Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a recently discovered DNA tumor virus that belongs to the γ-herpesvirus subfamily. Though numerous studies on KSHV and other herpesviruses, in general, have revealed much about their multilayered organization and capsid structure, the herpesvirus capsid assembly and maturation pathway remains poorly understood. Structural variability or irregularity of the capsid internal scaffolding core and the lack of adequate tools to study such structures have presented major hurdles to earlier investigations employing more traditional cryo-electron microscopy (cryoEM) single particle reconstruction. In this study, we used cryo-electron tomography (cryoET) to obtain three-dimensional reconstructions of individual KSHV capsids, allowing direct visualization of the capsid internal structures and systematic comparison of the scaffolding cores for the first time. We show that B-capsids are not a structurally homogenous group; rather, they represent an ensemble of “B-capsid-like” particles whose inner scaffolding is highly variable, possibly representing different intermediates existing during the KSHV capsid assembly and maturation. This information, taken together with previous observations, has allowed us to propose a detail pathway of herpesvirus capsid assembly and maturation.
Keywords: cryo-electron tomography, tumor herpesvirus, Kaposi’s sarcoma-associated herpesvirus, assembly, scaffolding, portal
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a DNA tumor virus and the most recently discovered human pathogen in the Herpesviridae family (Chang et al. 1994). It is the causative agent of all forms of Kaposi’s sarcoma and closely linked to AIDS-associated lymphomas, including primary effusion lymphoma and multicentric Castleman’s disease (Ganem 1998). KSHV and Epstein-Barr virus (EBV) are the two known human pathogens of the gammaherpesviruses subfamily of herpesviradae (Chang et al. 1994; Ganem 1997; Damania et al. 1999; Mueller 1999; Damania et al. 2000; Roizman and Pellett 2001; Carbone 2003). Like other herpesviruses, the KSHV virion consists of a double-stranded (ds) DNA genome enclosed within an icosahedral capsid shell, a thick, proteinaceous tegument compartment, and a lipid bilayer envelope spiked with glycoproteins (Rixon 1993; Steven and Spear 1997; Liu and Zhou 2005). Genetic and biochemical studies have suggested the existence of a bacteriophage-like DNA-packaging/ejection portal presumably located at one of the 12 vertices (Newcomb et al. 2001b), which has been confirmed recently by cryo-electron tomography (cryoET) studies of native KSHV capsids (Deng et al. 2007), although the exact orientation and radial placement of the portal remains controversial in herpes simplex virus (HSV-1) capsids (Trus et al. 2004; Cardone et al. 2007; Chang et al. 2007).
Since the 1970s, the scientific community has had biochemical evidence that herpesviruses form three distinct capsids during assembly in the host nucleus, designated A, B, and C-capsids, which at the time were only known as different bands, conveniently labeled alphabetically as “A”, “B” and “C”, according to the order of their positions on a density gradient. These particles were later discovered via protein analysis and negative stain electron microscopy to be an empty capsid shell (A-capsid), A-capsid containing scaffolding protein, and A-capsid containing the viral genome, respectively. In 1996, a fourth capsid category was discovered, termed the procapsid, that was different from A, B, and C-capsids in that it was spherical, rather than angular, and porous, rather than sealed (Newcomb et al. 1996). Procapsids are assembled upon a skeleton comprised of large numbers of the internal scaffold protein, SCAF (assembly protein [AP] in HSV-1). The procapsid spontaneously angularizes shortly after it forms, but exactly when this angularization occurs and whether or not A and B-capsids are both possibly dead-end products resulting from angularization at an inappropriate time is still under debate. At an indeterminate time following capsid assembly, the viral protease, PRO (Pr in HSV-1 and HCMV), cleaves the inner SCAF proteins, resulting in capsid angularization. Little is currently known about the roles of PRO and its cleavage of SCAF in KSHV assembly, partly due to the lack of any structural information regarding their organization inside KSHV capsids. The protease also disassembles the scaffolding protein complex within the capsid. When exactly this occurs is unclear, but Newcomb et al. in 2000 found that HSV-1 procapsids contain more scaffolding protein than B-capsids (Newcomb et al. 2001a), indicating perhaps that some scaffolding is lost during angularization. Either concurrent with or immediately after the cleaved scaffolding proteins leave the capsid, the viral genome is packaged into the capsid. The mechanism by which this packaging occurs likewise remains unclear, though the linear double stranded genome likely enters through the portal, a structurally unique vertex in the capsid (Newcomb et al. 2001b; Trus et al. 2004; Cardone et al. 2007; Chang et al. 2007; Deng et al. 2007).
With the advent of cryo-electron microscopy (cryoEM) in the late 1980s, it became possible to reconstruct the A, B, and C-capsids and procapsids and in three dimensions via single-particle reconstruction and icosahedral averaging, in which hundreds to thousands of particles are averaged together to overcome the low signal-to-noise ratio typical of cryoEM (Wu et al. 2000; Zhou et al. 2000; Trus et al. 2001; Heymann et al. 2003; Yu et al. 2003). Such studies have succeeded in generating clear reconstructions of the highly iterative and ordered capsid shell, but in the case of B-capsids, signal from the inner scaffolding was averaged out in reconstructions that failed to reveal any ordered internal structure, presumably due to either lack of icosahedral symmetry or variable numbers of scaffolding core structures among the individual capsids. Gammaherpesviruses have provided even more of a challenge as both EBV and KSHV are extremely difficult to grow in cell culture; KSHV, for example, requires chemical treatment to induce lytic replication (Renne et al. 1996). Nealon et al. were able to provide biochemical evidence that the amount of scaffolding protein in B-capsids seems to be variable, but there was still no definitive evidence clarifying the organization of scaffolding protein within the B-capsids (Nealon et al. 2001). In light of the above considerations, it became clear that cryoEM is inadequate for revealing the internal structures of the B-capsid, and thus we turned to cryoET.
Where cryoEM requires different particles for 3D reconstruction, cryoET can reveal 3D structures of an individual particle by reconstructing it using a tilt-series, or a series of micrographs taken at different tilt angles. The advantage cryoET holds over cryoEM is that it can extract structural information from even one single particle, structures unique to that particle will not get averaged out as they would by cryoEM after integrating signals from a population of particles analyzed. In this study, we used cryoET to reconstruct individual KSHV capsids, and show that B-capsids are actually a heterogeneous collection of B-capsids-like, or “B-type” capsids, with highly variable inner scaffolding formations that form hollow, spiky spheres. These spheres are sometimes situated in the center of the capsid, and sometimes adjacent to the inner wall opposite to the portal complex. Other times, the scaffolding appears disorganized and lacks any visible sphere-shape. Integrating this information, we have developed an updated model of herpesvirus capsid assembly and maturation.
Results and Discussion
KSHV A, B, and C-capsids
The difficulties in isolating human tumor herpesvirus capsids for structural studies have been well recognized (Wu et al. 2000; Nealon et al. 2001). In order to establish statistical significant observations, we collected a total of 33 cryoET tilt series containing 297 ice-embedded KSHV capsids. One example is provided Fig. 1. A total of 297 capsids were examined. 96 were A-capsids, 198 were B-capsids, and only 3 were C-apsids. Five high quality tomograms with 51 KSHV capsids were obtained using the marker-free alignment program developed by Winkler and Taylor (see details in Materials and Methods) (Winkler and Taylor 2006) as judged by the clear resolution of the unique portal complex in one of the twelve vertices (Fig. 1c). In addition, the eleven pentons and 150 hexons making up the capsid shell are clearly resolved and are similar to those obtained by cryoEM icoshaedral reconstruction (Fig. 1c) (Wu et al. 2000; Trus et al. 2001).
Figure 1.
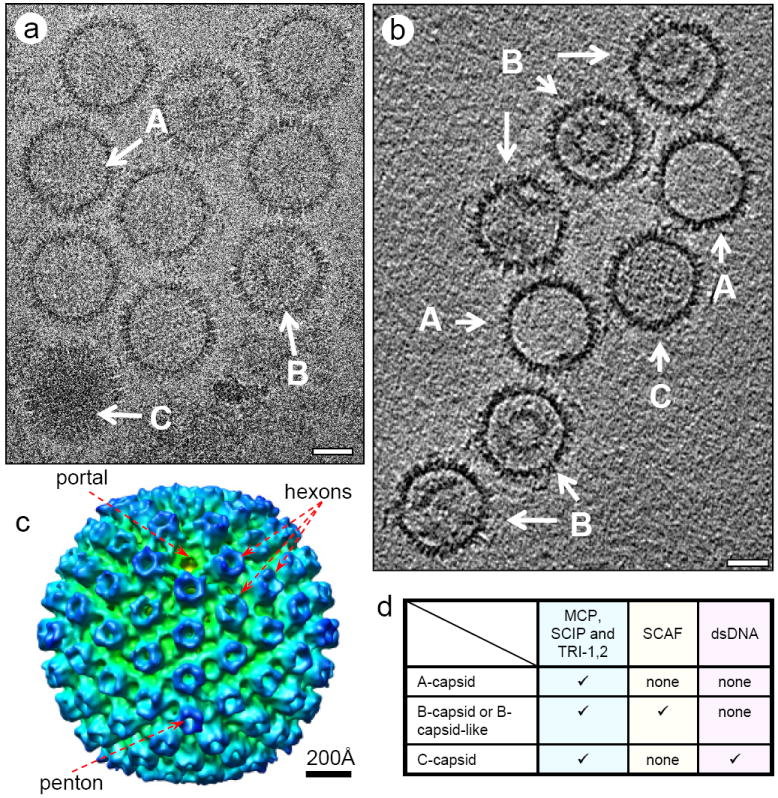
KSHV A, B and C-capsid. (a) electron micrograph and (b) central slice of a tomogram showing the KSHV A, B and C-capsid. (c) Radially colored surface representation of an averaged KSHV capsid showing the characteristic herpesvirus capsomers, including an “umbilical” portal, 11 pentons and 150 hexons. (d) components of KSHV A, B and C-capsid. Scale bar: 500 Å.
Our 3D tomograms reveal the existence of at least three classes of KSHV capsids. The capsid shell is practically identical from one class to the next, and the classes are only distinguished by their inner contents. One is completely empty; another has some density that sometimes forms a sphere structure and sometimes not; and the last is filled with a packed, uniform density (Fig. 1b). These classes are named A, B, and C-capsids, in ascending order of molecular density and corresponding to those established by density gradient and negative stain EM observation (Nealon et al. 2001). The characterizing chemical differences between these capsids are summarized in the table of Figure 1d. Biochemical and mass spectrometry studies have shown that all three capsid forms contain the proteins that make up the capsid shell, including major capsid protein (MCP), small capsomer interacting protein (SCIP), triplex monomer protein (TRI-1) and triplex dimer protein (TRI-2) (Nealon et al. 2001; O’Connor and Kedes 2006). It is generally believed that the A-capsid contains only capsid shell proteins, indicating it is an empty shell; B-capsids contain capsid proteins, SCAF (scaffolding protein) and PRO, and C-capsids contain capsid shell proteins and dsDNA only but no SCAF. The density slice from the 3D cryoET tomograms of KSHV capsid preparation clearly confirms the existence of all three types of capsid forms (Fig. 1b).
Individual A, B, and C-capsids were computationally extracted from the 3D tomogram volumes in order to examine them interactively in greater detail (Fig. 2). Most notably, the scaffolding core density in the B-capsid resembled the shape of a gear, with an inner shell diameter of 420 Å decorated by spiky outer densities (Fig. 2b). The spherically averaged density profiles plotted as a function of particle radius (Fig. 2d-f) show that A-capsids have one density peak between 480 Å and 630 Å, corresponding to the capsid shell. B-capsids have two additional peaks that range from ~190Å to ~400Å and represent the scaffolding protein inner sphere and outer spikes. C-capsids have one density peak as A-capsids do, but their density distribution between zero and ~400 Å is markedly higher than that of A-capsids. Due to the inherently limited resolution of current cryoET technology and small spacing between adjacent double helices, the details of the organization of dsDNA within C-capsids remain elusive, but our tomograms and reconstructions clearly show that the dsDNA is evenly distributed within the KSHV capsid shell. In a sample size of 51 particles, 15 were A-capsids, 34 were B-capsids, and only 2 were C-capsids, which resembles the ratio Nealon et al. reported (Nealon et al. 2001).
Figure 2.

CryoET of the KSHV A, B and C-capsid. (a-c) Serial density slices of a 3D cryoET reconstruction of KSHV A-capsid (a), B-capsid (b) and C-capsid (c). (d-f) Radial density profiles of KSHV A, B and C-capsid. Radial density profiles are calculated by spherical averaging of the 3D density maps. KSHV A, B and C-capsids share a major peak between a radii of 480-630Å. A-capsid (d) does not have apparent density inside. B-capsid (e) has two additional peaks, the higher of which is between a radii of 190-320Å, and the lower one is between a radii of 320-400Å. C-capsid (f) has continuous density distribution inside. (g-i) cut-away shaded surface view of KSHV A, B, and C-capsid. In (h) and (i), the internal structure of a B (scaffolding protein) and a C-capsid (dsDNA) are put into the capsid shell of an averaged A-capsid (g), respectively. Portal complex is highlighted in green. Scaffold is colored in golden (h) and dsDNA is colored in magenta (i). Scale bar: 500 Å
CryoET Reveals Dynamic or Variable Scaffolding Structures in “B-type” Capsids
Neither A nor C-capsids revealed much structural variation, consistent with prior data. In contrast, capsids with internal scaffolding cores exhibited great variation in these internal structures within the population. Thus, the B-capsids are, in reality, a heterogeneous collection of similar but non-identical particles, each potentially containing different amounts and configurations of SCAF. Figure 1b illustrates five B-type capsid particles, only two of which contain the sphere (or ring) structure previously thought to be characteristic of B-capsids. In some examples of B-type capsids, the scaffolding proteins form large, centrally situated hollow spiky spheres (Fig. 3a); in other examples the spheres are small and tucked away near the capsid wall opposite the portal complex (Fig. 3b), and in yet another variation, the scaffolding protein forms no sphere at all (Fig. 3c). Based on the variety of scaffolding arrangement apparent in our tomograms, we have elected to divide these B-type capsids further into three sub-categories: central sphere, off-center sphere, and non-sphere (Fig 3).
Figure 3.
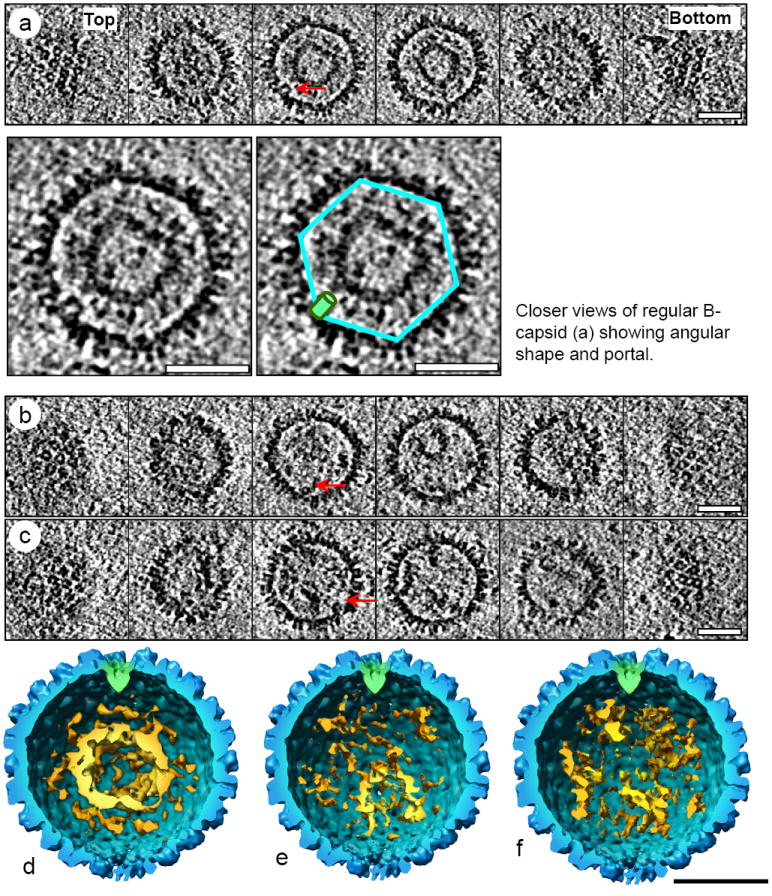
Three representative subgroups of KSHV “B-type” capsids. (a-c) Serial density slices of the 3D cryoET reconstruction KSHV capsids containing central sphere scaffolding core (a), or off-center sphere scaffolding core (b), or non-sphere scaffolding core (c). Arrows point to the portal complex in each capsid. (d-f) Cut-away shaded surface view of KSHV central sphere (d), off-center sphere (e) and non-sphere B-type capsid (f). The internal scaffolding core densities in each capsid are put into the capsid shell of an averaged capsid. Portal complex is highlighted in green. Scaffold is colored in golden. In the off-center sphere B-type capsids (b and e), the scaffold sphere is located away from the portal complex. Scale bar: 500 Å
The scaffolding protein in the traditionally defined B-capsids, or regular B-capsid, contains a core of SCAF in the form of a concentric spiky hollow sphere that is centered within the capsid. These inner spheres are approximately 75 Å thick and 200 Å in diameter (Fig. 3a and 3d). Another version of a SCAF-containing particle is the off-center sphere B-capsid that contains a smaller spiky, hollow sphere usually located eccentrically relative to the capsid shell and varying greatly in size (Fig. 3e). Interestingly, these spheres tend to be situated opposite of the portal complex, but the significance for this localization remains unclear. There is also additional density between the sphere and the capsid shell that may be unorganized or disintegrating scaffolding protein. The last group of B-type capsids has no discernable sphere-structure, but fills the capsid with densities that clump together in a way that suggests they may be SCAF sphere fragments (Fig. 3c and 3f). Out of our relatively small sample size of 34 B-type capsids, we found these sub-categories in a 16:14:4 (central: off-center: non-sphere) ratio. Because our current cryoET analysis only represents a snapshot of KSHV capsid distribution at a given time, it is not possible to distinguish whether the “irregular” B-capsids, i.e., the off-center sphere and the non-sphere-containing capsids, are precursors of regular B-capsids or subsequent capsid species originated from regular B-capsids. The fact that more regular B-capsids were found points to their abundance at the time of sample freezing and may also imply that they are a more stable intermediate capsid form if such observation is independent of time. It remains unclear whether they could extrude their inner contents even after angularization, as opposite to the view that angularization somehow “locks” the scaffolding protein in and prevents its escape. These important issues await further structural investigation of B-type capsids using time-resolved cryoET reconstructions similar to time-resolved cryoEM reconstructions of HSV-1 capsids (Heymann et al. 2003).
“Rope” Mechanism for Scaffolding
Our data showing a more or less fairly contiguous distribution of a fully-intact scaffold to a disintegrated, structureless conglomeration of protein lends support to the argument that B-capsids are an ensemble of assembly intermediates. The apparent sphere structure shows that the scaffolding protein is indeed structured, at least in a subset of B-capsids. However, the standard cryoEM fails to reconstruct this inner portion when integrating images from multiple similar sphere containing capsids, which indicates that these inner portions are not concretely fixed, or in register with, the icosahedral capsid shell. This new structural information, together with previously published data of herpesvirus capsid assembly, allows us to suggest a possible “rope” mechanism of herpesvirus scaffolding (Fig. 4a-b).
Figure 4.
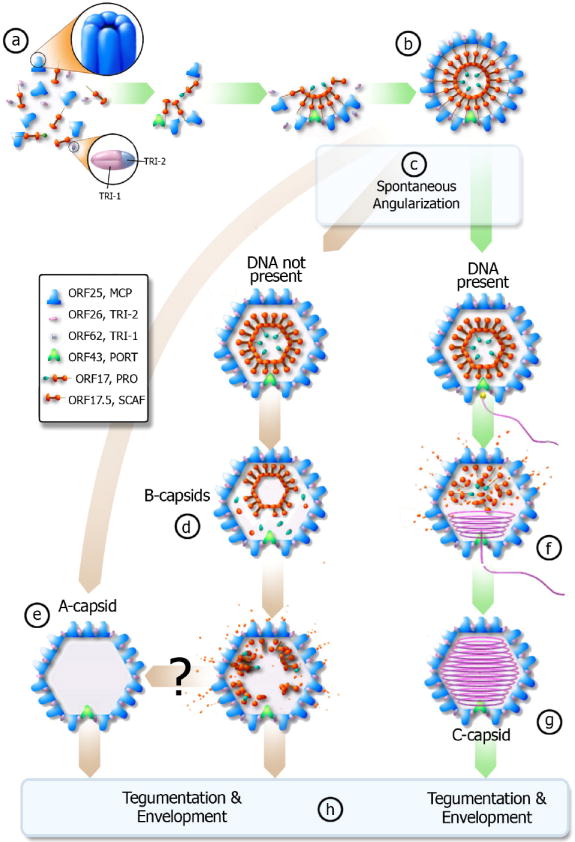
Proposed pathway of KSHV capsid assembly pathway – green arrows denote productive pathway (can create infectious virion), brown arrows denote unproductive pathway. (a) The scaffolding protein (orange) associates with MCP (blue). The scaffolding proteins then interact with each other and pull the MCP together through the “rope” mechanism (see text). Triplexes (pink and blue) bind the MCP together to form a spherical procapsid (b). If DNA is present and available when the procapsid spontaneously angularizes (c), then the DNA will enter the capsid via the portal complex (f) and the scaffold protein will evacuate the capsid to form a C-capsid (g). If the cleavage occurs prematurely and DNA is not present, the cleavage product can likely exit from the porous procapsid shell to result in A-capsid (e). If cleavage occurs slower than angularization, the scaffolding core will likely collapse and some scaffold protein will still evacuate the capsid while forming B-type capsid intermediates (d). It remains unclear whether B-capsid can eventually become an empty A-capsid (e). C-capsids will then obtain a tegument layer and an envelope to become an infectious virion while A and B-capsids lead to non-infectious enveloped particles (h).
Our cryoET tomograms clearly suggest that SCAF has an extended conformation extending a distance of approximately 200 Å (Fig. 2b and 2e). It has been shown that, in the absence of SCAF, MCP and triplex proteins can self assemble into capsid-like particles, but their size is much smaller than native capsids, suggesting that the role of SCAF is to define the size and curvature of assembled MCP intermediates (Saad et al. 1999; Newcomb et al. 2001a). Biochemical and structural data from other herpesviruses have shown that one end (C terminal) of SCAF interacts with MCP (Hong et al. 1996; Zhou et al. 1998). Our new cryoET data presented here demonstrates that the SCAF also, perhaps at its other end, has a tendency to self associate into a ring/sphere core, thus playing the role of a “rope”. In this “rope” model, scaffolding protein does not form a rigid, inflexible skeleton or template upon which capsid proteins assemble themselves, but instead acts as tethers to draw or pull MCP and TRI-1 and -2 within range of each other. The C-terminal end of SCAF reaches to the interface of MCP at the floor domain and they are drawn to close proximity to a critical concentration to promote inter- MCP interactions, thus ensuring the proper curvature and size of assembly intermediates and procapsid (Fig. 4a-b).
Plausible Pathway of Capsid Assembly and Maturation
We propose that KSHV capsid proteins originate as associated MCP-scaffold protein pairs that then bind together to serve as the basic building block of a procapsid (Fig. 4a). A quirk about herpesvirus capsid assembly is that studies have shown that the portal complex is not necessary for capsid assembly (Tatman et al. 1994; Thomsen et al. 1994; Zhou et al. 1998; Newcomb et al. 2001a; Newcomb et al. 2005). If assembling capsid proteins bind without preference to portal complexes, we should be able to observe capsids with a variable number of portal complexes, yet our data shows that each capsid still emerges with one and only one portal complex. Out of 49 capsids, we were able to identify a unique portal vertex in 40 of them. The remaining nine had vertices that fell within the “missing wedge” of information resultant from the cryoET technique. The reason for each capsid containing only one portal remains entirely unclear, but the supposition that the portal complex serves as a nucleating center in procapsid formation would be consistent with our data (Newcomb et al. 2005). In this case, portal-less capsid remains a possible outcome, but the predominant, energetically favorable, and non-abortive species in infected cells would be those with a single portal complex.
Therefore, in this model a portal complex initiates the capsid assembly through its interaction with MCP that, in turn, interacts with the C terminal helix of SCAF (Zhou et al. 1998). SCAF then brings together additional MCP molecules and ensures the proper curvature and capsid size through a “rope” mechanism as described above. Heterotrimers of ORF26 and ORF62 then clamp the shell together to form the porous, spherical procapsid (Fig. 4b). At this stage, the procapsid angularizes (Fig. 4c) and encounters a crossroad in its development with its fate dependent on the availability of nascent viral DNA and the timing of scaffolding cleavage as governed by the spontaneous assembly pathway first proposed in human cytomegalovirus (HCMV) capsid assembly (Yu et al. 2005). If there is no DNA present, protease is released and “cuts loose” the scaffolding protein from the capsid shell, causing the scaffolding protein to collapse on itself. The scaffolding protein then continues to degrade into sufficiently small peptides to allow free exit from the capsid via open channels in the capsid shell (Fig. 4d). The end result of this pathway is an empty A-capsid. In contrast, if there is DNA present at angularization, then the DNA enters via the portal complex (Fig. 4f) and most likely is condensed via a packaging mechanism in the portal complex to liquid crystal density to form a C-capsid (Fig. 4g). All three capsids (A, B, and C) can receive a layer of tegument protein and a glycoprotein-studded envelope, but only the C-capsid, has the potential to become a mature, infectious virion. Enveloped A and B-capsids will become non-infectious enveloped particles (NIEP) that arise during infections as first described for HCMV (Gibson 1996).
Material and Methods
KSHV capsid isolation
KSHV capsids were purified from the media of lytically induced KSHV-infected BCBL-1 cells as previously described (Wu et al. 2000; Nealon et al. 2001). An aliquot of 3-μl of the purified capsids was placed onto a carbon-coated holey grid and quickly frozen to liquid-nitrogen temperature so that the capsid particles were embedded in a thin layer of vitreous ice.
Cryo-electron tomography and 3D structural analyses
Tilt series were recorded with an electron dose of ~1 electron/Å2/micrograph at a magnification of 38,200×, pixels size of 7.9A/pixel, and 8 μm under-focus in a FEI 300 kV G2 Polara cryo-electron microscope (FEI Co., Hillsboro, Oregon), which is equipped with a 16-megapixel CCD camera and the EMMENU data acquisition software (TVIPS GmbH, Gauting, Germany). Image data were recorded from -70° to 70° with a 2 degree interval. Data processing was carried out using a marker-free alignment approach and back-projection 3D reconstruction method (Winkler and Taylor 2006). Briefly, a preliminary tomogram was obtained from the course-aligned images. Translational and rotational alignment, and tilt axis refinement were subsequently performed using the preliminary tomogram. Several rounds of refinement of alignment and area matching were carried out until no significant improvement in the alignment parameters and the 3D tomogram was detectable. The final 3D map was then computed using weighted back projection algorithm (Radermacher 1992). The final 3D tomograms were low-passed filtered to 50-Å resolution using a Gaussian type of filter. For averaging of cryoET reconstructions, 3D alignment of individual capsid reconstructions was first manually estimated and subsequently refined computationally by using the Foldhunter program (Jiang et al. 2001). Surface and volume rendering were performed using the Chimera visualization software package (Pettersen et al. 2004).
Acknowledgments
We thank Mr. Ivo Atanasov for technical assistance in imaging and Ms. Xiaorui Zhang for graphical and editorial assistance. This research is supported in part by NIH grants CA094809 and AI069015 (ZHZ) and CA088768 (DHK). BD is supported by a training fellowship from the Keck Center Pharmacoinformatics Training Program of the Gulf Coast Consortia (NIH Grant No. 1 T90 DK070109-01 and 1 R90 DK071505-01).
Footnotes
Structure Deposition The cryoET density maps are being deposited to the Protein Data Bank and EBI under accession code XXXX and XXXX, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Carbone A. Emerging pathways in the development of AIDS-related lymphomas. Lancet Oncol. 2003;4:22–29. doi: 10.1016/s1470-2045(03)00957-4. [DOI] [PubMed] [Google Scholar]
- Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Damania B, Choi JK, Jung JU. Signaling activities of gammaherpesvirus membrane proteins. J Virol. 2000;74:1593–1601. doi: 10.1128/jvi.74.4.1593-1601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, Lee H, Jung JU. Primate herpesviral oncogenes. Mol Cells. 1999;9:345–349. [PubMed] [Google Scholar]
- Deng B, O’Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi’s sarcoma-associated herpesvirus capsid by cryoelectron tomography. J Virol. 2007;81:3640–3644. doi: 10.1128/JVI.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. KSHV and Kaposi’s sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- Ganem D. Human herpesvirus 8 and its role in the genesis of Kaposi’s sarcoma. Curr Clin Top Infect Dis. 1998;18:237–251. [PubMed] [Google Scholar]
- Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong AD. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhou ZH. Comparative virion structures of human herpesviruses. In: Arvin A, Campadelli-Fiume G, Moore P, et al., editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge, UK: Cambridge University Press; 2005. In Press. [PubMed] [Google Scholar]
- Mueller N. Overview of the epidemiology of malignancy in immune deficiency. J Acquir Immune Defic Syndr. 1999;21:S5–10. [PubMed] [Google Scholar]
- Nealon K, Newcomb WW, Pray TR, Craik CS, Brown JC, Kedes DH. Lytic replication of Kaposi’s sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J Virol. 2001;75:2866–2878. doi: 10.1128/JVI.75.6.2866-2878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Brown JC. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J Virol. 2005;79:10540–10546. doi: 10.1128/JVI.79.16.10540-10546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Thomsen DR, Brown JC. In vitro assembly of the herpes simplex virus procapsid: formation of small procapsids at reduced scaffolding protein concentration. J Struct Biol. 2001a;133:23–31. doi: 10.1006/jsbi.2001.4329. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001b;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Kedes DH. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J Virol. 2006;80:1574–1583. doi: 10.1128/JVI.80.3.1574-1583.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Electron Tomography - Three-dimensional imaging with the Transmission Electron Microscopy. In: Frank J, editor. Weighted back-projection methods. New York: Plenum Press; 1992. pp. 91–115. [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Rixon FJ. Structure and assembly of herpesviruses. Seminars in Virology. 1993;4:135–144. [Google Scholar]
- Roizman B, Pellett PE. Fields Virology. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Herpesviridae: A Brief Introduction. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2381–2398. [Google Scholar]
- Saad A, Zhou ZH, Jakana J, Chiu W, Rixon FJ. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. Journal of Virology. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Spear PG. Structural Biology of Viruses. In: Chiu W, Burnett RM, Garcea R, editors. Herpesvirus capsid assembly and envelopment. New York: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- Tatman JD, Preston VG, Nicholson P, Elliott RM, Rixon FJ. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- Thomsen DR, Roof LL, Homa FL. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. Journal of Virology. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Heymann JB, Nealon K, Cheng N, Newcomb WW, Brown JC, Kedes DH, Steven AC. Capsid structure of Kaposi’s sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J Virol. 2001;75:2879–2890. doi: 10.1128/JVI.75.6.2879-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Wu L, Lo P, Yu X, Stoops JK, Forghani B, Zhou ZH. Three-dimensional structure of the human herpesvirus 8 capsid. J Virol. 2000;74:9646–9654. doi: 10.1128/jvi.74.20.9646-9654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Trang P, Shah S, Atanasov I, Kim YH, Bai Y, Zhou ZH, Liu F. Dissecting human cytomegalovirus gene function and capsid maturation by ribozyme targeting and electron cryomicroscopy. Proc Natl Acad Sci U S A. 2005;102:7103–7108. doi: 10.1073/pnas.0408826102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XK, O’Connor CM, Atanasov I, Damania B, Kedes DH, Zhou ZH. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J Virol. 2003;77:13182–13193. doi: 10.1128/JVI.77.24.13182-13193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Macnab SJ, Jakana J, Scott LR, Chiu W, Rixon FJ. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci U S A. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]


