Abstract
We have previously shown that Singleminded-2s (SIM2s), a member of the basic helix-loop-helix Per-Arnt-Sim (bHLH/PAS) family of transcription factors, is downregulated in breast cancer samples and has tumor suppressor activity. However, the mechanism by which SIM2s is repressed in breast cancer cells has not been determined. In this study, we show that transformation of MCF10A cells by Harvey-Ras (Ha-Ras) induces CCAAT/enhance binding protein β (C/EBPβ) and activates the NOTCH signaling pathway to block SIM2s gene expression. NOTCH-mediated repression acts through a C-repeat binding factor 1 (CBF1)-independent mechanism, as introduction of CBF1 had no effect on SIM2s expression. Consistent with C/ebpβ-dependent inhibition of SIM2s, C/ebpβ−/−mouse mammary glands express high levels of SIM2s and reestablishment of C/ebpβ isoforms decreased SIM2s mRNA levels in C/ebpβ immortalized mammary epithelial cell lines. These studies illustrate a novel pathway of tumor suppressor gene silencing in Ha-Ras-transformed breast epithelial cells and identify SIM2s as a target of C/EBPβ and NOTCH signaling.
Keywords: Singleminded-2s, NOTCH, C/EBP-beta, mammary gland
Introduction
Singleminded-2 (SIM2) is a member of the basic helix-loop-helix Per-Arnt-Sim (bHLH/PAS) family of transcription factors (Kewley et al., 2004). SIM2 is one of two mammalian orthologs of the Drosophila single-minded (sim) gene, which regulates central midline development (Crews et al., 1992). Previous studies have demonstrated that SIM2 is differentially expressed in prostate, colon and pancreatic cancers (Deyoung et al., 2002; Aleman et al., 2005; Halvorsen et al., 2007). We have shown that a short splice variant of SIM2, SIM2s, is expressed in human breast epithelial cells and is downregulated in primary human breast cancer samples (Kwak et al, 2007). Reestablishment of SIM2s in highly invasive breast cancer cells significantly inhibited growth and motility, suggesting that SIM2s is a breast tumor suppressor gene. Indeed, loss of SIM2s causes aberrant mammary ductal development with features suggestive of malignant transformation including increased proliferation, changes in polarity, downregulation of E-cadherin and invasion into the surrounding stroma (Laffin et al., 2008). Furthermore, short-hairpin RNA (shRNA)-mediated reduction of SIM2s (SIM2i) in MCF7 human breast cells promotes an epithelial-mesenchymal transition (EMT) and increases tumorigenic potential. These results suggest that SIM2s is required for normal mammary ductal development and inhibition of SIM2s is permissive for acquisition of an invasive mesenchymal phenotype; however, the mechanism by which SIM2s is suppressed during breast cancer progression has not been determined.
The Ras proto-oncogene is an important effector molecule that is essential for growth-dependent induction of numerous growth factors and cytokines. Although mutations in Ras are rare in breast cancers (Khleif et al., 1999), Ras is inappropriately activated in 50% of breast tumors and positively correlates with early neoplasia and poor prognosis (von Lintig et al., 2000). Ras activation is also associated with ERBB2-overexpressing breast tumors, which represent 30% of breast cancer cases (Slamon et al., 1989). Harvey-Ras (Ha-Ras)-induced tumors are characterized by activation of mitogen-activated protein kinase signaling and are cyclin D1 dependent (Yu et al., 2001; Dunn et al., 2005). In addition, by activating oncogenic pathways, Ha-Ras overexpression, driven by the mouse mammary tumor virus promoter, induces mammary adenocarcinomas (Sinn et al., 1987; Nielsen et al., 1991). Two Ha-Ras targets associated with breast cancer are the NOTCH signaling pathway and the CCAAT/enhance binding protein β (C/EBPβ) transcription factor (Nakajima et al., 1993; Weijzen et al., 2002). Increased NOTCH signaling has been demonstrated in a variety of human breast carcinomas (Stylianou et al., 2006), whereas C/EBPβ has been identified as an important mediator of Ras-induced tumorigenesis in a mouse skin carcinogenesis model known to cause Ha-Ras mutations (Zhu et al., 2002; Shuman et al., 2004; Shim et al., 2005). In the studies presented here, we show that Ha-Rasmediated transformation of MCF10A normal breast epithelial cells leads to a decrease in SIM2s expression resulting from C/EBPβ induction and NOTCH signaling.
Results and discussion
Ha-Ras transformation of MCF10A decreases SIM2s gene expression
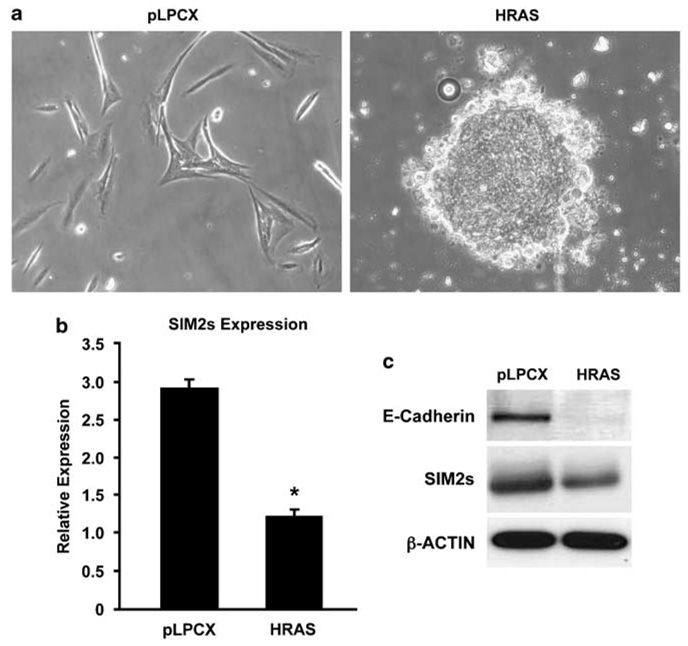
To define the mechanisms by which SIM2s is silenced in breast cancer, normal immortalized breast MCF10A cells were stably transduced with control vector or Ha-Ras expression constructs. Ha-Ras-MCF10A cells displayed hallmarks of transformation and EMT including a significant increase in invasion, anchorage-independent growth (data not shown), focus formation (Figure 1a) and downregulation of E-cadherin protein levels (Figure 1c) as compared to control cells. Evaluation of SIM2s expression in Ha-Ras-MCF10A cells, using real-time PCR and western blot analysis, showed a significant decrease in SIM2s mRNA (Figure 1a) and protein levels (Figure 1b), respectively. These results suggest that SIM2s is downregulated during Ha-Rasdependent transformation and pathways downstream of Ha-Ras suppress SIM2s gene expression.
Figure 1. Downregulation of Singleminded-2s (SIM2s) expression in HRAS-transformed MCF10A cells.
(a) Altered morphology of Harvey-Ras (Ha-Ras)-transformed MCF10A cells. Cells were transduced with either empty vector (pLPCX, left panel) or a vector containing the Ha-Ras coding sequence (HRAS, right panel). Differential contrast imaging of cells following 2 days of selection in medium containing 1 µg/ml puromycin, cells, showed that Ha-Ras-transduced cells lost their normal ovoid shape and displayed abnormal cellular organization. Ha-Ras construct was provided by Brian and Alana Welm and subcloned into EcoRI sites in pLPCX (Clontech). (b, c) SIM2s and E-cadherin expression is decreased in Ha-Ras-overexpressing MCF10A cells. Total RNA and protein were isolated from control (pLPCX) and Ha-Ras-overexpressing cells (HRAS) and analysed for SIM2s expression by quantitative real-time RT-PCR and western analysis (Chemicon) as previously described (Laffin et al., 2008). The results show that SIM2s and E-cadherin expression are significantly reduced in Ha-Ras-transduced MCF10A cells in comparison to control cells (mean ± s.e. for three replicates; *P=0.0002).
Reduction of SIM2s in MCF10A cells by shRNA induces an invasive EMT-like phenotype
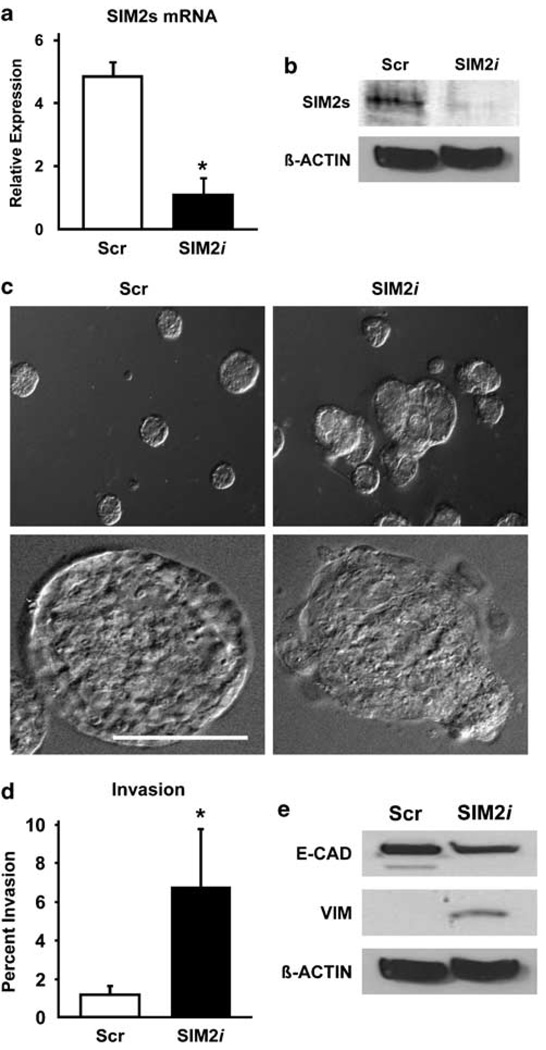
To determine if downregulation of SIM2s contributes to Ha-Ras-mediated transformation of MCF10A cells, we stably silenced SIM2s in MCF10A cells using a previously confirmed SIM2s-specific lentivirus shRNA construct (Laffin et al., 2008) (Figures 2a and b). When grown in three-dimensional cultures, control MCF10A cells formed regular, spherical acini, whereas a majority of SIM2i MCF10A acini were misshapen with cells budding into the surrounding matrix (Figure 2c). Loss of SIM2s also correlated with significantly increased in vitro invasive potential (Figure 2d). Furthermore, western blots showed that SIM2i-MCF10A cells had decreased E-cadherin and increased vimentin expression (Figure 2e). Together, these results show that reduction of SIM2s in MCF10A cells to levels observed in Ha-Ras-transformed MCF10A cells induces an invasive EMT-like phenotype.
Figure 2. Knockdown of Singleminded-2s (SIM2s) in MCF10A mammary epithelial cells by short-hairpin RNA (shRNA) disrupts three-dimensional acinar morphogenesis and induces a partial epithelial-mesenchymal transition (EMT).
(a, b) Inhibition of SIM2s expression in MCF10A cells by shRNA. MCF10A cells transduced with a confirmed SIM2-specific construct (SIM2s-sequence 3116; SIM2i) showed a significant reduction in SIM2s mRNA and protein as compared to a nonspecific control shRNA retroviral construct (Scr) (Laffin et al., 2008). (c) Low- and high-power differential interference contrast images of MCF10A acini grown on matrigel demonstrate that acinar morphogenesis is disrupted in SIM2i MCF10A cells as compared to Scr control. (d) SIM2i MCF10A cells display an increased ability to invade through a matrigel matrix using transwell invasion assays (Kwak et al., 2007). (e) Loss of SIM2s induces a partial EMT. Western blot analysis of Scr and SIM2i cells found that E-cadherin expression is reduced and vimentin expression is increased in SIM2i MCF10A cells as compared to Scr controls and is consistent with an EMT-like phenotype (Laffin et al., 2008). Data are represented as mean±s.e.m., asterisk (*) indicates P<0.05; Bar=100 µm.
Activation of NOTCH in Ha-Ras-MCF10A cells represses SIM2s through a CBF1-independent mechanism
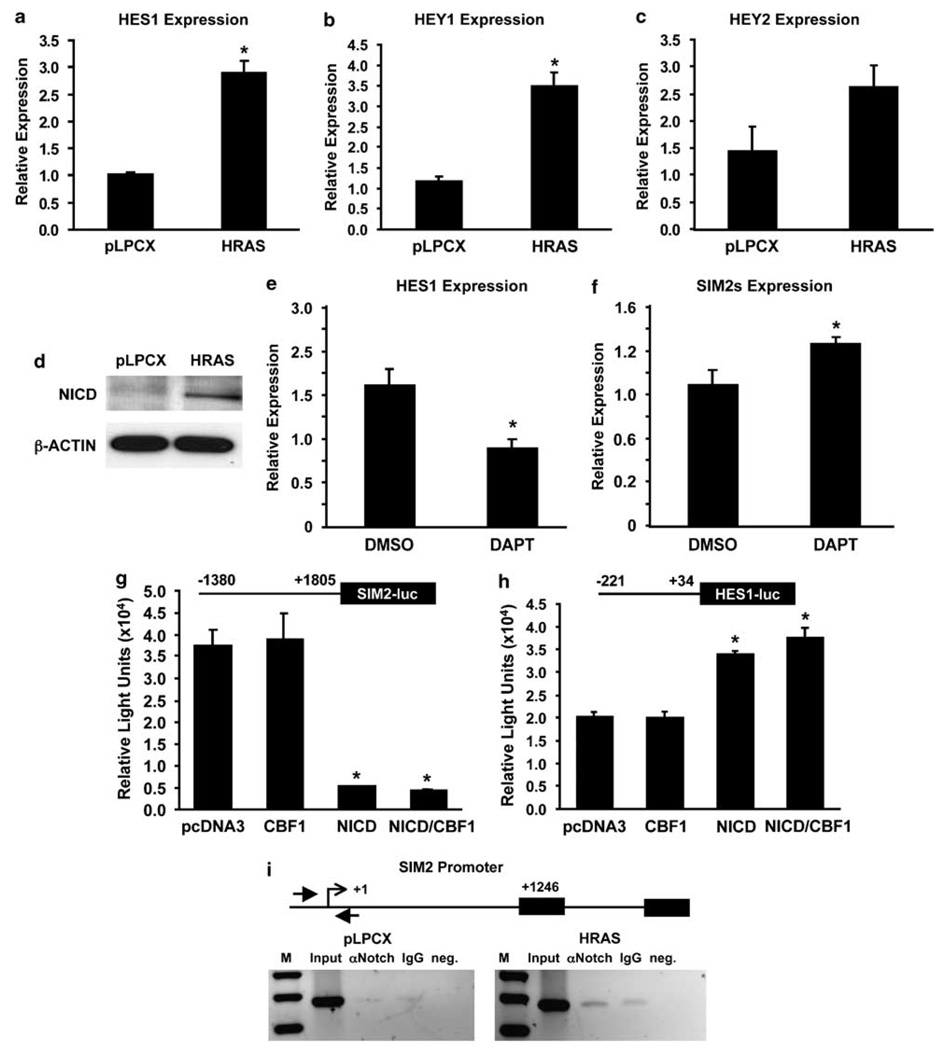
Increased NOTCH signaling has been demonstrated in a variety of human breast carcinomas (Stylianou et al., 2006). Studies have shown that Ha-Ras activates NOTCH, which is required to maintain the transformed phenotype in human cells (Weijzen et al., 2002). Activation of NOTCH signaling is an important mediator of Ras-induced tumorigenesis in the mouse mammary gland and is also implicated in stem cell maintenance (Weijzen et al., 2002; Kiaris et al., 2004; Sansone et al., 2007). Indeed, in Ha-Ras-MCF10A cells we observed a significant increase in the expression of known NOTCH target genes HES1, HEY1 and HEY2 by real-time PCR (Figures 3a–c) and detection of the NOTCH intracellular domain (NICD) by western blot analysis (Figure 3d), indicating that the NOTCH pathway is active in these cells. Furthermore, treatment with the γ-secretase inhibitor, DAPT (N-[N-(3,5-diffluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester), significantly blocked NOTCH-induced expression of HES1 and suppression of SIM2s gene expression (Figures 3e and f), implying that Ha-Ras-activated NOTCH signaling targets SIM2s.
Figure 3. Activation of NOTCH in Harvey-Ras (Ha-Ras)-MCF10A cells.
(a, b, c) Expression of NOTCH target genes in control and Ha-Ras-transduced MCF10A cells. Analysis of gene expression by real-time RT-PCR found that NOTCH target genes HES1, HEY1 and HEY2 are significantly higher in Ha-Ras-transduced cells compared to control MCF10A cells (*P<0.005). (d) Western blot analysis of Ha-Ras-MCF10A cells using a NOTCH intracellular domain (NICD)-specific antibody (Cell Signaling Technology) shows a significant increase in the active form of NOTCH. Together, these results suggest that the NOTCH pathway is activated by overexpression of RAS in MCF10A cells. (e, f) Treatment of Ha-Ras-MCF10A cells with 1 µm of the γ-secretase inhibitor, DAPT (N-[N-(3,5-difiuorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester) (Calbiochem), significantly blocked NOTCH-induced expression of HES1 and suppression of SIM2 gene expression (*P<0.05). (g, h) Effect of NICD and C-repeat binding factor 1 (CBF1) on HES1 and SIM2 promoter activity. Promoter reporter assays using either SIM2 or HES1 promoter constructs were (diagrammed above) cotransfected with vector control, NICD, CBF1 or both in HEK293T cells as previously described (Metz et al., 2006). The results show that HES1 promoter activity was significantly increased by NICD and NICD/CBF1 cotransfection compared to vector control, whereas SIM2 luciferase activity was suppressed by the NICD, independent of CBF1 (*P≤0.001). Data are expressed as the mean±s.e. for three wells per condition. The SIM2 promoter was amplified in two pieces with primers 5-ATCTGGGTAATCCCTTTCAAGCC-3 and 5-CCTGAGCTCCGAGCAACC-3, and 5-GTGGACAGCGGAGGTGCT-3 and 5-CCAAACCAAACCAGAATGC-3. Each piece was cloned onto pCR2.1 TOPO. The former fragment was subcloned onto pGL2 Basic into KpnI and XhoI restriction sites. The latter fragment was then subcloned into SacI and XhoI restriction sites to obtain the full-length SIM2 promoter on pGL2 Basic. (i) Binding of NOTCH to the SIM2 promoter. Chromatin immunoprecipitation (ChIP) analysis of the SIM2 promoter (Laffin et al., 2008) using a NOTCH1 antibody (Upstate) showed increased binding of NOTCH to the SIM2 promoter in MCF10A-HRAS cells as compared to control (pLPCX) cells (SIM2 primers FP: 5-GCCCACCCTGTGACCCTG-3, RP: 5-AAGTGACCCTTCTGCCCTTTC-3).
Previous studies have shown that the canonical NOTCH signaling pathway acts through C-repeat binding factor 1 (CBF1) to govern normal developmental processes (Bray, 2006). In addition, CBF1-independent mechanisms of NOTCH signaling that contribute to cell fate determination have also been identified (Martinez Arias et al., 2002). To evaluate the function of NOTCH signaling in regulation of SIM2s, a SIM2 promoter-luciferase reporter construct was cotransfected in the presence or absence of NICD or CBF1 expression vectors. NICD significantly repressed SIM2 promoter activity (P<0.0004); however, CBF1 had no effect on SIM2 promoter activity, suggesting a CBF1-independent mechanism of SIM2s repression by the NOTCH signaling pathway (Figure 3g). NICD function was confirmed using an HES1 promoter-controlled reporter. As expected, reporter activity was increased in cells cotransfected with either NICD or NICD plus CBF1, but not in CBF1 cells alone (Jarriault et al., 1995; Figure 3h). To determine whether SIM2 was a direct target of NICD, chromatin immunoprecipitation (ChIP) assays were employed to evaluate the region near the transcriptional start site of the SIM2 promoter (Figure 3i). The results show increased binding of NICD to the endogenous SIM2 promoter in Ha-Ras-over-expressing cells, suggesting that activation of NOTCH and release of the NICD by Ha-Ras contributes to inhibition of SIM2s expression.
C/EBPβ-dependent regulation of SIM2s in vitro and in vivo
C/EBPβ is a member of the basic leucine zipper family of transcription factors, which has an important function in mammary development and has recently been shown to be a critical mediator of Ras-induced tumorigenesis (Seagroves et al., 1998; Zhu et al., 2002; Grimm and Rosen, 2003). The C/EBPβ gene encodes a single 1.4 kb mRNA that, through alternative translation start sites, leads to production of three isoforms: LAP1, LAP2 and the dominant negative isoform, LIP. Both LAP isoforms contain the DNA binding and dimerization domains, whereas LIP is missing the transactivation domain and antagonizes LAP-mediated gene transcription by competing for LAP binding sites or by forming LIP/LAP heterodimers (Descombes and Schibler, 1991). The ratio of LAP to LIP has been proposed to have an important function in breast cancer (Zahnow, 2002; Grimm and Rosen, 2003). A low LIP/LAP ratio, indicating an increase in LIP expression, is associated with higher histological grading (Zahnow et al., 1997; Milde-Langosch et al., 2003) and has recently been shown to point toward a loss of transforming growth factor-β-cytostatic responses (Gomis et al., 2006). Indeed, overexpression of LIP in mouse mammary glands causes an increase in proliferation resulting in hyperplasias and invasive carcinoma (Zahnow et al., 2001). Although C/EBPβ is associated with breast cancer, its specific function in tumorigenesis is not well understood.
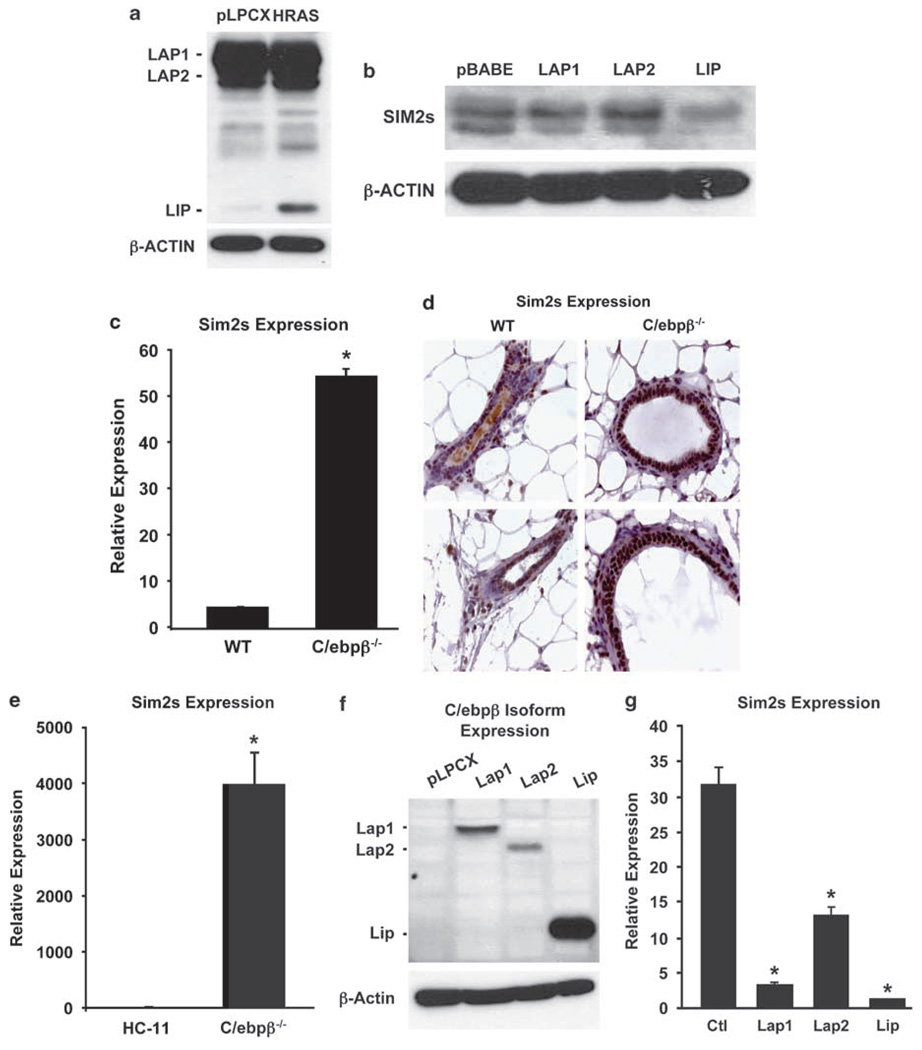
To determine the effect of Ha-Ras transformation on C/EBPβ, we used western blot analysis to evaluate levels of LAP1, LAP2 and LIP in Ha-Ras-MCF10A cells. These data revealed similar levels of LAP1 and LAP2; however, there was a significant increase in the truncated dominant negative isoform (LIP) in the Ha-Ras-MCF10A cells (Figure 4a). This is consistent with recent studies showing Ha-Ras-dependent induction of EGFR, which induces LIP (Martinez-Lacaci et al., 2000; Baldwin et al., 2004). To confirm if the increase in LIP alone alters SIM2s, MCF10A cells were stably transduced with control (pBABE), LAP1, LAP2 or LIP vectors. Western blot analysis revealed that LAP1 and LAP2 did not affect SIM2s levels, whereas overexpression of LIP leads to a reduction in SIM2s protein (Figure 4b).
Figure 4. Ha-Ras and developmental regulation of SIM2s by C/EBPβ.
(a) C/EBPβ isoform expression in Ha-Ras-MCF10A and control (pLPCX) MCF10A cells. Western blot analysis of Ha-Ras-MCF10A and control cell shows an Ha-Ras-dependent increase in C/EBPβ-LIP isoform expression (C/EBPβ antibody; Santa Cruz). (b) C/EBPβ-LIP-mediated suppression of SIM2s expression in MCF10A cells. Western blot for SIM2s (Chemicon) in MCF10A cells stably transduced with pBABE (vector control) or the indicated C/EBPβ isoforms shows a significant reduction of SIM2s protein levels in the LIP-transduced cells. Vectors (pBABEpuro or pEFIRESpuro backbones) containing human and mouse C/EBPβ isoforms were graciously provided by C Zahnow. (c, d) SIM2s expression in wild-type and C/ebpβ−/− mammary glands. Elevated SIM2s gene expression and protein levels in C/ebpβ−/− mammary glands as determined by real-time RT-PCR and immunohistochemistry, respectively (Laffin et al., 2008). (e) SIM2s expression in a C/ebpβ−/− mouse mammary epithelial cell line. Similar to the C/ebpβ−/− mammary glands, SIM2s levels are increased in mouse mammary epithelial cells derived from C/ebpβ−/− mammary glands (Lamb et al., 2003) by real-time RT-PCR as compared to normal HC11 cells (*P≤0.001). (f, g) Reestablishment of mouse C/ebpβ isoforms in the C/ebpβ−/− cells as determined by western blot analysis correlates with a decrease in SIM2s mRNA levels (*P≤0.001). Data are expressed as the mean±s.e. for three plates per condition.
C/ebpβ−/− mice are viable, but display immune system defects, abnormal brown adipose tissue function, skin irregularities and female infertility (Grimm and Rosen, 2003). In addition, mammary glands from C/ebpβ−/− mice have impaired ductal outgrowth and differentiation due to a disruption in cell fate determination (Seagroves et al., 1998). We have previously shown that SIM2s is expressed in luminal mammary epithelial cells and is required for proper ductal morphogenesis (Laffin et al., 2008). Using real-time PCR, we found that SIM2s is significantly increased in C/ebpβ−/− mammary glands when compared to wild-type mice (Figure 4c). The increase in SIM2s was confirmed at the protein level by immunohistochemistry and was found uniformly in the luminal epithelial cells of C/ebpβ null glands, compared to the more punctate expression pattern seen in wild-type glands (Figure 4d). To define further the relationship between C/ebpβ and SIM2s, we used an immortalized cell line established from mammary glands of C/ebpβ null virgin females. When compared to control cells, SIM2s expression was significantly increased in this in vitro model, consistent with the in vivo data. Furthermore, reestablishing each of the three C/ebpβ isoforms by lentiviral transduction (Figure 4f) led to a reduction in SIM2s expression (Figure 4g). Together, these results show that SIM2s is differentially regulated by C/EBPβ under both transforming and normal developmental conditions.
In summary, we have identified a pathway connecting Ha-Ras activation to SIM2s silencing through NOTCH and C/EBPβ. Ha-Ras has been shown to promote cleavage of NOTCH, releasing the active NICD (Weijzen et al., 2002), and to increase the LIP/LAP ratio (Nakajima et al., 1993). In addition, we also found that activation of NOTCH and C/EBPβ by Ha-Ras independently regulate SIM2s expression. We observed no changes in the LIP/LAP isoforms in Ha-Ras-MCF10A cells following treatment with a γ-secretase inhibitor (Supplementary Figure 1a), which we had previously shown to block RAS-mediated activation of HES1 and partially alleviated SIM2s suppression (Figures 3e and f), nor upregulation of NOTCH or NOTCH target genes HES1 and HEY1 in LIP-transduced MCF10A cells (Supplementary Figure 1b and c). We do not suggest that these are the only mediators of SIM2s repression in breast cancer cells, because Ras signaling affects many other pathways including phosphatidylinositol 3-kinase. Also, it will be important to evaluate SIM2s expression in other epigenetic and oncogene-driven models of breast cancer, such as MYC and WNT. Because NOTCH and C/EBPβ are differentially expressed in differing tumor types, these results may also explain the oncogenic function SIM2s has in pancreatic and prostate cancers (Sundaram, 2005; Gomis et al., 2006). It is of interest that two proteins involved in cell fate determination independently regulate SIM2s (Liu et al., 2005), suggesting that SIM2s has a function in mammary gland cell fate, possibly directing cells to differentiate. Future studies using gain- and loss-of-function mouse mammary gland models will elucidate the function of SIM2s in this process. We believe that downregulation of SIM2s expression is a requirement for breast cancer progression, and that elucidating pathways involved in SIM2s silencing will aid in the development of targeted therapies for treatment of this disease.
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Acknowledgements
We thank Bryan and Alana Welm (Huntsman Cancer Institute, Salt Lake City, UT, USA) for the Ha-Ras parent construct, Jeff Rosen (Baylor College of Medicine, Houston, TX, USA) for providing the C/ebpβ−/− mammary glands and Michael Lewis (Baylor College of Medicine, Houston, TX, USA) for critical reading of the paper. We also thank Keelan Anderson for technical assistance. This work was supported by awards from the National Cancer Institute RO1CA111551 (WWP), R01CA113795 (CAZ) and Howard Hughes Medical Institute Predoctoral Fellowship (TG).
References
- Aleman MJ, DeYoung MP, Tress M, Keating P, Perry GW, Narayanan R. Inhibition of Single Minded 2 gene expression mediates tumor-selective apoptosis and differentiation in human colon cancer cells. Proc Natl Acad Sci USA. 2005;102:12765–12770. doi: 10.1073/pnas.0505484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin BR, Timchenko NA, Zahnow CA. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPbeta-LIP in mammary epithelial cells. Mol Cell Biol. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Crews S, Franks R, Hu S, Matthews B, Nambu J. Drosophila single-minded gene and the molecular genetics of CNS midline development. J Exp Zool. 1992;261:234–244. doi: 10.1002/jez.1402610303. [DOI] [PubMed] [Google Scholar]
- Descombes P, Schibler U. A liver enriched transcriptional activator protein, LAP, and a transcriptional inhibitors protein, LIP, are transcribed from the same mRNA. Cell. 1991;3:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Deyoung MP, Scheurle D, Damania H, Zylberberg C, Narayanan R. Down’s syndrome-associated single minded gene as a novel tumor marker. Anticancer Res. 2002;22:3149–3157. [PubMed] [Google Scholar]
- Dunn KL, Espino PS, Drobic B, He S, Davie JR. The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 2005;83:1–14. doi: 10.1139/o04-121. [DOI] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Grimm SL, Rosen JM. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:191–204. doi: 10.1023/a:1025900908026. [DOI] [PubMed] [Google Scholar]
- Halvorsen OJ, Rostad K, Oyan AM, Puntervoll H, Bo TH, Stordrange L, et al. Increased expression of SIM2-s protein is a novel marker of aggressive prostate cancer. Clin Cancer Res. 2007;13:892–897. doi: 10.1158/1078-0432.CCR-06-1207. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Khleif SN, Abrams SI, Hamilton JM, Bergmann-Leitner E, Chen A, Bastian A, et al. A phase I vaccine trial with peptides reflecting ras oncogene mutations of solid tumors. J Immunother. 1999;22:155–165. doi: 10.1097/00002371-199903000-00007. [DOI] [PubMed] [Google Scholar]
- Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, et al. Modulation of Notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;2:259–266. doi: 10.1093/carcin/bgl122. [DOI] [PubMed] [Google Scholar]
- Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, et al. Loss of Singleminded-2s in the mouse mammary gland induces an epithelial mesenchymal transition associated with up-regulation of Slug and MMP2. Mol Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- Martinez-Lacaci I, Kannan S, De Santis M, Bianco C, Kim N, Wallace-Jones B, et al. RAS transformation causes sustained activation of epidermal growth factor receptor and elevation of mitogen-activated protein kinase in human mammary epithelial cells. Int J Cancer. 2000;88:44–52. doi: 10.1002/1097-0215(20001001)88:1<44::aid-ijc7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Metz RP, Kwak HI, Gustafson T, Laffin B, Porter WW. Differential transcriptional regulation by mouse single-minded 2S. J Biol Chem. 2006;281:10839–10848. doi: 10.1074/jbc.M508858200. [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K, Loning T, Bamberger AM. Expression of the CCAAT/enhancer-binding proteins C/EBPalpha, C/EBPbeta and C/EBPdelta in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat. 2003;79:175–185. doi: 10.1023/a:1023929504884. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LL, Discafani CM, Gurnani M, Tyler RD. Histopathology of salivary and mammary gland tumors in transgenic mice expressing a human Ha-ras oncogene. Cancer Res. 1991;51:3762–3767. [PubMed] [Google Scholar]
- Sansone P, Storci G, Giovannini C, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Seagroves TN, Krnacik S, Raught B, Gay J, Burgess-Beusse B, Darlington GJ, et al. C/EBPbeta, but not C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 1998;12:1917–1928. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim M, Powers KL, Ewing SJ, Zhu S, Smart RC. Diminished expression of C/EBPalpha in skin carcinomas is linked to oncogenic Ras and reexpression of C/EBPalpha in carcinoma cells inhibits proliferation. Cancer Res. 2005;65:861–867. [PubMed] [Google Scholar]
- Shuman JD, Sebastian T, Kaldis P, Copeland TD, Zhu S, Smart RC, et al. Cell cycle-dependent phosphorylation of C/EBPbeta mediates oncogenic cooperativity between C/EBPbeta and H-RasV12. Mol Cell Biol. 2004;24:7380–7391. doi: 10.1128/MCB.24.17.7380-7391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- von Lintig FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR. Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;62:51–62. doi: 10.1023/a:1006491619920. [DOI] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Zahnow CA. CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 2002;4:113–121. doi: 10.1186/bcr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnow CA, Cardiff RD, Laucirica R, Medina D, Rosen JM. A role for CCAAT/enhancer binding protein beta-liver-enriched inhibitory protein in mammary epithelial cell proliferation. Cancer Res. 2001;61:261–269. [PubMed] [Google Scholar]
- Zahnow CA, Younes P, Laucirica R, Rosen JM. Over-expression of C/EBPbeta-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J Natl Cancer Inst. 1997;89:1887–1891. doi: 10.1093/jnci/89.24.1887. [DOI] [PubMed] [Google Scholar]
- Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci USA. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)