Abstract
We investigated the role of the Notch signaling pathway in regulating several transcription factors that stabilize a neural fate and expand the neural plate. Increased Notch signaling in a neural lineage via a constitutively activated form (NICD) up-regulated geminin and zic2 in a cell autonomous manner, and expanded the neural plate domains of sox11, sox2, and sox3. Loss- and gain-of-function assays show that foxD5 acts upstream of notch1 gene expression. Decreasing Notch signaling with an anti-morphic form of a Notch ligand (X-Delta-1STU) showed that the foxD5-mediated expansion of the sox gene neural plate domains requires Notch signaling. However, geminin and zic2 appear to be dually regulated by foxD5 and Notch1 signaling. These studies demonstrate that: 1) Notch signaling acts downstream of foxD5 to promote the expression of a subset of neural ectodermal transcription factors; and 2) Notch signaling and the foxD5 transcriptional pathway together maintain the neural plate in an undifferentiated state.
Introduction
The neural ectoderm (NE) forms on the dorsal side of the Xenopus embryo in response to factors secreted from the blastula Chordin- and Noggin-expressing (BCNE) signaling center and the Organizer (De Robertis and Kuroda, 2004; Levine and Brivanlou, 2007). By gastrula stages, the NE expresses a large number of transcription factors whose expression domains widely overlap, and which coordinately promote a neural fate, expand the neural plate and regulate the onset of neural differentiation (Fig. 1; Sasai, 1998; Moody and Je, 2002). These include: 1) foxD5, a forkhead/winged helix gene (Sölter et al., 1999; Fetka et al., 2000; Sullivan et al., 2001); 2) geminin (gem), which interacts with the SWI/SNF complex (Kroll et al., 1998; Seo and Kroll, 2006); 3) the high-mobility group (HMG)-box genes sox2, sox3, soxD and sox11 (Uwanogho et al., 1995; Mizuseki et al., 1998a, b; Kishi et al., 2000; Hyodo-Miura et al., 2002; Wegner and Stolt, 2005; Dee et al., 2008); 4) the zinc-finger genes zic1, zic2 and zic3 (Brewster et al., 1998; Kuo et al., 1998; Mizuseki et al., 1998a; Nakata et al., 1997, 1998); and 5) the Iroquois genes Xiro1, Xiro2, and Xiro3 (Bellefroid et al., 1998; Gomez-Skarmeta et al., 1998, 2001). However, the interactions between these several genes during the establishment of the neural plate are largely unknown. Recently, we proposed a regulatory network, based on loss- and gain-of-function assays, that places foxD5 in an upstream position. It appears to directly regulate gem, sox11 and zic2, which together regulate the expression patterns of the other NE transcription factors (Fig. 1; Yan et al., 2009). This model, however, does not account for signaling pathways that might mediate the interactions.
Figure 1.
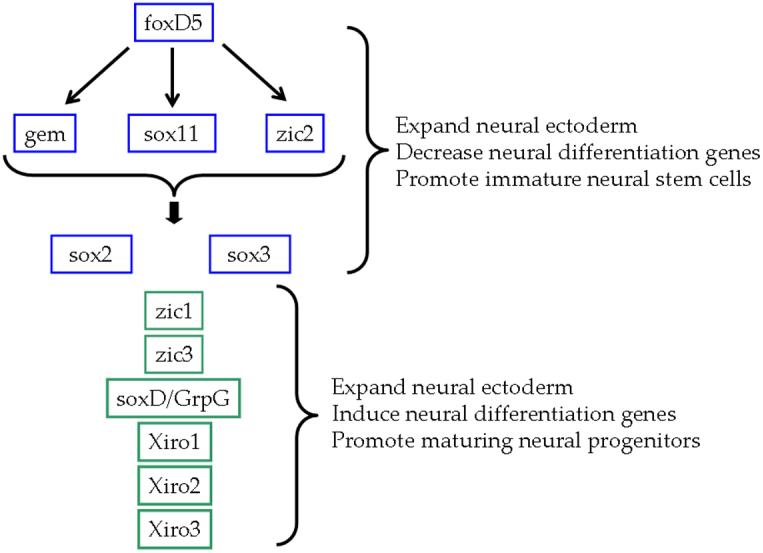
Previous studies indicate that foxD5 acts upstream of the other neural ectodermal transcription factors that expand the neural plate. Some of these decrease neural differentiation genes (blue boxes) and some promote the onset of neural differentiation (green boxes). foxD5 directly activates the transcription of gem, sox11 and zic2, and these three genes coordinately regulate the remaining NE transcription factors. (Based on Yan et al., 2009)
Notch signaling, which is highly conserved from Drosophila to humans, plays essential roles in many processes during development including neurogenesis (Artavanis-Tsakonas et al., 1999; Lai, 2004; Bray, 2006; Chitnis, 2007). When the Notch transmembrane receptor on the cell surface is bound by DSL (Delta/ Serrate/ Lag-2) ligands on the neighboring cells, the Notch intracellular domain (NICD) is cleaved, released into the cytoplasm and translocated into the nucleus. There, it combines with its transcriptional cofactor CSL (CBF1/ Suppressor of Hairless/ Lag-1) to activate downstream target genes, mainly members of the HES (Hairy/ Enhancer of Split) family of basic helix-loop-helix (bHLH) transcriptional regulators (Mumm and Kopan, 2000; Weinmaster, 2000). During early vertebrate development, Notch is expressed broadly throughout the neural plate, and its signaling pathway plays an essential role in maintaining the neural progenitor state and regulating the diversification of cell fate (Yoon and Gaiano, 2005; Louvi and Artavanis-Tsakonas, 2006; Chitnis, 2007). The classical view is that increasing Notch signaling within a cell up-regulates HES genes that subsequently repress neural differentiation bHLH genes (such as ngn and neuroD) and Notch DSL ligand genes.
The preponderance of work on Notch signaling during neural development focuses on the onset of neurogenesis, when progenitor cells become post-mitotic and acquire a particular neural cell fate. For example, at Xenopus and zebrafish neurula stages, increasing Notch signaling by expressing a constitutively activating form of Notch1 prevents expression of the neural differentiation genes and formation of primary neurons, whereas decreasing Notch signaling by interfering with Delta promotes neurogenesis (Chitnis et al., 1995; Itoh et al., 2003). However, Notch signaling also affects the size of the NE during Xenopus gastrulation (Coffman et al., 1993). Because Notch is expressed throughout the neural plate at a similar time as the NE transcription factors (Coffman et al., 1990, 1993), and several of those factors both expand the neural plate and inhibit neural differentiation (Fig. 1), we investigated whether Notch signaling has a role in regulating the expression of these genes. We show that: 1) foxD5 acts upstream of notch1; 2) increasing Notch signaling phenocopies the effects of foxD5 on gem, sox11, sox2, sox3 and zic2, but it does not alter the expression of NE genes that are inhibited by foxD5 (soxD, zic1, zic3, Xiro1-3); and 3) Notch signaling is required for the foxD5-mediated expansion of sox2, sox3 and sox11 expression domains but not for the up-regulation of gem and zic2. These studies demonstrate that: 1) Notch signaling acts downstream of foxD5 to promote the expression of a subset of NE transcription factors; and 2) the Notch signaling pathway and the foxD5 transcriptional pathway together maintain the neural plate in an undifferentiated state.
Results
Our previous work shows that foxD5 is required for the normal expression of gem, sox2, sox3, sox11, soxD, zic1-3 and Xiro1-3 in the nascent neural ectoderm (NE), and that increased levels of foxD5 induce gem, sox11, and zic2, expand the sox2 and sox3 NE domains and repress zic1, zic3 and Xiro1-3 (Fig. 1; Yan et al., 2009). To assess whether Notch signaling is involved in the regulation of any of these effects, we tested whether: 1) foxD5 acts upstream of the notch1 gene; 2) increased Notch signaling phenocopies foxD5-mediated effects on the NE genes that inhibit neural differentiation; and 3) decreased Notch signaling interferes with foxD5 regulation of these NE genes.
foxD5 acts upstream of notch1 gene expression
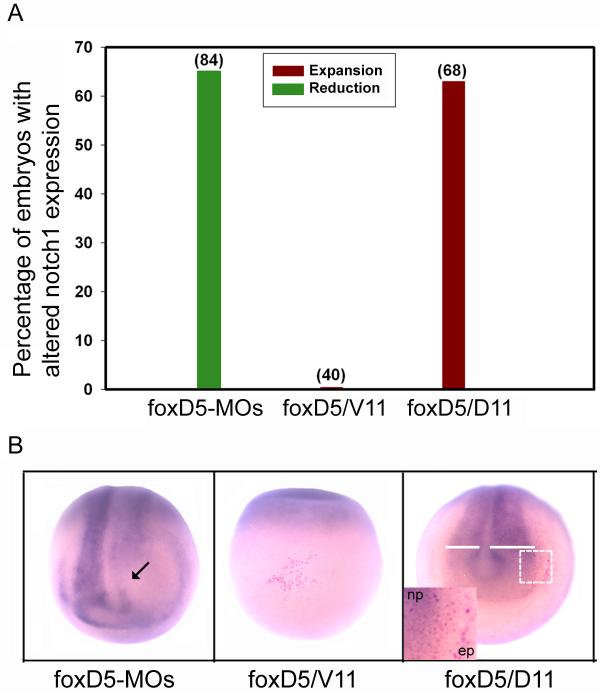
Endogenous foxD5 and notch1 mRNAs both are expressed diffusely throughout the early NE (Coffman et al., 1990, 1993; Sullivan et al., 2001). To assess whether foxD5 regulates notch1 expression, we microinjected into the precursor blastomere of the neural ectoderm (D11) either foxD5 anti-sense morpholino oligonucleotides (foxD5-MOs) to locally decrease FoxD5 levels, or foxD5 mRNA to locally increase them. Reducing endogenous foxD5 levels with foxD5-MOs blocked notch1 expression in the neural plate, whereas increased foxD5 significantly expanded the neural plate expression domain of notch1 (Fig. 2). The level of notch1 expression in the foxD5-expressing cells in the neural plate (tagged red by a βGal lineage tracer) was not increased above that of neighboring cells. Furthermore, expression of foxD5 in the epidermis adjacent to the neural plate or in a ventral epidermis precursor blastomere (V11) did not induce ectopic notch1 (Fig. 2B). These results demonstrate that foxD5: 1) is required for notch1 expression in the neural plate; 2) expands the notch1 domain in the neural plate in a non-cell autonomous manner; and 3) is not sufficient to activate ectopic notch1 transcription in the dorsal or ventral epidermis. Finally, expression of the constitutively signaling Notch1 construct (NICD) did not alter the expression of foxD5 in the neural ectoderm nor induce it in the ventral epidermis (Fig. 3A). Together, these results indicate that foxD5 acts upstream of notch1 during the formation of the neural ectoderm, but does not activate it in the epidermis.
Figure 2.
foxD5 acts upstream of notch1. A. Percentage of the embryos with altered notch1 expression after injection of foxD5-MOs in a neural plate precursor, or injection of foxD5 mRNA in either a ventral epidermis precursor (V11) or a neural plate precursor (D11). Numbers in parentheses indicate sample sizes. B. Embryos at neural plate stages processed for in situ hybridization detection of notch1 mRNA. Local depletion of FoxD5 by foxD5-MOs causes a loss of notch1 expression (arrow). Locally increasing FoxD5 by mRNA injection in the ventral epidermis (red cells) does not induce ectopic notch1 expression (middle panel), but doing so in the neural plate (right panel) expands the notch1 expression domain (compare white bars on injected [right] versus uninjected [left] sides). The inset shows that the foxD5-expressing cells (red nuclei) do not express notch1 at levels greater than surrounding cells in the neural plate (np), nor induce it in the adjacent dorsal epidermis (ep).
Figure 3.
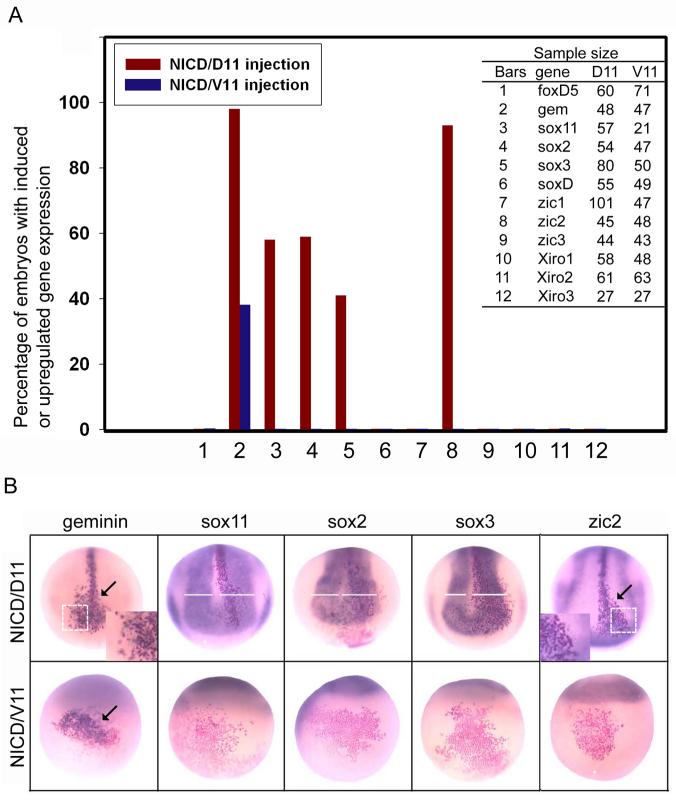
Effects of increased Notch1 signaling on the NE genes. A. Percentage of embryos in which NICD, expressed in either the neural plate (D11) or ventral epidermis (V11) lineages, altered the expression of twelve NE genes. The table to the right reports the numbers of embryos examined for each experiment. B. Examples of the in situ hybridization assays for those NE genes altered by NICD expressed in the indicated lineages. geminin and zic2 are up-regulated in a cell autonomous manner (arrows); insets show that NICD-expressing cells (red nuclei) express elevated levels of the respective genes compared to adjacent cells. NICD causes the expansion of the neural plate domains of the three sox genes (compare white bars on injected [right] versus uninjected [left] sides). NICD induces the ectopic expression of geminin in the ventral epidermis (arrow), but does not induce any of the other NE genes in this tissue (red cells; see also 3A).
Notch signaling up-regulates a subset of NE transcription factors
Because Notch signaling is proposed to expand an undifferentiated precursor population in the neural plate independent of regulating the cell cycle (Coffman et al., 1993), we tested whether expression of NICD would up-regulate those NE transcription factors, previously reported to also expand the NE, that function downstream of foxD5 (Fig. 1). First, expression of NICD in a single neural plate precursor did not alter the expression levels of soxD, zic1, zic3 or Xiro1-3 (Fig. 3A); this result is consistent with previous reports that these genes promote the onset of neural differentiation and are inhibited by foxD5 (Sasai 1998; Moody and Je, 2002; Yan et al., 2009). Second, gem and zic2, both of which maintain NE cells in an undifferentiated state (Brewster et al., 1998; Kroll et al., 1998), were up-regulated in the neural plate at high frequency, and their expression was elevated above endogenous levels in a cell autonomous manner (Fig. 3). Third, the neural plate expression domains of sox11, sox2 and sox3 were expanded, but their expression was not elevated above endogenous levels (Fig. 3); further, the expansion of the neural plate extended beyond the NICD-expressing clone indicating involvement of cell-to-cell signaling. The effects of increased Notch signaling on the NE genes partially phenocopied those of foxD5 (Sullivan et al., 2001; Yan et al., 2009): those genes expanded by foxD5 were also expanded by NICD, and two genes up-regulated cell autonomously by foxD5 (gem, zic2) were also up-regulated cell-autonomously by NICD in the neural plate. However, some effects of increased Notch signaling were not the same as those of foxD5. First, those genes inhibited by foxD5 (soxD, zic1-3, Xiro1-3) were unaffected by NICD. Second, the expression of sox11 was increased by both genes but not in the same way: foxD5 both up-regulated sox11 levels cell-autonomously in the neural plate and expanded its neural plate domain, whereas NICD only expanded the sox11 neural plate domain.
We also tested whether NICD can induce ectopic neural tissue by injecting the mRNA into a ventral epidermis precursor blastomere (V11). Consistent with a report that a different form of activated Notch does not convert epidermis to neural tissue based on a histological analysis (Coffman et al., 1993), we observed that of all the NE transcription factors tested, only gem was ectopically induced (Fig. 3). This is in contrast to foxD5, which ectopically induces gem, zic2, sox11, and sox3 at high frequency, and sox2, zic1 and Xiro2 at low frequency (Yan et al., 2009). Taken together, these results indicate that Notch signaling acts downstream of foxD5 to up-regulate or expand the expression of a subset of transcription factors in the NE, but unlike foxD5, it does not convert non-neural ectoderm to a neural fate.
Notch signaling is required for increased expression of some NE transcription factors
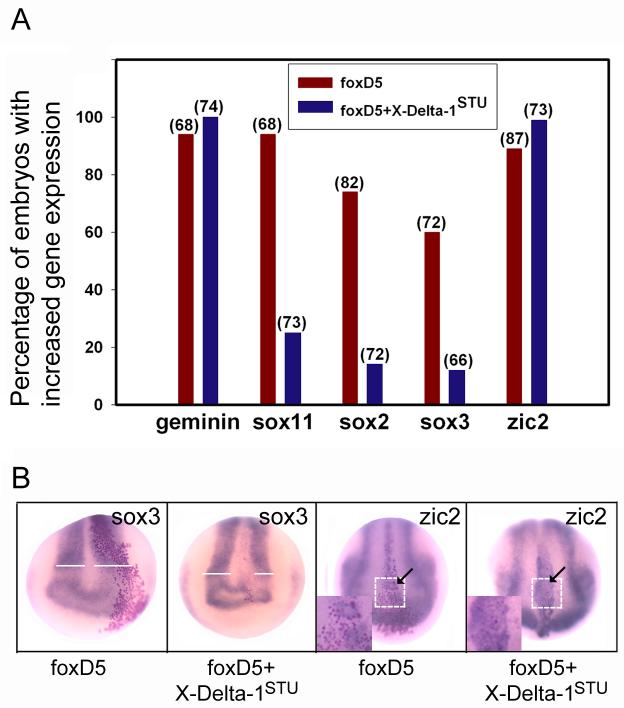
To determine whether Notch signaling is required for the increased expression of gem, zic2, sox11, sox2 or sox3, a dominant-interfering form of the ligand X-Delta-1 (X-Delta-1STU; Chitnis et al., 1995) was expressed in the neural precursor blastomere. This construct, which lacks the intracellular domain (ICD), is thought to prevent Notch signaling by preventing the cisendocytosis of endogenous Delta-1 and being unable to promote the Notch1 intracellular cleavage that releases the active NICD fragment (reviewed in Kiyota and Kinoshita, 2004). This construct has been used extensively to show that it blocks: 1) the ability of NICD to activate neuronal differentiation genes (Chitnis et al., 1995; Chitnis and Kintner, 1996; Ma et al., 1996; Chalmers et al., 2002); 2) lateral inhibition (Papalopulu and Kintner, 1996); 3) formation of ciliated epidermal cells (Deblandre et al., 1999); and 4) NICD expansion of the neural crest domain (Glavic et al., 2004; Kuriyama et al., 2006). Herein we show that X-Delta-1STU inhibits the expression of Hes1, a direct transcriptional target of NICD (Fig. 4B, 83.3%, n=36), thus demonstrating its efficacy in blocking Notch signaling in our assay. In contrast to NICD, X-Delta-1STU significantly decreased the expression of sox11, sox2 and sox3 (Fig. 4). sox11 expression was suppressed in the X-Delta-1STU –expressing cells in a cell autonomous manner without affecting the width of its neural plate domain. sox2 and sox3 expression also were suppressed within the clone at low frequencies (17%, 14%, respectively), but most notably the neural plate domain was smaller on the injected side (Fig. 4). We next co-expressed foxD5 and X-Delta-1STU mRNAs to test whether inhibition of Notch signaling blocked the effects of foxD5 on these genes. X-Delta-1STU inhibited the foxD5-mediated expansion of the sox11, sox2 and sox3 neural plate domains (Fig. 5). These results indicate that the expansion of the neural plate domains of the sox genes by foxD5 requires Notch signaling. Interestingly, the cell-autonomous up-regulation of sox11 by foxD5 is independent of Notch signaling.
Figure 4.
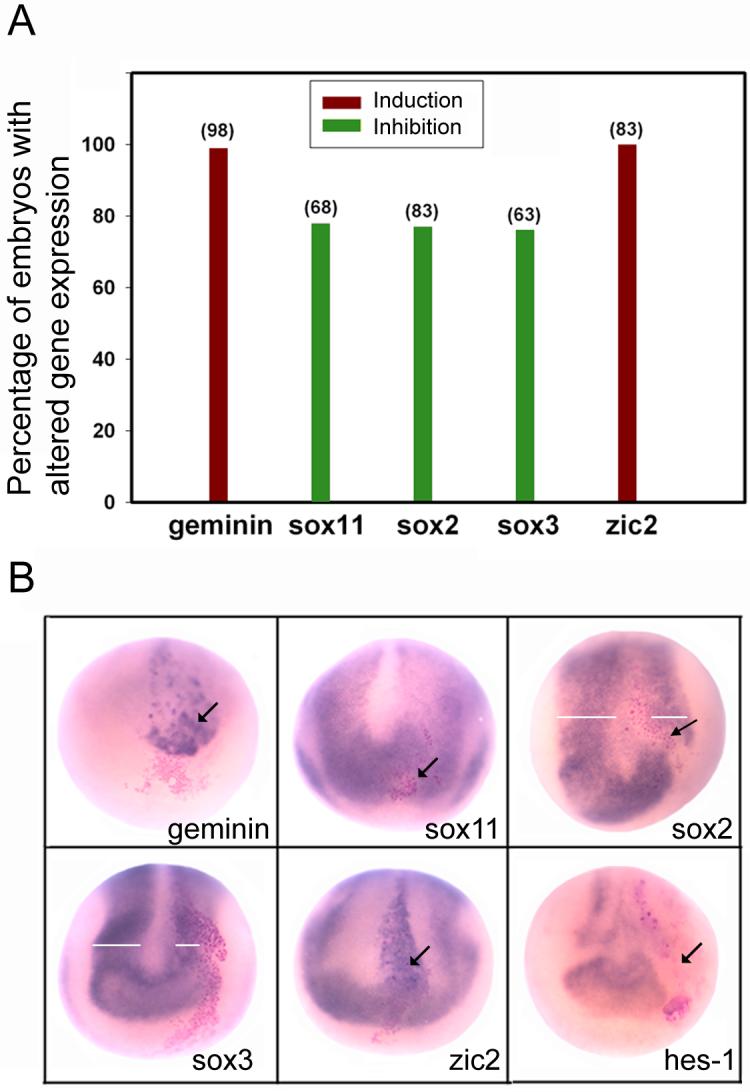
Decreased Notch signaling reduces the expression of the sox genes. A. Percentage of the embryos with altered NE gene expression after injection of X-Delta-1STU mRNA in the D11 lineage. The numbers in parentheses indicate sample sizes. B. Examples of the in situ hybridization assays showing cell autonomous up-regulation of geminin and zic2 (arrows) in the neural plate, cell autonomous repression of sox11, sox2 and hes1 (arrows) and reduction in width of the sox2- and sox3-expression domains in the neural plate (white bars).
Figure 5.
Decreased Notch signaling reverses foxD5-mediated effects on sox genes. A. Either foxD5 mRNA alone or foxD5 plus X-Delta-1STU mRNAs were injected into the D11 lineage. The numbers in parentheses indicate sample sizes. X-Delta-1STU interfered with foxD5-mediated expansion of the sox gene neural plate domains. The up-regulation of geminin and zic2 were not affected. B. Examples of the in situ hybridization assays showing reversal of foxD5 expansion of the sox3 domain and lack of reversal of the up-regulation of zic2 in the neural plate. zic2 expression is normally absent from the midline of the st14 neural plate (Brewster et al., 1998). When foxD5 is expressed in the midline by injecting mRNA into blastomere D11, zic2 is ectopically expressed (arrow). This ectopic expression is also observed when X-Delta-1STU is co-expressed (arrow). Insets show the effect is cell autonomous; nearly all of the mRNA-expressing cells (red nuclei) show ectopic zic2 expression in both samples.
Unexpectedly, X-Delta-1STU cell autonomously up-regulated the expression of gem and zic2 in the neural plate (Fig. 4), at frequencies very similar to the effects of NICD and foxD5 (Fig. 3; Yan et al., 2009). Co-expression of both foxD5 and X-Delta-1STU mRNAs did not alter these phenotypes, indicating that the foxD5 effects on gem and zic2 do not require Notch signaling in the same cell. Thus, gem and zic2 appear to be dually regulated by foxD5 and Notch signaling (Fig. 6). It is seemingly contradictory that gem and zic2 can be up-regulated by both increasing (by NICD) and decreasing (by X-Delta-1STU) Notch signaling within the same cell. Although we do not know the mechanism by which X-Delta-1STU promotes gem and zic2 expression, previous studies showed that Delta1 and Serrate1 are both expressed and available in their proteolytically cleaved, active forms (the ICDs) in the neural ectoderm, and both induce Notch target genes (Kiyota et al., 2001; Wettstein et al., 1997; Kiyota and Kinoshita, 2004). Furthermore, while the ICD-truncated forms of Serrate1 and Delta1 both cause increased numbers of primary neurons, they have distinctly different activities. The Serrate ICD acts in both the canonical Notch pathway and in an independent manner to activate genes that repress neuronal differentiation, and it can rescue the neurogenic effect of X-Delta-1STU (Kiyota and Kinoshita, 2004). Therefore, we predict that in our assay Serrate activity is intact in X-Delta-1STU —expressing cells, leading to induction of gem and zic2 expression. Alternatively, the X-Delta-1STU —expressing cells may lack a recently described interaction with the TGF-β/Smad signaling pathway (Hiratochi et al., 2007) that would normally repress gem and zic2. While our data indicate that gem and zic2 are up-regulated by both the NICD pathway and a pathway that is revealed in the absence of Delta1 function, it will be important to identify this additional pathway in order to understand the full scope of Notch signaling in the regulation of NE genes.
Figure 6.
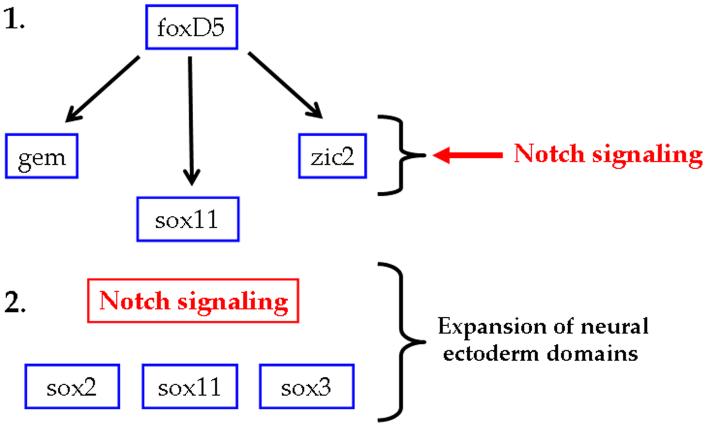
Notch signaling impacts the expression of NE transcription factors in two ways. 1. foxD5 directly up-regulates the expression of geminin (gem), sox11 and zic2 in both the neural plate and the ventral epidermis. Notch signaling, acting downstream of foxD5, co-regulates gem and zic2 expression levels, but only in the neural plate; it does not up-regulate sox11 expression levels. 2. Downstream of foxD5, Notch signaling is required for the expansion of the neural plate domains of the sox genes.
Discussion
During embryogenesis, Notch signaling is required for the maintenance and expansion of the neural stem and progenitor cells, and it inhibits neurogenesis by activating Hes1 and Hes5, the homologues of Drosophila hairy and Enhancer of split, which in turn repress the expression of the bHLH neural differentiation genes (Jarriault et al., 1995; Kageyama and Nakanishi, 1997; Lai, 2004; Chiba, 2006; Louvi and Artavanis-Tsakonas, 2006; Chitnis, 2007). Previous studies identified a large number of transcription factors that maintain neural stem and progenitor cells, but the integration of Notch signaling with the regulation of these factors is mostly unknown. In this study, we show that Notch signaling acts downstream of foxD5 to affect a subset of NE transcription factors that promote an undifferentiated neural state. Notch signaling is required for the expanded neural plate expression domains of sox11, sox2 and sox3, and it up-regulates gem and zic2 expression in the neural plate (Fig. 6).
Notch signaling expands the neural plate domains of sox genes downstream of foxD5
sox2 and sox3, which belong to the SoxB1 subgroup of HMG-box transcription factors, are highly expressed in the NE and in the endogenous neural stem cells in the adult central nervous system. Functionally, they both maintain neural stem and progenitor cells and inhibit neuronal differentiation (Avilion et al., 2003; Bylund et al., 2003; Graham et al., 2003; Ferri et al., 2004; Pevny and Placzek, 2005; Wegner and Stolt, 2005; Wang et al., 2006; Cavallaro et al., 2008; Dee et al., 2008; Kim et al., 2008). There are some conflicting data regarding how Notch signaling and the functions of SoxB1 genes interface. Some reports indicate that Sox2 acts upstream of Notch signaling because over-expressed Sox2 up-regulates the expression of Notch1, Notch ligands and its downstream target genes (Bani-Yaghoub et al., 2006). Moreover, Sox2 can directly regulate notch1 gene expression by binding to its 13th intron, which has a biological function in retinal neural progenitors (Taranova et al., 2006). However, there also is evidence that Sox2 responds to Notch signaling: 1) as we also report herein, expression of NICD expands the sox2 expression domain in the neural plate (Glavic et al., 2004); and 2) in avian neural crest cells, sox2 inhibition of differentiation is placed downstream of Notch signaling (Wakamatsu et al., 2004). Finally, some studies indicate that Notch signaling and SoxB1 work in parallel to inhibit neuronal differentiation (Bylund et al., 2003; Holmberg et al., 2008). Our studies provide an additional step in the pathway by showing that the expansion of sox2 and sox3 achieved by foxD5 during neural plate formation requires Notch signaling (Fig. 6).
sox11, a member of the SoxC subgroup, also is strongly expressed in the NE and functions in both neural induction and neural differentiation (Uwanogho et al., 1995; Hargrave et al., 1997; Hyodo-Miura et al., 2002; Bergsland et al., 2006). Although there is evidence that sox11 is involved in neuronal differentiation downstream of the bHLH proneural genes, our previous work indicates that in the embryonic NE sox11 functions in tandem with gem and zic2 to expand the neural plate expression of sox2 and sox3 (Fig. 1; Yan et al., 2009). To our knowledge there has not been a previous study of the interface between Notch signaling and sox11. Herein we show that Notch signaling is required for the foxD5-mediated expansion of sox11 neural plate expression, but it does not up-regulate the levels of sox11 in the NE.
Notch signaling up-regulates some NE transcription factors in parallel with foxD5
foxD5, gem and zic2 are well characterized as inhibitors of neural differentiation. foxD5 expands the neural plate and inhibits the expression of markers of axial differentiation in the NE (en2, Krox20) and of neural differentiation (ngnr1, neuroD, n-tubulin), leading to the interpretation that it functions to maintain the NE in an immature state (Sullivan et al., 2001). This is supported by the observations that foxD5 transcriptionally activates gem and zic2 and transcriptionally represses NE genes that promote neural differentiation (Yan et al., 2009). gem expands the neural plate, is expressed in neural stem cells and antagonizes Brg1, a protein that is required for activation of the bHLH neural differentiation genes (Kroll et al., 1998; Seo et al., 2005; Seo and Kroll 2006; Spella et al., 2007). zic2 expands the neural plate, is expressed in neural progenitor cells and inhibits the onset of neurogenesis (Brewster et al., 1998; Nakata et al., 1998; Aruga, 2004; Merzdorf, 2007).
The role of Notch signaling in the expression of these three NE genes has not been previously explored. We show by loss- and gain-of-function assays that notch1 expression is downstream of foxD5. Interestingly, strong Notch signaling, accomplished by expressing NICD, up-regulated both gem and zic2 in the neural ectoderm. Thus, in addition to being direct transcriptional targets of foxD5 (Yan et al., 2009), these genes appear to be either independently or co-regulated by Notch signaling. An important future goal will be to discern between these two possibilities. The fact that X-Delta-1STU also up-regulates gem and zic2 suggests that these two genes may be additionally regulated by another Notch ligand, such as Serrate, or another interacting pathway. It will be important to elucidate the involvement of the different Notch ligands in this signaling pathway (e.g., D’Souza et al., 2008).
In conclusion, elucidating the interactions between Notch signaling and the transcription factors involved in neural differentiation have been very important for understanding how this pathway regulates neuronal fate acquisition. We show that Notch signaling also interfaces with a subset of transcription factors that regulate the expansion of the neural plate and prevent neural differentiation. Further study of the role of Notch signaling in the earliest steps of neural fate acquisition should provide insights into the molecular mechanisms controlling the expansion of neural stem and progenitor cells (Gaulden and Reiter, 2008).
Experimental procedures
Embryo manipulation and mRNA injection
Fertilized Xenopus laevis eggs were obtained by gonadotropin-induced natural mating of adult frogs and prepared for injection as described (Moody, 2000). mRNAs for injection were synthesized by in vitro transcription with Ambion mMessage mMachine kits (Austin, TX, USA). The mRNAs of foxD5 (150pg, Sullivan et al., 2001), an activated form of notch1 (XNICD, 500pg, Coffman et al., 1993) or X-Delta-1STU, a truncated ligand lacking the intracellular domain (500pg, Chitnis et al., 1995) were mixed with β-gal mRNA (100pg; as a lineage tracer) and microinjected into either a dorsal animal (D11; neural plate precursor) or a ventral animal (V11; ventral epidermis precursor) blastomere at the 16-cell stage (Moody, 1987).
Morpholino antisense oligonucleotides
Two foxD5 morpholino antisense oligonucleotides (foxD5-MO) were synthesized to recognize the translational start site of all three foxD5 paralogues found in Xenopus laevis (5′-CAGACTCCTGGCTAAAGCTCATTGT-3′; 5′-TATACTCTGATGCTGGGTTTGTAGC-3′) (GeneTools, Philomath, OR). An equimolar mixture of the two foxD5-MOs was microinjected (16ng) into blastomere D11. foxD5-MO knock-down efficacy was assessed previously (Yan et al., 2009).
Whole mount in situ hybridization
Embryos were fixed and processed for whole mount in situ hybridization according to standard protocols (Sive et al., 2000) using digoxigenin-labeled RNA probes as following: foxD5 (Sullivan et al., 2001), geminin (Kroll et al., 1998), notch1 (Coffman et al., 1993), hes1 (Open BioSystems, BC041261) sox2 (Mizuseki et al., 1998a ), sox3 (Penzel et al., 1997), sox11 (Hiraoka et al., 1997), soxD (Mizuseki et al., 1998b), Xiro1 (Gomez-Skarmeta et al., 2001), Xiro2 (Gomez-Skarmeta et al., 1998), Xiro3 (Bellefroid et al., 1998), zic1 (Mizuseki et al., 1998a), zic2 (Brewster et al., 1998) and zic3 (Nakata et al., 1997). Embryos were analyzed for whether the expression domain was expanded or decreased in size compared to the uninjected side of the same embryo and for changes in staining intensity compared to adjacent uninjected cells.
Acknowledgements
We thank E. Bellefroid, E. Silva Casey, A. Chitnis, J. L. Gomez-Skarmeta, R. Grainger, T. Grammer, K. Kroll, R. Mayor, A. Ruiz i Altaba, T. Sargent and Y. Sasai for plasmids. We also thank Himani Datta Majumdar and Lynne Mied for technical assistance. This work was supported by NIH grant NS23158 and the George Washington University School of Medicine and Health Sciences.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny L, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 1998;17:191–203. doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signaling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, Wanke E, Brunelli S, Favaro R, Ottolenghi S, Nicolis SK. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135:541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev. Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Chiba S. Notch signaling in stem cell systems. Stem Cells. 2006;24:2437–2447. doi: 10.1634/stemcells.2005-0661. [DOI] [PubMed] [Google Scholar]
- Chitnis A. Notch signaling: a versatile tool for the fine patterning of cell fate in development. In: Moody SA, editor. Principles of Developmental Genetics. Academic Press; San Diego: 2007. pp. 316–340. [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Kintner C. Sensitivity of proneural genes to lateral inhibition affects the pattern of primary neurons in Xenopus embryos. Development. 1996;122:2295–2301. doi: 10.1242/dev.122.7.2295. [DOI] [PubMed] [Google Scholar]
- Coffman C, Harris W, Kintner C. Xotch, the Xenopus homolog of Drosophila notch. Science. 1990;249:1438–1441. doi: 10.1126/science.2402639. [DOI] [PubMed] [Google Scholar]
- Coffman CR, Skoglund P, Harris WA, Kintner CR. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–4728. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee CT, Hirst CS, Shih YH, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Fetka I, Doederlein G, Bouwmeester T. Neuroectodermal specification and regionalization of the Spemann organizer in Xenopus. Mech Dev. 2000;93:49–58. doi: 10.1016/s0925-4773(00)00265-3. [DOI] [PubMed] [Google Scholar]
- Gaulden J, Reiter JF. Neur-ons and neur-offs: regulators of neural induction in vertebrate embryos and embryonic stem cells. Hum Mol Genet. 2008;17(R1):R60–66. doi: 10.1093/hmg/ddn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004;131:347–359. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Glavic A, de la Calle-Mustienes E, Modolell J, Mayor R. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development at the neural plate. EMBO J. 1998;17:181–190. doi: 10.1093/emboj/17.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta J, de La Calle-Mustienes E, Modolell J. The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and downregulates Bmp4. Development. 2001;128:551–560. doi: 10.1242/dev.128.4.551. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hargrave M, Wright E, Kun J, Emery J, Cooper L, Koopman P. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Komatsu N, Sakai Y, Ogawa M, Shiozawa M, Aiso S. XLS13A and XLS13B: SRY-related genes of Xenopus laevis. Gene. 1997;197:65–71. doi: 10.1016/s0378-1119(97)00242-4. [DOI] [PubMed] [Google Scholar]
- Hiratochi M, Nagase H, Kuramochi Y, Koh CS, Ohkawara T, Nakayama K. The Delta intracellular domain mediates TGF-beta/Activin signaling through binding to Smads and has an important bi-directional function in the Notch—Delta signaling pathway. Nucleic Acids Res. 2007;35:912–922. doi: 10.1093/nar/gkl1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Hansson E, Malewicz M, Sandberg M, Perlmann T, Lendahl U, Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135:1843–1851. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- Hyodo-Miura J, Urushiyama S, Nagai S, Nishita M, Ueno N, Shibuya H. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells. 2002;7:487–496. doi: 10.1046/j.1365-2443.2002.00536.x. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–-358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Nakanishi S. Helix-loop-helix factors in growth and differentiation of the vertebrate nervous system. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M, Schöler HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Jono H, Kuriyama S, Hasegawa K, Miyatani S, Kinoshita T. X-Serrate-1 is involved in primary neurogenesis in Xenopus laevis in a complementary manner with X-Delta-1. Dev Genes Evol. 2001;211:367–376. doi: 10.1007/s004270100165. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Kinoshita T. The intracellular domain of X-Serrate-1 is cleaved and suppresses primary neurogenesis in Xenopus laevis. Mech. Dev. 2004;121:573–585. doi: 10.1016/j.mod.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- Kuo JS, Patel M, Gamse J, Merzdorf C, Liu X, Apekin V, Sive H. Opl: a zinc finger protein that regulates neural determination and patterning in Xenopus. Development. 1998;125:2867–2882. doi: 10.1242/dev.125.15.2867. [DOI] [PubMed] [Google Scholar]
- Kuriyama S, Lupo G, Ohta K, Ohnuma S, Harris WA, Tanaka H. Tsukushi controls ectodermal patterning and neural crest specification in Xenopus by direct regulation of BMP4 and X-delta-1 activity. Development. 2006;133:75–88. doi: 10.1242/dev.02178. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev. Biol. 2007;308:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998a;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Shiota K, Nakanishi S, Sasai Y. SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron. 1998b;21:77–85. doi: 10.1016/s0896-6273(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Moody SA. Fates of the blastomeres of the 16-cell-stage Xenopus embryo. Dev Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Moody SA. Cell lineage analysis in Xenopus embryos. Methods Mol Biol. 2000;135:331–347. doi: 10.1385/1-59259-685-1:331. [DOI] [PubMed] [Google Scholar]
- Moody SA, Je HS. Neural induction, neural fate stabilization, and neural stem cells. ScientificWorldJournal. 2002;2:1147–1166. doi: 10.1100/tsw.2002.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc Natl Acad Sci USA. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. A posteriorising factor, retinoic acid, reveals that anteroposterior patterning controls the timing of neuronal differentiation in Xenopus neuroectoderm. Development. 1996;122:3409–3418. doi: 10.1242/dev.122.11.3409. [DOI] [PubMed] [Google Scholar]
- Penzel R, Oschwald R, Chen Y, Tacke L, Grunz H. Characterization and early embryonic expression of a neural specific transcription factor xSOX3 in Xenopus laevis. Int. J Dev Biol. 1997;41:667–677. [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Sasai Y. Identifying the missing links: genes that connect neural induction and primary neurogenesis in vertebrate embryos. Neuron. 1998;21:455–458. doi: 10.1016/s0896-6273(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Kroll KL. Geminin’s double life: chromatin connections that regulate transcription at the transition from proliferation to differentiation. Cell Cycle. 2006;5:374–379. doi: 10.4161/cc.5.4.2438. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis, a Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Sölter M, Köster M, Hollemann T, Brey A, Pieler T, Knöchel W. Characterization of a subfamily of related winged helix genes, XFD-12/12′/12″ (XFLIP), during Xenopus embryogenesis. Mech. Dev. 1999;89:161–165. doi: 10.1016/s0925-4773(99)00195-1. [DOI] [PubMed] [Google Scholar]
- Spella M, Britz O, Kotantaki P, Lygerou Z, Nishitani H, Ramsay RG, Flordellis C, Guillemot F, Mantamadiotis T, Taraviras S. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience. 2007;147:373–387. doi: 10.1016/j.neuroscience.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Sullivan SA, Akers L, Moody SA. foxD5a, a Xenopus winged helix gene, maintains an immature neural ectoderm via transcriptional repression that is dependent on the C-terminal domain. Dev. Biol. 2001;232:439–457. doi: 10.1006/dbio.2001.0191. [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Endo Y, Osumi N, Weston JA. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Dev Dyn. 2004;229:74–86. doi: 10.1002/dvdy.10498. [DOI] [PubMed] [Google Scholar]
- Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J Comp Neurol. 2006;497:88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Yan B, Nelson KM, Moody SA. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.02.019. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]