Abstract
The veA gene positively regulates sterigmatocystin production in Aspergillus nidulans and aflatoxin production in A. parasiticus and A. flavus. Whether veA homologs have a role in regulating secondary metabolism in other fungal genera is unknown. In this study, we examined the role of the veA homolog, FvVE1, on production of two mycotoxin families, fumonisins and fusarins, in the important corn pathogen F. verticillioides. We found that FvVE1 deletion completely suppressed fumonisin production on two natural substrates, corn and rice. Furthermore, our results revealed that FvVE1 is necessary for the expression of the pathway-specific regulatory gene FUM21 and structural genes in the fumonisin biosynthetic gene (FUM) cluster. FvVE1 deletion also blocked production of fusarins. The effects of FvVE1 deletion on the production of these toxins were found to be the same in two separate mating types. Our results strongly suggest that FvVE1 play an important role in regulating mycotoxin production in F. verticillioides.
Keywords: Fusarium verticillioides, fumonisin, fusarins, FvVE1, veA, secondary metabolism
INTRODUCTION
The filamentous fungus Fusarium verticillioides (syn. F. moniliforme, teleomorph Gibberella moniliformis) is one of the most common causes of corn ear rot worldwide and can produce multiple families of mycotoxins (1, 2). Consequently, F. verticillioides mycotoxins are commonly detected in corn (maize, Zea mays L.) and often contaminate corn-based human food and animal feed (1, 3). Fumonisins are currently considered the most agriculturally significant F. verticillioides mycotoxins because they can cause several animal diseases and are epidemiologically associated with some human diseases (4, 5). Fumonisins are polyketide-derived metabolites that can inhibit ceramide synthase, a key enzyme in sphingolipid metabolism, and induce apoptosis (1, 5). Fumonisin B1 (FB1) is typically the most abundant fumonisin in contaminated corn and accounts for approximately 70% of the total fumonisin content. Fumonisin B2 (FB2) and fumonisin B3 (FB3) are also common in corn but typically comprise 10−20% of the total fumonisin content (1).
In F. verticillioides, fumonisin biosynthetic genes (FUM) are clustered. The cluster consists of seventeen genes, designated as FUM1 through FUM3 and FUM6 through FUM21 (6, 7, 8). Disruption of FUM1, FUM6, and FUM8 has been shown to abolish fumonisin production (7, 8). In many cases, genes responsible for the synthesis of fungal secondary metabolites such as sterigmatocystin, gibberellins, aurofusarin, trichothecenes, and lovastatin, are also found clustered and specific regulatory genes are located within these gene clusters (9-11). FUM21, a predicted a Zn(II)2Cys6 DNA-binding transcription factor located in the FUM cluster, positively regulates FUM gene expression and is required for fumonisin synthesis (12). Nevertheless, the regulatory mechanism controlling fumonisin biosynthesis is poorly understood. Among the genes involved in fumonisin gene regulation are FCC1, PAC1, and ZFR1 (13-15). FCC1 encodes a cyclin-like protein (Fcc1) that positively regulates fumonisin biosynthesis and conidiation (13) and interacts with FCK1, a cyclin-dependent kinase (Fck1) (16). PAC1 is required for growth at alkaline pH and may act as a repressor of fumonisin biosynthesis (14). ZFR1 encodes a zinc binuclear cluster-type protein (Zfr1) which functions as a positive regulator of fumonisin biosynthesis (15). Studies by Flaherty and Woloshuk (15) indicated that Fcc1 is required for Zfr1 function. On the other hand, Pac1 and Fcc1 seem to act independently of each other in regulating fumonisin biosynthesis (15).
In addition to fumonisins, F verticillioides produces other mycotoxins. Among them are the polyketide compounds fusarins (17-19). Fusarins have been reported to induce mutagenesis in mammalian cells in vitro (17) and to cause immunosupression (18). Although a polyketide synthase gene required for fusarin biosynthesis has been identified in several Fusarium species (20, 21), nothing is known about how fusarin biosynthesis is regulated.
In Aspergillus spp., the velvet gene (veA) regulates the biosynthesis of several secondary metabolites, including the polyketide toxins sterigmatocystin and aflatoxin (22-24). Whether veA homologs have a similar role in regulation of toxin production in other fungal genera has not been investigated. Previously, we identified FvVE1, a veA homolog in F. verticillioides, and demonstrated that it functions in regulation of morphogenesis (25). In this study, we investigate the role of FvVE1 in secondary metabolism in F. verticillioides, specifically in the biosynthesis of fumonisin and fusarins. Our results suggest that FvVE1 regulates biosynthesis of both fumonisin and fusarins in this important plant pathogenic fungus.
MATERIALS AND METHODS
Strains and media
The strains used in this study are: M-3125 (MAT1−1, FvVE1); M-3120 (MAT1−2, FvVE1); M312501 (MAT1−1, ΔFvve1::HygB); M31206 (MAT1−2, ΔFvve1::HygB); M312501C1 (MAT1−1, ΔFvve1::HygB, FvVE1::GenR); M31206C5 (MAT1−2, ΔFvve1::HygB, FvVE1::GenR). MAT1−1 and MAT1−2 are the two different mating type idiomorphs (alleles) in F. verticillioides. M-3120 and M-3125 are strain designations from the Fusarium Research Center culture collection (Pennsylvania State University, University Park, PA). The FvVE1 deletion strains and complementation strains were generated in both mating types as described by Li et al (25). In brief, the ΔFvve1 mutant strains were generated by gene replacement via double homologous recombination events using the hygromycin B resistance gene (HygB) as selectable marker (25). Complementation strains were obtained by transformation of the ΔFvve1 mutants with wild-type FvVE1 allele using the geneticin resistant gene, GenR, as a selectable marker (25).
V8 agar medium (10% V8 juice, 0.1% CaCO3, and 1.5% agar) was used for production of conidia. Corn medium and rice medium were prepared as previously described (26, 27) with some modifications. In this study we mixed 25 g of corn kernels and 40 mL distilled water in 250 mL flasks and 50 g of long-grain rice and 60 mL distilled water in 250 mL flasks. For RNA experiments, cracked corn kernel cultures were prepared by thoroughly mixing 250 g cracked corn kernels and 100 mL water and autoclaving. After cooling, the moistened kernels were combined with 25 ml of a suspension of F. verticillioides conidia (1 × 107 conidia per ml water) prepared from seven-day-old, V8 agar cultures of the fungus. This mixture was then distributed among eight 100- mm plastic Petri plates and incubated in the dark at 22 °C.
RNA Preparation and Northern Blots
At 36, 48 and 72 hours of incubation, 10 g of cracked corn culture was frozen in liquid nitrogen, placed at −80 °C until the nitrogen evaporated, and then lyophilized. Total RNA was isolated from the lyophilized material with TRIzol (Invitrogen Life Technologies, Carlsbad, California) using the protocol for samples with high polysaccharide content as described by the manufacturer. For Northern blot analysis, 5 μg of total RNA for each sample was subjected to electrophoresis in a 1.3% agarose gel containing 1.8% formaldehyde and then transferred to nylon membrane following standard protocols (28). 32P-labaled hybridization probes were prepared with the Ready-to Go DNA labeling kit (Amersham Biosciences, Little Chalfont, Buckinghamshire), and the hybridization, wash, and autoradiography procedures followed standard protocols (28). Templates for hybridization probes corresponding to FUM1, FUM8, and TEF1 were prepared by PCR that employed genomic DNA from wild-type F. verticillioides strain M-3120 and following primer pairs: The primers used to amplify DNA templates for Northern blot hybridization probes were: for FUM,1 rp405 (5’-TGGGACACAGTTCTCAAGGAGA-3’) and rp408 (5’-CAAGCTCCTGTGACAGAGATAC-3’); for FUM8, rp679 (5’-CGTAGTAGGAATGAGAAGGATG-3’) and rp680 (5’-GCAAGCTTTGTGGCTGATTGTC-3’); and for TEF1, rp992 (5’-ATGGGTAAGGARGACAAGAC-3’) and rp993 (5’-GGARGTACCAGTSATCATGTT-3’). TEF1, encoding the transcription elongation factor 1α, was used as loading control.
Reverse transcription PCR
Total RNA was treated with Turbo DNA-free DNase (Applied Biosystems, Carlsbad, California) following manufacturers recommended protocol. RNA from DNase treated samples was quantified on a Nanodrop spectrophotometer (Thermo Scientific, Delaware) and diluted to a concentration of 40 ng/μL. A total of 60 ng RNA was used per reverse transcription (RT) PCR reaction. RT-PCR was accomplished with the Easy-A One-tube RT-PCR System (Stratagene, La Jolla, California) following manufacturers recommended protocol. Primers used are: for TEF1, rb291 (5’-ATGGGTAAGGAGGACAAGAC-3’) and rb292 (5’-GGAAGTACCAGTGATCATGTT-3’); for FUM21, rb373 (5’-TAAATGCGAGACAGCATTTGCGGG-3’) and rb374 (5’-TGCATCTTGCCCTACTCAATCGGA-3’); for FUM8, rb379 (5’-TCCATGTTTACGGGCGCATTTGTC-3’) and rb380 (5’-TCGTGAAACCTAGACGCTTGCTGA-3’); for ZFR1, rb384 (5’-ATCCACGAAGGAGGCATGTTGGTA-3’ and rb385 (5’-AGGCGGATACAAAGAACGACAGGT-3’); and for FCC1, rb391 (5’-AATGTTTCCGCTTCCGCA-3’) and rb394 (5’-TGCCGCTTCTCCTTAGGTTCT-3’). When possible, primers were designed to amplify different size fragments from genomic DNA and cDNA. TEF1 was used as control reference to indicate amounts of total RNA. Primers for TEF1 amplify a 771-bp fragment from genomic DNA and a 324-bp fragment from cDNA. FUM21 primers amplify 920-bp and 707-bp fragments from genomic DNA and cDNA respectively. FUM8 primers amplify 789-bp and 638-bp fragments from genomic DNA and cDNA respectively. Primer pairs for ZFR1 and FCC1 amplify the same size fragments from genomic DNA and cDNA (566 bp and 725 bp, respectively due to the absence of introns in these genes).
Fumonisin analysis
A plug (1.6 cm diameter) containing mycelia and conidia from a 7-day old V8 agar culture was used as inoculum. In each case, the cultures were mixed twice during the first three days of incubation by shaking for 30 sec to ensure homogenization. Cultures were incubated for a total period of 2 weeks. After that time, the samples were lyophilized and ground to powder. Fumonisin analysis was performed as previously described by Abbas et al. (29). Samples were analyzed by liquid chromatography/electrospray ionization/mass spectrometry (LC/ESI/MS). Samples (10 g) from the ground corn and rice cultures were extracted with 50 mL of 70% methanol and filtered through No. 1 Whatman filter paper. An aliquot (10 mL) was applied to a SAX clean-up column (Varian, Harbor City, CA). The sample was reconstituted in 1 mL of acetonitrile:water (1:1) and diluted if necessary. The LC/ESI/MS analysis was performed on a Thermo Finnigan LCQ Advantage, coupled to a Thermo Finnigan Surveyor MS, and a Thermo Finnigan Surveyor MS Pump (Thermo Electron Corp., West Palm Beach, FL). A 10 μL aliquot was injected and each sample was evaluated in full-scan mode, using the appropriate mass ranges, fumonisin B1; 722 (M+H), fumonisin B2 and B3; 706 (M+H), fumonisin B4; 690 (M+H), fumonisin (FA1, FA2, FA3); 764, 748, 748 (M+H), and fumonisin C1; 708 (M+H). MS/MS was performed on 722 (M+H) for further confirmation of FB1. The column used for fumonisin analysis was a 3.0 mm × 150 mm i.d, 5 μm, Intersil ODS-3 column (MetaChem Technologies Inc., Torrance, CA). The mobile phase at initial elution starting condition consisted of water: 1% acetic acid in methanol (65:35) at 300 μL per min, followed by water:1% acetic acid in methanol:methanol (5:35:65) at 10 min. The gradient was held constant for 10 min, and returned to the initial starting conditions for 4 min for column equilibration. Quantitation of FB1, FB2, and FB3 was carried out by the external standard method where FB4 was calculated as a percentage of FB1. Other derivatives of fumonisin were monitored for qualitative purposes only.
Fumonisin and fusarins analysis was also carried out on crack-corn cultures used for RNA analysis to further investigate the correlation between fumonisin production and gene expression levels. Ten grams of cracked corn kernel culture were extracted in 25 ml acetonitrile:water (1:1, v/v) on a rotary shaker set at 250 rpm. After 2.5 hours of shaking the mixture was centrifuged at 500 g for 5 minutes and the supernatant was recovered for analysis by reversed phase liquid chromatography-mass spectrometry (LC-MS) in electrospray mode as previously described (30). Briefly, 10 grams of cracked corn kernel culture were extracted in 25 ml acetonitrile:water (1:1, v/v) on a rotary shaker set at 250 rpm. After 2.5 hours of shaking the mixture was centrifuged at 500 g for 5 minutes and the supernatant was recovered for analysis by reversed phase liquid chromatography-mass spectrometry (LC-MS) in electrospray mode. The LC-MS system consisted of a ThermoFinnigan LCQ Deca mass spectrometer coupled to a ThermoSpectraPhysics HPLC with a C18 column. The column used for fumonisin analysis was the same as the one described above (Intersil ODS-3 column MetaChem Technologies Inc., Torrance, CA). Samples were run on a gradient of 35 to 95% (v/v) methanol over 35 minutes at a flow rate of 0.3 ml per minute. Fumonisins and fusarins were detected by monitoring masses of 240 through 1000. The identities of fumonisins and fusarins were confirmed by retention time and the presence of appropriate [M+H]+ ions. Quantification was accomplished by comparison of the integrated intensity of ions corresponding to fumonisin and fusarin standards (30)
RESULTS and DISCUSSION
Fumonisin production on natural substrates
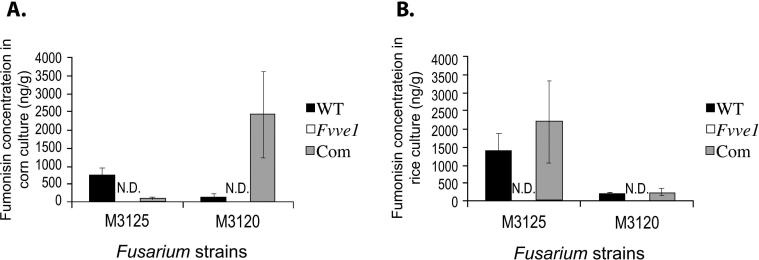
Fumonisins are currently considered the most economically important mycotoxins produced by the corn pathogen F. verticillioides because of their widespread occurrence in corn and their potential health effects on humans and animals (1, 2, 4, 5). Our current studies indicated that fumonisin production is affected in the ΔFvve1 mutants. In corn and rice cultures (Figure 1), FB1 was produced by the wild-type strains as well as by the complemented strains (Figure 2). Trace amounts of FB2 and FB3 were also detected in corn and rice cultures of the wild-type and complemented strains. In contrast, neither FB1, FB2, nor FB3 was detected in rice or corn cultures of the ΔFvve1 mutants (Figure 2). These results were consistent in both mating-type genetic backgrounds. Our findings reveled that the novel regulatory factor FvVe1 encoded by the FvVE1 gene is required for fumonisin production when the fungus grows on the natural substrates corn and rice (Figures 1 and 2).
Figure 1.
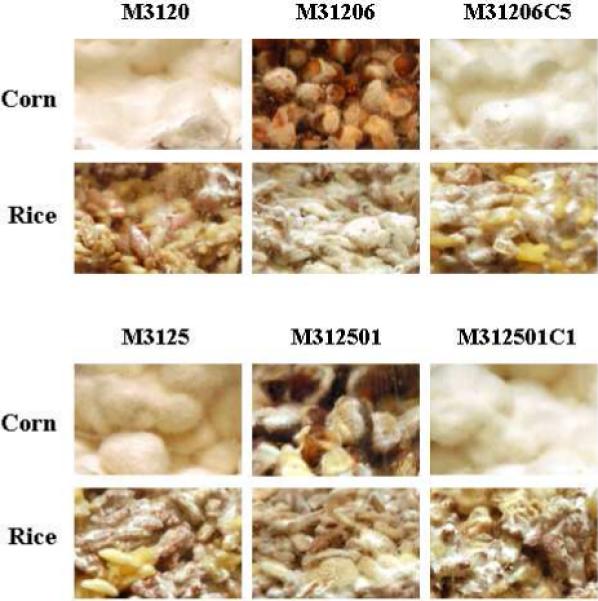
Photographs of wild-type (WT, M-3120 and M-3125), FvVE1deletion mutant (ΔFvve1, M31206 and M312501) and complemented (Com., M31206C5 and M312501C1) strains of MAT1−1 and MAT1−2 respectively, in corn and rice cultures.
Figure 2.
Production of fumonisins in wild-type (WT), FvVE1deletion mutant (ΔFvve1) and complemented (Com) strains of MAT1−1 and MAT1−2 in corn (A) and rice (B) cultures. Total fumonisins (B1, B2 and B3 combined) were analyzed at 2 weeks after inoculation. Bars indicate standard errors of 3 independent cultures. N.D. = not detected.
Expression of fumonisin biosynthetic genes
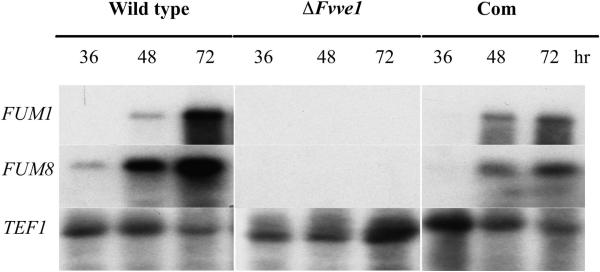
Previous studies showed that VeA, the FvVe1 homolog in Aspergillus species, is required for expression of sterigmatocystin/aflatoxin biosynthetic genes and for concomitant production of the toxins (22-24). However, the possible role of VeA homologs in activation of mycotoxin biosynthetic genes in other fungal genera was not known until now. To investigate whether the expression of fumonisin biosynthetic genes is regulated by the FvVE1 gene, transcription levels of FUM1 and FUM8 (essential fumonisin biosynthetic genes) were examined in the wild-type, ΔFvve1 mutant and complemented strains grown on cracked-corn medium. The Northern analysis in Figure 3 shows that transcription of FUM1 was first detected at 48 h after inoculation in both wild-type and complemented strains. FUM8 expression followed a similar pattern where transcripts started to accumulate slightly earlier in the wild-type strain. However, FUM1 and FUM8 transcripts were not detected in the ΔFvve1 mutant (Figure 3). The lack of fumonisin biosynthetic gene expression in the FvVE1 deletion mutant is most likely responsible for the lack of fumonisin production in this strain.
Figure 3.
Northern analysis of FUM1, and FUM 8 gene expression from wild-type M-3120, FvVE1 deletion mutant M31206 (ΔFvve1), and complementation (Com) strain M31206C5. Total RNA was isolated from mycelial tissue grown on cracked-corn medium at 36, 48 and 72 h after inoculation. TEF1 was used as loading control.
Expression of FUM21, FCC1 and ZFR1
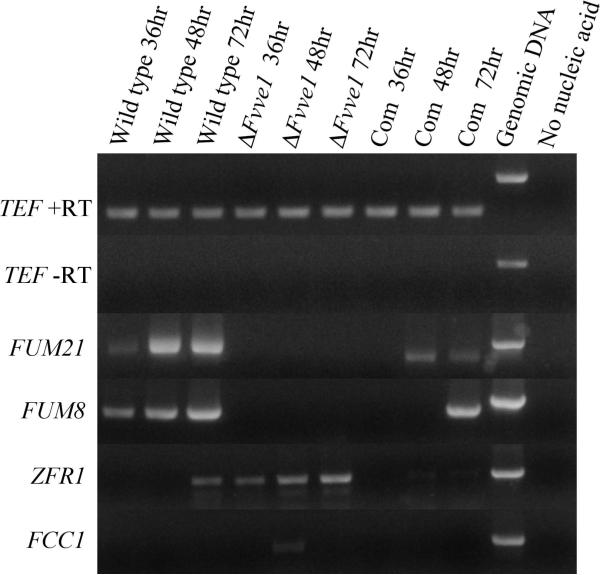
With the goal of further elucidating the mechanism through which FvVE1 regulates the expression of the FUM gene cluster and the concomitant fumonisin production, we investigated whether the expression of FUM21 was altered by the FvVE1 deletion. FUM21 gene encodes a putative Zn(II)2Cys6 DNA-binding transcriptional activator that is likely specific for FUM cluster genes (12). Furthermore, a functional FUM21 is necessary for fumonisin production in F. verticillioides. Genes encoding cluster-specific regulatory proteins have been previously found in other secondary metabolism gene clusters. Well known examples of these regulators are aflR and aflJ, demonstrated to govern the expression of sterigmatocystin/aflatoxin gene clusters in Aspergillus spp. (31-33). We have previously shown that veA is necessary for the expression of aflR and aflJ in Aspergillus (22-24). In the present study, in order to investigate whether FvVE1 plays a role in regulating the expression of FUM21 we chose RT-PCR analysis because of its ability to detect transcripts that are present at low levels. FUM21 transcripts were absent in the ΔFvve1 mutant cultures under conditions that allow expression of this gene in the wild type and complementation strains (Figure 4). FUM8 was also included in the RT-PCR analysis as an internal control for comparison of the RT-PCR and Northern experiments, which yielded essentially the same results. The absence of FUM21 transcripts in the ΔFvve1 mutant indicates that a functional FvVE1 is necessary for FUM21 expression.
Figure 4.
RT-PCR analysis of FUM21, FUM 8, ZFR1 and FCC1 gene expression from wild-type M-3120, FvVE1 deletion mutant M31206 (ΔFvve1), and complementation (Com) strain M31206C5. Total RNA was isolated from mycelial tissue grown on cracked-corn medium at 36, 48 and 72 h after inoculation. TEF1 was used as loading control. A no reverse transcriptase control reaction is shown for TEF1 primers indicating no genomic DNA remained after DNase treatment.
Genes outside the FUM cluster can also regulate fumonisin production in F. verticillioides. For example, deletion of the C-type cyclin-like gene, FCC1 , abolished fumonisin production on corn kernels and in a defined medium at high pH (14). To test whether FvVE1 controls this regulatory gene, the transcription levels of FCC1 were examined in wild-type, ΔFvve1 mutant and complemented strains grown on cracked-corn medium (Figure 4). Our results showed that FCC1 expression was very low and only detected in the FvVE1 deletion mutant at 48 h after inoculation. Although this result differs from those previously reported (13), where FCC1 expression was detected at higher levels in cracked-corn cultures, under the experimental conditions assayed in our study fumonisin production was detected as well as FUM1 and FUM8 expression, indicating that our culture system yielded reliable results. Fumonisin production in the fcc1 mutant is not blocked in the defined medium at low pH (14). Changes in pH did not rescue fumonisin production in the ΔFvve1 mutant (data not shown) as in the case of the FCC1 mutant (14). FCC1 is required for function of the Zfr1, a putative Zn(II)2Cys6 transcription factor postulated to control fumonisin production by regulating genes involved in the perception or uptake of carbohydrates (15, 34). In our study, ZFR1 transcripts were detected in the wild type, ΔFvve1 mutant and complementation strain, particularly in the ΔFvve1 mutant where ZFR1 transcription occurred earlier and was more abundant than in strains with a functional FvVE1 (Figure 4). This suggests that FvVE1 negatively influences ZFR1 expression. Further studies will focus on elucidating possible interactions between ZFR1 and FvVE1, and whether FvVE1 has a role in carbohydrate metabolism in F. verticillioides.
Cultures utilized for RNA studies were also analyzed for the fumonisins. In agreement with previous experiments (Figures 1 and 2), wild-type and complementation strains produced fumonisins but the ΔFvve1 mutant did not (Table 1). Even when incubation time was increased to 144 hours after inoculation of the crack-corn medium, fumonisin were not detected in ΔFvve1 cultures.
Table 1.
Fumonisin and Fusarin Analysis of Cracked-corn Cultures.
| Time | Strain | Fumonisinsa | Fusarinsb |
|---|---|---|---|
| 36 hours | Wild type | 0 | 0 |
| ΔFvve1 | 0 | 0 | |
| Comp | 0 | 196 | |
| 48 hours | Wild type | 1 | 0 |
| ΔFvve1 | 0 | 0 | |
| Comp | 0 | 864 | |
| 72 hours | Wild type | 98 | 330 |
| ΔFvve1 | 0 | 0 | |
| Comp | 127 | 1709 | |
| 96 hours | Wild type | 178 | 673 |
| ΔFvve1 | 0 | 0 | |
| Comp | 316 | 2066 | |
| 144 hours | Wild type | 281 | 1945 |
| ΔFvve1 | 0 | 0 | |
| Comp | 334 | 2246 |
μg fumonisins B1, B2 and B3 combined per gram of cracked corn culture
μg fusarins C1, C2 and C3 combined per gram of cracked corn culture
Fusarin analysis
The VeA homologs in Aspergillus regulate not only production of sterigmatocystin and aflatoxin, but also production of other secondary metabolites such as aflatrem, cyclopiazonic acid, and penicillin (22-24). In addition to fumonisins, F. verticillioides produces an array of other secondary metabolites, including the mycotoxins fusarins. These toxins have been reported to be mutagenenic as well as immunosuppressive (17-19). In order to evaluate the role of FvVE1 in fusarin biosynthesis, we examined the production of this compound in the wild-type, FvVE1 deletion mutant and complementation strains on cracked-corn medium. Fusarins were detected in extracts from cultures of wild-type and complementation strains, but were not detected in extracts of ΔFvve1 cultures (Table 1), indicating that FvVE1 is also necessary for fusarin biosynthesis in F. verticillioides. To our knowledge, this is the first report of a gene described to regulate fusarin production in F. verticillioides. Interestingly, a recent report by Estrada and Avalos (35) showed that the white-collar gene wcoA modulates fusarin production in a light-dependent manner in F. fujikuroi. In Aspergillus nidulans, VeA forms a nuclear protein complex that includes light-sensing proteins, such as the red phytochrome-like FphA and the white-collar LreA and LreB proteins responsive to blue light (36). In future studies we will investigate if a similar protein complex that includes FvVe1 also exists in F. verticillioides.
As in the case of veA regulation of secondary metabolism in Aspergillus (22-24), the effect of FvVE1 on secondary metabolism in F. verticillioides could also be broad. The differences in pigmentation observed in the natural substrate cultures (Figure 1, and data not shown) indicate that the synthesis of other unknown metabolites is also regulated by FvVE1.
In conclusion, we demonstrated that F. verticillioides FvVE1 is required for fumonisin and fusarin production on the natural substrates corn and rice. We also showed that FvVE1 is necessary for the expression of the fumonisin biosynthetic enzyme-encoding genes FUM1 and FUM8 as well as the transcription factor gene FUM21. This study also revealed that FvVE1 affects production of a second family of F. verticillioides secondary metabolites, namely fusarins. The consistent blockage of fumonisin on natural substrates, particularly in corn, suggests that FvVE1 could be a potential target to control fumonisin contamination of food commodities. We are currently exploring this possibility by investigating whether the FvVE1 deletion affects the ability of F. verticillioides to produce fumonisins in living corn plants.
ACKNOWLEDGMENT
We thank Bobbie Johnson and Jennifer Tonos for their help in analyzing fumonisins and Marcie Moore for the Northern blot analysis. We also thank Dapeng Bao for his technical support. This work was funded by NIH GM074267−01A1.
Abbreviation Used
- FB1
fumonisin B1
- FB2
fumonisin B2
- FB3
fumonisin B3
- veA
Velvet gene
- FvVE1
Fusarium verticillioides veA
LITERATURE CITED
- 1.Nelson PE, Desjardins AE, Plattner RD. Fumonisins, mycotoxins produced by Fusarium species: biology, chemistry, and significance. Annu. Rev. Phytopathol. 1993;31:233–252. doi: 10.1146/annurev.py.31.090193.001313. [DOI] [PubMed] [Google Scholar]
- 2.Rheeder JP, Marasas WFO, Vismer HF. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002;68:2101–2105. doi: 10.1128/AEM.68.5.2101-2105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezuidenhout SC, Gelderblom WCA, Gorst-Allman CP, Marthinus Horak R, Marasas WFO, Spiteller G, Vleggaar R. Structure elucidation of the fumonisins, mycotoxins from Fusarium moniliforme. J. Chem. Soc., Chem. Commun. 1988:743–745. [Google Scholar]
- 4.Gelderblom WCA, Jaskiewicz K, Marasas WFO, Thiel PG, Horak MJ, Vleggaar R, Kriek NPJ. Fumonisins-novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microbiol. 1988;54:1806–1811. doi: 10.1128/aem.54.7.1806-1811.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai K, Sullards MC, Allegood J, Wang E, Schmelz EM, Hartl M, Humpf HU, Liotta DC, Peng Q, Merrill AH., Jr. Fumonisins and fumonisin analogs as inhibitors of ceramide synthase and inducers of apoptosis. Biochim. Biophys. Acta. 2002;1585:188–192. doi: 10.1016/s1388-1981(02)00340-2. [DOI] [PubMed] [Google Scholar]
- 6.Proctor RH, Brown DW, Plattner RD, Desjardins AE. Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 2003;38:237–249. doi: 10.1016/s1087-1845(02)00525-x. [DOI] [PubMed] [Google Scholar]
- 7.Bojja RS, Cerny RL, Proctor RH, Du L. Determining the biosynthetic sequence in the early steps of the fumonisin pathway by use of three gene-disruption mutants of Fusarium verticillioides. J. Agric. Food Chem. 2004;52:2855–2860. doi: 10.1021/jf035429z. [DOI] [PubMed] [Google Scholar]
- 8.Seo JA, Proctor RH, Plattner RD. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 2001;34:155–165. doi: 10.1006/fgbi.2001.1299. [DOI] [PubMed] [Google Scholar]
- 9.Hohn TM, Krishna R, Proctor RH. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 1999;26:224–235. doi: 10.1006/fgbi.1999.1122. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 11.Malz S, Grell MN, Thrane C, Maier FJ, Rosager P, Felk A, Albertsen KS, Salomon S, Bohn L, Schafer W, Giese H. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 2005;42:420–433. doi: 10.1016/j.fgb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Brown DW, Butchko RAE, Busman M, Proctor RH. The Fusarium verticillioides FUM Gene Cluster Encodes a Zn(II)2Cys6 Protein That Affects FUM Gene Expression and Fumonisin. Euk. Cell. 2007;6:1210–1218. doi: 10.1128/EC.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shim WB, Woloshuk CP. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 2001;67:1607–1612. doi: 10.1128/AEM.67.4.1607-1612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaherty JE, Pirttila AM, Bluhm BH, Woloshuk CP. PAC1, a pH-regulatory gene from Fusarium verticillioides. Appl. Environ. Microbiol. 2003;69:5222–5227. doi: 10.1128/AEM.69.9.5222-5227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaherty JE, Woloshuk CP. Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster-type gene, ZFR1. Appl. Environ. Microbiol. 2004;70:2653–2659. doi: 10.1128/AEM.70.5.2653-2659.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bluhm BH, Woloshuk CP. Fck1, a C-type cyclin-dependent kinase, interacts with Fcc1 to regulate development and secondary metabolism in Fusarium verticillioides. Fungal Genet. Biol. 2006;43:146–154. doi: 10.1016/j.fgb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Cheng SJ, Jiang YZ, Li MH, Lo HZ. A mutagenic metabolite produced by Fusarium moniliforme isolates in Linxian County, China. Carcinogenesis. 1985;6:903–905. doi: 10.1093/carcin/6.6.903. [DOI] [PubMed] [Google Scholar]
- 18.Dong ZY, Zhan YH. Inhibitory effect of a mycotoxin, fusarin C, on macrophage activation and macrophage mediated cytotoxicity to tumor cells in mice. J. Exp. Clin. Cancer Res. 1987;6:31–38. [Google Scholar]
- 19.Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 2007;119:47–50. doi: 10.1016/j.ijfoodmicro.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Gaffoor I, Brown DW, Plattner R, Proctor RH, Qi W, Trail F. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell. 2005;4:1926–1933. doi: 10.1128/EC.4.11.1926-1933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song ZS, Cox RJ, Lazarus CM, Simpson TJ. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. ChemBioChem. 2004;5:1196–1203. doi: 10.1002/cbic.200400138. [DOI] [PubMed] [Google Scholar]
- 22.Kato N, Brooks W, Calvo AM. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell. 2003;2:1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duran RM, Cary JW, Calvo AM. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007;73:1158–1168. doi: 10.1007/s00253-006-0581-5. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Myung K, Guse D, Donkin B, Proctor RH, Grayburn WS, Calvo AM. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol Microbiol. 2006;62:1418–32. doi: 10.1111/j.1365-2958.2006.05447.x. [DOI] [PubMed] [Google Scholar]
- 26.Abbas HK, Mirocha CJ, Shier WT. Mycotoxins produced from fungi isolated from foodstuffs and soil: Comparison of toxicity in fiberblasts and rat feeding test. Appl. Environ. Microbiol. 1984;48:654–661. doi: 10.1128/aem.48.3.654-661.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbas HK, Cartwright RD, Xie W, Mirocha CJ, Richard JL, Dvorak TJ, Sciumbato GL, Shier WT. Mycotoxins production by Fusarium proliferatum isolates from rice with Fusarium sheath rot disease. Mycopathologia. 1999;147:97–104. doi: 10.1023/a:1007147813326. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: [Google Scholar]
- 29.Abbas HK, Cartwright RD, Xie W, Shier WT. Aflatoxin and fumonisin contamination of corn (maize, Zea mays) hybrids in Arkansas. Crop Protection. 2006;25:1–9. [Google Scholar]
- 30.Plattner RD, Weisleder D, Poling SM. Analytical determination of fumonisins and other metabolites produced by Fusarium moniliforme and related species on corn. Adv. Exp. Med. Biol. 1996;392:57–64. doi: 10.1007/978-1-4899-1379-1_5. [DOI] [PubMed] [Google Scholar]
- 31.Yu JH, Butchko RA, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
- 32.Payne GA, Nystrom GJ, Bhatnagar D, Cleveland TE, Woloshuk CP. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang PK. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genomics. 2003;268:711–719. doi: 10.1007/s00438-003-0809-3. [DOI] [PubMed] [Google Scholar]
- 34.Bluhm BH, Kim H, Butchko RA, Woloshuk CP. Involvement of ZFR1 of Fusarium verticillioides in kernel colonization and the regulation of FST1, a putative sugar transporter gene required for fumonisin biosynthesis on maize kernels. Mol. Plant Pathol. 2008;9:203–211. doi: 10.1111/j.1364-3703.2007.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estrada AF, Avalos J. The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet. Biol. 2008;45:705–718. doi: 10.1016/j.fgb.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]