I. Introduction
Dynamins play a crucial role in numerous membrane remodeling events throughout eukaryotic cells and have a relatively low nucleotide affinity and high rate of GTP hydrolysis. The propensity of dynamins to self-assemble and stimulate their own GTPase activity distinguishes them from other GTPases. The founding member, dynamin, regulates vesicle scission at the plasma membrane, endosome, and trans-Golgi network during endocytosis and caveolae internalization (Hinshaw, 2000). The dynamin-related protein (Drp1/Dnm1/ADL2B) is involved in mitochondrial fission, while mitofusins (Mfn1 and Mfn2) and OPA1/Mgm1 control fusion of the outer and inner mitochondrial membranes, respectively (Hoppins et al., 2007). Other dynamin family members control peroxisome (Vps1/Drp1) division as well as chloroplast division and cell wall formation in plants (ARC5/ADLs/Phragmoplastin) (Hong et al., 2003; Otegui et al., 2001; Praefcke and McMahon, 2004).
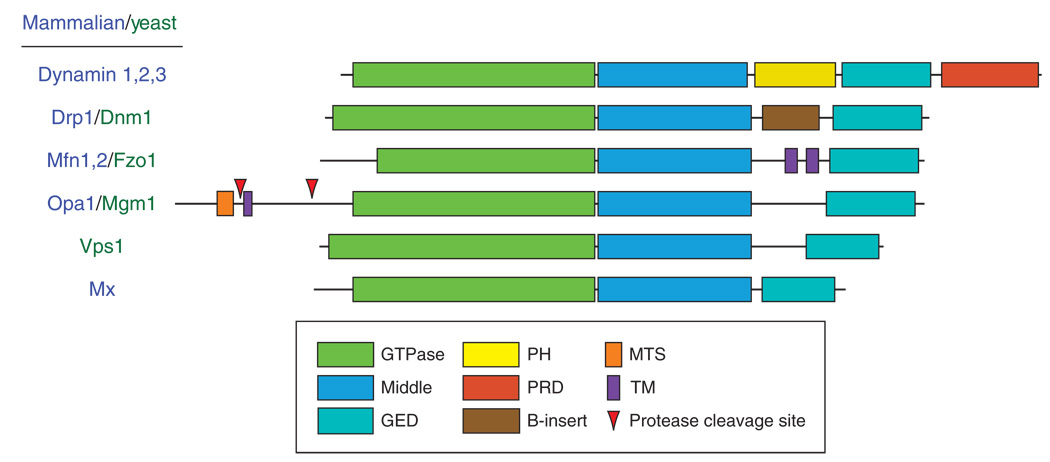
To achieve these varied tasks, all dynamins contain three conserved domains essential for function: a highly conserved GTPase domain, a middle domain, and a GTPase effector domain (GED) (Fig. 1). Each domain is required for self-assembly of dynamins into functional, oligomeric structures (Ingerman et al., 2005; Ramachandran et al., 2006; Smirnova et al., 1999; Song et al., 2004). In addition to these conserved motifs, dynamins contain other functional domains specific to the cellular mechanism associated with each protein (Fig. 1).
Fig. 1.
A schematic alignment of mammalian and yeast dynamins is presented and highlights the domain organization for each protein. The GTPase, middle, and GED topology is conserved among all family members. GED, GTPase effector domain; PH, pleckstrin homology; PRD, proline-rich domain; MTS, mitochondria targeting sequence; TM, transmembrane. (For Arabidopsis dynamins see Hong et al., 2003).
Dynamin, the family member studied most extensively, has an additional pleckstrin-homology (PH) domain and a proline-rich domain (PRD) (Fig. 1). The PH domain serves to target dynamin to negatively charged lipids (Klein et al., 1998; Tuma et al., 1993; Zheng et al., 1996), which may concentrate dynamin at the necks of invaginating pits during endocytosis (Achiriloaie et al., 1999; Artalejo et al., 1997; Lee et al., 1999). The PRD interacts with SH3-domain containing proteins, including endophilin, amphiphysin, intersectin, and cortactin. These dynamin partners all serve to help regulate vesicle endocytosis (Dawson et al., 2006; Schmid et al., 1998). Other dynamin family members contain transmembrane (TM) domains (mitofusin, Opa1/Mgm1), a mitochondrial targeting sequence (MTS; OPA1/Mgm1) and additional inserts whose functions remain unknown (see B-insert in Dnm1/Drp1) (Fig. 1). All of these domains are tailored to the cellular function associated with the individual proteins while maintaining the conserved GTPase, middle, and GED topology. For mitofusins, the TM domains anchor the protein in opposing membranes and likely act as tethers during mitochondrial fusion (Koshiba et al., 2004) in a mechanism believed to be similar to SNARE fusion events (Choi et al., 2006). The MTS found in Mgm1/OPA1 is essential for targeting the protein to the intermembrane space in mitochondria, where it is responsible for fusion events at the inner mitochondrial membrane and regulating cristae structure (Frezza et al., 2006; Meeusen et al., 2006; Meeusen and Nunnari, 2005). Some of the smallest dynamin-related proteins are the Mx proteins, which are involved in viral resistance (Haller and Kochs, 2002). The GTPase, middle, GED topology is maintained with little added sequence and no additional domains. Of all the dynamin family members studied to date, MxA contains the minimal set of domains essential for the function of dynamins.
Large oligomers of dynamins formed upon self-assembly, are amenable to visualization using various microscopic techniques. Specifically, transmission electron microscopy (TEM), atomic force microscopy (AFM), and scanning transmission electron microscopy (STEM) have been used to examine dynamins. To quantify the assembly state of the entire sample, biochemical techniques are also an essential tool. For dynamins, sedimentation assays provide a measure of the oligomeric state, while light scattering experiments provide a measure of conformational changes in dynamin structures due to GTP hydrolysis. When combined with high-resolution imaging techniques, these methods provide a complementary representation of dynamin self-assembly and structural properties, giving a more complete interpretation.
In this chapter, we will focus on three dynamin family members: human dynamin 1, yeast Dnm1, and human MxA. Despite differences in sequence, all three proteins contain similar structural features that can be attributed to the conserved GTPase–middle–GED topology. Each protein oligomerizes in low-salt conditions or with nucleotide analogs and forms helical arrays in the presence of lipid. In the absence of lipid, both dynamin and Dnm1 assemble into spirals while MxA forms curved filaments and rings (Fig. 2). For dynamin and Dnm1, the oligomeric state is tailored to its function: dynamin forms structures with sizes comparable to the size of necks at budding vesicles (~50 nm) (Hinshaw and Schmid, 1995), while Dnm1 forms significantly larger structures required for mitochondrial fission with sizes comparable to diameters observed at mitochondrial constriction sites (~100 nm) (Ingerman et al., 2005). Furthermore, both Dnm1/Drp1 and MxA proteins have an apparent affinity for lipid despite lacking a PH domain. Therefore, the polymers of dynamins may preferentially interact with lipid bilayers due to their inherent curvature. Comparing the similarities and differences in dynamin family members using a combination of biochemical and imaging techniques provides the opportunity to understand the relationship between conserved and unique protein domains associated with varied cellular functions.
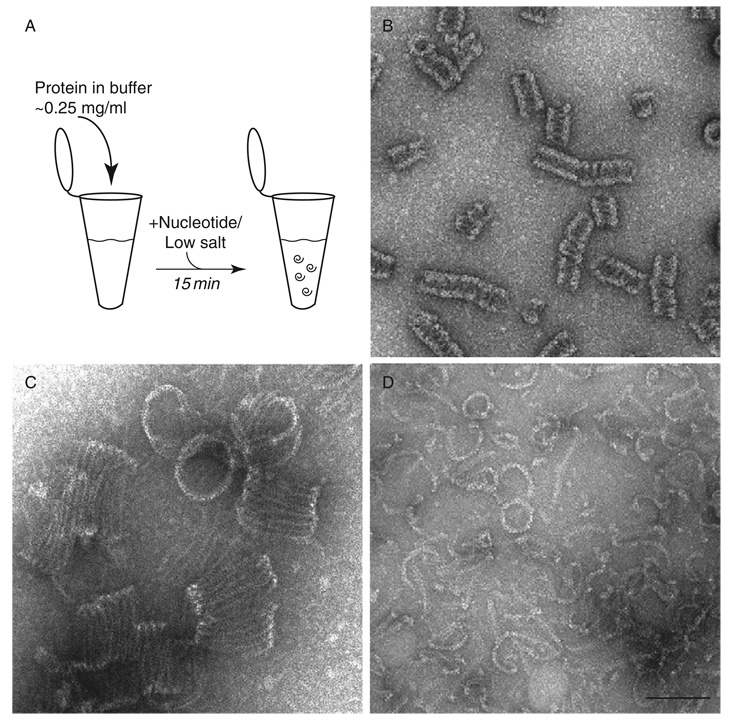
Fig. 2.
Oligomeric structures of dynamins are visualized using negative stain TEM. (A) Oligomers are generated under low salt conditions or in the presence of nonhydrolyzable nucleotides. (B) Dynamin spiral structures are shown that were dialyzed in the presence of GDP/BeF. (C) Larger spiral structures are observed for Dnm1 in the presence of GMP-PCP. (D) MxA curved filaments and occasional rings are presented after incubation with GMP-PCP. All protein structures were generated in HCB100. Scale bar, 100 nm.
II. Methods and Materials
A. Self-Assembly of Dynamins
Purified dynamin in high salt exists as a tetramer/monomer (Binns et al., 1999) and dilution into low salt conditions (<50 mM NaCl) forms ring and spiral structures (Hinshaw and Schmid, 1995). In addition, incubation with GDP/BeF, under physiological salt conditions, results in dynamin rings and spirals (Carr and Hinshaw, 1997). To make spirals, dynamin (~0.2 mg/ml) in HCB100 (Hepes Column BuVer (20 mM Hepes, pH 7.2, 1 mM MgCl2, 2 mM EGTA, 1 mM DTT) with 100 mM NaCl) is incubated with 1 mM GDP, 5 mM NaF, and 500 µM BeCl2 for 15 min at room temperature (Fig. 2A and B). Alternatively, AlF3 can be used in place of BeF2 by combining 5 mM NaF and 500 µM AlCl2. Dialysis of dynamin into HCB25 (25 mM NaCl) overnight at 4 °C also results in dynamin spirals, though not as consistent as with GDP/XFx.
Dnm1 oligomerizes into curved filaments in the absence of nucleotide (HCB100 alone) and forms spirals only in the presence of GMP-PCP (Fig. 2C) (Ingerman et al., 2005). These structures are much larger than dynamin spirals (~100 nm vs. ~50 nm). The addition of GTP (1 mM) to GMP-PCP–Dnm1 spirals caused the disassembly of highly ordered rings into curved filaments that are similar to those seen in the absence of nucleotides. For GMP-PCP spirals, Dnm1(~1.0 mg/ml) is dialyzed into 1 mM GMP-PCP in HCB150 (150 mM NaCl) overnight at 4 °C.
MxA protein also oligomerizes under certain conditions (Kochs et al., 2002, 2005), but not as well as other dynamins. Upon dialysis of MxA (~1 mg/ml) in HCB25–150 and the presence of 1 mM GMP-PCP, ring structures form (Fig. 2D) (Kochs et al., 2005). Additionally, long, straight assemblies are generated by dialysis of MxA in HCB50 and the presence of 1 mM GDP, 5 mM NaF, and 500 µM BeCl2 in 5% ethylene glycol for 20 h at 4 °C (Kochs et al., 2005).
B. Dynamin–Lipid Tubes
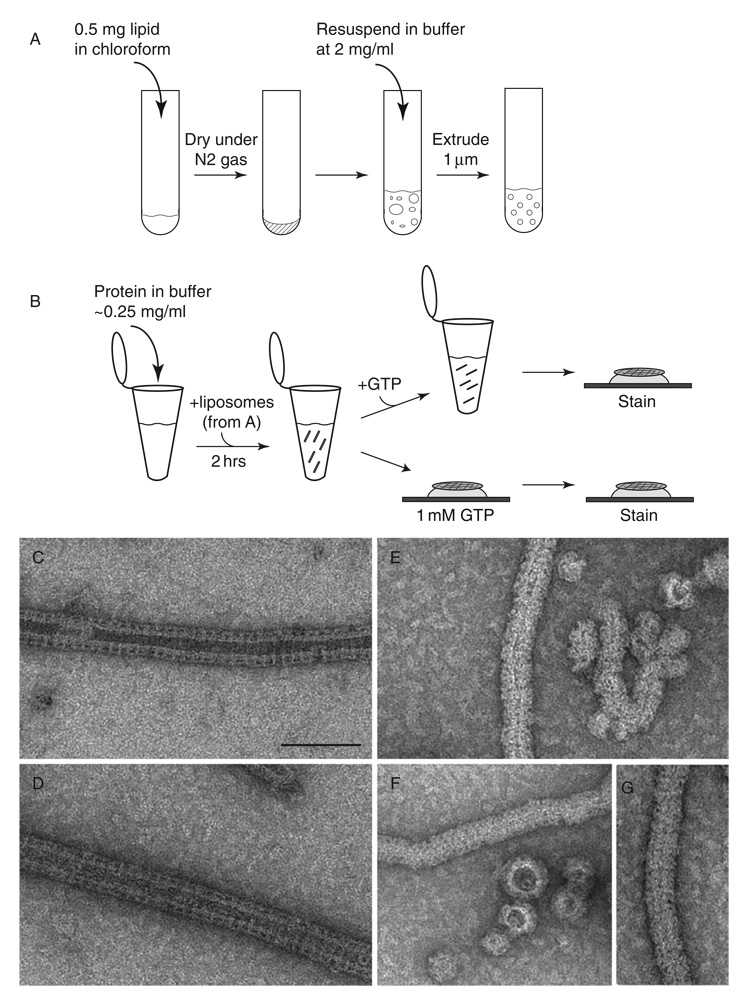
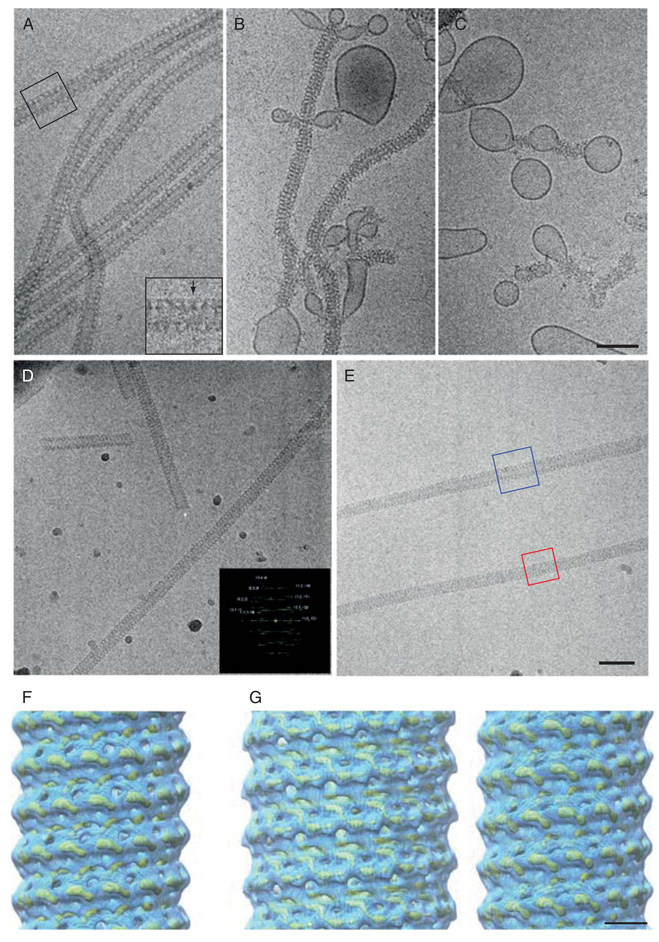
Dynamin interacts with lipid in vitro with a specific preference for negatively charged bilayers (Sweitzer and Hinshaw, 1998; Zheng et al., 1996). To generate dynamin oligomers on lipid bilayers, dynamin is incubated with liposomes for 1–2 h at room temperature. Liposomes are made by drying 50 µl of lipid in chloroform (100% synthetic phosphatidyl serine (PS), Avanti Polar Lipids) under nitrogen gas, keeping the lipid in vacuum overnight and resuspending to a final concentration of 2 mg/ml in buffer with physiological salt conditions (HCB100). The lipid is then extruded 11–15 times through a 1 µm polycarbonate membrane (Avanti Polar Lipids) to generate unilamellar vesicles of uniform size (Fig. 3A). Adding PS liposomes to dynamin at a final concentration of 0.2 mg/ml (protein and lipid) and incubating at room temperature for ~2 h results in the formation of long helical arrays of dynamin–lipid tubes (Fig. 3B–D). Dynamins form the best decorated tubes with 100% PS or 90% PS, 10% phosphoinositol-4,5-bisphophate (PI4,5P2). We have also had some success with total brain lipid (Avanti Polar Lipids), PS plus cholesterol (up to 10%), and a mixture of 70% PS and 30% galactosylceramide (GalCer). Upon addition of GTP (1 mM final), all but the GalCer tubes constrict and under certain conditions fragment (Fig. 3E–G) (Danino et al., 2004; Sweitzer and Hinshaw, 1998).
Fig. 3.
Dynamin-lipid tubes generated with extruded liposomes constrict and fragment upon addition of GTP. (A) To create extruded liposomes, lipid in chloroform solvent is dried under nitrogen gas in a glass tube and stored under vacuum overnight. The lipid is then resuspended in buffer and extruded through a 1 µm polycarbonate membrane (Avanti). (B) Liposomes are added to dynamin in HCB100 and incubated for ~2 h to generate dynamin–lipid tubes, which are observed using negative stain TEM (panels C and D). GTP is added either directly to the sample in the tube or by placing the grid with sample adhered to its surface on a drop of GTP in solution. (C, D) Negative stain EM of dynamin–lipid tubes prior to GTP addition are 50 nm in diameter. (E–G) In the presence of GTP, dynamin–lipid tubes fragment and constrict to 40 nm in diameter. Scale bar, 100 nm.
Well-ordered structures of dynamin tubes are formed with a dynamin mutant lacking the PRD (ΔPRD). ΔPRD dynamin forms tubes in the same manner as described earlier, but unlike wild-type dynamin, a constricted structure is observed in the presence of nonhydrolysable GTP analogs (GMP-PCP, GMP-PNP, and GTPγS). To form constricted tubes, ΔPRD dynamin (0.2 mg/ml in HCB100) was preincubated with GMP-PCP (1 mM final) for 15 min at room temperature prior to the addition of liposomes (~0.2 mg/ml) for 1–2 h at room temperature (Zhang and Hinshaw, 2001).
Dnm1 also forms helical arrays upon the addition of negatively charged liposomes despite lacking the PH domain. Unlike dynamin, a greater abundance of tubes are formed if nonextruded lipid is used in the sample, possibly due to the larger surface area required for the larger helical structures. As with dynamin, PS liposomes work well, but we also find a combination of 90% phosphatidylethanol-amine (PE; Avanti Polar Lipid) and 10% PI4,5P2 results in tubes with a more regular diameter and length. Addition of GMP-PCP to these tubes does not constrict the lipid; however, the tubes are more ordered. GMP-PCP was added to a final concentration of 1 mM and allowed to incubate for 15 min.
MxA forms protein–lipid tubes only with nanotubes made with GalCer, a lipid that makes extended lipid tubes without protein. This suggests that MxA may not be able to deform liposomes to lipid tubes, but it is capable of binding to a lipid tube with the correct diameter. As with dynamin, incubating MxA (0.2 mg/ml) with GalCer tubes (0.2 mg/ml comprised of 70% PS and 30% GalCer synthetic lipids) at 37 °C for 1 h results in protein helical oligomerization on the lipid substrate.
C. Quantifying Dynamin Oligomerization
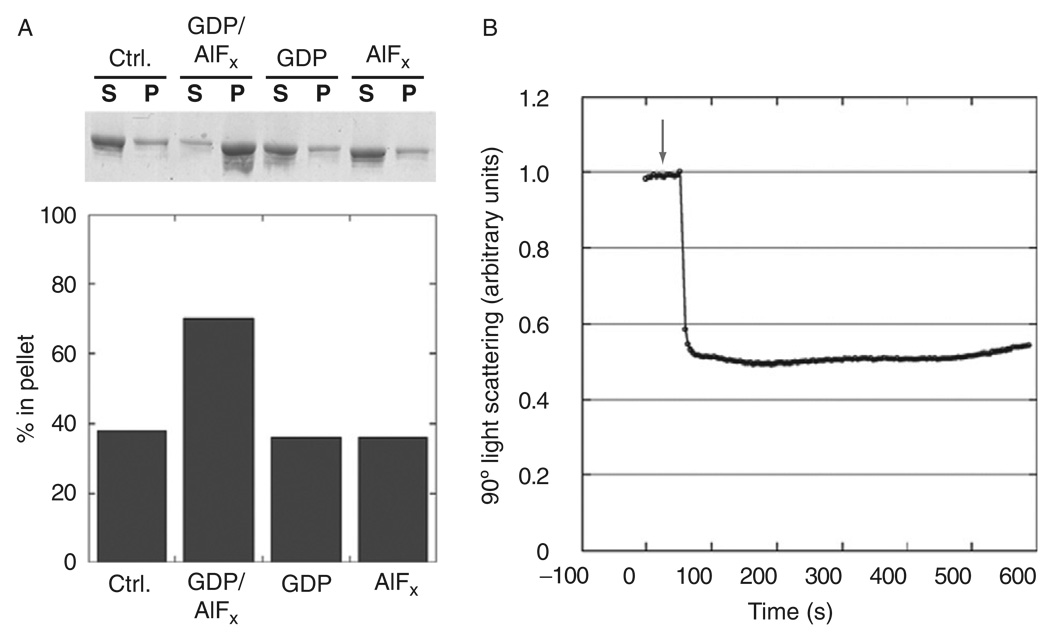
A common method used to quantify the amount of oligomer formation in solution is a sedimentation assay (Carr and Hinshaw, 1997; Danino et al., 2004; Hinshaw and Schmid, 1995). Samples incubating at room temperature are transferred to an ice bath to prevent any additional reactions. Samples are then centrifuged at 100,000×g for 15 min at 4 °C in a Beckman ultracentrifuge (TLA 100 rotor). The supernatant and pellet fractions can then be separated and loaded onto a 4–12% SDS-PAGE gel (Invitrogen) and stained with Coomassie to determine the amounts of protein. Assembled dynamin is found in the pellet (Fig. 4A). The relative densities of dynamin in each fraction can be quantified using gel-imaging software.
Fig. 4.
Biochemical assays quantify dynamin assembly and conformational changes upon addition of GTP. (A) The sup/pellet assay quantifies dynamin assembly under different conditions by centrifuging the sample at 100,000g and running the supernatant and pellet fractions on a gel (top). Imaging software quantifies the relative amount of protein in the pellet (bottom). (B) 90° light scattering measures the relative change in tube conformation. Addition of GTP (indicated with an arrow) leads to a rapid and dramatic decrease in scattering.
Ninety degree light scattering has been used to assess conformational changes in dynamin–lipid tubes treated with GTP. Dynamin tubes are prepared as previously described and diluted 1:10 with HCB100 (dynamin at 0.02 mg/ml). A PC1 Spectrofluorometer (ISS, Champaign, IL) was used at 350 nm with a 4% screen for excitation and 355 nm for emission with excitation and emission slit widths set at 5 nm with an OD1 filter. To begin, measurements of 90° light scattering were made at 0.1-s intervals up to 15 min. After obtaining a stable background, GTP is added to a final concentration of 1 mM, and is stirred gently with a pipette tip. The scattering curves are normalized, and arbitrary units are presented. Upon GTP addition to wild-type dynamin tubes, an immediate drop in light scattering is detected (Fig. 4B), which correlates with membrane constriction and supercoiling observed by electron microscopy (Fig. 3C and D).
D. Transmission Electron Microscopy
While sedimentation assays provide a measure of protein assembly, electron microscopy (EM) provides a means to visualize large oligomeric structures. Negative stain EM is a rapid, qualitative method to observe macromolecular structures with high contrast, although heavy atom stains introduce some structural artifacts, such as flattening. Cryo-EM overcomes many of the negative staining disadvantages by flash freezing the sample in a thin film of vitreous ice and imaging without stain. However, cryo-EM is significantly more time-consuming, and therefore, preliminary imaging of the sample by negative staining is commonly used to provide a simple, quick assessment of sample quality and structural homogeneity.
1. Negative Stain EM
To prepare a negative stain sample, the protein is adhered to carbon-coated mesh grids. To increase hydrophilicity, the grids can be glow discharged or plasma cleaned (Fischione Instruments) prior to adding sample. A small drop of sample (5–10 µl) is placed on a clean surface (i.e., parafilm) and the grid is placed, carbon-side down, onto the drop. After incubating for 0.5–2 min, the grid is washed in either buffer or 2% uranyl acetate (UA) solution in dH2O and blotted with filter paper and washed again before placing the grid on a drop of UA for 2 min, blotted again and dried. The entire time needed to generate a negative stain sample is less than 5 min.
Dynamin spirals and tubes are readily seen using a TEM operated at 100 kV (Fig. 2B and Fig 3C and D) and imaged at magnifications ranging from ~3000× to 35,000× with a bottom mount 1K × 1K CCD camera, which increases the magnification by a factor of ~1.3. To examine the effects of GTP, dynamin–lipid tubes are adhered to a grid and then incubated facedown on a drop of GTP in HCB100 (1–5 min) followed by subsequent staining and fixing with 2% UA. EM images of dynamin tubes before and after GTP treatment (Fig. 3C–F) show that the tubes constrict and fragment upon GTP hydrolysis.
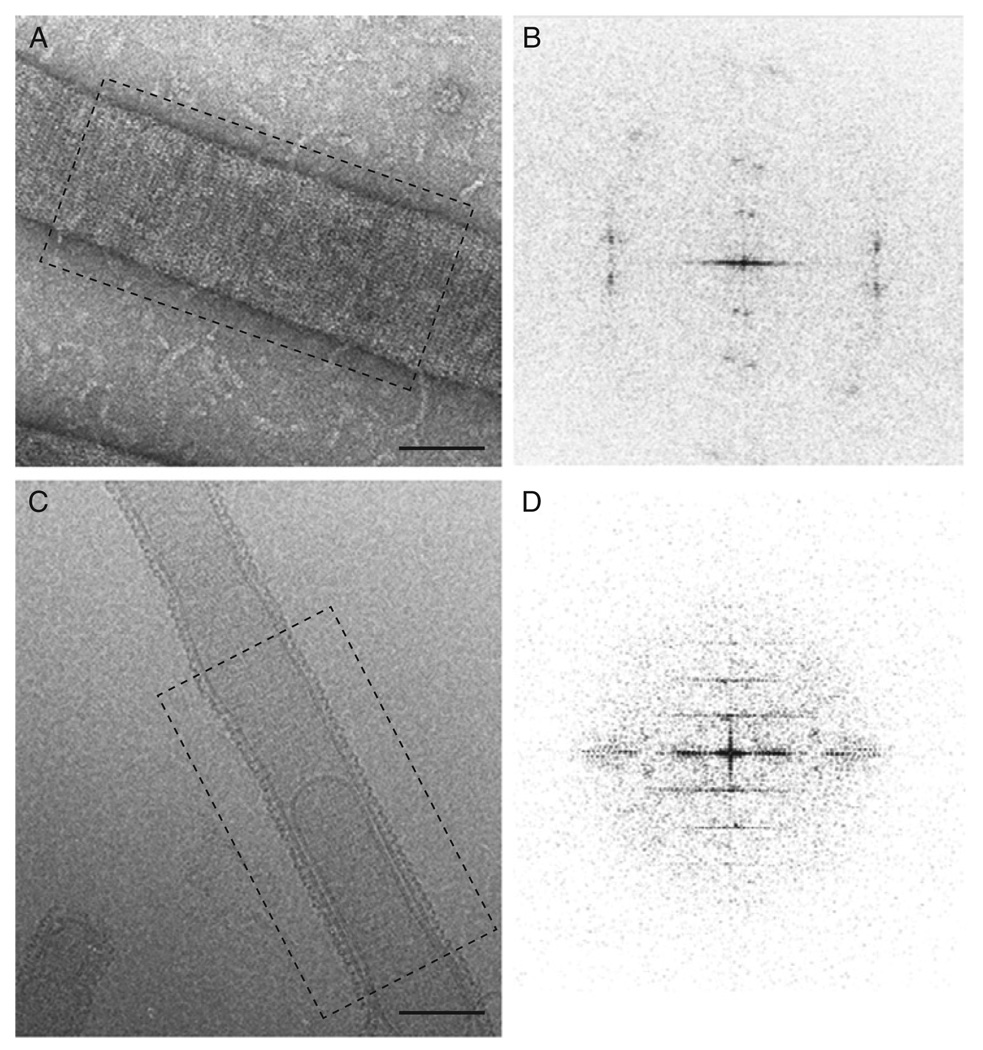
Dnm1 spirals generated by incubation with GMP-PCP are easily visualized by negative stain EM (Fig. 2C). These structures are a great deal larger than homologous dynamin spirals. Similarly, Dnm1–lipid tubes are significantly larger than are dynamin tubes (Fig. 5A), and because of the size of these tubes, negative stain flattens the tubes. The flattening is apparent when examining the Fourier transform of these tubes (Fig. 5B), which show diffraction spots consistent with a 2D lattice as opposed to layer lines observed for well-ordered helical structures (Fig. 5D). Adding 1 mM GMP-PCP to preformed Dnm1–lipid tubes for 15 min at room temperature improves the overall order of the helical structure, but some flattening is still observed.
Fig. 5.
Dnm1–lipid tubes are imaged using negative stain and cryo-EM. (A) Negative stain imaging of Dnm1–lipid tubes suggests flattening occurs due to stain and sample dehydration (B) Corresponding Fourier transform of image in A shows spots indicating a 2D lattice, indicative of tube flattening. (C) Cryo-EM image of Dnm1–lipid tube. (D) Fourier transform of image in (C) provides a regular helical pattern with layer lines suggesting a helical structure.
2. Cryo-EM
Cryo-EM eliminates the flattening effect of negative stain EM observed on Dnm1 tubes (Fig. 5C and D). In addition, cryo-EM allows for direct examination of the sample in its native state without staining. To control humidity and temperature, a Vitrobot (FEI Co.) system is used for sample vitrification. A 3–5 µl drop of sample is placed onto a holey carbon grid (Quantifoil R 3.5/1 with copper mesh) and blotted with filter paper to create a thin film of solution. The grid is immediately plunged into liquid ethane, which rapidly freezes the sample in noncrystalline, vitreous ice with the protein in its native state. The time and pressure of the blotting and subsequent freezing is computer controlled in the Vitrobot. Other manual devices work equally well with an experienced user. The advantage of the Vitrobot system is a novice can obtain good ice thickness and homogeneity. After freezing, the grid is stored under liquid nitrogen prior to examining the sample in the microscope. Using low-dose electron microscopy to prevent destruction of the sample, we are able to see ordered helical structures of the Dnm1–lipid tubes (Fig. 5C) with promising layer-line data in the Fourier transform (Fig. 5D). Cryo-electron tomography, a relatively new technique used to determine the structure of large cellular complexes, was also used to examine Dnm1 tubes. For this experiment, the vitreous sample is prepared as described earlier with the addition of gold particles applied to the grid prior to applying the sample. The sample is tilted to obtain a series of images at various angles (±70°) with the Serial-EM software (Mastronarde, 2005). The images are aligned and a three-dimensional reconstruction is obtained using IMOD software (Kremer et al., 1996). Tomographic reconstruction of Dnm1 tubes confirms that the flattening seen with negative stain on a carbon surface is no longer observed when cryo-EM methods are used.
Cryo-EM has also been used to visualize dynamin structures in their native states (Chen et al., 2004; Danino et al., 2004; Sweitzer and Hinshaw, 1998; Zhang and Hinshaw, 2001). Vitreous samples of dynamin–lipid tubes can be generated as described earlier. As observed with negative stain EM, a T-shape structure is seen at the lipid interface (Fig. 6A; see insert); a common feature observed among all dynamin family members examined to date, regardless of differences in protein circumference (Ingerman et al., 2005; Kochs et al., 2005; Low and Lowe, 2006). When GTP is added to the dynamin tubes, a constriction in the helical structure occurs that changes the overall diameter of the helical array from 50 to 40 nm (Fig. 6B and C). The major difference between cryo and negative stain samples is the lack of tube fragmentation in cryo-EM. The fragmentation observed in negative stain is due to the stain and sample drying. Another advantage of cryo-EM is that it allows for shorter time points to be observed after substrate addition. After the dynamin–lipid sample is added to the holey grids in the Vitrobot apparatus, GTP is added separately to the drop of sample on the grid and allowed to incubate for a specified amount of time before blotting and plunging the sample in liquid ethane. This procedure reveals that within 5 s after addition of GTP, most of the dynamin tubes are constricted and supercoiled with undecorated lipid bulges between constricted segments (Danino et al., 2004).
Fig. 6.
Cryo-EM imaging and reconstructions of dynamin are presented in the nonconstricted and constricted states. (A) Dynamin–lipid tubes formed as in Fig. 3 and imaged using cryo-EM. (B, C) When GTP is added, the tubes constrict the lipid bilayer, leading to bulges in regions of undecorated lipid. (D) ΔPRD dynamin forms well-ordered tubes in the presence of lipid and GMP-PCP, which diffract to ~20 Å resolution (Fourier transform in inset). Such images are averaged using helical reconstruction methods to generate a three-dimensional reconstruction (panel F). (E) ΔPRD dynamin–lipid tubes in both the nonconstricted and constricted states are boxed (blue and red boxes respectively) for image processing using the IHRSR reconstruction method (Egelman, 2006). (F) 3D map of dynamin–lipid tube solved by helical reconstruction methods (Zhang & Hinshaw, 2001) represented at both high (yellow mesh) and low (blue) thresholds. (G) IHRSR image reconstructions of the nonconstricted (left) and constricted (right) ΔPRD dynamin–lipid tubes (high-yellow mesh and low–blue thresholds) (Chen et al., 2004). Scale bar for A–E, 100 nm. Scale bar for F and G, 10 nm.
Cryo-EM also provides higher resolution images with less background noise since the sample is preserved in a thin layer of ice without stain or carbon support. Conversely, the low electron dose required for preventing sample damage results in low contrast and a higher background noise. Therefore, averaging numerous images to enhance the signal-to-noise ratio is necessary for subsequent image reconstructions. For dynamin, it was determined that removing the PRD favors the formation of well-ordered tubes that are ideal for image reconstruction (Zhang and Hinshaw, 2001). Additionally, constricted ΔPRD dynamin tubes in the presence of GMP-PCP are straight with a consistent helical pitch (Fig. 6D). Using helical reconstruction techniques, a three-dimensional structure for ΔPRD dynamin tubes was obtained for the constricted (nucleotide-bound) state (Fig. 6F) (Zhang and Hinshaw, 2001). The nonconstricted ΔPRD dynamin tubes (Fig. 6E, a section is boxed in blue) are more curved and flexible with a varied pitch, and therefore the traditional helical reconstruction techniques could not be applied. Instead, the Iterative Helical Real Space Reconstruction (IHRSR) method (developed by Edward Egelman at the University of Virginia) was used to overcome these problems. The IHRSR method uses a single-particle approach to generate three-dimensional reconstructions (Egelman, 2006), and was used to generate reconstructions for ΔPRD dynamin in both the nonconstricted and constricted states (Fig. 6G) (Chen et al., 2004).
3. Docking Structures to Cryo-EM Maps
Cryo-EM with image reconstruction methods allows for visualization of protein structures at resolutions greater than ~0.5 nm. Only recently has subnanometer resolution been achieved in part due to improved optics and sample stabilization in the microscope. To overcome the limited resolution, a comprehensive approach is used that combines X-ray crystallographic data with cryo-EM structures. There are numerous ways to dock X-ray structures into a cryo-EM map, including manually fitting crystal structures into cryo-EM density. Several visualization programs are available for manual fitting, including O (Jones et al., 1991), Pymol (http://pymol.sourceforge.net/), and Chimera (http://www.cgl.ucsf.edu/chimera). Using O, structures of the GTPase domain from a related dynamin family member and the PH domain from human were manually fit to the constricted ΔPRD dynamin structure determined by helical reconstruction (Zhang and Hinshaw, 2001).
The disadvantage of manual fitting is that the final model is subject to user prejudice. To prevent bias, automated fitting of X-ray structures provides a superior alternative. There are traditionally two methods used to match X-ray structures to the cryo-EM density: topology comparison and atom–voxel matching. For topology comparison, a density representation of the high-resolution structure is used to search orientations that best match the cryo-EM structure based on cross-correlation of the surfaces. For atom–voxel matching methods, the premise is to exhaustively search orientations in real or reciprocal space and find a best fit. To sample conformations with the cryo-EM density, various algorithms are available, including Monte Carlo simulations (see YAMMP/YUP (Tan et al., 1993; Tan et al., 2006) developed in group of Stephen C. Harvey). Monte Carlo simulations stochastically refine the structure(s) within the conformational space until a best fit is determined. This method requires a significant amount of user intervention but also allows a great deal of flexibility, because the force field for the simulation is user-defined. Consequently, bonds, nonbonds, volume exclusion, multidomain fitting, and other terms can be introduced along with simple rigid-body docking (Mears et al., 2006). The GTPase and PH crystal structures of dynamin were docked into the 3-dimensional density maps of dynamin using YAMMP/YUP and revealed a possible corkscrew mechanism of constriction (Mears et al. , 2007). Another docking package, SITUS, uses vector quantization to fit high-resolution structures into low-resolution density (Wriggers et al., 1999). This method allows for fast and exhaustive docking of rigid-body atomic structures to the cryo-EM structure, and new methods allow for flexible fitting as well (Wriggers and Birmanns, 2001).
E. Rotary Shadowing
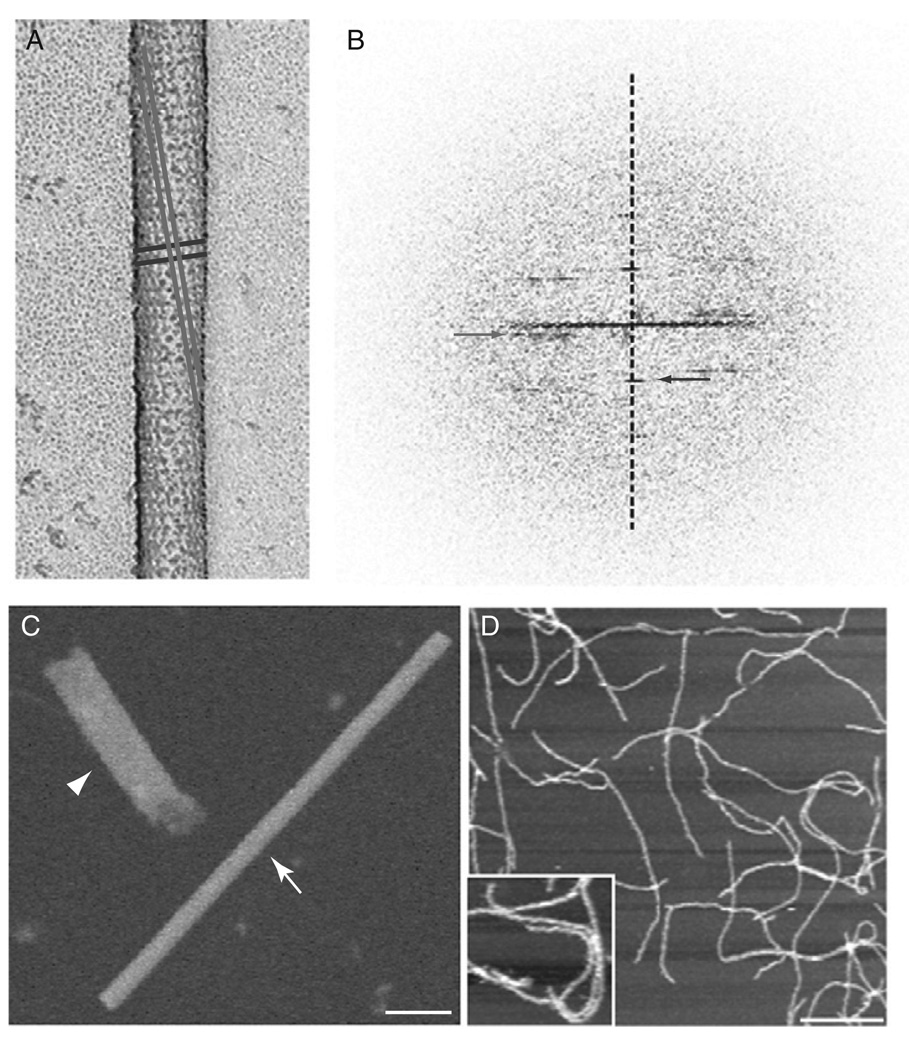
Rotary shadowing is commonly used to examine the shape of molecules and complexes. A thin layer of metal (usually Pt) applied at a low angle, while the sample is rotating, provides a clear replica of the top surface of the sample. This method is therefore useful for determining the hand of a helical array since only one side of the tube is imaged. With negative stain or cryo-EM the hand would be indiscernible because both the top and bottom layers of the helix are reflected equally. To determine the handedness of the ΔPRD dynamin tubes, the sample was freeze-dried and rotary-shadowed with Pt and carbon (Fig. 7A). A drop (10 µl) of ΔPRD dynamin tubes were directly added to clean mica (1 cm × 1 cm), washed with HCB0 (no NaCl) for 2 min and blotted from the back and side of the mica before freezing in liquid ethane. The sample was then stored in liquid nitrogen prior to freeze-drying and rotary shadowing. Later, the sample is placed quickly into a freeze-fracture machine (Balzers) that had been pumped down for several hours to a vacuum of 4 × 10−6 m bar and cooled to −150 °C. The fracture blade from the machine is placed over the sample to act as a cold trap as the temperature is raised to −95 °C. The sample is etched for ~30 min, and the temperature is returned to −150 °C prior to shadowing. While the sample is rotating (speed 9) Pt was applied for 6–8 s at a 6° angle followed by ~10 s of carbon. The sample is then warmed before removing from the machine. The replicas are floated off on deionized water and picked up on copper grids, and they are then examined by TEM. The hand of the long pitch and short pitch helices can be observed by eye (Fig. 7A) or determined by the Fourier transform (Fig. 7B).
Fig. 7.
Dynamin examined by several diverse image techniques. (A) Rotary shadowing of dynamin–lipid tubes reveals the surface of the tube and subsequently the hand of the helices. (B) Fourier transform of the shadowed image confirm the long (gray) pitch helix is left-handed and the short (black) pitch helix is right-handed. (C) STEM analysis of a dynamin spiral (highlighted by arrowhead) provides an accurate measure of mass over a defined length when compared with a TMV particle (arrow). Scale bar, 50 nm. (D) AFM imaging of dynamin tubes was used to examine conformational changes triggered by GTP hydrolysis. A zoomed image of dynamin tubes is shown in the inset. Scale bar, 2 µm.
F. Scanning Transmission Electron Microscopy
STEM analysis determines the mass of the specimen based on image intensity. With calibration of a specimen of known mass (usually tobacco mosaic virus (TMV); arrow in Fig. 7C), intensity in the STEM can be integrated over an isolated particle and converted to a molecular weight. STEM was used in this way to determine the number of dynamin molecules per turn of the helix (Fig. 7C). For STEM preparations, the ΔPRD dynamin spirals, made in the presence of GMP-PCP, were sent to Brookhaven National Laboratory (BNL) for imaging (Fig. 7C; arrowhead). The specimen is freeze-dried on holey carbon grids in the presence of TMV (as described on the BNL web page, http://www.biology.bnl.gov/stem/stem.html). The final concentration of dynamin spirals was 25–100 µg/ml. The images were processed using the PIC program on a VMS Dec Alpha; however, BNL currently has an updated program called PCMass25 available on the web that runs in Windows 95 or higher. The mass per length of TMV is known and acts as an internal control for the specimen. Dividing the length of the spiral by the helical pitch of 94.8 Å(determined by the diffraction pattern) and then dividing by the relative molecular mass of ΔPRD dynamin (87,100) determines the number of molecules per helical turn.
G. AFM
Dynamin tubes were examined by AFM in collaboration with Dr. Jan Hoh (Johns Hopkins University) to determine if the GTP-induced conformational change of the tubes could be observed in real time (Fig. 7D). To prepare the sample, dynamin tubes were diluted 1:1 with 20 mM NaCl prior to adding to clean mica. After 30 min, the sample was washed ten times with 20 mM NaCl and then imaged by AFM in a thin layer of liquid. Unfortunately, the conformational changes were not observed when GTP was added to tubes adhered to the mica; however, addition of GTP in solution prior to applying the sample to mica revealed supercoiled, shorter, and fewer tubes.
III. Discussion
In vitro studies of any protein require that the protein behave in a manner similar to in vivo preparations. For example, dynamin spirals and dynamin–lipid tubes observed in vitro are similar to dynamin structures observed at the necks of invaginating pits in nerve synapses (Evergren et al., 2004; Koenig and Ikeda, 1989; Takei et al., 1995). Also the large Dnm1 structures seen in vitro coincide with the mitochondrial constriction sites seen in wild-type yeast (Bleazard et al., 1999; Ingerman et al., 2005). In contrast, the oligomers of MxA still remain to correlate with structures seen in vivo; however, human MxA may also be membrane-associated as evidence suggests it interacts with the smooth endoplasmic reticulum (Accola et al., 2002).
The intrinsic quality of dynamins to self-assemble makes them amenable for structural studies as illustrated in this chapter. Dynamins have a strong propensity to self-assemble because of strong interactions between the GTPase, middle and GED domains. Even in 400 mM salt, dynamin exists as a tetramer (Binns et al., 1999). A decrease in salt concentration (<50 mM NaCl) leads to oligomerization of dynamin (rings/spirals) (Hinshaw and Schmid, 1995), Dnm1 (curved filaments) (Ingerman et al., 2005), and MxA (curved filaments) (Kochs et al., 2002). Additionally, nucleotide analogs interact with dynamins to form ring/spiral structures (Fig. 2). Dynamin forms spirals in the presence of GDP/BeF, a transition state analog, while nonhydrolysable GTP analogs have no observed affect. In contrast,Dnm1 and MxA form rings/spirals, albeit with different dimensions, in the presence of GTP analogs(GMP-PCP,GTPγS). The difference in nucleotide-dependent assembly between the dynamins may be directly related to the PRD region. ΔPRD dynamin behaves similar to Dnm1 and MxA and readily forms ring/spiral structures in the presence of GMP-PCP. In addition, ΔPRD dynamin–lipid tubes constrict when GMP-PCP is present (Fig. 6D and E). Therefore, the PRD may help regulate GTPase substrate affinity and activity.
The PH domain of dynamin is important for interactions with negatively charged lipids (Tuma et al., 1993; Zheng et al., 1996), and deleting this domain results in the loss of endocytosis as measured by transferrin uptake (Vallis et al., 1999). Moreover, the addition of negatively charged lipid to dynamin in solution stimulates GTP hydrolysis due to increased self-assembly of dynamin on lipid bilayers (Tuma et al., 1993). Dnm1 and MxA both form helical arrays in the presence of lipid, despite lacking a PH domain. The inherent curvature of the assembled oligomer may predispose the proteins to interact with membrane bilayers. However, Dnm1 favors lipid bilayers with less curvature because of the increased diameter of the oligomer.
When GTP is added, dynamin rapidly constricts and then disassociates from the lipid bilayer. The conformational change of dynamin–lipid tubes has also been examined in real time by fluorescence microscopy. In this work, Roux et al. observe a twisting motion resulting from GTP hydrolysis of dynamin (Roux et al., 2006). If the ends of the tubes are tethered, the twisting leads to fragmentation of the tubes. The tethered dynamin tubes are similar to the negative stain results, which also show dynamin tubes fragment upon GTP hydrolysis when attached to a support. Dynamin tubes free in solution twist and supercoil as observed by cryo-EM and as untethered tubes in light microscopy (Roux et al., 2006). In vivo, dynamin decorated necks of coated pits may be tethered by actin and the plasma membrane through numerous mediator proteins containing an SH3 domain to interact with the PRD in dynamin.
Examining the 3D structures of dynamin in the constricted and nonconstricted states show conformational changes in the “stalk” region of the T structure during lipid constriction (Chen et al., 2004). The middle domain and GED, which are conserved throughout the dynamin family and are responsible for driving self-assembly, are likely contained in this region, which undergoes a dramatic change from a relatively straight pattern to a kinked, zigzag pattern when GMP-PCP is added to the ΔPRD dynamin tube. Therefore, these conserved domains not only drive assembly but also regulate conformational changes that are essential for dynamin function.
It is unclear at this time what role cofactor proteins will have on assembly and constriction of dynamins. In the case of dynamin, many SH3 domain-containing binding partners (endophilin, amphiphysin, cortactin, etc.) are involved in endocytosis (Schmid et al., 1998), and many of these proteins contain lipid-binding motifs (Dawson et al., 2006). Still, the PH domain effectively targets dynamin to membranes in vitro (Tuma et al., 1993; Zheng et al., 1996) and is essential for endocytosis (Achiriloaie et al., 1999; Lee et al., 1999). Dnm1 is largely cytoplasmic in yeast cells, and other cofactor proteins (Fis1, Mdv1, and Caf4) are responsible for recruiting Dnm1 to the outer membrane of the mitochondria (Hoppins et al., 2007). Similarly, MxA is largely cytoplasmic, but some protein is associated with smooth ER (Accola et al., 2002). No binding partners have been identified for MxA, although it is able to interact directly with certain viral nucleocapsid proteins (Kochs and Haller, 1999).
In the future, examining additional dynamin family members will further define the fundamental mechanism of action of the dynamins and more importantly reveal the differences that are unique to the function of each protein. For example, Opa1/Mgm1 and mitofusins are required for mitochondrial fusion and understanding how these proteins exploit dynamins’ distinctive self-assembly property will provide great insight into this poorly understood mechanism. In addition, Vps1 and Mx proteins only contain the GTPase, middle, and GED domains with little additional sequence, suggesting these proteins are regulated by yet to be identified binding partners. Determining the process of how dynamins reach their functional sites is also crucial. For dynamin, the PRD has been shown to be essential for the localization of this protein to the plasma membrane while the inner membrane fusion protein (Opa1/Mgm1) contains a mitochondria targeting sequence. However, no comparable targeting sequence has been found for either Vps1 and Mx. Overall, comparison of dynamin family members provides an excellent example of modular protein domains organized around a conserved topology that dictate the function(s) for that protein.
IV. Summary
The tools presented in this chapter have been used to characterize the structural and biochemical properties of dynamins. The versatility in microscopic techniques allows for visualization of dynamins with varied shapes, including ring, spiral, and helical oligomers. Negative stain allows for structures to be examined quickly; however, larger structures may flatten, as was observed with Dnm1. Cryo-EM helps eliminate flattening, as shown with Dnm1, allows the specimen to be viewed in a more native state and freezing the sample rapidly following substrate addition (i.e., GTP) allows conformational changes to be observed in seconds. In addition, AFM, fluorescence microscopy, and light scattering assays can be used to observe structural rearrangements that occur upon GTP hydrolysis. Rotary shadowing reveals the surface structure of the sample and can be used to determine the hand of the helical array. The accuracy of STEM analysis provides a means to calculate atomic mass over a defined area or length of helix. Together, these methods provide a comprehensive approach for visualizing dynamins.
Acknowledgment
The authors thank Ye Fang and Dr. Jan Hoh (JHU) for assistance in acquiring AFM results and Dr. Blair Bowers (NHLBI/NIH) for work with rotary shadowing. We also thank Dr. Dan Sackett (NICHD/NIH) for help with light scattering and Dr. Edward Egelman (UVa) for his collaboration on image reconstruction of ΔPRD dynamin tubes using the IHRSR method.
References
- Accola MA, Huang B, Al Masri A, McNiven MA. The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 2002;277:21829–21835. doi: 10.1074/jbc.M201641200. [DOI] [PubMed] [Google Scholar]
- Achiriloaie M, Barylko B, Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol. Cell. Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artalejo CR, Lemmon MA, Schlessinger J, Palfrey HC. Specific role for the PH domain of dynamin-1 in the regulation of rapid endocytosis in adrenal chromaffin cells. EMBO J. 1997;16:1565–1574. doi: 10.1093/emboj/16.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns DD, Barylko B, Grichine N, Atkinson MA, Helms MK, Jameson DM, Eccleston JF, Albanesi JP. Correlation between self-association modes and GTPase activation of dynamin. J. Protein. Chem. 1999;18:277–290. doi: 10.1023/a:1021083211267. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JF, Hinshaw JE. Dynamin assembles into spirals under physiological salt conditions upon the addition of GDP and gamma-phosphate analogues. J. Biol. Chem. 1997;272:28030–28035. doi: 10.1074/jbc.272.44.28030. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat. Struct. Mol. Biol. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell. Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM. Bar domain proteins: A role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006;16:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J. Struct. Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Evergren E, Tomilin N, Vasylieva E, Sergeeva V, Bloom O, Gad H, Capani F, Shupliakov O. A pre-embedding immunogold approach for detection of synaptic endocytic proteins in situ. J. Neurosci. Methods. 2004;135:169–174. doi: 10.1016/j.jneumeth.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G. Interferon-induced mx proteins: Dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE. Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hong Z, Bednarek SY, Blumwald E, Hwang I, Jurgens G, Menzel D, Osteryoung KW, Raikhel NV, Shinozaki K, Tsutsumi N, Verma DP. A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant. Mol. Biol. 2003;53(3):261–265. doi: 10.1023/b:plan.0000007000.29697.81. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell. Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J. Biol. Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- Kochs G, Haener M, Aebi U, Haller O. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 2002;277:14172–14176. doi: 10.1074/jbc.M200244200. [DOI] [PubMed] [Google Scholar]
- Kochs G, Haller O. GTP-bound human MxA protein interacts with the nucleocapsids of Thogoto virus (Orthomyxoviridae) J. Biol. Chem. 1999;274:4370–4376. doi: 10.1074/jbc.274.7.4370. [DOI] [PubMed] [Google Scholar]
- Kochs G, Reichelt M, Danino D, Hinshaw JE, Haller O. Assay and functional analysis of dynamin-like Mx proteins. Methods Enzymol. 2005;404:632–643. doi: 10.1016/S0076-6879(05)04055-3. [DOI] [PubMed] [Google Scholar]
- Koenig JH, Ikeda K. Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J. Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lee A, Frank DW, Marks MS, Lemmon MA. Dominant-negative inhibition of receptor-mediated endocytosis by a dynamin-1 mutant with a defective pleckstrin homology domain. Curr. Biol. 1999;9:261–264. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC. A structural model for the large subunit of the mammalian mitochondrial ribosome. J. Mol. Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Meeusen SL, Nunnari J. How mitochondria fuse. Curr. Opin. Cell Biol. 2005;17:389–394. doi: 10.1016/j.ceb.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2006;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2007;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Schmid SL, McNiven MA, De Camilli P. Dynamin and its partners: A progress report. Curr. Opin. Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Song BD, Yarar D, Schmid SL. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol Cell. 2004;15:2243–2252. doi: 10.1091/mbc.E04-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Tan RK-Z, Harvey SC. Yammp: Development of a molecular mechanics program using the modular programming method. J. Comp. Chem. 1993;14:455–470. [Google Scholar]
- Tan RK-Z, Petrov AS, Harvey SC. YUP: A molecular simulation program for coarse-grained and multi-scale models. J. Chem. Theory Comput. 2006;2:529–540. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma PL, Stachniak MC, Collins CA. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J. Biol. Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Birmanns S. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat. Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. Identification of the binding site for acidic phospholipids on the pH domain of dynamin: Implications for stimulation of GTPase activity. J. Mol. Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]