Abstract
Hybrid FTMS instruments, such as the LTQ-FT and LTQ-Orbitrap, are capable of generating fast duty cycle linear ion trap MS/MS data along with high resolution information without compromising the overall throughput of measurements. Combined with online LC separations, these instruments provide powerful capabilities for proteomics research. In the present work, we explore three alternative strategies for high throughput proteomics measurements using hybrid FTMS instruments. Our accurate mass and time tag (AMT tag) strategy enables identification of thousands of peptides in a single LC-FTMS analysis by comparing accurate molecular mass and LC elution time information from the analysis to a reference database. An alternative strategy considered here, termed accurate precursor mass filter (APMF), employs linear ion trap (low resolution) MS/MS identifications generated by an appropriate search engine, such as SEQUEST, refined with high resolution precursor ion data obtained from FTMS mass spectra. The APMF results can be additionally filtered using the LC elution time information from the AMT tag database, which constitutes a precursor mass and time filter (PMTF), the third approach implemented in this study. Both the APMF and the PMTF approaches are evaluated for coverage and confidence of peptide identifications and contrasted with the AMT tag strategy. The commonly used decoy database method and an alternative method based on mass accuracy histograms were used to reliably quantify identification confidence, revealing that both methods yielded similar results. Comparison of the AMT, APMF and PMTF approaches indicates that the AMT tag approach is preferential for studies desiring a highest achievable quantity of identified peptides. In contrast, the APMF approach does not require an AMT tag database and provides a moderate level of peptide coverage combined with acceptable confidence values of ∼99%. The PMTF approach yielded a significantly better peptide identification confidence, >99.9%, that essentially excluded any false peptide identifications. Since AMT tag databases that exclude incorrect identifications are desirable, this study points to the value of a multi-pass APMF approach to generate AMT tag databases, which are then validated using the PMTF approach. The resulting compact, high quality databases can then be used for subsequent high-throughput, high peptide coverage AMT tag studies.
Introduction
Liquid chromatography coupled to mass spectrometry (LC-MS) and tandem mass spectrometry (LC-MS/MS) has become a broadly applied technique for analyzing complex peptide mixtures to determine protein composition and relative abundance [1-5]. Modern mass spectrometry instrumentation can provide exceptional mass measurement accuracy (MMA), extending to 1 ppm or better. The most advanced MS technologies are Fourier transform (FT) ion cyclotron resonance (ICR) and Orbitrap, both employing FT-generated spectra of characteristic ion frequencies, which provide the best currently available resolutions and mass measurement accuracies [6-11]. The hybrid-FTMS instruments, such as the LTQ-FT and LTQ-Orbitrap (Thermo Electron, San Jose, CA), are capable of generating fast duty cycle linear ion trap MS/MS data along with the high resolution information, without compromising the overall throughput of measurements. By coupling these instruments with online LC separations, they become powerful and flexible tools for proteomics [10-12]. As would be expected, the high quantity and better quality of information used to produce a peptide identification, the more confident the results [13-19].
Several proteomics approaches have combined complementary results from multiple LC-MS and LC-MS/MS analyses to provide improved throughput with comprehensive and accurate results [4, 20-23]. As an example, using the accurate mass and time tag (AMT tag) strategy it is generally feasible to identify thousands of peptides in a single LC-FTICR measurement [2-5] based upon accurate molecular mass and normalized LC elution time information matched to data compiled in an AMT tag database derived from previous shotgun LC-MS/MS measurements. Once created, the AMT tag database can be utilized in multiple studies involving the corresponding biological system, using higher-throughput LC-FTICR measurements. In the present study, we further refined this strategy by extending it to the hybrid-FT instruments that are capable of concurrently generating both LC-MS/MS information and the high mass accuracy LC-MS data used for AMT tag measurements.
LC-MS proteomics data involving a statistically large number of identifications can be characterized using a mass accuracy histogram to evaluate the mass measurement accuracy and precision and to guide an optimal choice of mass accuracy tolerance values used for subsequent peptide identification steps [16-19]. Such datasets can be recalibrated using either the least square fitting procedure [17] or more advanced procedures such as multidimensional recalibration based upon the histogram maximization approach and nonparametric regression models [18, 23, 24]. The recalibration efficiently removes systematic mass measurement errors and reduces the mass error spread; i.e. it improves both the mass measurement accuracy and precision [13, 14-19]. In addition to the mass measurement accuracy characterization, the mass accuracy histograms can be used to evaluate the confidence of identifications derived from accurate mass measurements [18]. In this work, we further explore this approach and evaluate it for various levels of confidence, comparing it with an alternative approach based upon decoy databases [25, 26]. The number of peptides sufficient to confidently identify a protein is evaluated for various measurement strategies.
A recent report, published after this manuscript was prepared, investigated three approaches for shotgun protein identification by combining MS and MS/MS information obtained in MudPIT experiments using LTQ-Orbitrap [27], and showed improved protein identification sensitivity for low-abundance proteins when the results from MS and MS/MS analyses are combined. In the present study the two approaches used previously, based upon AMT tag and low resolution MS/MS methods, are integrated at the level of measurements and data processing, in the context of high throughput LC-MS measurement. Parameters critical for obtaining improved coverage and confidence for peptide identifications are explored. These efforts are aimed at the development of an improved approach, such as one which incorporates the empirical distributions of true and false identifications so as to optimize the approach for specific applications.
Methods
To evaluate the strategies under consideration, we have used a Shewanella oneidensis global cell lysate tryptic digest that we commonly employ for quality control evaluation of our high-throughput LC-MS systems [17]. An LTQ Orbitrap XL MS instrument was coupled to a 75 μm i.d. capillary LC column using 100 min LC separations as previously described [28], providing 2940 high resolution, high accuracy full MS spectra (400-2000 m/z, resolution of 100,000 each) along with lower resolution MS/MS spectra for the 6 most abundant precursor ions, obtained in the linear ion trap in parallel with acquisition of the high resolution spectra. The MS/MS data were analyzed with SEQUEST to provide peptide identifications for further filtering according to a data processing strategy described below. The AMT tag database for S. oneidensis has been used extensively in multiple studies [29-35]. In this study, we use subsets of approximately 20K to 50K peptide AMT tags, filtered according to the peptide tag quality metrics. The LC-MS data were processed using the PNNL developed software Decon2LS (http://omics.pnl.gov/software/) that uses a version of the THRASH algorithm [36] to detect features (and their monoisotopic masses) in the individual mass spectra. The AMT tag data processing was done as described previously [5, 17, 18, 37].
Results
The AMT tag strategy
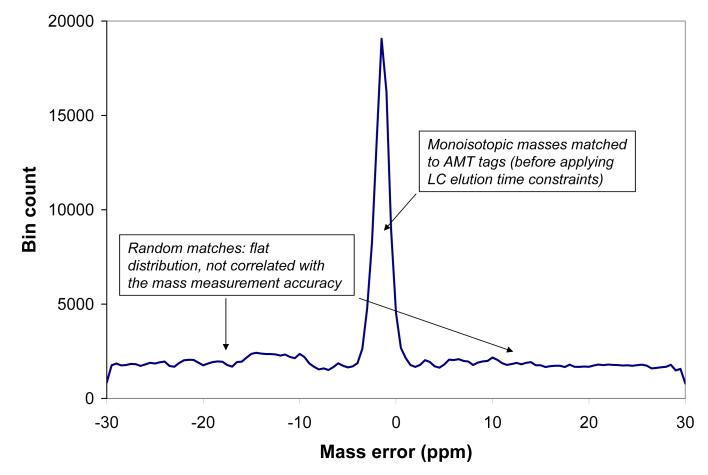
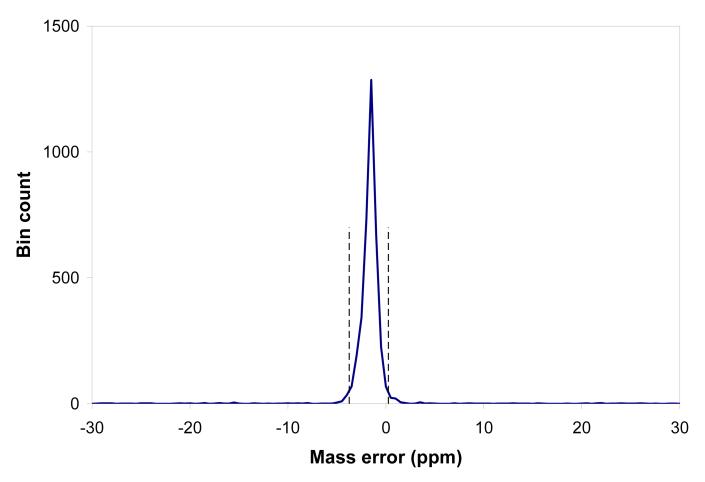
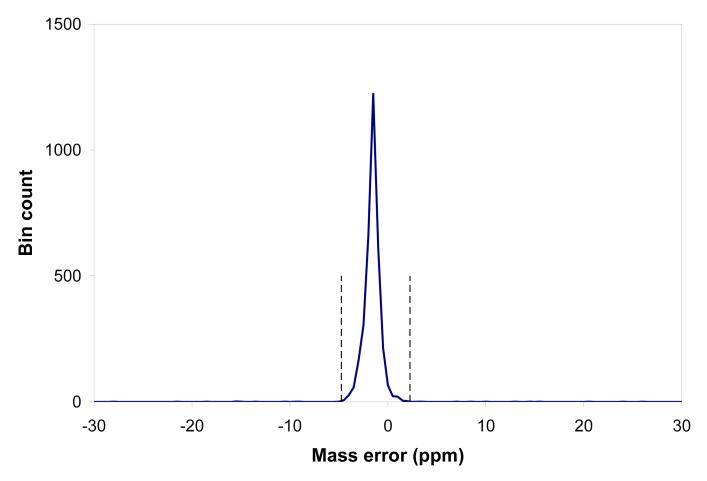
As one benchmark for this study, we used the AMT tag processing pipeline developed at our laboratory [2-5]. The AMT tag approach considers the MS and MS/MS portions of the experimental data (in this case acquired with a hybrid-FT instrument) separately. Thus, with hybrid instruments, the MS/MS data contributes to the population of peptides and related metrics in the AMT tag database, while the MS data is independently matched with the AMT tag database for subsequent peptide identifications and quantitation. All full MS spectra obtained during an LC separation are subjected to a deisotoping procedure, resulting in a set of up to ∼106 isotopic envelopes. The global collection of experimental monoisotopic masses can be matched to theoretical masses from the AMT tag database using only a mass constraint; Figure 1a shows a histogram of mass residuals that results (note that instead of a bar chart, as commonly used for histograms, we are connecting points by linear segments, i.e. linear interpolation, in order to obtain a better representation of the distribution profile, as discussed below). This step is commonly applied in our laboratory for quality control purposes to assess the mass measurement accuracy and precision independent of LC separation performance. The well-defined peak of the histogram corresponds to matches between experimentally observed masses and peptide masses from the AMT tag database. The position and width of the peak give a measure of the mass measurement accuracy and precision of the whole collection of the high resolution mass spectra comprising the LC separation. Knowing the range of the expected mass measurement errors it is now possible to make the next processing step where repeatedly observed peaks are grouped into elution features.
Figure 1.
Histograms characterizing statistics of AMT tag identifications for a single LC-LTQ Orbitrap dataset for Shewanella oneidensis tryptic digest peptide mixture.
a). Mass accuracy histogram obtained at the first stage of the AMT tag process, prior to applying elution time constraints. Each monoisotopic mass observed in the high resolution LC-MS data is matched against theoretical peptide masses from the AMT tag list; the statistics of matches is plotted in the histogram in terms of the number of matches per 0.5 ppm bin (Y axis) vs. the mass difference in ppm (X axis).
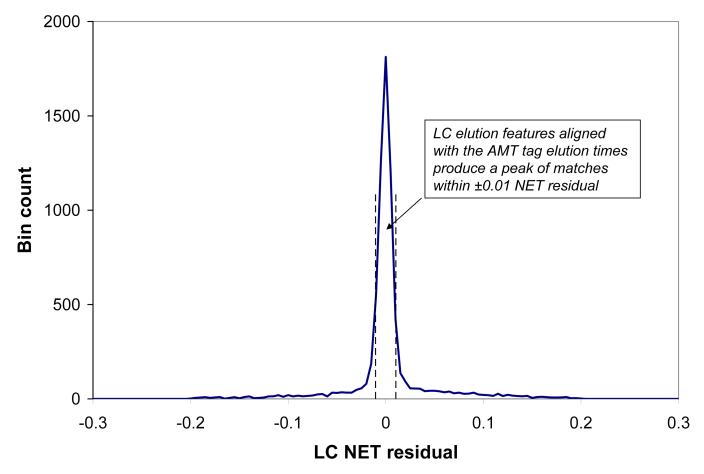
b). Histogram of normalized LC elution time (NET) residuals: the number of LC-MS features per 0.005 NET bin vs. the LC NET residual; vertical dashed lines show automatically determined NET tolerance margins, -0.0105 and 0.0103.
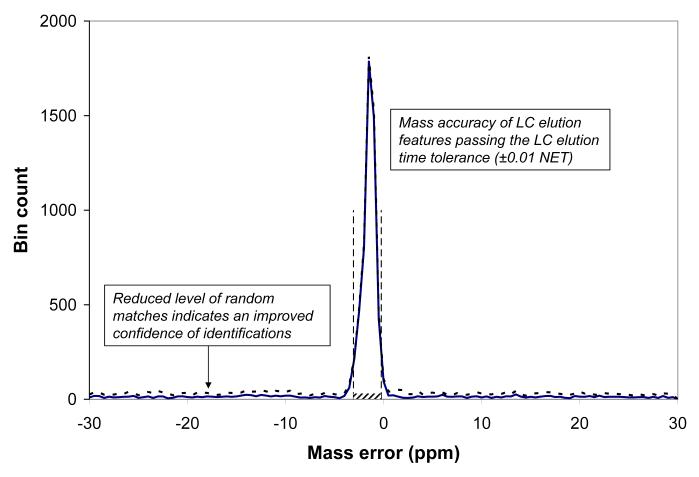
c). Mass accuracy histogram for LC-MS features passing the NET tolerance margins; the number of LC-MS features per 0.5 ppm bin (Y axis) vs. molecular mass difference (X axis); vertical lines show automatically determined mass tolerance margins, -3.1 ppm and -0.2 ppm, used in the final AMT tag matching process. The shaded rectangular area at the bottom of the peak corresponds to the portion of accidental, or false, matches. The dashed curve shows a histogram obtained for the decoy AMT tag database with doubled number of tags, see text. Note that the bin count is approximately 10-fold smaller in (c) than (a) largely due to the number of individual spectral features that collapse to single LC-MS features.
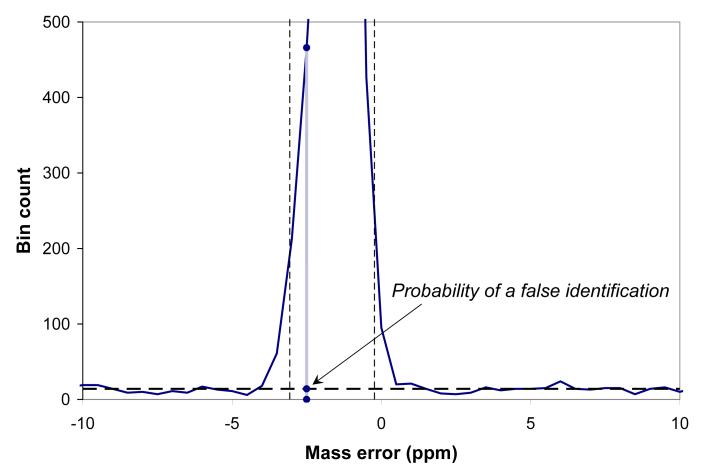
d). Expanded view of the previous histogram for FDR estimation; the horizontal dashed line shows interpolated level of the flat component that represents a probability of purely random matches; the vertical segment shows an example of a match with a mass difference within the limit of mass precision; the portion of it below the random level represents the relative probability of the identification being false; the decoy results are not shown. The vertical segment shows estimation of a specific FDR value for a peptide identification based on the mass error value.
The distribution of putative identifications occurs against a pronounced background of random (i.e. false) matches, evident as the nearly flat component of the mass accuracy histogram in Figure 1a. We will later show how the ratio of the “true” identifications under the peak versus this flat “false” (i.e. random match) component is improved with refinement of the identification process.
In the next processing step isobaric LC-MS “features” are determined, each one consisting of isotopic envelopes observed repeatedly in a number of sequential mass spectra; here we required at least 2 sequential observations to define such a feature, in this case a set of approximately ∼105 features are detected. An ion intensity corresponding to the elution profile maximum is used as a measure of abundance for each particular ion species and the elution time maximum is taken as the characteristic elution time of the LC-MS feature. The measured LC elution time (ET) is then converted to a normalized elution time (NET) by means of a calibration procedure that aligns experimentally obtained elution times with the NET values from the AMT tag database [5, 29, 38]. The 2-term linear NET calibration expression is used to convert the “physical” elution time ET into dimensionless NET:
where C1 is a scaling factor and C0 accounts for an uncertainty in the time axis origin. The NET calibration can be followed by a NET-alignment process [23] that accounts for non-linear deviations in a particular LC separation. The NET calibration procedure uses elution features that match AMT tags with mass residuals passing mass accuracy values determined using the histogram peak in Figure 1a, in this case approximately -4 to +1 ppm. The quality of the NET alignment is characterized by the histogram of NET residuals, shown in Figure 1b. A well defined peak of matching NET values is indicative of a successful NET calibration and a width less than ±∼0.01 NET full width at half maximum (FWHM) is obtained regularly [23]. The baseline width of the peak can be used to determine NET tolerance margins used for peptide identification, as shown by vertical dashed lines in Figure 1b.
In the AMT tag matching procedure the LC-MS features are typically filtered using these LC NET tolerance margins. All passing features contribute to the LC-MS feature mass accuracy histogram, as shown in Figure 1c. The histogram shows a peak of matching masses with the apex position and peak width similar to the peak of the histogram of single matches (Figure 1a), since both the histograms reflect the mass accuracy and precision of the same LC-FTMS dataset. The LC-MS feature masses in this analysis are based upon the median of all monoisotopic masses for each respective LC-MS feature. We have found that this use of the median mass typically produces somewhat narrower LC-MS feature histograms compared to the use of an average mass, since lower abundance species having less precise isotopic masses, give a more limited contribution. The baseline width of the histogram peak can be interpreted as the range of mass residuals characteristic of the mass precision for the experimental dataset. The vertical dashed lines in Figure 1c show the mass error tolerance margins of -3.1 ppm and -0.2 ppm that are determined by default in our software analysis. The default data processing procedure also includes a mass correction step that re-centers the histogram peak at 0 ppm mass error, as well as processing steps that reduce the baseline width of the peak [18, 23, 24]. These steps are omitted here to show the pre-processed mass accuracy distributions for all the alternative strategies under consideration.
At this stage both the LC elution time and mass accuracy tolerances are determined, and the AMT tag matching procedure can be performed to identify a set of peptides within the selected LC NET and mass tolerances; in the particular case of Figure 1c we obtain a set of 5180 unique peptide identifications. The level of random “false” matches (i.e. the flat component of the histogram) in Figure 1c is much smaller than in Figure 1a primarily since the features contributing to Figure 1c are filtered according to both mass and elution time. However, another “refining” factor is our requirement that each LC-MS feature must include a number of isotopic features repeatedly observed in at least two sequential mass spectra (user selected). This filter rejects noisy isotopic features such as artifacts of the deisotoping procedure, etc. It follows that in addition to the mass accuracy characterization, the mass accuracy histogram can be used to evaluate the effectiveness of the matching results in terms of a projected percentage of random matches [18]. The mass accuracy based approach for estimating the false discovery rate (FDR) evaluation is considered in more detail below.
The total area of the histogram Figure 1c is equal to the total number of matches with mass residuals within the full x-range of the plot, in this case ± 30 ppm. Obviously, matches with large mass errors, far beyond the mass error peak characteristic for a particular dataset, can be considered as random matches; the probability of such random matches is determined by the probability distribution of peptide masses, and can be well approximated by a flat distribution within the range ± 30 ppm used here [18, 39, 40]. (In fact, the background of random matches can also be skewed in cases when there is a significant systematic mass difference between experimental and theoretical mass lists. Here it is sufficient to consider a model of the flat background over the relevant mass range). Thus, the ratio of histogram areas below and above the level of random matches gives an estimate of the ratio of false and true identifications. Generally only matches with small mass differences are accepted, e.g. within the mass tolerance margins corresponding to the baseline width of the histogram peak so as to minimize the FDR. In this case the false matches are represented by a rectangular area Ar between the two margins, below the interpolated level of the flat component, as shown by a rectangle at the bottom of the histogram Figure 1c. The FDR estimated from the histogram area (subscript A) can be defined as follows:
| (1) |
here At is the total area of the histogram between the chosen mass tolerance margins; the value is equal to the total number of matches passing the matching constraints, At = Nmatch. The FDR value is a measure of the identification confidence C:
| (2) |
In the case of Figure 1c, FDRA = 0.02, and the overall confidence of identifications is C = 0.98, or 98%. It is not possible at this stage to distinguish which are the true and false peptide identifications based simply upon the LC-MS data; each has an approximately 98% probability of being true and 2% probability of being false. However, it is possible to further extrapolate this logic to the level of a single identification, as illustrated in Figure 1d for a particular match with a mass difference dm = -2.5 ppm, which is close to the chosen right tolerance margin of -3.1 ppm. One can use the ratio of the random match amplitude (horizontal line) to the interpolated histogram value for a particular dm value, (vertical segment) to compute the individual (specific peptide) FDR as FDRi ≈ 0.03 (subscript “i” stands for “individual”). This corresponds to an identification confidence Ci ≈ 97%. The closer a particular match is to the histogram apex position, the better is the individual identification confidence; from this perspective the value FDRA, eq. 1, can be considered as the weighted average of all individual FDRi values. Globally tightening the NET and mass tolerance cut-offs improves the overall FDRA, but at the cost of rejecting identifications having lower FDRi. Additionally, beyond a certain limit of tighter cut-offs this FDRA reduction produces diminishing returns, e.g. an FDRA ≈ 0.01 for NET tolerance ±0.005 and mass tolerance 0.74 ppm (i.e. the apex position ±0.37 ppm), and where the number of identified peptides is reduced from ∼5000 to less than ∼2000. Thus, the size of the AMT tag database and the overall mass and NET accuracy quality combine to set an upper limit on the confidence of any peptide identification obtained in this analysis. We note that further FDR improvements can be achieved using data refining approaches [41]. It should be noted that the peptide level FDR of ∼1% corresponds to much lower FDR for identified proteins, when multiple peptides are used for protein identification [27, 42 - 46].
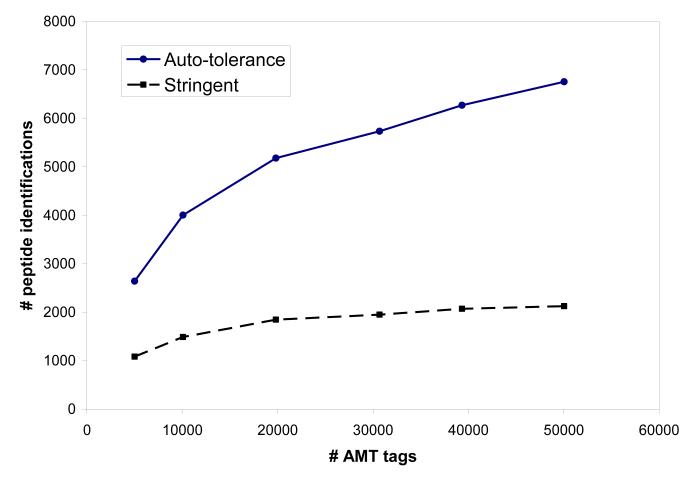
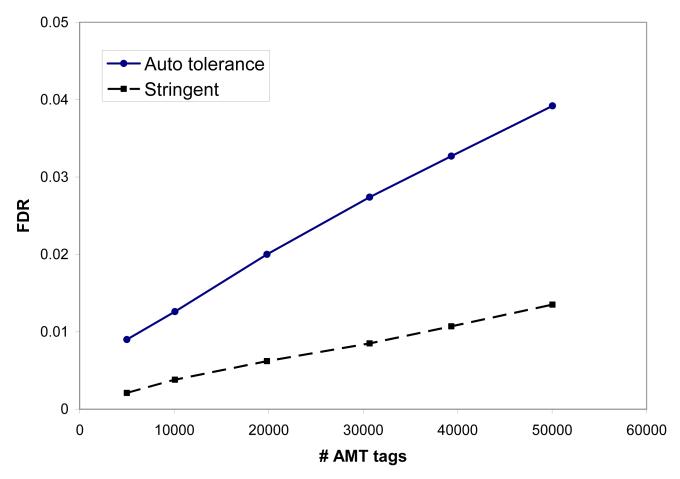
As indicated above, an important factor contributing to the level of FDR obtained with the AMT tag approach is the size of the AMT tag database used, NMT (Table 1). Filters applied to AMT tags to obtain reduced subsets included the number of times an AMT tag was observed in shotgun LC-MS/MS measurements, nobserved, and the high discriminant score HDS [38]. AMT tag sets used had from 5000 to approximately 50,000 AMT tags; smaller NMT correspond to higher nobserved and HDS thresholds, i.e. “higher quality” sets, which generally include higher abundance peptides. The column Nmatch shows a number of peptide identifications obtained with a corresponding set of AMT tags; a ratio of Ctag = Nmatch / NMT is listed in the rightmost column. The smaller AMT tag sets correlate better with the experimental LC-MS data, e.g. 53% of AMT tags (Ctag = 0.53) were matched to LC-MS features for the smallest dataset having 5000 AMT tags. The number of peptide identifications obtained vs. NMT is plotted in Figure 2a; corresponding FDR values are given in Figure 2b. Using the most restrictive rules used here, it can be seen that peptide identifications level off for AMT tag sets having approximately 20,000 tags, while FDR increases approximately linearly with NMT. The AMT tag database having NMT ≈ 20,000 tags provided a low FDR of 0.02 for the automated (less stringent) data analysis, along with coverage of >5000 peptides, approaching the saturation level, thus this configuration is chosen here as a benchmark AMT tag processing scenario.
Table 1.
Filters applied to AMT tags to obtain reduced subsets. n-observed is the number of times an AMT tag was observed in shotgun LC-MS/MS measurements used to compile the AMT tag database; HDS, high discriminant score [38]; NMT, the number of AMT tags passing the filters; Nmatch is the number of unique peptides matched to the AMT tag subset; Ctag is a fraction of AMT tags that matched to experimentally observed features, Ctag = Nmatch / NMT
| n-observed | HDS | NMT | Nmatch | Ctag |
|---|---|---|---|---|
| 180 | 0.99 | 5000 | 2639 | 0.53 |
| 70 | 0.95 | 10079 | 4004 | 0.40 |
| 20 | 0.95 | 19819 | 5180 | 0.26 |
| 8 | 0.95 | 30684 | 5735 | 0.19 |
| 6 | 0.8 | 39332 | 6269 | 0.16 |
| 4 | 0.7 | 50039 | 6756 | 0.14 |
Figure 2.
Number of peptide identifications (a) and FDR (b) obtained using the AMT tag processing of Shewanella tryptic digest LC-MS data from the LTQ-Orbitrap, for AMT tag databases of various sizes, # AMT tags, Table 1. Solid line, circles: mass and NET tolerance determined by default in our software analysis; dashed curve, squares: stringent mass and NET tolerance criteria: ±0.005 NET and ±0.37 ppm mass accuracy, see text.
The accurate precursor mass filter approach
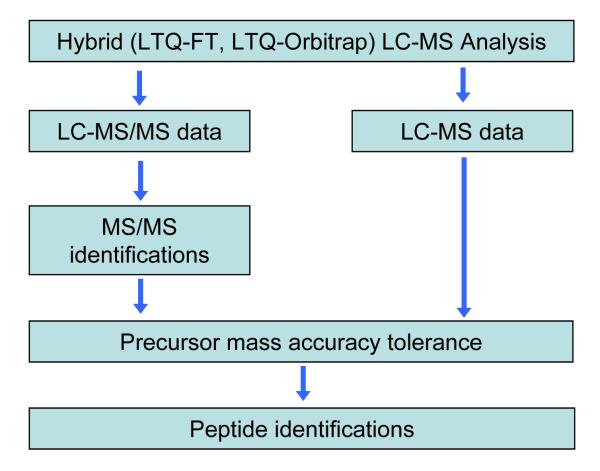
In this section, we apply a new peptide identification processing approach, outlined in Figure 3, to the same Shewanella oneidensis LC-LTQ Orbitrap dataset considered in the preceding section. We now incorporate use of the linear trap MS/MS spectra and their SEQUEST peptide identification scores [47] as well as the accurate mass precursor measurement capability. Similar approaches have been recently applied for proteomics studies using hybrid-FT MS instruments [13, 15]. The two types of information generated in one LC-Orbitrap MS analysis, peptide MS/MS identifications and LC-FT MS data, can be combined via the accurate mass information obtained from the LC-MS measurements or based upon the calculated mass of the peptide identified. In this analysis we applied somewhat relaxed filters compared to those typically used for the SEQUEST identifications: DelCn > 0.08, XCorr > 1.7, 2.0 and 2.8 for charge states 1, 2, and ≥ 3; the single highest XCorr identification for each MS/MS spectrum was used, i.e. RankXc = 1. Use of these relaxed criteria was reasonable because of the strong correlation between the quality of identification scores and mass measurement errors of the accurate m/z for precursor ions that is additionally used [2, 47]. Theoretical molecular masses were calculated for each peptide identified by SEQUEST, and corresponding Orbitrap high resolution mass spectra were searched for a matching peak. Additional information on the APMF procedure can be found in [42]. Figure 4a shows the histogram of mass residuals between the theoretical masses and monoisotopic masses of isotopic features found in the Orbitrap spectra. The histogram peak shape and apex position are similar to the AMT tag histogram in Figure 1c, since both histograms represent the mass measurement accuracy statistics of the same LC-MS dataset. Mass tolerance margins corresponding to the baseline width of the histogram peak represent the range of acceptable mass differences and can be used to “filter” the low resolution MS/MS identifications, similar to the final step of AMT tag matching procedure. This procedure is referred to as the accurate precursor mass filter (APMF).
Figure 3.
Outline of the accurate precursor mass filter (APMF) data analysis process.
Figure 4.
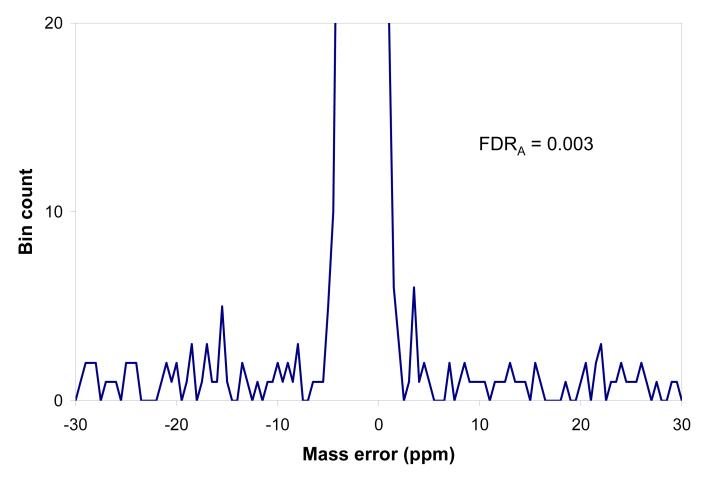
The histogram of mass residuals between precursor masses found in the high resolution precursor MS spectra and theoretical masses calculated for peptides produced by the SEQUEST search based upon the linear ion trap MS/MS spectra, obtained as a result of the accurate precursor mass filter (APMF) analysis. a). The histogram peak area corresponds to 3577 peptides passing tolerance range from -3.75 to 0.25 ppm, vertical segments b). Y-scale expanded to show the level of matches outside the range of mass measurement accuracy, |dM| > 10 ppm, on average 1.5 counts per 0.5 ppm bin, corresponding to 12 false positive identifications per 8 ppm tolerance range, FDRA ∼ 0.003.
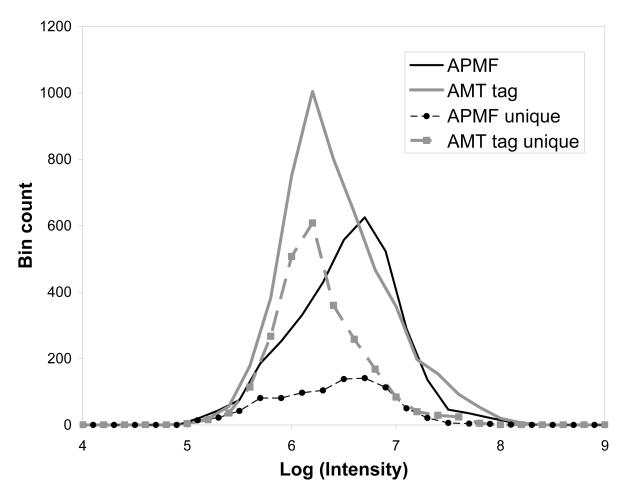
The APMF analysis provided 3577 peptide identifications, using the optimal tolerance range, -3.75 to 0.25 ppm, defined as the base-line interval of the histogram peak, Figure 4a. Compared to 5180 peptides found using the AMT tag strategy, 2659 of the peptides were identified by both approaches. Interestingly, 918 peptides were not identified by the AMT tag approach as implemented for this analysis. A major reason for this was the stringent filtering that was used to generate the limited size AMT tag database used, 20K AMT tags out of more than 100K peptide putative identifications typically used. The reduced size of the AMT tag database was selected in an attempt to increase the overall confidence of identifications (i.e. improve the FDR) at the expense of increasing the overall number of false negatives. Analysis of the ion abundances for precursor ions revealed that higher abundance peptides are generally identified by both approaches, Figure 5. It should be noted that the intensities used for this comparison come from different sources. The precursor ion intensity from full MS high resolution spectra is used as the ion abundance measure for the APMF identifications. The intensity of AMT tag identifications is determined as the maximum intensity in a corresponding elution profile. These different sources means that for the vast majority of peptides in common the AMT tag approach will have a higher measured abundance versus the APMF. Intensity ratios for 2659 peptides identified by both approaches produced on average the AMT tag identifications had a higher measured intensity by 3.2 fold, indicating that precursor ions for LIT MS/MS were chosen before the ions reached the elution peak maximum. To compensate for this bias, the factor of 3.2 was applied to the measured intensities of all MS/MS identifications to produce the values in the histogram Figure 5.
Figure 5.
Intensity distribution of Shewanella oneidensis peptides identified from accurate mass FTMS spectra and low resolution LIT MS/MS spectra of the same LC-MS run using LTQ-Orbitrap. Horizontal axis: ion intensity, common logarithm; vertical axis: number of identifications per 0.2 bin. Black solid line: MS/MS identifications filtered using APMF; gray solid line: AMT tag identifications from the accurate mass full MS Orbitrap spectra; circles: peptides identified by APMF and not identified by AMT tag; squares: peptides identified by AMT tag and not identified by APMF.
The 2521 peptides identified by the AMT tag approach but not identified by the APMF approach (squares) are biased towards peptides with lower abundances relative to all AMT tag identifications (gray curve in Figure 5), which indicates a higher sensitivity of the AMT tag results for this particular experiment. Along with the insufficient ion abundance, the reduced number of APMF identifications can be attributed to the undersampling inherent in “shotgun” proteomics, i.e. an insufficient number of MS/MS spectra were acquired over the time peptides elute. The 100 min long LC separation provided 2900 high resolution Orbitrap spectra and approximately 18,000 linear ion trap (LIT) MS/MS spectra; the time interval when most of the peptides eluted was ∼2/3 of the total separation time. Given ∼12,000 MS/MS spectra and ∼2000 full MS useful spectra, the number of APMF identifications per MS/MS spectrum is ∼1/3, compared to ∼3 identifications per high resolution MS spectrum, as obtained with the AMT tag approach.
For low intensity ions who’s MS peaks appear in two or three MS scans, the errors in mass accuracy tend to increase. The use of a tight mass tolerance would result in biased removal of these identifications. This might be one of reasons that the ‘APMF-only’ peptides tend to be of larger intensity, along with the undersampling issue. However we do not expect this to be a major issue since the APMF mass tolerance, -3.75 to +0.25 ppm, is in fact less stringent than the mass tolerance used for the AMT tag results, -3.1 to -0.2 ppm.
It should be noted that we consider results from a single LC-MS analysis compared with the AMT tags from multiple MS/MS datasets that have been rolled up and filtered. It can be expected that repeated APMF analysis on multiple datasets would result in virtually full overlap between the two approaches.
While fewer, the APMF identifications have improved confidence levels, as seen from the suppressed level of the flat component of the mass accuracy histogram, Figure 4. The expanded view in Figure 4b shows the low level of random matches (1 to 3 occurrences per 0.5 ppm mass error bin). Using eq. 1 we obtain FDRA = 0.0027, i.e. identification confidence C = 0.997, suggesting that ∼10 peptide identifications are false. The improved confidence is obtained despite using relaxed SEQUEST score filters, since the APMF filter is a powerful method to eliminate most false identifications in the AMT tag database. The coverage and FDR of the APMF procedure most likely can be optimized by using MS/MS scoring distributions with mass and NET distributions to quantify the total uncertainty for each individual identification. This optimization could even include a “garbage collection” step that could check the vicinity of an MS/MS spectrum for deisotoping errors, similar to approaches used elsewhere [48].
Our instrument settings aim to maximize the total number of high mass accuracy MS scans and then acquire the highest number of LIT MS/MS scans to maintain those measurements, but there are alternative practical approaches. Mann and coworkers [15] used LTQ FT programmed to perform survey scans of the whole peptide mass range, select the three most abundant peptide signals and perform limited mass range SIM scans for high mass accuracy measurements; simultaneously with the SIM scans, the linear ion trap was used to obtain an MS/MS spectrum and peptide fragment ions were further isolated and fragmented to yield the MS3 spectrum. Such an approach provides even higher identification confidence [15] due to improved mass accuracy of the SIM scans and additional information from MS3 scans. However the number of MS/MS scans is significantly reduced since high resolution mass spectra, the survey scan, and three SIM scans are acquired for each three MS/MS scans. As shown above, the total number of MS/MS identifications is limited by the number of MS/MS spectra over the LC separation, thus we expect that higher numbers of identifications with the analogous approaches yields increased numbers of MS/MS spectra. A key point in all such approaches are the relative trade offs in the numbers of peptides ultimately identified and the level of confidence in these identifications, and the “best” choice depends on the details of the specific application. In addressing this issue we next consider a strategy that tries to combine the high coverage and throughput of the AMT tag approach with further improved identification confidence, provided by the APMF approach.
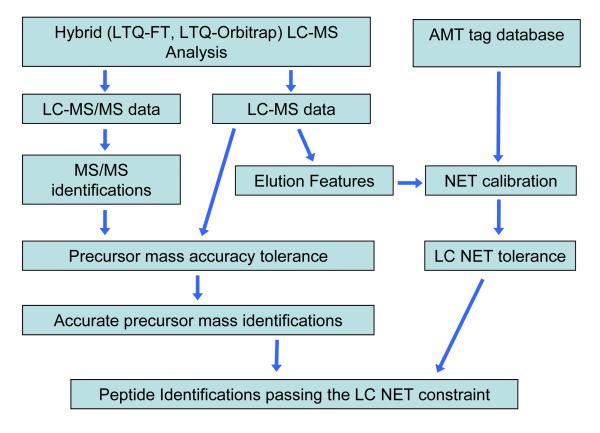
A Precursor Mass and Time Filter approach
To fully explore the capabilities of hybrid-FT instruments, we have initially considered alternative strategies combining both LC-MS/MS and LC-MS accurate mass information, and developed a “Precursor Mass and Time Filter” (PMTF) workflow, as shown in Figure 6. The approach is a combination of the two approaches described above: it starts with the low resolution MS/MS identifications, then the APMF filter is applied, and finally all identifications are filtered according to the elution times tabulated in the AMT tag database. An extended subset of AMT tags can be used, as discussed below; here we used an expanded set of 50K peptide AMT tags filtered according to quality scores, compared to 20K AMT tags used for the above AMT tag processing. The details of the AMT tag filters provided in Table 1 were considered above. The LC-MS features are determined based on the high mass accuracy LC-MS data and are used for NET alignment in order to establish the correspondence between observed physical elution times and dimensionless NET values from the AMT tag list. The NET residuals histogram is used to determine the LC NET tolerance margins for filtering MS/MS identifications (as in Figure 1b).
Figure 6.
Flow chart for the Precursor Mass and Time Filter (PMTF) approach. The approach starts with the low resolution MS/MS identifications, then the APMF filter is applied, and finally all identifications are filtered according to the elution times tabulated in the AMT tag database.. LC-MS elution time data are aligned to AMT tag LC NET values to improve specificity of the LC elution time constraint. As a result of multiple filters the peptide ID FDR <∼ 0.1% is achieved, even with increased AMT tag sets (e.g., here 50K).
Unlike the AMT tag strategy, in this approach LC-MS features are not used directly for peptide identification. Each LC-MS feature consists of a series of the “same” ion species observed during an interval of the LC elution time [2-5] (i.e. as the LC peak elutes). As with the AMT tag process, an ion that is observed only once, i.e. in a single full MS spectrum, is generally not counted as an elution feature. Such a “repeated observation” requirement greatly improves the confidence of identifications, since it rejects most “noisy” ion species. The APMF procedure produced substantially improved identification confidence, so that no such a “repeated observation” filter was necessary. Similarly, the PMTF approach did not employ the elution features in the matching process, rather, single precursor ion occurrences were used. This improved the dynamic range and coverage due to inclusion of species that failing the “repeated observation” filter. (We note that LC-MS elution features were still used for the NET calibration and alignment steps).
Under the PMTF process, identifications obtained from the linear trap MS/MS data are filtered both using the accurate (precursor) mass and NET constraints; the described procedure used the APMF step prior to AMT tag NET filtering, but results are identical if the filtering sequence is reversed. Either relaxed or stringent SEQUEST score filters can be used; for comparison we apply the same relaxed filters as for the APMF processing described above.
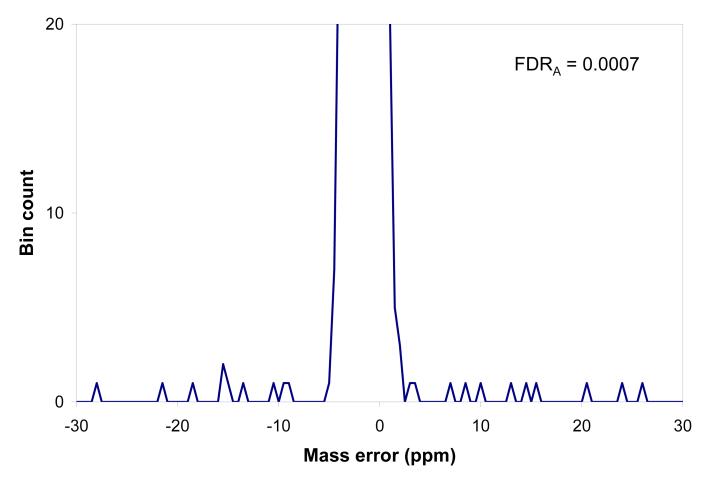
Figure 7a shows the mass accuracy histogram corresponding to MS/MS peptide identifications passing NET constraints, before applying the accurate mass filter. The histogram represents statistics of mass measurement accuracy of a single LC-FTMS dataset, in this case the Shewanella oneidensis LTQ-Orbitrap dataset used in the previous sections. Not surprisingly, the peak shape and position are similar to Figures 1c and 4a. The mass tolerance margins corresponding to the baseline width of the histogram peak are used as mass constraints; the peak area corresponds to 3301 peptides passing a mass accuracy range from -3.75 to 0.25 ppm. The confidence of identifications can be estimated using the mass accuracy histogram. The expanded view of the random match component, Figure 7b, shows sporadic single counts of matches in the area beyond the instrument precision (±10 ppm). There are 14 accidental matches per 40 ppm range (-30 to -10 and 10 to 30 ppm), corresponding to 1.4 false matches across the 4 ppm range used for mass filtering. Because of the very low level of random matches, the tolerance range can be maximized to include the whole peak; using the expanded range -4.75 to 2.25 ppm we obtain 3387 matches within 7 ppm range, corresponding to 2.45 false matches. The corresponding FDRA = 2.45/3387 ≈ 7.2×10-4, or C ≈ 0.9993.
Figure 7.
Mass accuracy histogram corresponding to PMTF peptide identifications passing NET constraints, before applying the accurate mass filter. a) full scale view: the histogram peak corresponds to 3387 matches using the expanded mass tolerance range -4.75 to 2.25 ppm, vertical segments; b). Y-scale expanded to show the level of accidental matches, 14 matches in the range beyond the instrument mass precision, ±10 ppm, corresponding to projected ∼2 false matches within the 7 ppm mass tolerance range.
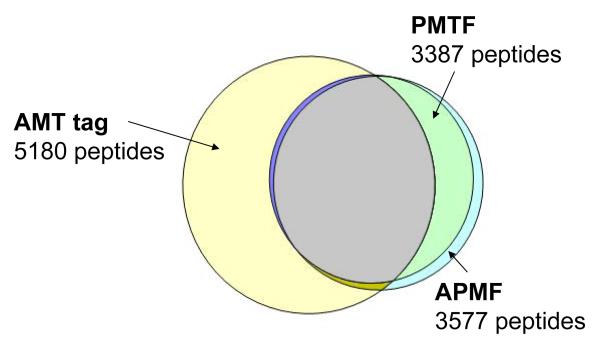
The number of identified peptides, 3387, is smaller than the 3577 peptides identified using the APMF approach. 3301 peptide identifications are found within each list; 86 peptides unique to the PMTF approach can be attributed to the wider mass accuracy tolerance used. 2702 peptides observed with the PMTF approach were also reported in the AMT tag results; 2478 peptides matched to AMT tags and not observed with the PMTF approach generally have relatively lower abundance, similarly to the APMF vs. AMT tag comparison, Figure 5. Figure 8 shows the diagram for peptides identified by each approach; 685 peptides identified with the PMTF approach and not reported in the AMT tag results can be attributed to a larger database used, 50K vs. 20K AMT tags, see discussion section below.
Figure 8.
Peptides identified with the three alternative strategies, the AMT tag (5180 peptides), APMF (3577 peptides) and the PMTF approach (3387 peptides). The 3387 high confidence (FDR ≈ 0.0007) peptides identified with the PMTF approach include 3301 peptides also found by the AMT tag approach and 2702 peptides found by the APMF approach. 685 peptides identified with the PMTF approach and not reported in the AMT tag results can be attributed to a larger database used, 50K vs. 20K AMT tags. 86 peptides identified with the hybrid approach and not identified by the APMF approach are due to the wider mass accuracy tolerance used for the PMTF approach. 918 peptides identified by APMF and not reported by AMT tag are due to the reduced subset of AMT tags used (20K), LC elution time constraints and the elution feature constraint. 276 APMF peptides were not reported by the PMTF approach since they did not pass the AMT tag filter and/or the LC elution time filter.
The mass accuracy tolerance reduced to 2 ppm range, -2.75 to -0.75 ppm, produces 2798 peptide identifications, with the projected number of random matches equal to 0.7: 1 or 0 probable false identifications. The high identification confidence is obtained using relaxed XCorr and DelCn filters. The confidence can be further improved using stricter filters, including more conservative signal to noise threshold and stricter deisotoping settings. It is concluded here that the filters based on measured quantities, such as accurate precursor mass and elution time, are very efficient in producing identifications with very high levels of confidence.
Discussion
The impact of these new peptide identification approaches on the AMT tag strategy can be seen in Table 2. The APMF and PMTF approaches are applied to the same experimental dataset as the AMT tag strategy and have resulted in quite different FDRs and peptide identification counts. The PMTF approach has produced an exceptionally high confidence level that nearly eliminates any false identifications. The confidence values estimated in terms of FDR based upon the mass accuracy histograms can be compared to that obtained using the decoy database approach [25, 26]. Each of the three approaches has been applied twice: first with a database that only contained the normal S. oneidensis sequences, and second with a composite database, which included the original “forward” database merged with the decoy database of the same size. The SEQUEST search used with the APMF and PMTF approaches used a composite reversed decoy database. In the case of the AMT tag strategy, we generated the decoy portion of the AMT tag list by adding an 11 Da shift to each AMT tag’s molecular mass [49]; the resulting composite decoy AMT tag database included NMT ≈ 40,000 AMT tags, which is twice the number of the original database. The mass accuracy histogram, illustrated by the dashed curve in Figure 1c, represents statistics of matches to the decoy AMT tag database. The histogram shows an increased magnitude of the flat component, Hr, produced by purely random matches. Using simple probability density estimations one can show that the random matches component is approximately proportional to the number of AMT tags used for matching, NMT:
| (3) |
Here Nobs is the number of experimentally observed features and kr is the proportionality coefficient; Figure 1c histograms have Hr ≈ 60 matches / ppm for the decoy and Hr ≈ 30 matches / ppm for the forward matching results, consistent with NMT = 20K and 40K, respectively. The value Hr determines the false matches component of the histogram Ar involved in the FDR evaluation, eq. 1:
| (4) |
Here ΔMt is the mass accuracy tolerance interval used as the matching constraint. Optimally, the value ΔMt reflects the precision of mass measurements, which can be determined as the baseline width of the histogram peak, i.e. the interval between the two tolerance margins, illustrated by vertical dashed lines in Figure 1c. We can now rewrite the equations for FDR and identification confidence C as follows:
| (5) |
| (6) |
Table 2.
Coverage and confidence of identified peptides for the alternative strategies; peptide counts for composite decoy searches are corrected according to eq. 11
| Strategy | AMT tag | APMF | PMTF |
|---|---|---|---|
| #peptides found with regular database |
5180 | 3577 | 3387 |
| #peptides, composite decoy database, corrected |
5159 | 3507 | 3337 |
| # decoy peptides: Ndecoy | 144 | 11 | 1 |
| FDR from histogram areas: FDRA | 0.020 | 0.0027 | 0.00072 |
| FDR based on Ndecoy : FDRD | 0.028 | 0.0031 | 0.00030 |
An important conclusion from these expressions is that the identification confidence is directly related to the precision of mass measurements, that determines the tolerance range ΔMt.
It is useful to note that the kr value is largely independent of the nature of the two lists matched to each other. In particular, kr does not depend on any quality score filters used to compile the AMT tag sets; it also does not depend on any metrics of quality of experimental features involved in matching. The following example gives an order-of-magnitude evaluation of factors contributing to the level of accidental matches Hr. Consider a list of experimentally observed ion masses, Nobs = 10,000, covering m/z range 500 to 1500, on average 10,000/1000 = 10 feature observations per unit mass (for simplicity we disregard the non-uniformity of the m/z-distributions of each list, which can be accounted for by applying an additional factor). Thus the probability of a random match to an AMT tag at m/z 1000 can be roughly evaluated as ∼10-2 per 1 mDa range, or ∼10-2 per 1 ppm bin, considering 1000 such bins populated by 10 different ion species. In the case of an AMT tag list having NMT = 10,000 theoretically accurate m/z values, the matching process generates Hr∼1×10-2 × 104 = 100 random matches per 1 ppm bin. Importantly, this level of false matches is created regardless of how well the two lists correlate to each other; e.g. in the extreme case of matching an AMT tag list to itself we still obtain a similar level of noisy identifications, assuming an AMT tag list of similar size. As can be seen from this example, Hr is proportional to Nobs × NMT, in agreement with eq. 3. The AMT tag matching process includes an additional step of filtering using LC NET constraints, which gives >10-fold reduction in the level of random hits, kNET∼0.1. Using this correction, we obtain an order of magnitude estimation kr ≈ 1 ×10-7 /ppm. The dataset used for Figure 1c has 15,086 experimentally observed LC-MS features matched to 19,819 forward, then 39,638 composite forward + shifted AMT tags. Using eq. 3 and kr estimated above we arrive at Hr = 30 hits/ppm for the forward database and 60 hits/ppm for the composite database, in agreement with experimental results (Figure 1c). It follows that the matching procedure produces a number of random matches that is approximately proportional to the product of the number of peptides on each list, times the tolerance range ΔMt (nominator in eq. 6), showing the significance of the mass measurement precision for the identification confidence.
An additional factor that influences the confidence of identifications is related to the quality of experimental and theoretical lists involved in the matching process. The overall quality of the AMT tag list with respect to a particular experimental dataset can be characterized by the following ratio:
| (7) |
In other words, 26% of the AMT tags will have matches in the experimental dataset. It follows that the ideal case scenario with 100% correspondence between two lists can improve FDR and identification confidence by a factor of 1÷0.26 ≈ 4. In practice, more sophisticated filtering approaches can be applied to improve the quality of the AMT tag list, often involving assumptions on distribution density of SEQUEST Xcorr and/or other quality scores [41]. Similar considerations apply to the quality of experimentally observed LC-MS features:
| (8) |
Given the variability of experimental conditions, Qobs is always smaller than 1. Assuming a limit Qobs < ∼0.5 and a similar limit for QAMT we estimate that refinement of the quality would likely be limited to ∼2 fold improvement of identification confidence in this approach, and be obtained at the cost of a reduction of coverage. More sophisticated mass and NET alignment algorithms can further improve FDR by reducing ΔMt and kNET [18, 23, 24]. Thus, the AMT tag matching confidence is defined by the quality and size of the matching data, as well as the mass measurement precision (ΔMt) and chromatographic separation quality.
It should be noted that the values estimated above, eq. 7 and 8, are obtained for the LC-MS data set chosen as a benchmark for this study. To the best of our knowledge, >5000 peptides identified in a single LC-MS measurement (i.e. 26% of AMT tags being searched), is larger than quantities reported in similar studies elsewhere, even when multiple LC-MS measurements are involved. Thus we consider this a demonstration of the high coverage and high throughput operation. Furthermore, at the protein level, >∼20% of the whole proteome of Shewanella was identified in a single LC-MS analysis, again representing high coverage. Further increased proteome coverage can be obtained by applying fractionation methods and additional analyses of the resulting fractions. Since the time this manuscript was first prepared, the technology has improved to the point that we are now reproducibly obtaining 10,000 to 15,000 peptides in a single Shewanella LC-MS QC measurement [50].
Table 2 summarizes the AMT tag results for the target database, NMT = 20k, and the composite decoy database, NMT = 40k. As expected from eq. 3, the level of random hits has increased proportionally by NMT, and FDRA evaluated from the mass accuracy histogram (eq. 1) has also doubled. The number of true hits is similar for both cases:
| (9) |
The composite decoy search has produced Ndecoy = 144 decoy AMT tags, consistent with the proportion of expected false positives based on FDRA = 0.02, which is ∼100 random matches. An alternative way to estimate FDR is based on the count of decoy AMT tags obtained from the composite decoy AMT tag matching:
| (10) |
This equation effectively assumes that the conventional search (using 20k AMT tags) should have the total number of matches reduced by Ndecoy and a number of random matches ∼Ndecoy. FDRD ≈ 0.028 agrees quite well with FDRA≈ 0.02, which serves as validation of both approaches for FDR estimation.
The second strategy we are characterizing is based upon MS/MS identifications subjected to the accurate precursor mass filter, a strategy that does not require a pre-generated AMT tag database. The data processing discussed above has been repeated using composite reversed decoy databases for SEQUEST searching, and results are summarized in the APMF column of Table 2. The number of identified peptides, Nmatch = 3577, is reduced compared to 5180 peptides found with AMT tag approach. However, the FDR obtained from the mass accuracy histogram improved, with FDRA decreasing from 0.02 to 0.003. The composite decoy search has produced a similar number of matches, Nmatch = 3518, with a small number of decoy peptides Ndecoy = 11 (the scrambled decoy search produced similar quantities). Considering that the number of random false matches is similar to Ndecoy, we estimate FDRD using equation 10, FDRD = 0.0031. This value is in a good agreement with FDRA = 0.0027, estimated above based on mass accuracy histogram, which confirms that the both FDR evaluation approaches produce consistent results.
Finally, we have conducted a composite decoy processing method with the PMTF approach. The same decoy sequence searches were used as in the previous case; the APMF results were additionally filtered using the AMT tag database with NET constraints, as described above. The decoy search produced 1 decoy peptide out of 3337 identifications, close to the expected ∼2 false matches estimated above using the mass accuracy histogram approach.
The improved peptide identification confidence generally translates into fewer peptides needed for the confident identification of a protein [27, 42-46]. In a study currently in progress, we investigate the quantity and quality of protein identifications produced by the alternative strategies. The decoy database approach is extended to the level of protein identifications and the number of proteins identified with decoy peptides, i.e. false protein identifications, is used to quantify the protein level FDR. The results [42] show the conventional search to yield 977 proteins from 3387 peptides identified with PMTF. The composite reversed decoy search produced 970 proteins (1 decoy) based on 3337 peptides. The total number of PMTF-detected proteins is reduced compared to the AMT tag approach if all protein identifications are considered (1177), including proteins identified with a single peptide. With PMTF, 565 proteins are identified with two ore more peptides, with 0 decoy proteins. Given the insufficient decoy protein statistics, the probability of false protein identification can be approximately estimated using extrapolation to higher levels n:
| (11) |
Here Nmatch is the total number of identified peptides, as defined above, n is the number of peptides used for the protein identification, Np1 is the number of peptides giving protein identifications with n =1, and FDR is the probability of false peptide identification. 565 proteins identified by PMTF with n ≥ 2 have FDRP2 ∼ 4×10-5, estimated using eq. 11, compared to 774 proteins identified by two or more peptides (n ≥ 2) with the AMT tag approach, which have decoy-based FDRP2 ∼ 0.04 (0.07 using eq.11). The AMT tag protein identifications attain a high confidence level of < 0.01 for n ≥ 4 peptides per protein, 432 proteins total. The two strategies provide a stratification of identifications, and a choice of higher number of proteins identified with a reasonable confidence (∼0.95), vs. a conservative subset of proteins identified with further increased confidence.
Conclusions and Future Directions
We have explored three peptide identification strategies for bottom-up proteomics measurements using as an example Shewanella oneidensis LC-MS (MS/MS) dataset from an LTQ-Orbitrap. The AMT tag strategy uses high mass accuracy LC-MS data; observed LC-MS features are matched to a pre-generated AMT tag database (from previous LC-MS/MS analyses), using accurate mass and LC elution time constraints. The APMF strategy uses peptide (SEQUEST) identifications from the linear trap MS/MS portion of the same LC-MS dataset, filtered using the accurate precursor masses observed in the accurate mass Orbitrap spectra to significantly increase the peptide identification confidence. The confidence of peptide identifications with the APMF approach is >0.99; the approach can be utilized with hybrid-FT MS instruments (e.g. LTQ-FT or LTQ-Orbitrap) and does not require a pre-generated AMT tag database. A disadvantage of the APMF approach is that it does not benefit from the use of previously obtained information, e.g. as contained in an AMT tag database, that effectively includes information related to whether a peptide should be observed, and when it would be observed in an LC separation, and that can significantly improve the confidence level for identifications. Thus, we evaluated a hybrid strategy that combines together the APMF and AMT tag database, starting with the linear ion trap MS/MS identifications subjected to the accurate precursor mass filter, and in which all identifications are then matched to the AMT tag database and filtered according to the corresponding elution times. The PMTF approach was found capable of producing an improved confidence of peptide identifications (> 0.999). The total numbers of peptides identified with the AMT tag approach was higher than for each of the two MS/MS based approaches, which can be attributed to under-sampling for LC-MS/MS measurements, an issue that can be addressed by combining the results from multiple analyses.
The various strategies produced peptide identification FDR values ranging from ∼3% to well below one percent. In particular, the PMTF approach can produce ∼3000 peptide identifications with FDR ∼0.00025, corresponding to projected 0 or 1 false identifications. In order to reliably quantify the identification confidence we have applied two alternative approaches: the commonly used one that involves the decoy database, and the newer approach based on the mass accuracy histogram. The two approaches have produced consistent results, supporting the validity of the FDR estimates. It should be emphasized that the both approaches estimate the false discovery rate related to the probability of random attribution and do not address other sources of incorrect identifications, which can be elucidated by careful examination involving additional sources of information, such as the high resolution MS/MS and de-novo peptide sequencing [51].
The mass accuracy-based approach was capable of quantifying very low FDR values, e.g. ∼10-4, whereas the decoy approach is inaccurate due to insufficient statistics of decoy matches. Another advantage of the mass accuracy-based FDR calculation was a more straightforward implementation that did not require duplicate processing using the decoy database. It was found possible to attribute an individual FDR value to each identification based on its mass accuracy, an option not available with the decoy approach.
The mass accuracy histogram analysis revealed that the level of random matches is determined by the number of experimental identifications multiplied by the number of theoretically exact masses being searched. The portion of random matches passing the mass accuracy constraints is proportional to the mass tolerance range, meaning that the overall confidence of identifications is determined by the precision of mass measurements. Constraints arising from additional measured quantities, e.g. elution time, further improve the identification confidence. A substantial confidence improvement was achieved when MS/MS data was incorporated into the accurate mass and elution time identification process.
We note that even with a perfect peptide ID (0% FDR in the peptide MS/MS database), the peak matching FDR is a function of the database size, so smaller high confidence databases are attractive in many cases. Future work will implement multi-pass APMF approach to generate high confidence AMT tag databases, which can be then validated using PMTF approach, allowing compact high quality databases to be used for subsequent high-throughput, high coverage studies.
Generally, we note that this work clearly illustrates how peptide identification criteria, mass accuracy, or LC NET cut-offs, etc. impact the quality of the results achieved, in terms of both coverage and FDR. Indeed, we explicitly point out how the size of the AMT tag database used impacts both the number of peptide identifications as well as the resulting FDR. This inter-dependence of parameters presents a significant challenge for proteomics applications: which is better, a large set of proteins identified with modest FDR, or a smaller set identified with lower FDR? The answer often also depends on details of the application (e.g. biomarker discovery). While it is clear that the PMTF approach is advantageous, there are numerous variations that differ somewhat in details. We believe that the best approach will be one that makes optimal use of the data sets generated by both parts of hybrid MS instruments, considers the grdations in data quality/confidence, as well as the knowledge encapsulated from previous analyses (e.g. AMT tags).
For these reasons we believe an optimal methodology will include a statistically based approach that optimizes the overall data analysis pipeline in a manner that effectively accounts for the quality of AMT tags (e.g. scores for peptide identifications from MS/MS data), as well as uncertainties in mass and LC NET measurements, etc. In such an approach no cut-offs or “hard” selection criteria are used, but rather overall optimized probability values are obtained, based upon global optimization of the data analysis. Indeed, we are presently evaluating such an approach for application to the AMT tag data analysis pipeline [41] and plan to further refine this approach to incorporate key aspects of the PMTF approach described here. The present work points the way to further improved statistically grounded approaches that optimize both the coverage and quality of identifications in proteomics measurements.
ACKNOWLEDGEMENTS
Portions of this work were supported by the National Center for Research Resources (RR 018522), the National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01), the National Institute of General Medical Sciences (NIGMS, R01 GM063883). Work was performed in the Environmental Molecular Science Laboratory, a DOE national scientific user facility located on the campus of Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Smith RD, Anderson GA, Lipton MS, Paša-Tolic’ L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Pasa-Tolic L, Masselon C, Barry RC, Shen Y, Smith RD. Proteomic analyses using an accurate mass and time tag strategy. Biotechniques. 2004;37(4):621–639. doi: 10.2144/04374RV01. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs JM, Monroe ME, Qian WJ, Shen Y, Anderson GA, Smith RD. Ultra-sensitive, high throughput and quantitative proteomics measurements. Int. J. Mass Spectrom. 2005;240:195–212. [Google Scholar]
- 5.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom. Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comisarow MB, Marshall AG. Fourier transform ion cyclotron resonance spectroscopy. Chem. Phys. Lett. 1974;25:282–283. [Google Scholar]
- 7.Marshall AG. Milestones in Fourier transform ion cyclotron resonance mass spectrometry technique development. Int. J. Mass Spectrom. 2000;200:331–356. [Google Scholar]
- 8.Marshall AG, Hendrickson CL. Fourier transform ion cyclotron resonance detection: principles and experimental configurations. Int. J. Mass Spectrom. 2002;215:59–75. [Google Scholar]
- 9.Makarov A. Electrostatic Axially Harmonic Orbital Trapping: A High-Performance Technique of Mass Analysis. Anal. Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 10.Makarov AA, Denisov E, Lange O, Horning SJ. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. Am. Soc. Mass Spectrom. 2006;17:977–982. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Makarov AA, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 12.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 13.Haas Wilhelm, Faherty Brendan K., Gerber Scott A., Elias Joshua E., Beausoleil Sean A., Bakalarski Corey E., Li Xue, Villen Judit, Gygi Steven P. Optimization and Use of Peptide Mass Measurement Accuracy in Shotgun Proteomics. Molecular & Cellular Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci USA. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilch Bartosz, Mann Matthias. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biology. 2006;7(5):R40, 1–10. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanofsky CM, Bell AW, Lesimple S, Morales F, Lam TT, Blakney GT, Marshall AG, Carrillo B, Lekpor K, Boismenu D, Kearney RE. Multicomponent Internal Recalibration of an LC-FTICR-MS Analysis Employing a Partially Characterized Complex Peptide Mixture: Systematic and Random Errors. Anal. Chem. 2005;77:7246–7254. doi: 10.1021/ac050640q. [DOI] [PubMed] [Google Scholar]
- 17.Tolmachev AV, Zhang R, Langley CC, Monroe ME, Qian W, Strittmatter E, Liu T, Shukla A, Udseth HR, Smith RD. Accurate mass proteomics measurements using capillary LC-FTICR optimized for sensitivity and dynamic range. 53rd ASMS Conf.; San Antonio, TX. June 2005; proceedings on CD ROM. [Google Scholar]
- 18.Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Mass measurement accuracy in analyses of highly complex mixtures based upon multidimensional recalibration. Anal. Chem. 2006;78(24):8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- 19.Zubarev R, Mann M. On the Proper Use of Mass Accuracy in Proteomics. Molecular & Cellular Proteomics. 2007;6:377–381. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. PEPPeR, a platform for experimental proteomic pattern recognition. Mol Cell Proteomics. 2006;5(10):1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, Wu CC, MacCoss MJ. Label-free comparative analysis of proteomics mixtures using chromatographic alignment of high-resolution muLC-MS data. Anal Chem. 2008;80(4):961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- 22.Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Müller M. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7(19):3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 23.Jaitly N, Monroe ME, Petyuk VA, Clauss TRW, Adkins JN, Smith RD. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 2006;78(21):7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
- 24.Petyuk VA, Jaitly N, Moore RJ, Ding J, Metz TO, Tang K, Monroe ME, Tolmachev AV, Adkins JN, Belov ME, Dabney AR, Qian WJ, Camp DG, 2nd, Smith RD. “Elimination of systematic mass measurement errors in liquid chromatography - mass spectrometry based proteomics using regression models and a priori partial knowledge of the sample content. Anal. Chem. 2008;80(3):693–706. doi: 10.1021/ac701863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 26.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 27.Lu Bingwen, Motoyama Akira, Ruse Cristian, Venable John, Yates John R., III Improving Protein Identification Sensitivity by Combining MS and MS/MS Information for Shotgun Proteomics Using LTQ-Orbitrap High Mass Accuracy Data. Anal. Chem. 2008;80:2018–2025. doi: 10.1021/ac701697w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manes NP, Estep RD, Mottaz HM, Moore RJ, Clauss TR, Monroe ME, Du X, Adkins JN, Wong SW, Smith RD. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J Proteome Res. 2008;7(3):960–968. doi: 10.1021/pr070432+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petritis K, Kangas LJ, Ferguson PL, Anderson GA, Pasa-Tolic L, Lipton MS, Auberry KJ, Strittmatter EF, Shen YF, R. Zhao, Smith RD. Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Anal. Chem. 2003 MAR 1;75(5):1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 30.Romine MF, Elias DA, Monroe ME, Auberry K, Fang RH, Fredrickson JK, Anderson GA, Smith RD, Lipton MS. Validation of Shewanella oneidensis MR-1 small proteins by AMT tag-based proteome analysis. Omics-A Journal Of Integrative Biology. 2004;8(3):239–254. doi: 10.1089/omi.2004.8.239. [DOI] [PubMed] [Google Scholar]
- 31.Elias DA, Monroe ME, Marshall MJ, Romine MF, Belieav AS, Fredrickson JK, Anderson GA, Smith RD, Lipton MS. Global detection and characterization of hypothetical proteins in Shewanella oneidensis MR-1 using LC-MS based proteomics. Proteomics. 2005;5(12):3120–3130. doi: 10.1002/pmic.200401140. [DOI] [PubMed] [Google Scholar]
- 32.Shen YF, Strittmatter EF, Zhang R, Metz TO, Moore RJ, Li FM, Udseth HR, Smith RD, Unger KK, Kumar D, Lubda D. Making broad proteome protein measurements in 1-5 min using high-speed RPLC separations and high-accuracy mass measurements. Anal. Chem. 2005;77(23):7763–7773. doi: 10.1021/ac051257o. [DOI] [PubMed] [Google Scholar]
- 33.Fang RH, Elias DA, Monroe ME, Shen YF, Mcintosh M, Wang P, Goddard CD, Callister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Molecular & Cellular Proteomics. 2006;5(4):714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Elias DA, Monroe ME, Smith RD, Fredrickson JK, Lipton MS. Confirmation of the expression of a large set of conserved hypothetical proteins in Shewanella oneidensis MR-1. Journal of Microbiological Methods. 2006;66(2):223–233. doi: 10.1016/j.mimet.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Norbeck AD, Callister SJ, Monroe ME, Jaitly N, Elias DA, Lipton MS, Smith RD. Proteomic approaches to bacterial differentiation. Journal Of Microbiological Methods. 2006;67(3):473–486. doi: 10.1016/j.mimet.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J. Am. Soc. Mass Spectrom. 2000;11(4):320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 37.Monroe ME, Tolić N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23(15):2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 38.Strittmatter EF, Kangas LJ, Petritis K, Mottaz HM, Anderson GA, Shen Y, Jacobs JM, Camp DG, 2nd, Smith RD. Application of peptide LC retention time information in a discriminant function for peptide identification by tandem mass spectrometry. J Proteome Res. 2004;3(4):760–769. doi: 10.1021/pr049965y. [DOI] [PubMed] [Google Scholar]
- 39.Mann M. Useful Tables of Possible and Probable Peptide Masses. Abstracts of the 43rd ASMS Conference on Mass Spectrometry and Allied Topics; Atlanta, GA. May 21-26, 1995. [Google Scholar]
- 40.Zubarev RA, Håkansson P, Sundqvist B. Accuracy Requirements for Peptide Characterization by Monoisotopic Molecular Mass Measurements. Anal. Chem. 1996;68:4060–4063. [Google Scholar]
- 41.Jaitly N, Adkins JN, Dabney AR, Monroe ME, Norbeck AD, Mottaz HM, Lipton MS, Anderson GA, Smith RD. A Statistical Approach to Quantifying Uncertainties in the AMT Tag Pipeline. 56th ASMS Conf.; Dencer, CO. June 2008; proceedings on CD ROM. [Google Scholar]
- 42.Tolmachev AV, Monroe ME, Moore RJ, Purvine SO, Adkins JN, Anderson GA, Smith RD. Strategies for obtaining confident identifications in high coverage, high throughput LC-MS proteomics measurements using hybrid FT instruments. 56th ASMS Conf.; Denver, CO. June 2008; proceedings on CD ROM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 45.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 46.Cociorva D, Yates JR., III DTASelect 2.0: Improving the Confidence of Peptide and Protein Identifications. 54th ASMS Annual Meeting Proceedings; Seattle, WA. May 2006. [Google Scholar]
- 47.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 48.Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24(7):1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petyuk VA, Qian WJ, Chin MH, Wang H, Livesay EA, Monroe ME, Adkins JN, Jaitly N, Anderson DJ, Camp DG, II, Smith DJ, Smith RD. Spatial mapping of protein abundances in the mouse brain by voxelation integrated with high-throughput liquid chromatography—mass spectrometry. Genome Res. 2007;17:328–336. doi: 10.1101/gr.5799207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clauss T, et al. manuscript in preparation.
- 51.Shen Y, Tolić N, Hixson KK, Purvine SO, Pasa-Tolić L, Qian WJ, Adkins JN, Moore RJ, Smith RD. Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal Chem. 2008;80(6):1871–882. doi: 10.1021/ac702328x. [DOI] [PMC free article] [PubMed] [Google Scholar]