Abstract
Combination treatment with pegylated-interferon-α and ribavirin, the current recommended therapy for chronic hepatitis C virus (HCV) infection, results in a sustained virological response (SVR) in only about half of patients. Because genes involved in the interferon-α pathway may affect anti-viral responses, we analyzed the relationship between variants in these genes and SVR among participants in the HALT-C trial. Patients had advanced chronic hepatitis C and had previously failed to respond to interferon-based treatment. Participants were treated with peginterferon-α2a and ribavirin during the trial. Subjects with undetectable HCV RNA at week 72 were considered to have had an SVR. Subjects with detectable HCV RNA at week 20 were considered non-responders. We used TaqMan assays to genotype 56 polymorphisms found in 13 genes in the interferon-α pathway. This analysis compares genotypes for participants with an SVR to non-responders. The primary analysis was restricted to European American participants because a priori statistical power was low among the small number (n=131) of African American patients. We used logistic regression to control the effect of other variables that are associated with treatment response. Among 581 European American patients, SVR was associated with IFNAR1 IVS1-22G (adjusted odds ratio [aOR], 0.57; p=0.02); IFNAR2 Ex2-33C (aOR, 2.09; p=0.02); JAK1 IVS22+112T (aOR, 1.66; p=0.04); and ADAR Ex9+14A (aOR, 1.67; p=0.03). For the TYK2 -2256A promoter region variant a borderline association was present among European American participants (OR, 1.51; p=0.05) and a strong relationship among African American patients; all 10 with SVR who were genotyped for TYK2-2256 carried the A variant compared to 68/120 (57%) non-responders (p=0.006). In conclusion, genetic polymorphisms in the interferon-α pathway may affect responses to antiviral therapy of chronic hepatitis C.
Keywords: epidemiology, hepatitis C/genetics, chronic, interferon-alpha/therapeutic use, gene frequency, pharmacogenetics, polymorphism, genetic
Background
Hepatitis C virus (HCV) is an important cause of cirrhosis and hepatocellular carcinoma in the United States and worldwide.(1) Combination therapy with ribavirin and pegylated-interferon-α is the current best therapy for patients with chronic HCV infection. This regimen produces sustained virologic response (SVR) in 80% of patients infected with HCV genotypes 2 or 3, but in less than 50% of patients who are infected with the much more common HCV genotype 1.(2) Other factors associated with a poor response to therapy include high baseline HCV RNA level, hepatic fibrosis, older age, male gender and African ancestry.(2–4) Relatively little is known about the role of host genetic factors in HCV treatment response.(5)
Because of the crucial role of the interferon-α pathway in achieving viral clearance, it is plausible that genes involved in this pathway may alter a patient’s response to interferon-α-based therapy. In response to viral infections, type I interferon-α is produced endogenously in epithelial cells, plasmacytoid dendritic cells and lymphocytes. By binding to its receptor, interferon-α activates the JAK-STAT pathway, which in turn induces the expression of host genes with direct antiviral properties.(6) In addition, interferon-α exerts indirect immunomodulatory effects through enhancement of antigen presentation and induction of the adaptive (cellular) immune response. (7, 8) An understanding of the role of genes involved in the interferon-α pathway in HCV treatment response could identify patients at increased risk of treatment failure and, potentially, lead to the development of new therapeutic strategies.
The Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) trial is a multi-center clinical trial designed to evaluate the safety and efficacy of long-term pegylated-interferon-α2a for treatment of advanced chronic hepatitis C infection in patients who did not respond to previous interferon therapy. (9) All HALT-C patients had evidence of bridging fibrosis or cirrhosis on liver biopsy. During the ‘lead-in’ phase of HALT-C, patients received treatment with pegylated-interferon-α2a and ribavirin. We are conducting studies of potential associations between variants in candidate genes and response to this course of treatment in HALT-C patients. (10, 11)The present analysis investigates the possible impact of polymorphisms in genes for interferon-α, interferon-α receptors, the interferon-α pathway and interferon-α induced elements on treatment response.
Methods
Subjects
The HALT-C Trial
The design of the HALT-C trial, along with the major clinical, biochemical, and virological responses to the ‘Lead-in’ phase of therapy have been published.(9, 12) In brief, patients with advanced chronic hepatitis C (Ishak fibrosis score ≥3) who had not previously responded to interferon treatment (either with or without ribavirin) and who had a Child-Turcotte-Pugh score <7 were invited to enroll. Patients with other forms of liver disease, HIV infection, active illicit drug use, or current alcohol abuse were excluded. All subjects had quantifiable levels of serum HCV RNA, according to the Roche Amplicor Monitor assay version 2.0 (lower limit of detection, 100 IU/ml).
Treatment during the ‘Lead-in phase’ of the trial (i.e., first 24 weeks) consisted of pegylated-interferon-α2a (Pegasys®) 180 mcg/week parenterally, plus ribavirin (Copegus®) 1.0–1.2 g orally per day. Subjects with detectable HCV RNA at week 20 were considered to be ‘non-responders’. Subjects with undetectable HCV RNA at week 20 remained on treatment with pegylated interferon and ribavirin through week 48 and were followed until week 72 (24 weeks after pegylated interferon and ribavirin treatment was stopped). Subjects were considered to have an SVR if they had undetectable HCV RNA at week 72. Patients with undetectable HCV RNA at week 20, but detectable HCV RNA by week 72 were termed “relapsers.” Overall, 18% of patients achieved an SVR on this regimen, including 20% of European Americans and 6% of African Americans.(9, 12)
Information on demographic variables and other potential covariates were assessed through patient interviews. Race was self- identified as White, Black, Asian/Pacific Islander, American Indian/Alaskan Native, other or unknown, and ethnicity as Latino/Hispanic or not. For purposes of the present analysis of genetic variants, subjects who reported that they were White and not of Latino/Hispanic ethnicity are considered ‘European Americans.’ Those who categorized themselves as Black are considered ‘African Americans’.
Subjects entering the HALT-C trial were offered participation in an ancillary study to evaluate genetic polymorphisms associated with hepatitis C disease severity and response to treatment. Of the 1145 subjects enrolled in the Lead-in phase of the HALT-C trial, 1051 (91.7%) provided written informed consent to participate in the genetics ancillary study. The demographic and clinical characteristics of the genetic study participants did not differ from the overall group of HALT-C Lead-in phase subjects in any meaningful way.(11)
Current Study
This analysis compares genotype results in HALT-C patients with SVR to non-responders. Patients with virologic response (absence of detectable HCV RNA) at week 20, but virologic breakthrough during treatment or relapse after therapy discontinuation were not genotyped for the sake of cost efficiency. The present study was restricted to European American and African American patients because there were too few members of other racial/ethnic groups who met the criteria for SVR or non-responder (n=84 among all groups combined) to permit a meaningful statistical analysis.
Laboratory
DNA Extraction
DNA was extracted at SeraCare (Gaithersburg, MD) from frozen whole blood using the Gentra Systems Puregene kit (n=117) or from either Epstein-Barr transformed B-lymphocytes (n=636) or frozen peripheral blood mononuclear cells (n=21) using Qiagen DNA purification columns (Qiagen Inc, Valencia, CA). The majority (62.4%) of patients who achieved SVR had DNA extracted by the Gentra Systems method, whereas 96.8% of patients who did not achieve SVR had DNA extracted by the Qiagen method.
Genotyping
Table 1 lists the single nucleotide polymorphisms (SNPs) investigated in this study. In choosing variants for investigation, we gave priority to polymorphisms deemed more likely to have functional significance because they cause non-synonymous changes in coding or are located in possible gene promoter or regulatory regions; nonetheless ‘synonymous SNPs’, SNPs in introns and SNPs in untranslated regions were also examined. This analysis was limited to variants for which minor allele frequency was ≥5% among European American patients in this study.
Table 1.
Genetic variants examined with gene name, gene symbol, dbSNP identifier, common variant alias, protein changes, observed minor allele frequency (MAF) and results for test of Hardy-Weinberg (HW) proportions.
Based on Human Genome Build 35.1 and dbSNP build 126.
| Genetic Information | European Americans | African Americans | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Name | Symbol | dbSNP Identifier† | Variant* | Protein | MAF‡ | PHW§ | MAF‡ | PHW§ |
| Interferon-α | ||||||||
| Interferon, alpha-1 | IFNA1 | |||||||
| rs28383793 | −90G>A | 5.3 | 0.64 | 1.2 | 0.89 | |||
| rs1758566 | Ex1+51T>C | 9.3 | 0.67 | 19.5 | 0.78 | |||
| Interferon-α Receptor | ||||||||
| Interferon (alpha, beta and omega) receptor 1 | IFNAR1 | |||||||
| rs2243592 | IVS1−22T>G | 38.2 | 0.72 | 34.7 | 0.76 | |||
| rs2257167 | Ex4−30G>C | V168L | 13.7 | 0.68 | 16.5 | 0.72 | ||
| Interferon (alpha, beta and omega) receptor 2 | IFNAR2 | |||||||
| rs3153 | IVS1−4640G>A | 29.3 | 0.80 | 12.6 | 0.46 | |||
| rs4986956 | Ex2−33T>C | F8S | 7.3 | 0.97 | 5.7 | 0.36 | ||
| rs7279064 | Ex2−28T>G | F10V | 31.4 | 0.63 | 17.7 | 0.23 | ||
| rs2834158 | IVS3−43T>C | 31.7 | 0.69 | 18.7 | 0.36 | |||
| rs2236757 | IVS6−50A>G | 29.4 | 0.96 | 20.2 | 0.93 | |||
| Signal transduction in response to Interferon-α - JAK-STAT- Pathway | ||||||||
| Janus kinase 1 | JAK1 | |||||||
| rs310204 | IVS2+867T>G | 11.2 | 0.11 | 47.3 | 0.90 | |||
| rs2274947 | IVS4−186G>C | 12.3 | 0.79 | 19.5 | 0.59 | |||
| rs2256298 | IVS6−27C>T | 25.3 | 0.25 | 40.8 | 0.96 | |||
| rs310214 | IVS10−720A>G | 31.1 | 0.06 | 36.3 | 0.77 | |||
| rs2230587 | Ex14+62C>T | S671S | 12.2 | 0.81 | 18.1 | 0.46 | ||
| rs3737139 | Ex14−19C>G | A687A | 5.3 | 0.61 | 3.1 | 0.72 | ||
| rs2230588 | Ex15−53A>G | P721P | 25.7 | 0.06 | 38.7 | 0.59 | ||
| rs2254002 | IVS22+112G>T | 37.0 | 0.13 | 19.2 | 0.91 | |||
| Tyrosine kinase 2 | TYK2 | |||||||
| rs12720217 | −2256A>T | 14.6 | 0.43 | 38.1 | 0.42 | |||
| rs12720222 | IVS2−69G>A | 8.0 | 0.04 | 14.5 | 0.59 | |||
| rs12720356 | Ex15+4T>G | I684S | 9.0 | 0.46 | 2.3 | 0.79 | ||
| Signal transducer | STAT1 | |||||||
| and activator of transcription 1 | rs2066802 | Ex3+64T>C | L21L | 6.7 | 0.72 | 6.9 | 0.60 | |
| rs2066795 | IVS12−103C>T | 16.3 | <0.001 | 28.1 | 0.003 | |||
| rs2066804 | IVS21−8C>T | 24.4 | 0.13 | 15.6 | 0.14 | |||
| Signal transducer and activator of transcription 2 | STAT2 | |||||||
| rs2066807 | Ex20+58 | M594I | 6.7 | 0.10 | 0.8 | 1.00 | ||
| Interferon-α induced genes with antiviral properties | ||||||||
| Adenosine deaminase, RNA-specific | ADAR | |||||||
| rs2229857 | Ex3−451A>G | K384R | 28.7 | 0.80 | 43.0 | 0.50 | ||
| rs1127309 | Ex10+14G>A | V849V | 27.2 | 0.77 | 33.6 | 0.38 | ||
| rs1127313 | Ex16+1095C>T | 49.6 | 0.17 | 21.9 | 0.37 | |||
| Eukaryotic translation initiation factor 2A-alpha kinase 2 | EIF2AK2 | |||||||
| rs2307466 | Ex2+87C>G | 6.1 | 0.41 | 0.8 | 0.93 | |||
| rs2254958 | Ex2+110C>T | 39.2 | 0.95 | 15.4 | 0.96 | |||
| rs2373114 | IVS4+746A>G | 7.6 | 0.44 | 35.5 | 0.04 | |||
| rs4648174 | IVS4−206A>G | 38.2 | 0.65 | 14.0 | 0.72 | |||
| rs2307483 | IVS6−70T>C | 7.4 | 0.50 | 18.3 | 0.13 | |||
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells | NFKB1 | |||||||
| rs3774936 | IVS1+11190A>T | 32.4 | 0.87 | 11.2 | 0.58 | |||
| rs230532 | IVS2−826A>T | 33.0 | 0.63 | 16.1 | 0.39 | |||
| rs230521 | IVS5+4216G>C | 38.9 | 0.06 | 40.4 | 0.31 | |||
| rs4648058 | IVS12−458G>C | 31.3 | 0.53 | 22.5 | 0.24 | |||
| rs230547 | IVS23−1330T>C | 10.3 | 0.00 | 18.2 | 0.01 | |||
| Myxovirus resistance 1, interferon-inducible protein | MX1 | |||||||
| rs2071430 | −5932G>T | 11.1 | 0.09 | 22.0 | 0.54 | |||
| rs2838035 | IVS3+673T>G | 12.3 | 0.10 | 13.8 | 0.27 | |||
| rs464397 | IVS3+725T>C | 48.8 | 0.46 | 19.7 | 0.54 | |||
| rs458582 | IVS5+404G>T | 49.3 | 0.28 | 18.7 | 0.16 | |||
| rs455599 | IVS5+577G>A | 49.2 | 0.32 | 18.7 | 0.16 | |||
| rs469270 | IVS11−198G>A | 41.7 | 0.88 | 39.7 | 0.05 | |||
| rs35870315 | IVS11−21T>C | 10.8 | <0.001 | 12.0 | 0.91 | |||
| rs469390 | Ex13+4G>A | V379I | 40.1 | 0.88 | 45.0 | 0.12 | ||
| rs2280807 | IVS14+43A>G | 12.0 | 0.15 | 21.4 | 0.61 | |||
| rs1050008 | Ex16+114A>G | A541A | 13.5 | 0.00 | 23.8 | 0.67 | ||
| rs469304 | Ex16−64G>A | Q565Q | 44.8 | 0.40 | 21.4 | 0.60 | ||
| 2′, 5′-oligoadenylate synthetase 1 | OAS1 | |||||||
| rs3741981 | Ex3+15G>A | G162S | 44.2 | 0.19 | 29.0 | 0.14 | ||
| rs2285934 | IVS3+2480T>G | 35.8 | 0.73 | 43.1 | 0.90 | |||
| rs3177979 | Ex6+16G>A | A352T | 36.0 | 0.28 | 8.5 | 0.94 | ||
| Interferon regulatory factor | 0.09 | |||||||
| Interferon regulatory factor 3 | IRF3 | |||||||
| rs2304204 | Ex1+95A>G | 24.8 | 0.27 | 32.4 | 0.09 | |||
| rs3204440 | Ex1−53T>C | 5.2 | 0.04 | 5.1 | 0.54 | |||
| rs2304205 | Ex1−40T>G | 24.9 | 0.18 | 39.8 | 0.73 | |||
| rs2304206 | IVS1+17C>T | 24.5 | 0.53 | 38.6 | 0.97 | |||
| rs7251 | Ex8−81G>C | S427T | 32.8 | 0.57 | 34.4 | 0.55 | ||
dbSNP database, http://www.ncbi.nlm.nih.gov/SNP
Common genetic variant nomenclature adapted from Antonarakis SE and the Nomenclature Working Group
Minor allele frequency (%)
P-values for Hardy-Weinberg proportions, Pearson c2 (1 degree of freedom)
Genotyping was performed at the National Cancer Institute’s Core Genotyping Facility (CGF) with one exception (see below). To validate the publicly available data on genetic variation (NCBI SNP) and determine population specific genotype frequencies, CGF sequences samples from 102 individuals (Coriell Biorepository), which include representatives from various racial/ethnic groups. (13) Decisions regarding the development of specific genotyping assays for this study were based on these verified sequences and the observed genotype frequencies. Genotyping was performed using optimized TaqMan™ assays and analyzed on the ABI 7900HT platform (ABI, Foster City, CA). These assays are publicly available (including primers, probes and conditions) on the SNP500Cancer website (http://snp500cancer.nci.nih.gov).(13) Nomenclature for the description of these sequence variations is described in http://snp500cancer.nci.nih.gov/terms_snp_region.cfm. Genotyping for STAT2 Ex20+58 (rs2066807) was performed at SAIC-Frederick using similar methods.
DNA Sequencing
Because results for TYK2 -2256A>T differed markedly between African American patients with SVR and non-responders, we performed DNA sequencing to validate the performance of the TaqMan assay. Selected specimens were prepared by using the M13 tagged PCR primers from the SNP500 Cancer website.(13) After checking the PCR product via an agarose gel, the PCR products were sequenced using an ABI 3730XL machine. Sequence tracings were analyzed with the Sequencher 4.1.4 program (Genecodes, Ann Arbor, MI).
Data analysis
Data were analyzed at the New England Research Institute’s Data Coordinating Center, Computer Sciences Corporation and at the National Cancer Institute using SAS software (SAS Institute, Inc, SAS/STAT® 9.1 User’s Guide, 2004, Cary, NC, SAS Institute, Inc.). Because the frequency of genetic polymorphisms and response to therapy may both vary by ancestry, we analyzed European Americans and African Americans separately. The a priori statistical power was low for the small number of African American participants, therefore we focused on the results among European American patients and viewed results among African American patients as potential confirmatory evidence. For European Americans and African Americans separately, we examined whether genotype distribution (non-responders plus SVR) deviated from the proportions expected under Hardy-Weinberg equilibrium (HWE) on the basis of the Pearson’s chi square test.
For European American participants we compared genotype frequency in patients with SVR to non-responders by calculating unadjusted odds ratios (OR) and 95% confidence intervals (CI) using logistic regression.(14) Patients who were homozygous for the major variant served as the referent group for comparisons to patients who were heterozygous, patients who were homozygous for the minor variant, and patients who were either heterozygous or homozygous for the minor variant (dominant genetic model). To perform adjusted analyses for the dominant genetic model, we created logistic regression models that included the following covariates: HCV RNA level at baseline, HCV genotype, fibrosis stage, dose of pegylated-interferon and ribavirin received in HALT-C, previous treatment with ribavirin, and the presence of the HFE H63D variant (which is associated with treatment response in a previous analysis of HALT-C data).(10) We considered confounding to be present in the analysis of a variant when the adjusted OR differed from the unadjusted OR by at least 15%(15). To investigate which variables accounted for observed confounding, we performed stepwise logistic regression analyses that included the six covariates listed above plus gender and age as candidate variables.
For variants in which the adjusted OR differed meaningfully from the unadjusted OR, we also examined potential statistical interaction in a series of stratified logistic regression analyses that considered HCV RNA level at baseline, HCV genotype, fibrosis stage, dose of pegylated-interferon and ribavirin received in HALT-C, previous treatment with ribavirin, and the presence of the HFE H63D variant. These models tested for the presence of multiplicative interaction on a log scale.
Results
Descriptive Data
Selected demographic and clinical information are summarized in Table 2. Among the 581 European American participants in this analysis, 138 had an SVR and 443 were non-responders. Those with SVR were younger (mean age, 48.1 versus 50.1 years), had lower fibrosis scores, were less likely to be infected with HCV genotype 1, had lower mean HCV RNA and were less likely to have received ribavirin previously. The number of African American participants was much smaller (11 with SVR and 120 non-responders). Among these patients, those with SVR were less likely to have been infected with HCV genotype 1.
Table 2.
Demographic and clinical characteristics of the study population, HALT-C Study
| European Americans (n=581) | African Americans (n=131) | |||||
|---|---|---|---|---|---|---|
| Characteristic | Sustained Virologic Responders (n=138) | Non-Responders (n=443) | P | Sustained Virologic Responders (n=11) | Non-Responders (n=120) | P |
| Age (mean, sd) | 48.1 (6.33) | 50.1 (7.60) | 0.002 | 52.7 (5.04) | 51.8 (6.92) | 0.68 |
| Male (n, %) | 106 (77%) | 330 (75%) | 0.58 | 4 (59%) | 71 (36%) | 0.14 |
| Ishak Fibrosis Score | ||||||
| 2–4 (n, %) | 108 (78%) | 253 (57%) | <0.0001 | 9 (82%) | 75 (63%) | 0.2 |
| 5–6 (n, %) | 30 (22%) | 190 (43%) | 2 (18%) | 45 (38%) | ||
| HCV Genotype 1 (n, %) | 100 (72%) | 422 (95%) | <0.0001 | 9 (82%) | 116 (97%) | 0.01 |
| HCV RNA level (log 10) (mean, sd) | 6.28 (0.68) | 6.49 (0.46) | 0.001 | 5.96 (0.86) | 6.38 (0.51) | 0.13 |
| Prior treatment: | ||||||
| Interferon alone (n, %) | 65 (47%) | 96 (22%) | <0.0001 | 2 (18%) | 26 (22%) | 0.79 |
| Interferon and ribavirin (n, %) | 73 (53%) | 347 (78%) | 9 (82%) | 94 (78%) | ||
Genotype Results
Our analysis examined results performed on 56 alleles (from 13 genes) for which the observed minor allele frequency was & ge;5% among European American patients (Table 1). For two assays (STAT1 IVS12-103C>T; NFKB1 IVS23-1330T>C), the genotype distribution (non-responders plus SVR or non-responders only) deviated from the proportions expected under HWE (P<0.01) in both European American and African American subjects. For two other assays (MX1 IVS11-21T>C; MX1 Ex16+114A>G), the genotype distribution deviated from the HWE proportions among European Americans alone.
Associations between genotype and treatment results among European American patients are presented in Table 3 and Figure 1. For IFNA1, the gene that codes for interferon-α, we genotyped a SNP in the promoter region and another located in exon 1, and found no association for either. The interferon-α receptor consists of two major subunits, which are encoded by IFNAR1 and IFNAR2. We examined seven SNPs in these two genes, including three that cause non-synonymous changes. Patients who carried the minor variant for the intronic IFNAR1 IVS1-22T>G SNP were less likely to achieve SVR when other variables were considered in the model. Results for a model with HCV genotype, fibrosis stage, and previous treatment that included ribavirin produced the same results as the full model (adjusted OR, 0.57; 95% CI, 0.36–0.90; p=0.02).
Table 3.
Genotype frequency for genetic variants in European American patients with a sustained virological response (SVR) or non-response (NR), odds ratio (OR) for SVR and 95% confidence interval (95% CI) - HALT-C Study
| Gene | Variant*/Protein | Genotype | SVR | % | NR | % | OR | 95% | CI | ORa** | 95% | CI | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interferon-α | |||||||||||||
| IFNA1 | −90G>A | AA | 122 | 90 | 379 | 89 | 1.00 | ||||||
| GA+GG | 13 | 10 | 45 | 11 | 0.90 | 0.47 | 1.72 | 0.87 | 0.41 | 1.84 | 0.72 | ||
| Ex1+51T>C | TT | 116 | 85 | 341 | 81 | 1.00 | |||||||
| CT+CC | 20 | 15 | 80 | 19 | 0.74 | 0.43 | 1.25 | 0.70 | 0.38 | 1.29 | 0.25 | ||
| Interferon-α Receptor | |||||||||||||
| IFNAR1 | IVS1−22T>G | TT | 61 | 45 | 160 | 37 | 1.00 | ||||||
| GT+GG | 76 | 55 | 277 | 63 | 0.72 | 0.49 | 1.06 | 0.57 | 0.36 | 0.90 | 0.02 | ||
| Ex4−30G>C | GG | 103 | 75 | 327 | 74 | 1.00 | |||||||
| V168L | GC+CC | 34 | 25 | 112 | 26 | 0.96 | 0.62 | 1.50 | 0.85 | 0.50 | 1.44 | 0.55 | |
| IFNAR2 | IVS1−4640G>A | GG | 63 | 46 | 216 | 51 | 1.00 | ||||||
| GA+AA | 73 | 54 | 209 | 49 | 1.20 | 0.81 | 1.76 | 1.35 | 0.85 | 2.13 | 0.20 | ||
| Ex2−33T>C | TT | 111 | 81 | 385 | 88 | 1.00 | |||||||
| F8S | TC+CC | 26 | 19 | 55 | 13 | 1.64 | 0.98 | 2.74 | 2.07 | 1.12 | 3.85 | 0.02 | |
| Ex2−28T>G | TT | 56 | 43 | 206 | 49 | 1.00 | |||||||
| F10V | TG+GG | 75 | 57 | 215 | 51 | 1.28 | 0.86 | 1.91 | 1.33 | 0.84 | 2.12 | 0.22 | |
| IVS3−43T>C | CC | 58 | 43 | 212 | 48 | 1.00 | |||||||
| TC+TT | 78 | 57 | 227 | 52 | 1.26 | 0.85 | 1.85 | 1.32 | 0.84 | 2.08 | 0.23 | ||
| IVS6−50A>G | GG | 63 | 47 | 213 | 51 | 1.00 | |||||||
| AG+AA | 72 | 53 | 205 | 49 | 1.19 | 0.81 | 1.75 | 1.36 | 0.86 | 2.16 | 0.19 | ||
| Signal Transduction Response (JAK-STAT Pathway) | |||||||||||||
| JAK1 | IVS2+867T>G | TT | 108 | 79 | 347 | 80 | 1.00 | ||||||
| GT+GG | 29 | 21 | 88 | 20 | 1.06 | 0.66 | 1.70 | 1.38 | 0.79 | 2.42 | 0.26 | ||
| IVS4−186G>C | GG | 108 | 79 | 332 | 76 | 1.00 | |||||||
| GC+CC | 29 | 21 | 104 | 24 | 0.86 | 0.54 | 1.37 | 1.04 | 0.61 | 1.78 | 0.88 | ||
| IVS6−27C>T | CC | 74 | 54 | 252 | 58 | 1.00 | |||||||
| TC+TT | 64 | 46 | 185 | 42 | 1.18 | 0.80 | 1.73 | 1.37 | 0.87 | 2.16 | 0.17 | ||
| IVS10−720A>G | AA | 62 | 46 | 220 | 50 | 1.00 | |||||||
| AG+GG | 73 | 54 | 218 | 50 | 1.19 | 0.81 | 1.75 | 1.50 | 0.94 | 2.37 | 0.09 | ||
| Ex14+62C>T | CC | 107 | 78 | 336 | 77 | 1.00 | |||||||
| S671S | CT+TT | 30 | 22 | 103 | 23 | 0.92 | 0.58 | 1.45 | 1.04 | 0.61 | 1.76 | 0.90 | |
| Ex14−19C>G | CC | 123 | 90 | 392 | 89 | 1.00 | |||||||
| A687A | CG+GG | 14 | 10 | 46 | 11 | 0.97 | 0.52 | 1.82 | 1.28 | 0.62 | 2.66 | 0.50 | |
| Ex15−53A>G | AA | 74 | 55 | 231 | 57 | 1.00 | |||||||
| P721P | GA+GG | 60 | 45 | 173 | 43 | 1.08 | 0.73 | 1.61 | 1.27 | 0.79 | 2.03 | 0.32 | |
| IVS22+112G>T | GG | 53 | 38 | 184 | 42 | 1.00 | |||||||
| GT+TT | 85 | 62 | 253 | 58 | 1.17 | 0.79 | 1.73 | 1.66 | 1.03 | 2.66 | 0.04 | ||
| TYK2 | −2256A>T | TT | 91 | 66 | 327 | 74 | 1.00 | ||||||
| AT+AA | 47 | 34 | 112 | 26 | 1.51 | 1.00 | 2.28 | 1.47 | 0.90 | 2.40 | 0.12 | ||
| IVS2−69G>A | GG | 114 | 83 | 369 | 84 | 1.00 | |||||||
| AA+AG | 24 | 17 | 68 | 16 | 1.14 | 0.69 | 1.90 | 1.21 | 0.65 | 2.26 | 0.54 | ||
| Ex15+4T>G | TT | 114 | 84 | 359 | 83 | 1.00 | |||||||
| I684S | TG+GG | 22 | 16 | 74 | 17 | 0.94 | 0.56 | 1.58 | 0.76 | 0.40 | 1.42 | 0.38 | |
| STAT1 | Ex3+64T>C | TT | 116 | 85 | 381 | 88 | 1.00 | ||||||
| L21L | CT+CC | 21 | 15 | 53 | 12 | 1.30 | 0.75 | 2.25 | 1.23 | 0.64 | 2.40 | 0.54 | |
| IVS12−103C>T | CC | 99 | 72 | 323 | 75 | 1.00 | |||||||
| TC+TT | 39 | 28 | 109 | 25 | 1.17 | 0.76 | 1.79 | 1.21 | 0.72 | 2.03 | 0.47 | ||
| IVS21−8C>T | CC | 83 | 61 | 252 | 57 | 1.00 | |||||||
| TC+TT | 53 | 39 | 187 | 43 | 0.86 | 0.58 | 1.28 | 0.81 | 0.51 | 1.28 | 0.36 | ||
| STAT2 | Ex20+58 | CC | 120 | 88 | 378 | 87 | 1.00 | ||||||
| M594I | CG+GG | 16 | 12 | 55 | 13 | 0.92 | 0.48 | 1.72 | |||||
| Interferon-α Induced Genes with Anti-Viral Properties | |||||||||||||
| ADAR | Ex2−451A>G | GG | 61 | 45 | 229 | 52 | 1.00 | ||||||
| K384R | AG+AA | 75 | 55 | 208 | 48 | 1.35 | 0.92 | 1.99 | 1.57 | 1.00 | 2.49 | 0.05 | |
| Ex9+14G>A | GG | 63 | 46 | 240 | 55 | 1.00 | |||||||
| V849V | AG+AA | 74 | 54 | 197 | 45 | 1.43 | 0.97 | 2.10 | 1.67 | 1.06 | 2.64 | 0.03 | |
| Ex15+1095C>T | TT | 33 | 24 | 121 | 28 | 1.00 | |||||||
| CT+CC | 103 | 76 | 316 | 72 | 1.20 | 0.77 | 1.86 | 1.35 | 0.80 | 2.28 | 0.26 | ||
| EIF2AK2 | Ex2+87C>G | CC | 121 | 90 | 385 | 88 | 1.00 | ||||||
| CG+GG | 14 | 10 | 55 | 13 | 0.81 | 0.44 | 1.51 | 0.63 0.23 |
0.30 | 1.31 | 0.22 | ||
| Ex2+110C>T | CC | 44 | 32 | 167 | 39 | 1.00 | |||||||
| CT+TT | 94 | 68 | 265 | 61 | 1.35 | 0.90 | 2.02 | 1.15 0.15 |
0.72 | 1.85 | 0.57 | ||
| IVS3+746A>G | GG | 121 | 89 | 366 | 84 | 1.00 | |||||||
| AG+AA | 15 | 11 | 70 | 16 | 0.65 | 0.36 | 1.18 | 0.70 | 0.34 | 1.43 | 0.33 | ||
| IVS4−206A>G | AA | 41 | 30 | 172 | 40 | 1.00 | |||||||
| GA+GG | 95 | 70 | 257 | 60 | 1.55 | 1.03 | 2.35 | 1.26 | 0.78 | 2.04 | 0.34 | ||
| IVS5−70T>C | TT | 121 | 89 | 367 | 85 | 1.00 | |||||||
| TC+CC | 15 | 11 | 67 | 15 | 0.68 | 0.37 | 1.23 | 0.74 | 0.36 | 1.52 | 0.41 | ||
| NFKB1 | IVS1+11190A>T | AA | 54 | 40 | 205 | 47 | 1.00 | ||||||
| AT+TT | 82 | 60 | 228 | 53 | 1.37 | 0.92 | 2.02 | 1.58 | 0.99 | 2.53 | 0.06 | ||
| IVS2−826A>T | AA | 52 | 38 | 199 | 46 | 1.00 | |||||||
| AT+TT | 85 | 62 | 229 | 54 | 1.42 | 0.96 | 2.11 | 1.61 | 1.00 | 2.57 | 0.05 | ||
| IVS5+4216G>C | GG | 47 | 34 | 156 | 36 | 1.00 | |||||||
| CG+CC | 90 | 66 | 280 | 64 | 1.07 | 0.71 | 1.60 | 1.08 | 0.68 | 1.74 | 0.74 | ||
| IVS12−458G>C | GG | 60 | 44 | 209 | 48 | 1.00 | |||||||
| CG+CC | 77 | 56 | 230 | 52 | 1.17 | 0.79 | 1.72 | 1.32 | 0.84 | 2.09 | 0.23 | ||
| IVS23−1330T>C | CC | 113 | 82 | 356 | 81 | 1.00 | |||||||
| CT+TT | 24 | 17 | 81 | 19 | 0.93 | 0.57 | 1.54 | 0.70 | 0.38 | 1.26 | 0.23 | ||
| MX1 | −5932G>T | GG | 112 | 82 | 346 | 79 | 1.00 | ||||||
| TG+TT | 25 | 18 | 91 | 21 | 0.85 | 0.52 | 1.39 | 0.78 | 0.44 | 1.39 | 0.39 | ||
| IVS3+673T>G | TT | 112 | 81 | 334 | 76 | 1.00 | |||||||
| TG+GG | 26 | 18 | 103 | 24 | 0.75 | 0.47 | 1.22 | 0.69 | 0.40 | 1.22 | 0.20 | ||
| IVS3+725T>C | CC | 34 | 25 | 104 | 75 | 1.00 | |||||||
| TC+TT | 100 | 75 | 324 | 76 | 0.94 | 0.60 | 1.48 | 0.89 | 0.52 | 1.53 | 0.68 | ||
| IVS5+404G>T | TT | 34 | 24 | 110 | 76 | 1.00 | |||||||
| TG+GG | 100 | 75 | 322 | 75 | 1.01 | 0.64 | 1.57 | 0.98 | 0.58 | 1.67 | 0.94 | ||
| IVS5+577G>A | AA | 35 | 24 | 110 | 76 | 1.00 | |||||||
| GA+GG | 102 | 74 | 327 | 75 | 0.98 | 0.63 | 1.52 | 0.99 | 0.58 | 1.67 | 0.96 | ||
| IVS11−198G>A | AA | 49 | 37 | 140 | 33 | 1.00 | |||||||
| GA+GG | 85 | 64 | 284 | 67 | 0.86 | 0.57 | 1.28 | 0.93 | 0.57 | 1.50 | 0.76 | ||
| IVS11−21T>C | TT | 113 | 83 | 348 | 81 | 1.00 | |||||||
| TC+CC | 23 | 17 | 84 | 19 | 0.84 | 0.51 | 1.40 | 0.80 | 0.45 | 1.45 | 0.47 | ||
| Ex13+4G>A | AA | 52 | 38 | 154 | 35 | 1.00 | |||||||
| V379I | GA+GG | 85 | 62 | 286 | 65 | 0.88 | 0.59 | 1.31 | 0.92 | 0.58 | 1.48 | 0.74 | |
| IVS14+43A>G | AA | 103 | 76 | 344 | 79 | 1.00 | |||||||
| AG+GG | 33 | 24 | 93 | 21 | 1.19 | 0.75 | 1.87 | 1.22 | 0.72 | 2.07 | 0.47 | ||
| Ex16+114A>G | AA | 100 | 74 | 327 | 77 | 1.00 | |||||||
| A541A | AG+GG | 36 | 26 | 97 | 23 | 1.21 | 0.78 | 1.89 | 1.23 | 0.73 | 2.06 | 0.44 | |
| Ex16−64G>A | AA | 44 | 32 | 135 | 31 | 1.00 | |||||||
| Q565Q | AG+GG | 92 | 68 | 301 | 69 | 0.94 | 0.62 | 1.42 | 1.09 | 0.67 | 1.79 | 0.72 | |
| OAS1 | Ex3+15G>A | AA | 53 | 39 | 127 | 30 | 1.00 | ||||||
| G162S | GA+GG | 83 | 61 | 291 | 70 | 0.68 | 0.46 | 1.02 | 0.65 | 0.41 | 1.05 | 0.08 | |
| GG | |||||||||||||
| IVS3+2480T>G | 65 | 47 | 173 | 40 | 1.00 | ||||||||
| TG+TT | 72 | 53 | 261 | 60 | 0.73 | 0.50 | 1.08 | 0.75 | 0.48 | 1.19 | 0.22 | ||
| Ex6+16G>A | AA | 65 | 47 | 177 | 40 | 1.00 | |||||||
| A352T | AG+GG | 73 | 53 | 262 | 60 | 0.76 | 0.52 | 1.12 | 0.76 | 0.48 | 1.20 | 0.24 | |
| Interferon Regulatory Factor | |||||||||||||
| IRF3 | Ex1+95A>G | AA | 73 | 53 | 255 | 59 | 1.00 | ||||||
| GA+GG | 64 | 47 | 179 | 41 | 1.25 | 0.85 | 1.84 | 1.44 | 0.91 | 2.27 | 0.12 | ||
| Ex1−53T>C | TT | 123 | 90 | 385 | 90 | 1.00 | |||||||
| CT+CC | 13 | 10 | 42 | 10 | 0.97 | 0.50 | 1.86 | 1.02 | 0.48 | 2.16 | 0.96 | ||
| Ex1−40T>G | TT | 75 | 55 | 249 | 58 | 1.00 | |||||||
| GT+GG | 61 | 45 | 179 | 42 | 1.13 | 0.77 | 1.67 | 1.36 | 0.86 | 2.14 | 0.19 | ||
| IVS1+17C>T | CC | 73 | 54 | 254 | 59 | 1.00 | |||||||
| CT+TT | 63 | 46 | 179 | 41 | 1.23 | 0.83 | 1.80 | 1.42 | 0.90 | 2.23 | 0.13 | ||
| Ex8−81G>C | GG | 57 | 41 | 206 | 47 | 1.00 | |||||||
| S427T | CG+CC | 81 | 59 | 232 | 53 | 1.26 | 0.86 | 1.86 | 1.48 | 0.93 | 2.35 | 0.10 | |
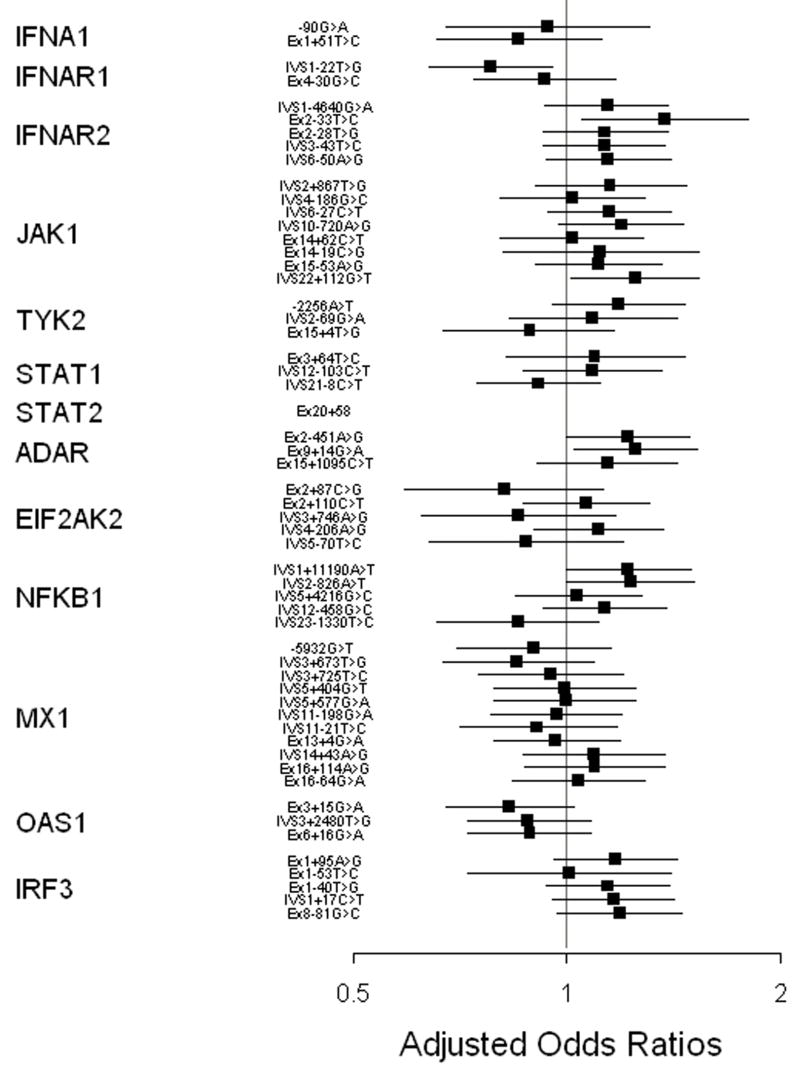
Figure 1.
Adjusted odds ratios and 95% confidence intervals for associations between genotype and treatment results among European American patients, HATL-C Trial.
Patients who carried the C variant for the IFNAR2 SNP found 33 bp before the end of exon 2 (IFNAR2 Ex2-33T>C) were more likely to have had an SVR (adjusted OR, 2.07; 95% CI, 1.12–3.85; p=0.02). This IFNAR2 variant changes phenylalanine to serine at amino acid position 8. To investigate which variables accounted for the confounding observed for IFNAR2 Ex2-33 (26% difference between adjusted OR and unadjusted OR), we performed stepwise logistic regression analyses. No single variable accounted for the observed confounding, but a model that included HCV genotype, fibrosis stage, baseline HCV RNA level, and whether previous treatment included ribavirin yielded results that were similar to the full model (adjusted OR, 2.09; p=0.02).
Binding of interferon-α to its receptor activates Janus kinase 1 and protein tyrosine kinase 2, which are encoded by JAK1 and TYK2, respectively. Among 8 SNPS in JAK1 examined, there was an association with IVS22+112G>T and SVR (adjusted OR, 1.66; 95% CI, 1.03–2.66; p=0.04) that, per the stepwise model, depended on all six covariates that comprised the full model. For TYK2 we examined three SNPs. For promoter region variant TYK2 -2256, SVR was more common among European American patients who carried the A allele (unadjusted OR, 1.51; 95% CI, 1.00–2.28). This relationship was stronger among the African American patients (Supplemental Table 1): among the 10 patients with an SVR who were successfully genotyped for this locus none were homozygous for major variant (T) compared to 52/120 (43%) of non-responders (0.006, Fishers exact test). To exclude the possibility that genotyping error explained these results among African American patients, we sequenced the region that includes TYK2 -2256 from these 10 African American patients with an SVR and a sample of African American non-responders. The sequence based results were fully concordant with those obtained by TaqMan genotyping. The frequency of the TYK2 -2256T allele among the 74 Human Diversity Panel subjects of African origin (63.5%; http://snp500cancer.nci.nih.gov/snp.cfm?ethnic=true&hdp=true&snp_id=TYK2-01) was much more similar to the frequency among HALT-C African American non-responders (64.2%) than those with an SVR (35.0%). In contrast, there was no association for either a TYK2 variant located in intron 2 or a non-synonymous variant in exon 15, and response to therapy.
TYK2, together with JAK1, phosphorylates signal transducer and activator of transcription 1 (STAT1), and STAT2. Phosphorylated STAT1 and STAT2 form heterodimers that translocate into the nucleus following interferon receptor activation. We examined three STAT1 SNPs and one SNP found on STAT2. None of these four SNPs were associated with SVR.
We also examined variants in genes that code for proteins with antiviral properties that are induced by interferon-α. RNA-specific adenosine deaminase (ADAR) exhibits its antiviral properties by RNA editing conducted through deamination of adenosine to produce inosine. We examined three exonic variants of ADAR and found that the synonymous Ex9+14G>A SNP was associated with SVR (adjusted OR, 1.67; 95% CI, 1.06–2.64; p=0.03). In the stepwise model, fibrosis stage, dose of pegylated-interferon and ribavirin received in HALT-C, and previous treatment with ribavirin accounted for higher OR compared to the unadjusted model. Eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) is a serine/threonine kinase that can inhibit translation by phosphorylation of eukaryotic protein synthesis initiation factor -2α. We examined two exonic and three intronic variants of EIF2AK2. Patients who carried EIF2AK2 IVS4-206G were more likely to have an SVR in the unadjusted analysis, but this association was lessened in the adjusted analysis (adjusted OR 1.26; 95% CI, 0.78–2.04; p= 0.34) due to consideration of viral genotype. EIF2AK2 activates the transcription factor NF-κB, which is essential for interferon-β expression.(16, 17) We examined five intronic variants of NFKB1but none had a statistically significant association with SVR.
Human cytoplasmic MxA, a highly conserved GTPase, can inhibit viral RNA synthesis. We examined 11 variants of its gene, MX1, some of which were in genetic linkage. In our analysis of MX1 -5932G>T (also known as MX1 -88), which was associated with treatment response in previous studies, we found no association (unadjusted OR, 0.85; 95% CI, 0.52–1.39). No association was observed for any of the ten other variants of MX1 that we examined. The 2′-5′ oligoadenylate synthetase system (2′-5′-OAS) mediates the degradation of viral and cellular RNA, mainly by activation of RNAse L, which cleaves single strand RNA and inhibits protein synthesis. We genotyped three variants of OAS1, including two SNPs that cause non-synonymous amino acid changes. Patients who carried the G variant for OAS1 Ex3+15G>A (G162S) were somewhat less likely to achieve SVR (unadjusted OR, 0.68; 95% CI, 0.46–1.02), although these results did not reach statistical significance.
Interferon-regulatory factor 3 (IRF-3) is a transcription factor activated in HCV infected cells. IRF-3 exerts antiviral effects by binding to interferon-stimulated response elements (ISREs) and inducing expression of type I interferon and numerous other antiviral genes. HCV NS3/4A can block IRF-3 phosphorylation and activation. Variation in the IRF3 gene could, therefore, have an impact on viral persistence and reduce response to interferon.(18, 19) We examined five IRF3 variants, but found no evidence that any affected response to HCV treatment.
We looked for evidence of a multiplicative statistical interaction by creating a series of stratified logistic regression models for these variants: IFNAR1 IVS1-22T>G; IFNAR2 Ex2-33T>C; JAK1 IVS2+867T>G; JAK1 IVS22+112G>T; ADAR Ex3-451A>G; and EIF2AK2 IVS4-206A>G. For IFNAR2 Ex2-33T>C, a strong association with SVR was present among participants who had a baseline HCV RNA level >6.5 log10 copies/ml (the median value; adjusted OR, 4.80; 95% CI, 1.87–12.35; p=0.001), but not among those with lower viral levels (adjusted OR, 1.07; 95% CI, 0.47–2.45; p=0.9; interaction p-value= 0.03). No other interactions yielded a p-value <0.05.
Discussion
We examined whether variants of genes related to interferon-α were associated with response to treatment with pegylated interferon alfa-2a plus ribavirin in patients with advanced chronic hepatitis C infection who had failed a previous course of interferon-based treatment. Among European American patients, variants associated with SVR were: the intronic IFNAR1 IVS1-22G variant (lower frequency in patients with SVR); IFNAR2 Ex2-33C (higher frequency with SVR), which changes an amino acid in an interferon-α receptor subunit; the intronic variant IVS22+112T (higher frequency with SVR) TYK2 -2256A (higher frequency with SVR), a promoter region SNP, and ADAR Ex9+14A (higher frequency with SVR), which is a synonymous variant. Statistical power was limited for our analysis of the relatively small number of African Americans enrolled in this study. Nonetheless, the observation that the TYK2 -2256A allele predicted SVR was supported by a very strong association in African American patients; whereas 43% of African American non-responders were homozygous for the major (T) variant, 0/10 African American patients with an SVR had this genotype. Although the number of African American patients with an SVR in this study is small, this finding does not appear to be due to chance and it is not due to genotyping error. The evidence that the negative predictive value for SVR for the TYK2 TT genotype is high among African American patients (100% in this small study) merits confirmation in a larger study.
There have been relatively few previous studies of interferon-α related genes and response to therapy for HCV infection (5). Virahep-C cohort investigators recently reported results for nine variants located in seven of the genes we examined.(20) For IFNA IVS1-22T>G [rs2243592] and IFNAR2 Ex2-33T>C [rs4986956]) we found an association with SVR after adjusting for variables that predict treatment response, but these associations were not present in the Virahep-C analysis, which did not adjust for these factors. Most of the variants we found to be associated with SVR had stronger OR after we controlled for known predictors of treatment response in logistic regression models and only one was statistically significant at the p=0.05 level without adjustment (table 3). For example, for IFNAR2 Ex2-33T>C the OR increased from 1.64 (p=0.06) without adjustment to 2.09 (p=0.02) in the adjusted model. Therefore, the consideration of other variables that predict treatment response in the present study could account for the difference with Virahep-C. In addition, the number of European American subjects in the present study (n=581) is greater than that in Virahep-C (n=194).
We know of no prior reports for the TYK2 -2256A polymorphisms that we can compare to our results or of any ‘functional’ studies of this genetic variant. However, consistent with our finding that a SNP in the promoter region of TYK2 affects response to treatment with interferon-α-based therapy, a recent report suggests that altered expression of TYK2 may cause interferon resistance in vitro. In a study of HCV replicon cell lines, those that continued viral replication in the presence of interferon-α displayed markedly reduced expression of the TYK-2 protein, as well as JAK-1.(21) Future studies of TYK2 -2256A should examine its effect in other populations of HCV-infected patients undergoing treatment with pegylated interferon plus ribavirin and investigate the functional consequences of this variant in liver tissue.
Previous studies of MX1 -5932G>T (also known as -88MXA) suggested that the T allele is associated with better response to interferon-α therapy. In separate Japanese populations, Hijikata et al and Suzuki et al reported that homozygosity for the G allele was significantly less frequent among patients who achieved SVR after interferon treatment compared to non-responders. (22, 23) In European patients, Knapp also observed a lower frequency of the GG genotype in non-responders to therapy, especially among patients with viral genotype 1.(24) In contrast, we found no evidence that carrying MX1 -5932T improved treatment response among the European American patients enrolled in the HALT-C trial. The reason for the different results reported from these studies is not clear. The studies differ with regard to factors that are related to response to interferon-based treatments, such as the degree of liver fibrosis in the populations, exclusion of HCV non-1 genotypes, or type of interferon used (pegylated vs. regular), and it is also possible that the differences between studies are due to chance.
Some strengths of our study include the uniform treatment of patients with the current ‘gold standard’ regimen (pegylated interferon plus ribavirin) and careful follow-up for treatment outcomes within the context of an NIH-sponsored clinical trial. However, potential limitations of this study should be considered as well. HALT-C trial participants have advanced fibrosis and failed to respond to prior anti-HCV treatment, raising the question of whether results from HALT-C can be generalized to other patients with chronic hepatitis C. It is reassuring that HALT-C investigators have found strong associations for risk factors for treatment failure (i.e., older age, HCV genotype 1, higher HCV RNA level, the presence of cirrhosis) that have been reported in previously untreated patients.(12) Among European American participants, the study had statistical power ≥80% to detect a two- fold association between the presence of an allele and SVR for variants with a minor allele frequency≥15%. Although this includes most of the variants that we examined, the study’s statistical power for less common variants was suboptimal. Finally, because we examined 56 genetic variants in this study, it is possible that some or all of the ‘statistically significant’ associations we observed are due to chance. Expert opinion differs regarding the necessity of adjusting for multiple comparisons(25) and in investigating candidate genes we favor presenting results without such adjustments, interpreting the results conservatively and seeking to replicate the findings in multiple populations. For that reason, we believe that the TYK2 -2256A variant, which was associated with treatment response in separate analyses of European American and African American patients, is the most promising result from this study. In vitro evidence of altered function would have strengthened our findings, but the extensive studies required to evaluate these polymorphisms for function were beyond the scope of this project. Evaluation of genetic polymorphisms for altered function is challenging because there are multiple levels at which alterations may produce functional changes (e.g., transcriptional, posttranscriptional, posttranslational differences) (26) and because these alterations are often tissue specific.(27, 28) Unfortunately, this project was not designed to assess potential functional mechanisms in liver tissue. Well-powered genome-wide association studies have yielded important advances in the understanding of a range of medical conditions. Unfortunately, the HALT-C study would have low statistical power to detect differences that are statistically significant at the genome-wide level; (26) a consortium may be required in order to bring together sufficient subjects for a genome-wide association study of HCV treatment response.
In summary, several interferon-α-related host genetic polymorphisms were associated with response to therapy for chronic hepatitis C in this study. If confirmed, these findings could provide insight into the variability in response to treatment for HCV and, possibly, provide prognostic information for responsiveness to therapy.
Supplementary Material
Acknowledgments
The HALT-C Trial was supported by the National Institute of Diabetes & Digestive & Kidney Diseases (contract numbers are listed below). Additional support was provided by the National Institute of Allergy and Infectious Diseases (NIAID), the National Cancer Institute, the National Center for Minority Health and Health Disparities and by General Clinical Research Center grants from the National Center for Research Resources, National Institutes of Health (grant numbers are listed below). Additional funding for HALT-C was supplied by Hoffmann-La Roche, Inc. through a Cooperative Research and Development Agreement (CRADA) with the National Institutes of Health. The present study was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Financial Disclosures: Financial relationships of the authors with Hoffmann-La Roche, Inc., are as follows: T. R. Morgan is a consultant, is on the speaker’s bureau, and receives research support; H. L. Bonkovsky receives research support; R. K. Sterling is a consultant, is on the speaker’s bureau, and receives research support; K. L. Lindsay is a consultant and receives research support; R. J. Fontana is on the speaker’s bureau; W. M. Lee receives research support; A. M. Di Bisceglie is a consultant and receives research support; R. T. Chung receives research support. Authors with no financial relationships to this project are T. M. Welzel, D. Naishadham, R. M. Pfeiffer, E. C. Wright, A. A. Hutchinson, A. T. Crenshaw, A. Bashirova, M. Carrington, M. Dotrang, M. G. Ghany, D. R. Gretch, S. J. Chanock, and T.R. O’Brien.
Footnotes
This is publication #32 of the HALT-C Trial.
In addition to the authors of this manuscript, the following individuals were instrumental in the planning, conduct and/or care of patients enrolled in this study at each of the participating institutions as follows: University of Massachusetts Medical Center, Worcester, MA: (Contract N01-DK-9-2326) Gyongyi Szabo, MD, Maureen Cormier, RN, Donna Giansiracusa, RN
University of Connecticut Health Center, Farmington, CT: (Grant M01RR-06192) Michelle Kelley, RN, ANP
Saint Louis University School of Medicine, St Louis, MO: (Contract N01-DK-9-2324) Bruce Bacon, MD, Brent Neuschwander-Tetri, MD, Debra King, RN
Massachusetts General Hospital, Boston, MA: (Contract N01-DK-9-2319, Grant M01RR-01066), Jules L. Dienstag, MD, Andrea E. Reid, MD, Wallis A Molchen, Loriana Di Giammarino
University of Colorado School of Medicine, Denver, CO: (Contract N01-DK-9-2327, Grant M01RR-00051) Gregory T. Everson, MD, Jennifer DeSanto, RN, Carol McKinley, RN
University of California - Irvine, Irvine, CA: (Contract N01-DK-9-2320, Grant M01RR-00827) John C. Hoefs, MD, M. Mazen Jamal, MD, MPH, Choon Park, RN
University of Texas Southwestern Medical Center, Dallas, TX: (Contract N01-DK-9-2321, Grant M01RR-00633) Janel Shelton, Rivka Elbein, RN, BSN
University of Southern California, Los Angeles, CA: (Contract N01-DK-9-2325, Grant M01RR-00043) Carol B. Jones, RN, Susan L. Milstein, RN
University of Michigan Medical Center, Ann Arbor, MI: (Contract N01-DK-9-2323, Grant M01RR-00042) Anna S.F. Lok, MD, Pamela A. Richtmyer, LPN, CCRC
Virginia Commonwealth University Health System, Richmond, VA: (Contract N01-DK-9-2322, Grant M01RR-00065) Mitchell L. Shiffman, MD, Charlotte Hofmann, RN, Paula Smith, RN
National Institute of Diabetes and Digestive and Kidney Diseases, Liver Disease Branch, Bethesda, MD: T. Jake Liang, MD, Yoon Park, RN, Elenita Rivera, RN, Vanessa Haynes-Williams, RN
National Institute of Diabetes and Digestive and Kidney Diseases, Division of Digestive Diseases and Nutrition, Bethesda, MD: Leonard B. Seeff, MD, Patricia R. Robuck, PhD, Jay H. Hoofnagle, MD
University of Washington, Seattle, WA: (Contract N01-DK-9-2318), Chihiro Morishima, MD, Minjun Chung Apodaca, BS, ASCP
New England Research Institutes, Watertown, MA: (Contract N01-DK-9-2328) Kristin K Snow, Sc.D., Teresa M. Curto, MPH, Linda J. Massey
Armed Forces Institute of Pathology, Washington, DC: Zachary D Goodman, MD
Data and Safety Monitoring Board Members: (Chair) Gary L. Davis, MD, Guadalupe Garcia-Tsao, MD, Michael Kutner, PhD, Stanley M. Lemon, MD, Robert P. Perrillo, MD
References
- 1.NIH Consensus Statement on Management of Hepatitis C. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, Brown RS, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Ferenci P. Predictors of response to therapy for chronic hepatitis C. Semin Liver Dis. 2004;24 Suppl 2:25–31. doi: 10.1055/s-2004-832925. [DOI] [PubMed] [Google Scholar]
- 5.Yee LJ. Host genetic determinants in hepatitis C virus infection. Genes Immun. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 6.Chung RT, Gale M, Jr, Polyak SJ, Lemon SM, Liang TJ, Hoofnagle JH. Mechanisms of action of interferon and ribavirin in chronic hepatitis C: Summary of a workshop. Hepatology. 2008;47:306–320. doi: 10.1002/hep.22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes E, Harcourt G, Brown D, Lucas M, Phillips R, Dusheiko G, Klenerman P. The dynamics of T-lymphocyte responses during combination therapy for chronic hepatitis C virus infection. Hepatology. 2002;36:743–754. doi: 10.1053/jhep.2002.35344. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann RM, Diepolder HM, Zachoval R, Zwiebel FM, Jung MC, Scholz S, Nitschko H, et al. Mapping of immunodominant CD4+ T lymphocyte epitopes of hepatitis C virus antigens and their relevance during the course of chronic infection. Hepatology. 1995;21:632–638. [PubMed] [Google Scholar]
- 9.Lee WM, Dienstag JL, Lindsay KL, Lok AS, Bonkovsky HL, Shiffman ML, Everson GT, et al. Evolution of the HALT-C Trial: pegylated interferon as maintenance therapy for chronic hepatitis C in previous interferon nonresponders. Control Clin Trials. 2004;25:472–492. doi: 10.1016/j.cct.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Bonkovsky HL, Naishadham D, Lambrecht RW, Chung RT, Hoefs JC, Nash SR, Rogers TE, et al. Roles of iron and HFE mutations on severity and response to therapy during retreatment of advanced chronic hepatitis C. Gastroenterology. 2006;131:1440–1451. doi: 10.1053/j.gastro.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 11.Morgan TR, Lambrecht RW, Bonkovsky HL, Chung RT, Naishadham D, Sterling RK, Fontana RJ, et al. DNA polymorphisms and response to treatment in patients with chronic hepatitis C: Results from the HALT-C trial. J Hepatol. 2008;49:548–556. doi: 10.1016/j.jhep.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiffman ML, Di Bisceglie AM, Lindsay KL, Morishima C, Wright EC, Everson GT, Lok AS, et al. Peginterferon alfa-2a and ribavirin in patients with chronic hepatitis C who have failed prior treatment. Gastroenterology. 2004;126:1015–1023. doi: 10.1053/j.gastro.2004.01.014. discussion 1947. [DOI] [PubMed] [Google Scholar]
- 13.Packer BR, Yeager M, Staats B, Welch R, Crenshaw A, Kiley M, Eckert A, et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:D528–532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosmer DW, Lemeshow S. Applied Logistic Regression. 2. John Wiley and Sons; New York, NY: 2000. [Google Scholar]
- 15.Rothman KJ, Greenland S. Modern Epidemiology. 2. Philadephia: Lippincott-Raven Publishers; 1998. p. 256. [Google Scholar]
- 16.Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 17.Tan SL, Katze MG. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J Interferon Cytokine Res. 1999;19:543–554. doi: 10.1089/107999099313677. [DOI] [PubMed] [Google Scholar]
- 18.Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 19.Nakaya T, Sato M, Hata N, Asagiri M, Suemori H, Noguchi S, Tanaka N, et al. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Yee LJ, Im K, Rhodes SL, Tang Y, Tong X, Howell C, et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. J Hepatol. 2008;49:184–191. doi: 10.1016/j.jhep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazari S, Taylor L, Haque S, Garry R, Florman S, Luftig R, Regenstein F, et al. Reduced expression of Jak-1 and Tyk-2 proteins leads to interferon resistance in Hepatitis C virus replicon. Virology Journal. 2007;4:89. doi: 10.1186/1743-422X-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Hijikata M, Ohta Y, Mishiro S. Identification of a Single Nucleotide Polymorphism in the MxA Gene Promoter (G/T at nt --88) Correlated with the Response of Hepatitis C Patients to Interferon. Intervirology. 2000;43:124–127. doi: 10.1159/000025035. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki F, Arase Y, Suzuki Y, Tsubota A, Akuta N, Hosaka T, Someya T, et al. Single nucleotide polymorphism of the MxA gene promoter influences the response to interferon monotherapy in patients with hepatitis C viral infection. Journal of Viral Hepatitis. 2004;11:271–276. doi: 10.1111/j.1365-2893.2004.00509.x. [DOI] [PubMed] [Google Scholar]
- 24.Knapp S, Yee LJ, Frodsham AJ, Hennig BJW, Hellier S, Zhang L, Wright M, et al. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 4:411–419. doi: 10.1038/sj.gene.6363984. [DOI] [PubMed] [Google Scholar]
- 25.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]
- 26.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 27.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 28.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, et al. Tissue-Specific Genetic Control of Splicing: Implications for the Study of Complex Traits. PLoS Biology. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.