Abstract
The p53 homologueΔNp63α is overexpressed and inhibits apoptosis in a subset of human squamous cell carcinomas (SCC). Here we report that in normal keratinocytes overexpressing ΔNp63α and in human squamous carcinoma cells, ΔNp63α physically associates with phosphorylated, transcriptionally active nuclear c-Rel, a NF-κB family member, resulting in increased c-Rel nuclear accumulation. This accumulation and the associated enhanced proliferation driven by elevated ΔNp63α are attenuated by c-Rel siRNA or overexpression of mutant IκBαM, indicating that c-Rel-containing complex formation is critical to the ability of elevated ΔNp63α to maintain proliferation in the presence of growth arresting signals. Consistent with a role in growth regulation, ΔNp63α:c-Rel complexes bind a promoter motif and repress the CDK inhibitor p21WAF1 in both human squamous carcinoma cells and normal keratinocytes overexpressing ΔNp63α. The relationship between ΔNp63α and activated c-Rel is reflected in their strong nuclear staining in the proliferating compartment of primary HNSCC. This is the first report indicating that high levels of ΔNp63α interact with activated c-Rel in keratinocytes and SCC, thereby promoting uncontrolled proliferation, a key alteration in the pathogenesis of cancers.
Keywords: Δ Np63α, c-Rel/NF-κB, IκB, head and neck squamous cell carcinoma
Introduction
Considerable debate has focused on the role of the p53 homologue, p63, in human cancer pathogenesis1. Dysregulation of p63 is observed in a majority of squamous cancers, with p63 gene amplification and/or overexpression reported in squamous cell cancers of the head and neck, lung, cervix and skin2–4. Functional determination of the consequences of p63 overexpression is complicated by the existence of multiple protein variants of p63, which demonstrate overlapping and opposing functions.
The p63 gene is transcribed as 2 classes: TA and ΔN5. TAp63 isoforms contain an NH2-terminal p53-like transactivation domain, and are capable of transactivating known p53-responsive genes, as well as distinct sequences5–7. In contrast, ΔN isoforms lack this domain due to alternate promoter usage and can block transactivation by either p53 or TAp63 isoforms, while still harboring direct transactivation potential8–10. p63 overexpression in human cancers has been predominantly associated with ΔNp63 isoforms1,2,4. Additional complexity within each class of isoform is derived from C-terminal alternative splicing, giving rise to TA- and ΔN-p63α, β and γ. Unlike p53, the p63 gene is critical for normal development of stratified squamous epithelium11,12, and several studies have indicated a requirement for temporal regulation of individual p63 isoforms in both development and maintenance of mature epidermis13–15. However, the specific contribution of each of the known isoforms remains a subject of active investigation.
Previously, we utilized primary murine epidermal keratinocytes and adenoviral vectors to mimic ΔNp63α overexpression observed in human squamous cell cancers (SCC). We demonstrated that overexpressed ΔNp63α maintains keratinocyte proliferation and blocks morphological and biochemical differentiation, despite the presence of signals that induce growth arrest and differentiation10,16. ΔNp63α overexpression was subsequently shown by others to promote survival in a subset of head and neck squamous cell carcinomas by physical association with and blockade of transcription of apoptosis genes by another p53 family member, p7317. To gain mechanistic insight into the altered growth regulation of murine keratinocytes associated with elevated ΔNp63α expression, we profiled extracts from keratinocytes overexpressing ΔNp63 or β-galactosidase (β-gal) for differential transcription factor binding, which provided evidence for a novel form of regulation of NF-κB by ΔNp63.
NF-κB is widely expressed, with effects that are cell type- and context-dependent. Dysregulation of NF-κB activity is associated with multiple human diseases including cancer18, and therapeutics targeting constitutive NF-κB activity are the subject of clinical trials in oncology19. The NF-κB family consists of 5 subunits, which function as homo- and heterodimers. Rel-A, Rel-B and c-Rel contain a transactivation domain, whereas p50/105 and p52/100 do not. Within the normal epidermis, NF-κB plays an important role in regulating homeostasis20,21. During the development and progression of squamous cell carcinoma, the NF-κB1-Rel-A (p50/p65) heterodimer has been implicated in promotion or repression of the malignant phenotype dependent on the context22,23.
Here, we show that murine keratinocytes overexpressing ΔNp63α accumulate transcriptionally active c-Rel in their nuclei, and that nuclear c-Rel accumulation is required to maintain ΔNp63α-mediated proliferation in the presence of signals that normally induce growth arrest. Accumulation of c-Rel is also seen in the nuclei of tumor specimens and cell lines of human head and neck squamous cell carcinomas (HNSCC) expressing endogenous ΔNp63α. Additionally, ΔNp63α and c-Rel physically interact. Their association is observed in vitro in both human and murine cells, and has been confirmed in murine cells in vivo on the promoter of the cyclin dependent kinase inhibitor p21WAF1. These findings provide a mechanism whereby c-Rel contributes to the altered growth regulation of ΔNp63α-overexpressing keratinocytes. This is the first report demonstrating ΔNp63α–mediated regulation of active c-Rel, which is known for its oncogenic propensity24,25, and implicates ΔNp63α–c-Rel complexes in human HNSCC.
Materials and Methods
Cell Culture
Primary keratinocytes isolated from C57Bl/6NCr mice were cultured in 0.05mM Ca2+-containing medium to maintain proliferation, and induced to differentiate by elevating Ca2+ levels to 0.12mM10. HNSCC cell lines 11A, 22B and 38 have been described previously26. N-ethylmaleimide (NEM), a thiol modifier, was added to the culture media following immediately following adenoviral transduction, to block in vivo phosphorylation27.
Gene Transfer
Adenoviruses (ΔNp63α, ΔNp63p40, IκBαM, β-gal) and transduction methodology were described previously10,16,2,28.
Reporter constructs, NF-κB30 or p21WAF131, were transfected using Lipofectamine Reagent System (Life Technologies, Gaithersburg, MD) or for co-transfections with siRNA, Lipofectamine 2000. Activity relative to protein concentration was determined via the Luciferase Assay System (Promega Corporation, Madison, WI).
Reporter assay-only transfections
Keratinocytes were transfected 17h post-adenoviral transduction with the NF-κB reporter construct (3μg) and harvested 24h post-transfection.
C-Rel siRNA and reporter assay transfections
Keratinocytes were transfected with a siRNA pool (c-Rel or non-targeting, 200pmol) (Dharmacon, Lafayette, CO) plus NF-κB reporter construct (1.5μg) 24h prior to adenoviral introduction of ΔNp63α or β-gal. Samples were harvested 24h later.
Transcription factor binding assay
Nuclear extracts from ΔNp63α, ΔNp63p40 or β-gal-overexpressing keratinocytes29 were used to screen the Panomics DNA Array I (Panomics, Fremont, CA).
Western Analysis
Primary antibodies used were: Rel-A (F-6), Rel-B (C-19), c-Rel (C), p100/52 (447), p105/50 (E-10), IκBα (C21), IκBβ (C20), IκBε (M121), p63 DNA-binding domain (4A4) and p63 α–domain (H129), all from Santa Cruz Biotechnology (Santa Cruz, CA); actin (AC-15, Sigma Immuno Chemicals, St. Louis, MO); keratin 10 and filaggrin (BABCO, Richmond, CA). Signal was detected using horseradish peroxidase linked anti-mouse, anti-goat or anti-rabbit secondary antibodies.
Phosphatase Assay
Nuclear extracts were incubated in 1X SAP buffer +/− 10U Shrimp Alkaline Phosphatase (SAP) (Promega, Madison, WI) for 3h at 37°C followed by inactivation at 65°C.
BrdU Incorporation Analysis
FACS analysis was performed as described previously16. 17 h post-adenoviral infection (ΔNp63α or β-gal) cells were maintained in fresh 0.05mM Ca2+-containing medium or switched to 0.12mM for 24 h, with addition of 10μM BrdU for the final 4h. siRNA experiments were performed as described except that keratinocytes were transfected with the siRNA pools as noted 12h prior to adenoviral transduction.
Co-immunoprecipitation analysis
Lysates were pre-cleared with appropriate antibody and beads and then incubated overnight at 4°C with c-Rel (4A4), p63 (H-129), Rel-A (F-6), or control antibody. Protein A/G Plus beads were added for the final hour, samples washed 4x with PBS, resolved by SDS-PAGE and analyzed. The ExactaCruz F reagent system (Santa Cruz Biotechnology) was employed for cases where the same species was used to generate the primary antibody for immunoprecipitation and western analysis.
RT-PCR
Primary murine keratinocytes were transfected with siRNA pools (c-Rel or non-targeting, 200pmol) 8 h prior to adenoviral transduction (ΔNp63α or β-gal). 17h later, cultures were exposed to 0.12mM Ca2+ for 15 hours, or maintained in 0.05mM Ca2+-containing medium. RNA was harvested via the Qiagen RNeasy Plus Mini (Qiagen, Hilden Germany) and reverse transcribed (1μg) using the Accuscript High Fidelity 1st Strand cDNA Synthesis Kit (Stratagene, Cedar Creek, TX) with an oligodT primer. Target sequences were amplified from cDNA pool aliquots in 1X reaction buffer (10mM Tris-HCl, pH8.3, 50mM KCl and 1.5mM MgCl2), 400μM of each dNTP, 5U AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA) and 250ng of each primer. Following a 3 minute hot start, the reaction profile was: denaturation 94°C, 30s; annealing 30s (p21WAF1 57°C, 22 cycles, 478 bp product; HPRT 51°C, 25 cycles, 526 bp product); elongation 72°C, 45s. The primer sequences were:
P21WAF1(F): 5′-AATCCTGGTGATGTCCGACCTGTT-3′
P21WAF1(R): 5′-AGACCAATCTGCGCTTGGAGTGAT-3′
HPRT(F): 5′-CGTCGTGATTAGCGATGATGA-3′
HPRT(R): 5′-TTCAAATCCAACAAACTCTGGC-3′
PCR products were quantified using Spot densitometry software on an Alpha Innotech imaging system (San Leandro, CA).
Electrophoretic mobility shift assays (EMSA)
The LightShift Chemiluminescent EMSA Kit was used (Pierce, Rockford, IL) with oligonucleotides from the p21WAF1 promoter p63 binding site #132: p63BS#1 forward: 5′-TGGCCATCAGGAACATGTCCCAACATGTTGAGCTCTGGCA- 3′ and p63BS#1 reverse: 5′-TGCCAGAGCTCAACATGTTGGGACATGTTCCTGATGGCCA-3′. Oligonucleotides were end-labeled using the 3′ Biotin end-labeling kit (Pierce, Rockford, IL), and incubated with nuclear extract (6μg/reaction) prior to resolution (4% acrylamide). For radioactive EMSAs, oligonucleotides were 5′ end-labeled using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and γ-32P-dATP. Nuclear extracts (6μg/reaction) were incubated at room temperature with 1μl of labeled probe (20,000 cpm) and resolved by gel electrophoresis.
Chromatin Immunoprecipitation
ΔNp63α or β-gal-overexpressing keratinocytes were fixed in 1% formaldehyde solution for 10 minutes. The reaction was stopped by the addition of 1X glycine buffer. Following washing, cells were scraped into PBS. Chromatin was isolated and sheared enzymatically for 10 minutes (ChIP-IT Express Kit, Active Motif, Carlsbad, CA). Samples were immunoprecipitated overnight at 4°C with antibodies to: c-Rel (sc-71), p63α H129 (sc-8344), or IgG control (sc-2027). The chromatin was eluted and cross-links reversed prior to proteinase K digestion. Following a 3 minute hot start, the PCR reaction profile was: denaturation 94°C, 30s; annealing 58°C, 30s; elongation 72°C, 30s for 35 cycles; 220 bp product. The primer sequences were:
P21BS#1(F): 5′-ACTAGCTTTCTGGCCTTCAGGAAC-3′
P21BS#1(R): 5′-CCTGATACATGTCACAAGATACATACCACC- 3′
Immunostaining
Patient matched carcinoma and normal stratified squamous epithelium biopsies were obtained under IRB-approved NIH protocol 04-C-0141 in the outpatient clinic. Frozen sections (10μm) on silanated glass were fixed with 4% paraformaldehyde/PBS at 4°C for 5 min. Nonspecific binding was blocked with 5.5% serum/TBS, and endogenous tissue peroxidase quenched with 0.6% H2O2/TBS prior to incubation with primary antibodies, c-Rel (C), ΔNp63 (N-16) or isotype control (diluted 1:100 in 3% BSA/TBS) overnight at 4°C. Samples were then incubated with biotinylated secondary antibody, then avidin-biotin complex (Vector Labs Vectastain Elite ABC Kit, Burlingame, CA) and 3, 3′-diaminobenzidine (1–5 min depending on target antigen) to reveal immune complexes. Sections were counterstained with Gill’s Formula hematoxylin (Vector Labs), dehydrated, cleared, and mounted using Permount (Fisher, Alanta, GA).
Results
ΔNp63α overexpression promotes nuclear c-Rel accumulation
ΔNp63α expression is associated with the proliferative compartment of normal stratified squamous epithelium and this protein is overexpressed in squamous cell carcinomas2,3,5. Previously we showed that elevated exogenous ΔNp63α maintains primary murine keratinocytes in an undifferentiated proliferative state in the presence of signals that normally induce growth arrest and differentiation10,16. To identify downstream targets of elevated ΔNp63α that may mediate these effects in squamous epithelium, nuclear extracts prepared from keratinocytes overexpressing ΔNp63 or β-gal were screened for differential transcription factor regulation. Differential binding to a NF-κB consensus sequence was observed in samples overexpressing ΔNp63 as compared to β-gal controls (not shown), indicating the potential involvement of NF-κB in the altered growth regulation associated with ΔNp63 overexpression.
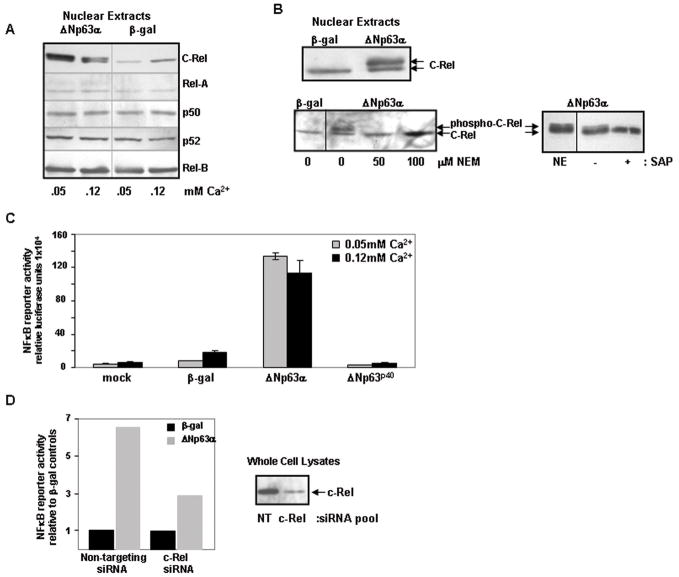
Five NF-κB subunits form hetero/homodimers that display differential binding affinity for multiple NF-κB consensus sequences33. To confirm the altered NF-κB binding activity observed with elevated ΔNp63 and to determine which subunits were involved, we profiled nuclear extracts from ΔNp63α vs β-gal-overexpressing keratinocytes by western blotting. C-Rel was enhanced in nuclei of keratinocytes overexpressing ΔNp63α relative to β-gal control, while nuclear levels of the other NF-κB subunits were unaffected (Figure 1A).
Figure 1. ΔNp63α overexpression results in increased nuclear levels of transcriptionally active phosphorylated c-Rel in murine keratinocytes.
A) Elevated Δ Np63α expression specifically enhances nuclear levels of the NF-κ B subunit c-Rel. Western blots of nuclear extracts from primary murine keratinocytes harvested 21h post-adenoviral introduction of human ΔNp63α or β-gal. Cultures were maintained in medium containing 0.05mM Ca2+ throughout or exposed to 0.12 mM Ca2+ for the final 4h. C-Rel levels are increased in the nuclei of ΔNp63α-overexpressing keratinocytes. B) c-Rel is phosphorylated in response to elevated ΔNp63α. Western analysis of nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal cultured under control conditions (top) or in the presence of the thiol modifier N-ethylmaleimide (NEM) at concentrations noted for 21h following adenoviral introduction to block in vivo phosphorylation (bottom left). The upper species is eliminated in the presence of NEM. Phosphorylation was confirmed by western analysis of nuclear extracts derived from keratinocytes overexpressing ΔNp63α incubated in the presence or absence of Shrimp Alkaline Phosphatase (SAP) for 3h at 37°C (bottom right). A non-incubated control nuclear extract (NE) is included for visual reference. SAP treatment results in the loss of the upper phosphorylated species. C) ΔNp63α overexpression results in NF-κ B-mediated transactivation; α-domain of ΔNp63α required. NF-κB responsive reporter gene activity in keratinocytes overexpressing ΔNp63α, ΔNp63p40 (ΔNp63 lacking α-domain) or β-gal. Samples were harvested 24h post-transfection. Results depict mean +/− s.d. of triplicate samples of a representative experiment performed thrice. D) c-Rel is required for NF-κ B-mediated transactivation following ΔNp63α overexpression. Keratinocytes were co-transfected with NF-κB reporter construct and c-Rel-targeting siRNA or control siRNA, 24h prior to adenoviral infection with ΔNp63α or β-gal. Samples were harvested 24h post adenoviral introduction. Right side: Western blot reveals depletion of c-Rel in whole cell lysates at time of harvest (NT: non targeting siRNA). Left side: Fold increase in NF-κB reporter gene activity in ΔNp63α overexpressing cultures compared to β-gal control cultures in the presence or absence of c-Rel siRNA. Triplicate wells were averaged and are presented as fold increase relative to β-gal controls that are normalized to 1.0. Experiment was performed twice with consistent results; representative experiment shown.
The c-Rel detected in nuclear extracts from ΔNp63α-overexpressing keratinocytes resolved into two species, potentially reflecting post-translational modifications (Figures 1A/B). To address whether the two species of c-Rel observed in the nuclei of ΔNp63α-overexpressing keratinocytes reflected differences in phosphorylation status, keratinocytes were incubated with N-ethylmaleimide (NEM), a thiol modifier that had been previously shown to block c-Rel phosphorylation in vivo27, for 21h immediately following adenoviral transduction. Culturing with NEM resulted in loss of the upper species, indicating that it is a phosphorylated form of c-Rel (Figure 1B). As confirmation, nuclear extracts isolated from keratinocytes overexpressing ΔNp63αwere treated with 10U Shrimp Alkaline Phosphatase (SAP) for 3h at 37°C. Incubation with SAP eliminated the upper species, confirming that it is a phosphorylated form of c-Rel (Figure 1B).
Nuclear c-Rel accumulated in response to elevated ΔNp63α expression in keratinocytes is transcriptionally active
Phosphorylation of c-Rel is known to positively impact its transactivation capacity34. To assess whether c-Rel enhancement resulting from ΔNp63α elevation impacts NF-κB transcriptional activity, keratinocytes were transfected with a NF-κB responsive luciferase reporter construct30 following adenoviral introduction of ΔNp63α orβ-gal. This revealed a 6 – 17 fold increase in NF-κB reporter activity in ΔNp63α-overexpressing keratinocytes relative to β-gal controls (Figure 1C). Using this assay, we also compared keratinocytes overexpressing ΔNp63p40, a truncated form of ΔNp63 lacking the entire α-COOH-terminus that is present in ΔNp63α (Figure 1C). In contrast to ΔNp63α, no reporter gene activity was observed in keratinocytes overexpressing ΔNp63p40, indicating a requirement for the α–tail of p63 in mediating this NF-κB transactivation activity. Repetition of the NF-κB reporter assay in keratinocytes in which c-Rel had been reduced using siRNA prior to the adenoviral introduction of ΔNp63α or β-gal revealed a > 50% reduction in fold increase of reporter activity relative to samples in which c-Rel was not targeted (Figure 1D). The siRNA silencing of c-Rel was incomplete (Figure 1D, western blot); therefore this degree of reduction in activity underscores the critical contribution of c-Rel to ΔNp63α-induced NF-κB-mediated transactivation in keratinocytes.
Enhanced nuclear NF-κ B levels are required for sustained proliferation mediated by Δ Np63α
C-Rel is critical for antigen dependent B-cell proliferation and TCR-mediated T-cell proliferation, and has been implicated in the maintenance of normal keratinocyte proliferation21,35,36. A substantial block in nuclear accumulation of c-Rel was achieved by hindering NFκB nuclear translocation through the introduction of an adenovirus encoding the IκBαM super-repressor (Figure 2A, western blot)28. This approach reduced ΔNp63α-induced nuclear accumulation of c-Rel to levels approximating those in β-gal control cultures. FACS analysis revealed that β-gal control cultures underwent normal Ca2+-induced growth arrest in both the absence and presence of the IκBαM super-repressor (note decrease in S-phase fraction, Figure 2A, histogram). Consistent with previous results10, ΔNp63α-overexpressing keratinocytes do not arrest in response to 0.12mM Ca2+ (histogram, - IκBαM). Blocking NF-κB subunit translocation with the IκBαM super-repressor restored responsiveness to Ca2+-induced growth arrest in ΔNp63α-overexpressing keratinocytes (histogram, +IκBαM).
Figure 2. Enhanced nuclear c-Rel is required for ΔNp63α-mediated loss of keratinocyte growth regulation, but not differentiation defects.
A–C: Flow cytometry analysis of BrdU incorporation in keratinocytes overexpressing ΔNp63α or β-gal under proliferating (0.05mM Ca2+) or differentiating (0.12mM Ca2+) conditions. A) Blocking NF-κB nuclear translocation with the IκBαM super-repressor restores normal Ca2+-mediated growth regulation to ΔNp63α-overexpressing keratinocytes. Primary murine keratinocytes (1.5 days post plating) were co- infected with adenovirus encoding ΔNp63α or β-gal in combination with IκBαM super-repressor or empty vector control. 17h post-infection the medium was changed; cells were maintained for a further 24h in 0.05mM or 0.12mM Ca2+ and pulsed with BrdU (10μM) for the final 4h. Results depict mean +/− s.d. of triplicate samples from a representative experiment performed thrice. Right: Western blot of corresponding nuclear extracts confirms that co-infection with the IκBαM super-repressor effectively reduces levels of nuclear c-Rel in ΔNp63α overexpressing keratinocytes. B) c-Rel siRNA knockdown partially restores normal growth arrest in ΔNp63α-overexpressing keratinocytes. Keratinocyte cultures were transfected with c-Rel or non-targeting siRNA 12 hours prior to introduction of ΔNp63α or β-gal by adenovirus. Decreasing c-Rel levels by siRNA results in an overall reduction of proliferation in all cultures (right panel vs left panel of histogram), and partially restores Ca2+-mediated growth arrest to ΔNp63α-overexpressing keratinocytes (right side, right panel of histogram). Results depict mean +/− s.d. of triplicate samples from a representative experiment. Right: Western blot of whole cell lysates confirms that c-Rel expression is reduced in keratinocytes transfected with c-Rel siRNA. C) Rel-A does not contribute to the aberrant growth arrest response observed in ΔNp63α overexpressing keratinocytes. Keratinocytes were transfected with Rel-A targeted siRNA to deplete Rel-A levels, or with non-targeting siRNA as control. Depleting Rel-A by siRNA has no effect on keratinocyte proliferation under these conditions. Results depict mean +/− s.d. of triplicate samples from a representative experiment. Right: Western blot of whole cell lysates confirms that Rel-A siRNA effectively reduces Rel-A expression in these cultures. D) Blocking NF-κB nuclear translocation does not restore induction of markers of terminal differentiation in ΔNp63α-overexpressing keratinocytes. Western blot of whole cell lysates from keratinocytes overexpressing ΔNp63α or β-gal +/− IκBαM super-repressor and exposed to 0.12mM Ca2+ for 24h. Blocking NF-κB nuclear translocation does not restore the Ca2+-mediated induction of the early marker of keratinocyte differentiation, keratin 10, or the late marker, filaggrin.
An siRNA approach was used to further dissect the requirement for specific NF-κB subunits in the aberrant proliferation observed in conjunction with ΔNp63α overexpression10. Overall levels of proliferation were decreased in both β-gal and ΔNp63α-overexpressing cultures following c-Rel depletion as compared to control cultures with non-targeting siRNA (Figure 2B). However, the relative growth arrest response in β-gal control cultures remained similar (48.9% S-phase reduction with non-targeting siRNA vs 43.8% S-phase reduction with c-Rel siRNA) (Figure 2B, histogram). These results suggest that normal constitutive levels of c-Rel do not significantly impact Ca2+-mediated growth arrest in normal cells. Consistent with previous findings10, ΔNp63α-overexpressing keratinocytes that had been transfected with non-targeting siRNA abnormally continued proliferating following exposure to 0.12 mM Ca2+ (0% reduction in the S-phase, Figure 2B). In contrast, transfecting c-Rel-targeting siRNA into ΔNp63α-overexpressing keratinocytes partially restored growth arrest response (26.4% reduction in the S-phase, Figure 2B, right side histogram). The use of c-Rel siRNA did not entirely block c-Rel protein expression (western blot), thus it is likely that some c-Rel protein remained available for nuclear accumulation and contributed to the remaining abnormal proliferation observed. As c-Rel was the only NF-κB subunit detectably altered in ΔNp63α-overexpressing keratinocytes (Fig. 1A), these siRNA results taken together with the IκBαM super-repressor data (Figure 2A), support a requirement for enhanced nuclear c-Rel in the mediation of enhanced proliferation by ΔNp63α.
Rel-A has been implicated in the maintenance of normal keratinocyte proliferation21, therefore we also used Rel-A-targeting siRNA to assess the contribution of Rel-A to growth regulation in ΔNp63α-overexpressing keratinocytes. In contrast to studies using c-Rel siRNA, depleting Rel-A did not restore normal growth arrest to ΔNp63α-overexpressing keratinocytes (Figure 2C). Based on these findings, and our observation that ΔNp63α overexpression does not alter nuclear levels of Rel-A (Figure 1A), we conclude that Rel-A does not participate in the aberrant growth arrest response in keratinocytes overexpressing ΔNp63α.
In conjunction with loss of Ca2+-mediated growth regulation, elevation of ΔNp63α protein expression blocks the onset of squamous morphology as well as the induction of keratinocyte differentiation-specific gene expression10,16. Blocking c-Rel translocation with the IκBαM super-repressor did not restore expression of differentiation markers to ΔNp63α-overexpressing keratinocytes, indicating NF-κB subunits do not participate in this aspect of ΔNp63α biological activity (Figure 2D).
Enhanced nuclear levels of c-Rel in response to elevated ΔNp63α result from altered intracellular localization without altering cytoplasmic Iκ B: c-Rel interactions
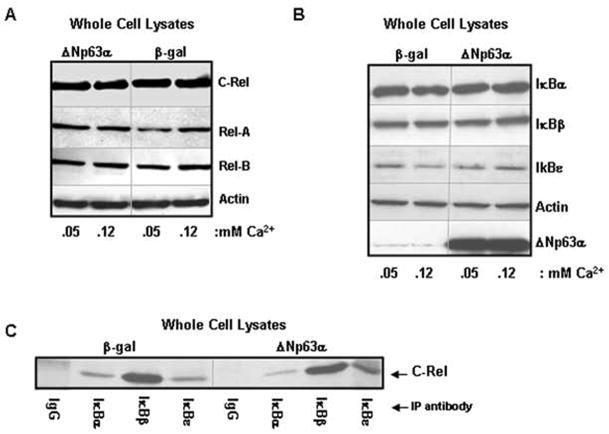
Both ΔN and TAp63 isoforms modulate gene transcription8,10,37. To address whether the enhanced nuclear NF-κB levels reflect ΔNp63α’s activity as a transcription factor, total levels of each subunit in whole cell lysates were assessed. No changes in total cellular expression of c-Rel, Rel-A or Rel-B were observed between ΔNp63α-overexpressing and control cultures (Figure 3A), indicating that enhanced nuclear c-Rel levels resulted from altered intracellular localization. p50/105 and p52/100 were undetectable under these conditions (not shown).
Figure 3. Mechanism of nuclear c-Rel enhancement is not dependent on disruption of cytoplasmic IκB:c-Rel interactions.
A) ΔNp63 overexpression does not alter total levels of cellular NF-κB. Western blots of whole cell lysates from primary mouse keratinocytes harvested 21h post-adenoviral introduction of human ΔNp63α or β-gal. Cultures were maintained in medium containing 0.05mM Ca2+ or exposed to 0.12 mM Ca2+ for the final 4h. B) Levels of IκB regulatory proteins are not decreased in ΔNp63α-overexpressing keratinocytes that accumulate nuclear c-Rel. Western blot of whole cell lysates of keratinocytes overexpressing β-gal or ΔNp63α. C) ΔNp63α overexpression does not inhibit normal cytoplasmic interactions between c-Rel and the IκB proteins. Co-immunoprecipitation analysis of whole cell lysates from keratinocytes overexpressing ΔNp63α or β-gal.
Control of NF-κB localization within the cell is largely mediated by the IκB family of proteins, which sequester NF-κB in an inactive state in the cytoplasm. Upon phosphorylation, the IκBs are degraded, freeing NF-κB hetero/homodimers to translocate to the nucleus38. Western analysis of whole cell lysates derived from keratinocytes overexpressing ΔNp63α vs β-gal showed no reduction in IκB protein levels in keratinocytes harboring elevated ΔNp63α(Figure 3B).
As IκB levels are maintained in ΔNp63α-overexpressing keratinocytes, we speculated that ΔNp63α might perturb normal cytoplasmic interactions between NF-κB and the IκBs. We focused on c-Rel, as this is the only subunit altered in ΔNp63α-overexpressing keratinocytes. Co-immunoprecipitation analyses of whole cell lysates revealed that the normal associations between IκBα, β or ε and c-Rel remain intact in the presence of overexpressed ΔNp63α (Figure 3C).
ΔNp63α and c-Rel physically associate in the nuclei of keratinocytes expressing high levels of Δ Np63α
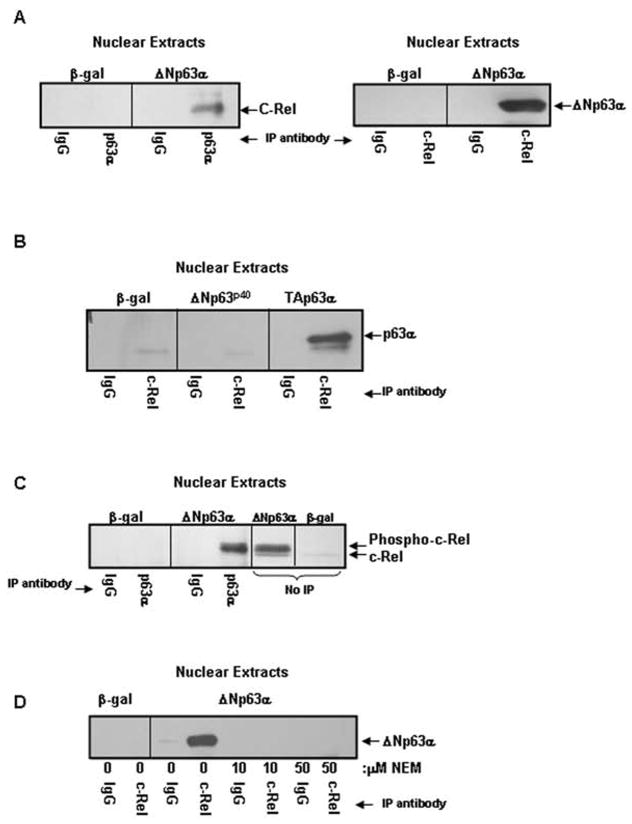
In addition to sequestering NF-κB in the cytoplasm, IκBα and IκBε can disrupt NF-κB: DNA interactions in the nucleus, and through nuclear export signals can actively shuttle NF-κB back into the cytoplasm39,40. IkBα/ε shuttling is mediated by CRM1, which when blocked results in enhanced nuclear NF-κB accumulation. A failure in shuttling, potentially due to a blocked association between c-Rel and IκBα/IκBε, could result in nuclear accumulation of c-Rel in ΔNp63α-overexpressing keratinocytes. Unlike our findings in whole cell lysates (Figure 3C), we detected no association between c-Rel and IκBα, β or ε in nuclear extracts derived from either β-gal or ΔNp63α-overexpressing keratinocytes (not shown). However, co-immunoprecipitation analyses revealed a physical interaction between ΔNp63α and c-Rel in nuclear extracts of ΔNp63α-overexpressing keratinocytes (Figure 4A). In contrast to c-Rel, interaction was not seen between ΔNp63α and Rel-A in ΔNp63α-overexpressing keratinocytes (not shown).
Figure 4. ΔNp63α and c-Rel physically interact in a phosphorylation, p63 α-domain dependent manner.
A) ΔNp63α and c-Rel physically interact in the nuclei of ΔNp63α overexpressing keratinocytes. Co-immunoprecipitation analysis of nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal. Nuclear extracts were immunoprecipitated with antibody to p63 (left panel) or c-Rel (right panel) and probed for c-Rel or p63, as noted. B) The α-domain but not the ΔN-domain of p63 is required for the interaction between p63 and c-Rel. Nuclear extracts from keratinocytes overexpressing ΔNp63p40 (a truncated form of ΔNp63 lacking the α-COOH-terminus), TAp63α, or β-gal were immunoprecipitated with antibody to c-Rel and probed for p63. C) ΔNp63α associates with phosphorylated c-Rel. Co-immunoprecipitation analysis of nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal resolved alongside ΔNp63α or β-gal nuclear extracts (no-IP). Co-migration and western blot reveals that the upper, phosphorylated species of c-Rel interacts with ΔNp63α. D) Protein phosphorylation is required for association between ΔNp63α and c-Rel. Co-immunoprecipitation analysis of nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal. Culture with NEM for one day following adenoviral infection to block in vivo phosphorylation eliminates the interaction between ΔNp63α and c-Rel.
Association between Δ Np63α and phospho-c-Rel requires the p63α-domain and phosphorylation
To assess if the α-COOH-terminus of ΔNp63α is required for the physical interaction between ΔNp63α and c-Rel, co-immunoprecipitation was performed with nuclear extracts from keratinocytes overexpressing ΔNp63p40, which lacks the α-tail, as well as, keratinocytes overexpressing TAp63α, which differs from ΔNp63α only at the NH2-terminus. No interaction was seen between ΔNp63p40 and c-Rel (Figure 4B), but interaction was observed between c-Rel and TAp63α, suggesting that the α-tail of p63 contributes to this interaction.
To address which c-Rel species interacts with ΔNp63α, co-immunoprecipitation reactions were resolved on a gel next to non-immunoprecipitated nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal (Figure 4C). This revealed that the upper, phosphorylated form of c-Rel is the predominant species that interacts with ΔNp63α in the nuclei of keratinocytes overexpressing ΔNp63α. To determine if phosphorylation is necessary for this physical interaction, keratinocytes overexpressing ΔNp63α were cultured in the presence of 10μM NEM and then subjected to co-immunoprecipitation. Blocking protein phosphorylation by NEM treatment abrogated the interaction between ΔNp63α and c-Rel (Figure 4D).
Overexpressed Δ Np63α physically associates with c-Rel on the p21WAF1 promoter
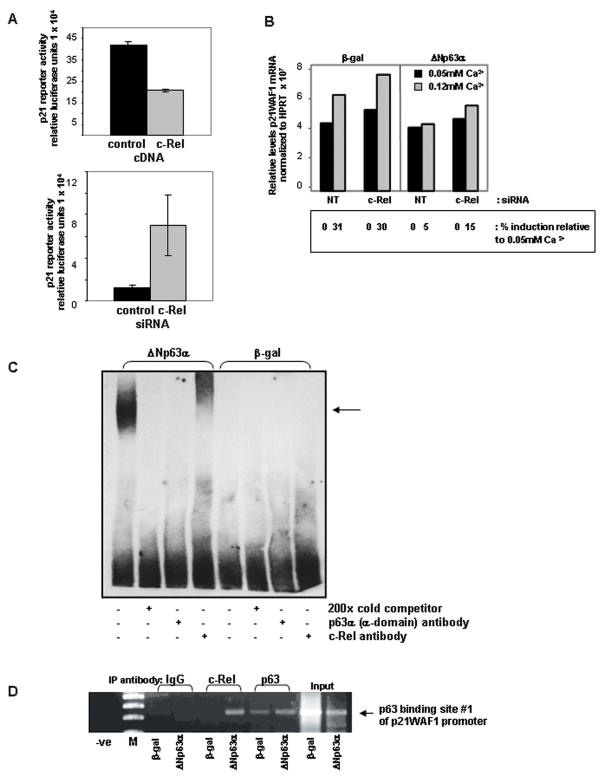
Previously we showed16 that overexpression of ΔNp63α in primary murine keratinocytes blocks induction of the cyclin dependent kinase inhibitor p21WAF1 in response to elevated extracellular Ca2+. Others have demonstrated that ΔNp63α binds to and acts as a transcriptional repressor for p21WAF132. Since enhanced levels of c-Rel are critical to ΔNp63α’s ability to maintain proliferation under conditions that normally induce growth arrest (Figure 2A/B), we asked whether c-Rel also regulates p21WAF1. Co-transfection assays using a luciferase reporter under control of the p21WAF1 promoter confirmed that, like ΔNp63α, c-Rel negatively regulates the p21WAF1 promoter (Figure 5A). Keratinocytes co-transfected with the p21WAF1 promoter construct in combination with a human c-Rel cDNA exhibited a ≥50% decrease in luciferase activity relative to the control samples (top), while use of siRNA to decrease endogenous c-Rel levels resulted in enhanced p21WAF1 promoter activity compared to control (bottom).
Figure 5. ΔNp63α and c-Rel both negatively regulate the cyclin dependent kinase inhibitor p21WAF1, and interact in vitro and in vivo at a p63 binding site on the p21WAF1 promoter.
A) Modulating c-Rel levels alters p21WAF1 reporter gene activity. p21WAF1 reporter gene activity following co-transfection in combination with a human c-Rel cDNA construct, c-Rel targeted siRNA, or controls. Overexpression of c-Rel represses p21WAF1 reporter gene activity (Figure 5A, top), while reducing c-Rel expression levels with targeted siRNA enhances expression of a p21WAF1 luciferase reporter construct (bottom). B) Incomplete silencing of c-Rel by targeted siRNA allows slight restoration of induction of endogenous p21WAF1. Semi-quantitative RT-PCR analysis of p21WAF1. Ca2+-mediated induction of p21WAF1 is unaffected by siRNA knockdown of c-Rel in control keratinocytes overexpressing β-gal. Consistent with previous results16, 32 Ca2+-mediated p21WAF1 induction is blocked by the overexpression of ΔNp63α. siRNA knockdown of c-Rel results in a small but reproducible induction of p21WAF1 in ΔNp63α overexpressing keratinocytes in response to 0.12mM Ca2+ (15% in c-Rel targeted vs 5% induction in non-targeted siRNA controls), as determined by spot densitometry analysis using an Alpha Innotech imaging system. Experiment was repeated with consistent results. C) Δ Np63α and c-Rel physically associate on the p21WAF1 promoter in vitro. EMSA analysis of nuclear extracts from keratinocytes overexpressing ΔNp63α or β-gal. The p63 binding site #1 from the p21WAF1 promoter used in the reporter gene assays was biotin-labeled and used in the binding reactions. A protein:DNA complex seen only in the presence of overexpressed ΔNp63α is supershifted with a c-Rel antibody, and interrupted with a p63-specific antibody. The experiment was performed 4 times with consistent results; representative experiment is presented. D) ΔNp63α and c-Rel physically associate on the p21WAF1 promoter in vivo. ChIP analysis was performed on samples derived from keratinocytes overexpressing ΔNp63α or β-gal using the antibodies noted. PCR primers were designed to flank the p63 binding site #1 from the p21WAF1 promoter. Association of the C-Rel:ΔNp63α complex with p63 binding site #1 is observed. Input DNA: PCR products generated using DNA template from total genomic DNA. Lane labeled “-ve” indicates an absence of DNA in the PCR reaction. M: molecular weight marker. Results shown are representative of 2 independent experiments.
We also evaluated the effect of depleting c-Rel on endogenous p21WAF1 induction. siRNA-mediated depletion of c-Rel in β-gal control keratinocytes did not impact the induction of p21WAF1 mRNA expression following exposure to 0.12mM Ca2+ (Figure 5B). Consistent with previous results, ΔNp63αoverexpression blocks normal Ca2+-mediated induction of p21WAF1. As silencing of c-Rel by siRNA transfection is incomplete (Figure 2B), some c-Rel remains available for nuclear accumulation. Despite this, depletion of c-Rel in keratinocytes overexpressing ΔNp63α resulted in a small but reproducibly detectable induction of p21WAF1 in response to 0.12mM Ca2+ (Figure 5B).
The p21WAF1 promoter contains two known p53 response elements, which have been previously shown to bind p6332. Electrophoretic mobility shift assays (EMSAs) performed using the “p63 binding site #1”32 which corresponds to the p53/p63 consensus site in the reporter constructs used in Figure 5A, revealed that nuclear extracts derived from ΔNp63α-overexpressing keratinocytes produced a DNA:protein complex that could be supershifted with a c-Rel antibody, demonstrating a physical association in vitro between ΔNp63α and c-Rel on the p21WAF1 promoter (Figure 5C) consistent with a role for c-Rel in regulating p21WAF1. In addition to the p53/p63 consensus binding sites, promoter analysis of the p21WAF1 promoter sequence revealed the presence of one potential c-Rel/p65 binding sequence (Drs. B Yan, Z Chen and C Van Waes, unpublished), but EMSAs performed with oligonucleotides to this sequence did not reveal binding (not shown). The association between ΔNp63α and c-Rel on p63 binding site #1 of the p21WAF1 promoter was confirmed in vivo by ChIP analysis of ΔNp63α-overexpressing vs β-gal control keratinocytes using antibodies to either c-Rel or p63. As shown in Figure 5D, a c-Rel:ΔNp63αcomplex in ΔNp63α-overexpressing keratinocytes occupies this p53/p63 consensus site in vivo.
Δ Np63α and c-Rel are strongly expressed thoughout head and neck squamous cell carcinomas
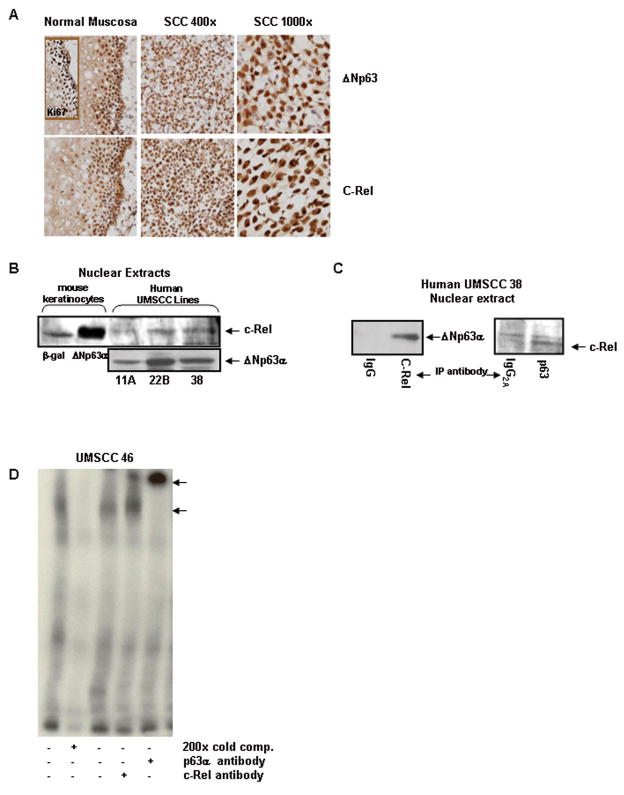
To determine whether the association between ΔNp63αand c-Rel extends to normal and malignant human squamous epithelia, immunostaining was performed on human squamous mucosa and HNSCC tumor samples. Nuclear expression of both p63 and c-REL is associated with the basilar proliferative compartment of normal human mucosa, as defined by Ki67 immunostaining (Figure 6A). Nuclear co-localization demonstrated by strong nuclear staining of both proteins is diffusely seen throughout squamous cell carcinoma tissue samples (Figure 6A). Increased, diffuse nuclear co-staining of ΔNp63 and c-REL was observed in the malignant squamous epithelia of 13/16 (81%) of HNSCC specimens examined, indicating such nuclear co-localization is common in HNSCC.
Figure 6. Endogenous ΔNp63α and c-Rel expression are correlated and expanded in primary human cancers, and associate in the nuclei of human squamous cell carcinoma cells.
A) Nuclear expression patterns of ΔNp63α and c-Rel are expanded and associated in primary human squamous cell carcinomas. Immunostaining of normal mucosa and squamous cell carcinoma tissue sections with p63 and c-Rel. The proliferative compartment of normal mucosa is identified by Ki67 immunoreactivity. B) Endogenous Δ Np63α and c-Rel are present in nuclei of human HNSCC lines. Western blots of nuclear extracts prepared from SCC squamous cell carcinoma lines. Mouse keratinocytes overexpressing ΔNp63α or β-gal are included as controls. C) Endogenous nuclear ΔNp63α and c-Rel physically interact in squamous cell carcinoma cells. Co-immunoprecipitation analysis of UMSCC-38 nuclear extracts. Nuclear extracts were immunoprecipitated with antibody to c-Rel (left panel) or p63 (right panel) and probed for c-Rel or p63, as noted. D) Endogenous nuclear ΔNp63α and c-Rel derived from squamous carcinoma cell lines are associated on the p21WAF1 promoter in vitro. EMSA analyses of nuclear extracts derived from the HNSCC cell line UMSCC 46. A 32P labeled probe using the p63 binding site #1 from the p21WAF1 promoter was used in these reactions. A protein:DNA complex is seen and can be partially supershifted with a c-Rel antibody. Use of the smaller 32P tag in the HNSCC experiments allowed for a supershift band to be seen with the p63 antibody as well.
Endogenous Δ Np63α and c-Rel physically associate in nuclei of HNSCC cell lines
Next we addressed whether a physical association between endogenous ΔNp63α and c-REL occurs in cells of human cancers known to express high levels of ΔNp63α. Western blotting of nuclear extracts from the UMSCC-11A, -22B and -38 squamous cell carcinoma lines revealed that all of these lines express both ΔNp63α and a form of c-REL that co-migrates with the phosphorylated species seen in keratinocytes with elevated ΔNp63α(Figure 6B). Co-immunoprecipitation analysis of nuclear extracts isolated from these cell lines revealed a physical association between ΔNp63α and c-REL (Figure 6C), consistent with our findings in primary mouse keratinocytes. EMSAs performed with nuclear extracts from the HNSCC line UMSCC-46 revealed that a protein:p63 binding site #1 DNA complex is also formed in this cell background that can be partially supershifted with a c-Rel antibody (Figure 6D). As in murine keratinocytes overexpressing ΔNp63α, ChIP assay confirmed association of both p63 and c-REL with the same p21WAF1 promoter site in UMSCC-46 (H. Lu, unpublished observations). This confirms the presence of endogenous ΔNp63α:c-REL complexes that exhibit DNA binding activity in HNSCC on a relevant target gene in vitro.
Discussion
We show that overexpressing ΔNp63α in primary murine keratinocytes leads to the nuclear accumulation of phosphorylated, transcriptionally active c-Rel, which is required to maintain aberrant proliferation mediated by overexpressed ΔNp63α. In these cells, and in human SCC cells endogenously expressing these proteins, ΔNp63α and phospho-c-Rel physically associate in the nuclei and on the p21WAF1 promoter.
c-Rel was originally identified as the cellular counterpart of the v-Rel oncogene, known to cause lymphomas. c-Rel plays an important role in normal cellular homeostasis21,35,36, including that of the epidermis21, and enhanced nuclear c-Rel has been associated with solid and hematopoietic cancers41,42. In contrast to numerous studies of the NF-κB heterodimer p50:p65, the role of c-Rel in transformation of squamous epithelium remains largely unexplored. However, several studies point to the oncogenic propensity of dysregulated c-Rel expression in other systems. Retroviral overexpression of full length wildtype c-Rel can transform primary spleen cells in vitro25. Furthermore, forced overexpression of c-Rel in vivo under control of the MMTV-LTR promoter resulted in mammary tumorigenesis, and correlated with induction of NF-κB target genes including c-myc and cyclin D124. Treatment of these c-Rel-transformed mammary tumor cells with dimethylbenzanthracene in vitro resulted in epithelial to mesenchymal transition43.
The transforming ability of c-Rel both in vitro and in vivo is dependent on the presence of its transactivation domain44,45. c-Rel’s transformation capacity can be enhanced by mutations and deletions within the transactivation domain, suggesting that the strength of transactivation activity can determine the potency of c-Rel44,46. The transactivation domain of c-Rel contains multiple phosphorylation sites and variable levels of phosphorylation have been shown to influence transactivation of distinct sets of target genes47. In this report we show that, in addition to being phosphorylated, the c-Rel that is modulated by ΔNp63α has transcriptional NF-κB reporter-enhancing and p21 gene-repressing activity. Future studies will aim to identify the impact of sustained ΔNp63α elevation on c-Rel target gene expression.
Regulation of NF-κB is a dynamic process38–40. In the classical paradigm of NF-κB regulation, cytoplasmic IκB proteins retain NF-κB in an inactive state, with NF-κB nuclear translocation following IκB degradation. Once within the nucleus, NF-κB induces resynthesis of IκBs, and IκBα and IκBε can dissociate NF-κB from DNA and usher it to the cytoplasm via their nuclear export functions39,40. IκBβ can function in its phosphorylated form to dissociate NF-κB from DNA, while unphosphorylated IκBβ forms a ternary complex with NF-κB and DNA and can protect it from dissociation by IκBα or IκBε48. Our data support a model whereby enhanced ΔNp63α expression results in nuclear accumulation of c-Rel without disrupting IkB:c-Rel cytoplasmic interactions or causing degradation of the IκBs (Figure 3). We have shown that c-Rel physically interacts with ΔNp63α in the cell nucleus and propose that this association inhibits nuclear, but not cytoplasmic, interaction of c-Rel with the IκB proteins by blocking binding. This results in enhanced nuclear accumulation of c-Rel due to the inability of IκBαand IκBε to interact with and remove c-Rel. The c-Rel that accumulates in the nuclei of ΔNp63α overexpressing cells is phosphorylated and transcriptionally active, as determined by reporter gene assay, and can interact in a complex with ΔNp63α on the p21WAF1 promoter to block promoter activity.
The physical association between ΔNp63 and phosphorylated c-Rel requires the α-COOH-terminus of ΔNp63α; like ΔNp63α, TAp63α also physically associates with c-Rel, while ΔNp63p40 does not (Figure 4B). Although less is understood about the role of TAp63 in cancer development, dysregulated TAp63α has been reported to influence the development and progression of chemically-induced skin tumors49. Whether the downstream effects of TAp63α in this context are mediated by c-Rel remains to be determined.
It was initially proposed that overexpression of ΔNp63 in human cancers blocks the tumor suppressor activity of p5350. It has recently been shown that the ability of ΔNp63α to repress p73-dependent apoptosis enhances the survival of a subset of squamous cell carcinoma cells17. The data presented herein support a novel mechanism whereby overexpression of ΔNp63α induces dysregulation of the proto-oncogene c-Rel via physical association, resulting in loss of normal keratinocyte growth regulation. Enhancement of transcriptionally active c-Rel and activation of downstream effectors could be a means whereby ΔNp63α influences the growth and phenotypic characteristics of human cancers. Consistent with our model, a recent clinical trial targeting constitutively active NF-κB in HNSCC via a proteasome inhibitor was found to block nuclear localization of Rel-A, but not c-Rel (Allen et al, manuscript submitted). The findings presented here suggest that distinct NF-κB complexes can promote proliferation of keratinocytes, act in concert with other NF-κB dimers to promote an aggressive cancer phenotype, and offer novel targets and useful biomarkers for optimizing therapeutic efficacy in this subset of poorly responsive cancers.
Acknowledgments
Intramural projects Z01 BO 04006-06 LIMB (CDER/FDA); Z01-DC-00016 (NIDCD/NIH)
IκBαM super-repressor and ΔNp63p40 adenoviruses were gifts of Drs. D. Guttridge and D. Sidransky. Thanks are extended to Dr. Bin Yan for p21WAF1 promoter analysis, Drs. M. Stacey Ricci and David Gius for critical reading and Drs. Christophe Cataisson and Stuart Yuspa for helpful discussions.
Abbreviations
- β-gal
β-galactosidase
- NEM
N-ethylmaleimide
- HNSCC
Head and Neck Squamous Cell Carcinoma
- EMSA
electrophoretic mobility shift assay
- SAP
shrimp alkaline phosphatase
- ChIP
chromatin immunoprecipitation
- TBS
Tris buffered saline
References
- 1.King KE, Weinberg WC. p63: defining roles in morphogenesis, homeostasis, and neoplasia of the epidermis. Mol Carcinog. 2007;46:716–24. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- 2.Hibi K, Trink B, Patturajan M, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000;97:5462–67. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wrone DA, Yoo S, Chipps LK, et al. The expression of p63 in actinic keratoses, seborrheic keratosis, and cutaneous squamous cell carcinomas. Dermatol Surg. 2004;30:1299–302. doi: 10.1111/j.1524-4725.2004.30403.x. [DOI] [PubMed] [Google Scholar]
- 4.Lin Z, Liu M, Li Z, et al. DeltaNp63 protein expression in uterine cervical and endometrial cancers. J Cancer Res Clin Oncol. 2006;132:811–16. doi: 10.1007/s00432-006-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 6.Osada M, Park HL, Nagakawa Y, et al. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25:6077–89. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006;580:4544–50. doi: 10.1016/j.febslet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2005;20:3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 9.Romano RA, Birkaya B, Sinha S. A functional enhancer of keratin14 is a direct transcriptional target of deltaNp63. J Invest Dermatol. 2007;127:1175–86. doi: 10.1038/sj.jid.5700652. [DOI] [PubMed] [Google Scholar]
- 10.King KE, Ponnamperuma RM, Yamashita T, et al. DeltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–44. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 11.Mills AA, Zheng B, Wang XJ, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–18. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 13.Koster MI, Kim S, Mills AA, et al. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Candi E, Rufini A, Terrinoni A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–47. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 15.Truong AB, Kretz M, Ridky TW, et al. A p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–97. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King KE, Ponnamperuma RM, Gerdes MJ, et al. Unique domain functions of p63 isotypes that differentially regulate distinct aspects of epidermal homeostasis. Carcinogenesis. 2006;27:53–63. doi: 10.1093/carcin/bgi200. [DOI] [PubMed] [Google Scholar]
- 17.DeYoung MP, Johannessen CM, Leong CO, et al. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–68. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- 18.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–43. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 19.Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res. 2007;13:1076–82. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- 20.Seitz CS, Lin Q, Deng H, et al. Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc Natl Acad Sci USA. 1998;95:2307–12. doi: 10.1073/pnas.95.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gugasyan R, Voss A, Varigos G, et al. The transcription factors c-rel and Rel-A control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms. Mol Cell Biol. 2004;24:5733–45. doi: 10.1128/MCB.24.13.5733-5745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loercher A, Lee TL, Ricker JL, et al. Nuclear factor-kappaB is an important modulator of the altered gene expression profile and malignant phenotype in squamous cell carcinoma. Cancer Res. 2004;64:6511–23. doi: 10.1158/0008-5472.CAN-04-0852. [DOI] [PubMed] [Google Scholar]
- 23.Dajee M, Lazarov M, Zhang JY, et al. A NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–43. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 24.Romieu-Mourez R, Kim DW, Shin SM, et al. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Mol Cell Biol. 2003;23:5738–54. doi: 10.1128/MCB.23.16.5738-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilmore TD, Cormier C, Jean-Jacques J, et al. Malignant transformation of primary chicken spleen cells by human transcription factor c-Rel. Oncogene. 2001;20:7098–103. doi: 10.1038/sj.onc.1204898. [DOI] [PubMed] [Google Scholar]
- 26.Ondrey FG, Dong G, Sunwoo J, et al. Constitutive activation of transcription factors NF-κB, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Mol Carcinog. 1999;26:119–29. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Glineur C, Davioud-Charvet E, Vandenbunder B. The conserved redox-sensitive cysteine residue of the DNA-binding region in the c-Rel protein is involved in the regulation of the phosphorylation of the protein. Biochem J. 2000;352(Pt 2):583–91. [PMC free article] [PubMed] [Google Scholar]
- 28.Guttridge DC, Albanese C, Reuther JY, et al. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–99. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber E, Matthias P, Muller MM, et al. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiDonato JA, Mercurio F, Karin M. Phosphorylation of I kappa B alpha precedes but is not sufficient for its dissociation from NF-kappa B. Mol Cell Biol. 1995;15:1302–11. doi: 10.1128/mcb.15.3.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 32.Westfall MD, Mays DJ, Sniezek JC, et al. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunsch C, Ruben SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–21. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Valdepenas C, Martin AG, Ramakrishnan P, et al. NF-kappaB-inducing kinase is involved in the activation of the CD28 responsive element through phosphorylation of c-Rel and regulation of its transactivating activity. J Immunol. 2006;176:4666–74. doi: 10.4049/jimmunol.176.8.4666. [DOI] [PubMed] [Google Scholar]
- 35.Hsia CY, Cheng S, Owyang AM, et al. c-Rel regulation of the cell cycle in primary mouse B lymphocytes. Int Immunol. 2002;14:905–16. doi: 10.1093/intimm/dxf055. [DOI] [PubMed] [Google Scholar]
- 36.Liou HC, Jin Z, Tumang J, et al. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–71. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- 37.Wu G, Nomoto S, Hoque MO, et al. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–57. [PubMed] [Google Scholar]
- 38.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, Hannink M. Characterization of the nuclear import and export functions of Ikappa B(epsilon) J Biol Chem. 2002;277:23358–66. doi: 10.1074/jbc.M111559200. [DOI] [PubMed] [Google Scholar]
- 40.Tam WF, Lee LH, Davis L, et al. Cytoplasmic sequestration of rel proteins by IkappaBalpha requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20:2269–84. doi: 10.1128/mcb.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sovak MA, Bellas RE, Kim DW, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100:2952–60. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodig SJ, Savage KJ, Nguyen V, et al. TRAF1 expression and c-Rel activation are useful adjuncts in distinguishing classical Hodgkin lymphoma from a subset of morphologically or immunophenotypically similar lymphomas. Am J Surg Pathol. 2005;29:196–203. doi: 10.1097/01.pas.0000149689.75462.ff. [DOI] [PubMed] [Google Scholar]
- 43.Shin SR, Sanchez-Velar N, Sherr DH, et al. 7,12-dimethylbenz(a)anthracene treatment of a c-rel mouse mammary tumor cell line induces epithelial to mesenchymal transition via activation of nuclear factor-kappaB. Cancer Res. 2006;66:2570–75. doi: 10.1158/0008-5472.CAN-05-3056. [DOI] [PubMed] [Google Scholar]
- 44.Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928–36. doi: 10.1038/sj.onc.1206801. [DOI] [PubMed] [Google Scholar]
- 45.Fan Y, Rayet B, Gelinas C. Divergent C-terminal transactivation domains of Rel/NF-kappa B proteins are critical determinants of their oncogenic potential in lymphocytes. Oncogene. 2004;23:1030–42. doi: 10.1038/sj.onc.1207221. [DOI] [PubMed] [Google Scholar]
- 46.Fan Y, Gelinas C. An optimal range of transcription potency is necessary for efficient cell transformation by c-Rel to ensure optimal nuclear localization and gene-specific activation. Oncogene. 2007;26:4038–43. doi: 10.1038/sj.onc.1210164. [DOI] [PubMed] [Google Scholar]
- 47.Starczynowski DT, Reynolds JG, Gilmore TD. Mutations of tumor necrosis factor alpha-responsive serine residues within the C-terminal transactivation domain of human transcription factor REL enhance its in vitro transforming ability. Oncogene. 2005;24:7355–68. doi: 10.1038/sj.onc.1208902. [DOI] [PubMed] [Google Scholar]
- 48.Tran K, Merika M, Thanos D. Distinct functional properties of IkappaB alpha and IkappaB beta. Mol Cell Biol. 1997;17:5386–99. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koster MI, Lu SL, White LD, et al. Reactivation of developmentally expressed p63 isoforms predisposes to tumor development and progression. Cancer Res. 2006;66:3981–86. doi: 10.1158/0008-5472.CAN-06-0027. [DOI] [PubMed] [Google Scholar]
- 50.Crook T, Nicholls JM, Brooks L, et al. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene. 2000;19:3439–44. doi: 10.1038/sj.onc.1203656. [DOI] [PubMed] [Google Scholar]