies SSO1545 as a member of AAA+ ATPase family 3,4. The structure of another uncharacterized ATPase, Paby2304 from Pyrococcus abyssi (PDB 2qen), was recently released. Paby2304 and SSO1545 share 36% sequence identity and can be superimposed with an RMSD 1.95 Å for 318 Cα [Fig. 1B], suggesting that these two proteins have similar functions.
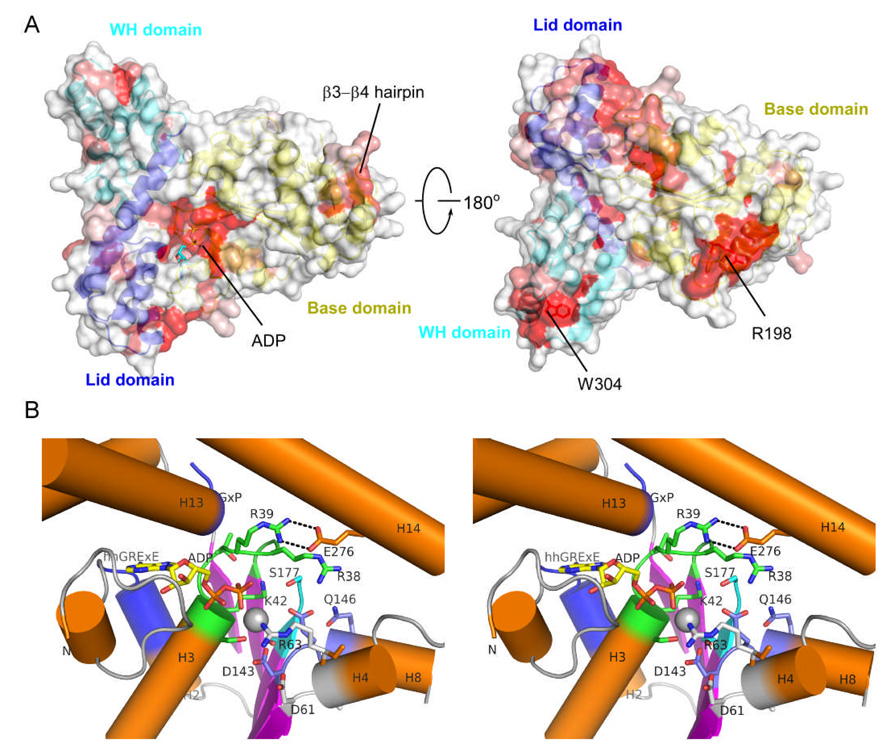
The three domains of the ADP-bound SSO1545 form a triangular shape of molecular dimensions 69 Å × 66 Å × 41 Å with each domain occupying one corner [Fig. 1A]. The base and lid domains interact like a typical AAA+ ATPase, with the nucleotide binding site located at the domain interface. The ADP is protected by the N-terminal region of the base domain (residues 1–43) and helices H11 and H13 of the lid domain. The WH domain is connected to the NTPase domain through an extended helix H14. This “bridging” helix (H14) is in contact with the base domain (H4, and H4–H5 loop region) through three salt bridges involving four residues, Lys275/Glu67 and Glu276/Arg39. Additionally, helix H17 of the WH domain (residues 333–336) also interacts with the helix H10 of the base domain (residues 177–184). The interactions between the bridging helix (H14) and the WH domain are mediated primarily through hydrophobic contacts and an additional salt bridge between Glu278 and Arg288. In addition, the C-terminal region of the WH domain (residues 348–350) also interacts weakly with the lid domain (residues 237–240).
Homologs of SSO1545 are widespread among archaea and are also found in bacteria, such as TM1011 from Thermotoga maritima. SSO1545 does not display significant overall sequence similarities beyond AAA+ motifs to three characterized Orc 1–3 proteins in Sulfolobus solfataricus 32, all of which are AAA+ ATPases with similar domain organizations consisting of the NTPase and WH domains. Analysis of sequence conservation of SSO1545 and its homologs identified several conserved sites on the molecular surface of SSO1545, including the nucleotide binding site and the arginine finger region [Fig. 2A]. All common AAA+ motifs, except sensor II, are present in SSO1545. The Walker A, Walker B, sensor I motifs are near the bound ADP and magnesium ion [Fig. 2B]. The Walker A motif (P-loop, residue 36–43, GhRRxGK[TS]) is located between the first strand β1 and the helix H3. An arginine from the P-loop (Arg 39) forms salt bridges with Glu276 of the helix H14. The strict conservation of these two residues in SSO1545 homologs indicates that this interaction is likely important for SSO1545 function. The Walker B (residues 143–146, DExD) and sensor I (residues 176–177, GS) motifs are located in the C-terminal loops following β5 and β6, respectively. Two STAND specific motifs, hhGRExE (residues 16–22) and GxP (residues 240–242), are involved in the interaction with the ADP adenosine base, but are not highly conserved among SSO1545 homologs.
Figure 2.
Surface conservation pattern and the nucleotide binding site of SSO1545. (A). Molecular surface of SSO1545 colored by sequence conservation. The most conserved residues are shown in red, the non-conserved residues in white. The three domains of SSO1543 are shown in ribbon representation and colored as yellow, blue and cyan respectively. The orientation of left panel is the same as in Fig. 1A. (B) Close-up stereo view of the ADP binding site. The bound ADP (yellow) and magnesium ion (silver) are shown in sticks and sphere respectively. Walker A (P-loop, green), Walker B (W-B, blue), sensor I (S-I, cyan), sensor II (S-II, white) are shown in cartoon and sticks. The STAND specific hhGRExE and GxP motifs are also highlighted in blue.
The sensor II motif is usually a conserved arginine residue from the N-terminus of the third helix of the lid domain in a prototypical AAA+ ATPase. However, sequence analysis indicates that STANDs generally do not have a corresponding conserved motif on the lid domain 1. In SSO1545, no corresponding positively charged residues are found on the equivalent helix (H13). The usual arginine of the sensor II motif is replaced by a strictly conserved Trp244 and no other nearby residues on the lid domain could fulfill this role. Therefore, we concluded that SSO1545 does not possess the typical AAA+ sensor II motif. The Apaf-1 structure suggests that an equivalent sensor II motif might reside on the winged helix (WH) domain 7. However, the bridging helix H14 (residues 258–284) of SSO1545 is located above the nucleotide binding site so that no residues of the WH domain are in the vicinity of the nucleotide [Fig. 2B]. Additionally, the potential sensor II of Apaf-1 (His438) is not conserved in SSO1545. Thus, the sensor II motif is also not likely to be located on the WH domain in SSO1545. Instead, a strictly conserved arginine residue (Arg63) on the base domain could potentially serve a similar role. Although Arg63, does not interact with ADP in the crystal structure, it resides on a short 310 helix (H4) between strand β2 and helix H5 and this region of the base domain interacts directly with the bridging helix (H14). A mechanism for the ATP-induced conformation change (activation) can be proposed based on this location of the sensor II motif. The interaction between Arg63 and γ-phosphate of ATP would induce conformational changes to the H4 helix and its surrounding region. The interaction between the bridge helix H14 and the base domain could be disrupted (e.g. Arg39/Glu276 salt bridges), resulting in a conformation switch of helix H14 and the associated WH domain. Thus, SSO154 may be a novel representative of the ATP-dependent activation process in AAA+ ATPases.
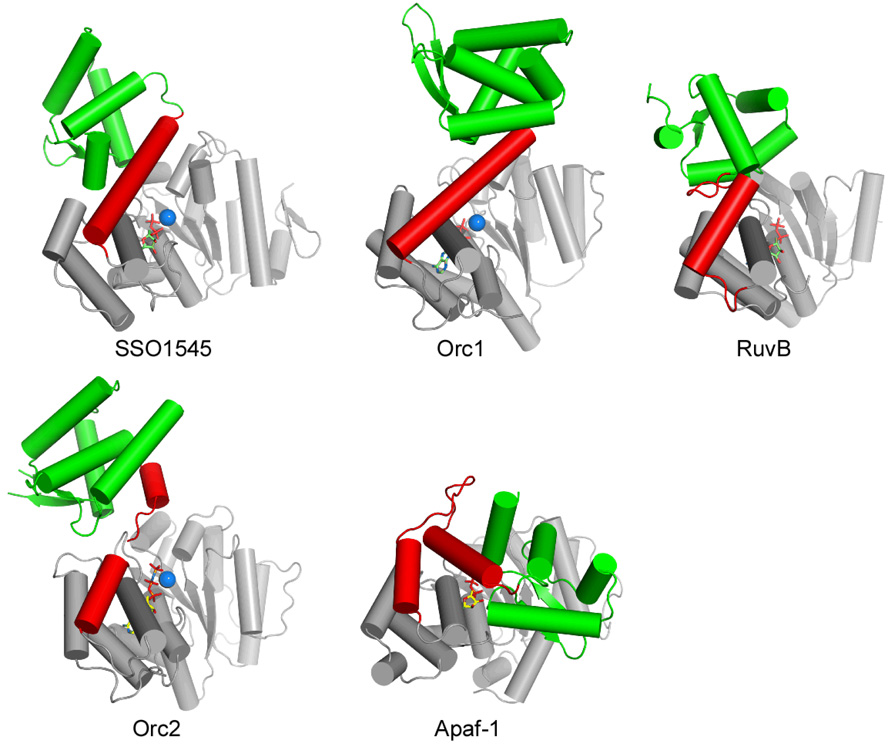
Other AAA+ ATPases have the same domain organization as SSO1545, including the Holliday junction helicase RuvB (PDB 1in5) 33, archaeal origin replication complex proteins Orc1 (PDBs 2qby and 2v1u) 30,34 and Orc2 (PDB 1w5t) 29, and apoptotic protease activating factor Apaf-1 (PDB 1z6t) 7. The overall structures of the NTPase domains in these proteins are similar [Fig. 3]. SSO1545 contains a novel β3–β4 β-hairpin insert between helices H6 and H7. A hairpin insertion, involved in binding RuvA, is also observed at a similar location in RuvB 35. It is plausible that this region of SSO1545 is also involved in protein/protein interaction. The main structural differences among the above proteins are in the linker region of the NTPase/WH domains, as well as the locations/orientation of the WH domain with respect to the NTPase domain. The linkers (the last helix of the lid domain is defined as a part of the linker) have different lengths and conformations [Fig. 3]. SSO1545 and Orc1 contain a single long helix linker, while the linkers of Orc2 and Apaf-1 consist of two helices jointed by a flexible loop. As a result, the WH domains are observed in significant different spatial arrangements with respect to the NTPase domains. Although SSO1545 and Apaf-1 are both classified as STANDs, the arrangement of their WH domains is significantly different. The WH domain of SSO1545 is located away from the wedge opening between the base domain and the lid domain, whereas the WH domain of Apaf-1 sits in front of the nucleotide binding site. The location of the WH domain in SSO1545 is more similar to those in Orc2 and RuvB [Fig. 3].
Figure 3.
Structural comparisons of SSO1545 with AAA+ ATPases with similar domain organizations: RuvB (PDB 1in5), Orc1 (PDB 2v1u), Orc2 (PDB 1w5t) and Apaf-1 (PDB 1z6t, residues 105–450). These structures were superimposed base on their respective NTPase domains (gray). The linker regions connecting the NTPase domain and the WH domain (green) are shown in red. The nucleotides and magnesium ions are shown in sticks and spheres respectively.
The physiological function of SSO1545 is currently unknown. However, the ADP bound form of SSO1545 is likely inactive in vivo by analogy to other AAA+ ATPases where the NTPase activity is used to regulate the conformation of the WH domain. The overall structural integrity of the WH domain is contributed by a disulfide bond (not conserved among SSO1545 homologs), between Cys301 and Cys345 (observed in monomer B), from the β8–β9 wing and the H15–H16 loop, respectively. A DALI search with the WH domain alone indicated similarity to a large number of WH domains, such as transcription repressor SmtB (PDB 1smt, Z=8.6, RMSD 1.6 Å for 63 aligned Cα) [Fig. 4A] 36, despite low sequence identity (<20% for approx. 60 aligned Cα atoms). The known roles for WH domains are to bind DNA or to mediate protein-protein interactions. The WH domains can bind DNA in at least two modes 37. In the majority of cases, the third helix (H17 of SSO1545) acts as a recognition helix and docks to the major groove of the DNA. A second mode, represented by the transcription factor RFX-1 38, is also observed in which the β hairpin wing is inserted into the major groove while the third helix makes contacts with the minor groove. Archaeal Orc1 proteins make use of both the β hairpin wing and the recognition helix to interact with the minor and major grooves of DNA, respectively 30,34. The wing of the SSO1545 WH domain (β8 and β9 hairpin) is very short [Fig. 4A]; therefore, it is unlikely to contribute significantly to DNA interaction due to steric consideration. If SSO1545 were to interact with DNA, it is likely through the canonical recognition helix mode (H17 in SSO1545) or yet another unknown mode. The electrostatic surface properties of the WH domain in SSO1545 differ from other DNA binding WH domains, such as SmtB 36. In contrast to the predominately positive surface observed in DNA binding WH domains 37, the positive electrostatic potential around the H3 is less extensive and is located near the domain interface between the WH and base domains [Fig. 4B] 26. The positive potential is even less significant when the WH domain is considered in the context of the electrostatic potential of the full-length protein [Fig. 4C]. Additionally, surface exposed residues on putative recognition helix H17, e.g. Ser322, Tyr325, Thr329 and Lys333, are not conserved among SSO1545 homologs, raising doubts on the role of H17 in DNA binding. Alternatively, the WH domain of SSO1545 may interact with some other component in the cell, as seen in RPA32 and Apaf-1 7,39. Trp304, located on the surface of helix H16, is strictly conserved in SSO1545 homologs and, together with several nearby highly conserved residues (Ser305, Lys308, Glu321, Glu342 and Tyr344), forms a small highly conserved surface area that may constitute a potential docking site [Fig. 2A]. Interestingly, a sequence similarity search of the Sulfolobus solfataricus genome indicated two other homologs of the SSO1545 WH domain: SSO1204 (114 residues) and SSO3257 (182 residues). The WH domains in these proteins may serve a similar function as SSO1545 based on sequence conservation.
The arginine finger motif (residues 194–198, PhhGR) is located on an extended loop between H10 and β7 with the Arg198 exposed to the solvent [Fig. 2A]. The presence of the absolutely conserved arginine finger indicates that SSO1545 is likely to form an oligomeric assembly when it binds to ATP, as do other AAA+ proteins. Typical AAA+ ATPases often form hexameric (or heptameric) rings. This assembly allows the arginine finger of one promoter to interact with the ATP bound to a neighboring promoter, forming an active configuration for ATP hydrolysis. Modeling studies suggested that hexamer or heptamer rings of SSO1545 can be constructed based on the HslU hexamer (PDB 1do0) or the NtrC heptamer (PDB 1ny6). In these models, the WH domains and the NTPase domains are stacked together like two co-axial donuts, as proposed in RuvB hexamer model 33. Conformational changes at the bridging helix H14 and the WH domain are required to avoid steric clashes. The CluPro webserver 40 predicts another, wind-wheel like, ring structure in which the base domains form the inner ring and the WH domains the outer ring. This ring is flat with a larger central channel (21 Å in diameter).
Further experiments are needed to elucidate the oligomerization state of SSO1545 in its ATP-bound form and the role of the WH domain, as well as its physiological function. The crystal structure and the information present here should be invaluable for these endeavors. The JCSG has developed The Open Protein Structure Annotation Network (TOPSAN), a wiki-based community project to collect, share, and distribute information about protein structures determined at PSI centers. TOPSAN offers a combination of automatically generated, as well as comprehensive, expert curated annotations, provided by JCSG personnel and members of the research community. Additional information about SSO1545 is available at http://www.topsan.org/explore?pdbID=2fna.
Acknowledgements
Portions of this research were performed at the APS Beamline ID-23-D of the GM/CA-CAT and SSRL. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). The SSRL is a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Genomic DNA from Sulfolobus solfataricus (DSM 1617; P2) (ATCC Number: 35092D-5) was obtained from the American Type Culture Collection (ATCC).
Grant Sponsor: National Institute of General Medical Sciences, Protein Structure Initiative; Grant Number: U54 GM074898.
References
- 1.Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 2.Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 4.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 5.Aravind L, Dixit VM, Koonin EV. The domains of death: evolution of the apoptosis machinery. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 6.Koonin EV, Aravind L. The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 7.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 8.Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, Xue D, Shi Y. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 9.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. J Appl Cryst. 2002;35:278–281. [Google Scholar]
- 12.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact samples cassettes and a small-scale robot. J Appl Cryst. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller MD, Deacon AM. An X-ray microsource based system for crystal screening and beamline development during synchrotron shutdown periods. Nuclear Instruments and Methods in Physics Research Section A. 2007;582:233–235. doi: 10.1016/j.nima.2007.08.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 15.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 16.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 17.Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, Lamzin VS, Kleywegt GJ, Perrakis A. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 20.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Tickle IJ, Laskowski RA, Moss DS. Error estimates of protein structure coordinates and deviations from standard geometry by full-matrix refinement of γB- and βB2-crystallin. Acta Crystallogr D. 1998;54:243–252. doi: 10.1107/s090744499701041x. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 23.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 25.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 27.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 30.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 31.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 32.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 33.Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- 34.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 35.Yamada K, Miyata T, Tsuchiya D, Oyama T, Fujiwara Y, Ohnishi T, Iwasaki H, Shinagawa H, Ariyoshi M, Mayanagi K, Morikawa K. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell. 2002;10:671–681. doi: 10.1016/s1097-2765(02)00641-x. [DOI] [PubMed] [Google Scholar]
- 36.Cook WJ, Kar SR, Taylor KB, Hall LM. Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J Mol Biol. 1998;275:337–346. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- 37.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 38.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 39.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 40.Comeau SR, Camacho CJ. Predicting oligomeric assemblies: N-mers a primer. J Struct Biol. 2005;150:233–244. doi: 10.1016/j.jsb.2005.03.006. [DOI] [PubMed] [Google Scholar]