Abstract
FIAT represses osteocalcin gene transcription by heterodimerizing with ATF4 and preventing it from binding to DNA. We report here the expression profiles of FIAT and ATF4 during osteoblastogenesis. Messenger RNA levels for the osteoblast transcriptional regulators Satb2, Runx2, Fiat and Atf4 were quantified using Real Time-reverse transcription PCR (RT-qPCR) and respective protein levels monitored by immunodetection in differentiating primary osteoblast cultures. Satb2, Fiat and Atf4 mRNA levels remained constant throughout the differentiation sequence, whereas Runx2 transcript levels were significantly increased by 12 days post-confluency. Using immunofluorescence, the SATB2, RUNX2, and ATF4 signals appeared to increase as a function of time in culture. FIAT protein expression was readily detected in early cultures, but signal intensity decreased thereafter. When immunoblotting was used to quantify the relative amounts of FIAT and ATF4 proteins, the expression levels of the two proteins were found to be inversely correlated. The decrease in FIAT protein levels coincided with increased binding of ATF4 to the osteocalcin gene promoter, and with increased osteocalcin expression measured by RT-qPCR or immunoblotting. Immunohistochemistry of long bones from mice at E16.5 and 2 days post-natal revealed that both proteins are initially expressed in osteoblasts. In adult bone, FIAT was detected in osteocytes, while ATF4 expression was observed in active osteoblasts and lining cells, but not in osteocytes. Taken together, these data support the idea that a stoichiometric excess of ATF4 over FIAT in mature osteoblasts releases ATF4 from sequestration by FIAT, thereby allowing ATF4 homodimerization and subsequent transactivation of the osteocalcin gene.
Keywords: ATF4, FIAT, leucine zipper transcription factors, osteoblasts, osteocytes, osteoblastogenesis, bone development
1. Results and discussion
The basic domain-leucine zipper (bZIP) transcription factor ATF4 (activating transcription factor 4) has recently been shown to mediate multiple aspects of bone formation, including osteoblast-specific gene expression, osteoblast differentiation, type I collagen synthesis, and osteoclast differentiation (Elefteriou et al., 2005; Yang et al., 2004). The ATF4 transcript is ubiquitously expressed but its protein mainly accumulates in osteoblasts where it is not subjected to ubiquitination-mediated degradation like in most other cell types (Yang and Karsenty, 2004). ATF4 dimerizes through its leucine domain to form a large variety of homodimers and/or heterodimers and bind to DNA via its basic region for target gene transactivation. Different dimerization partners confer different DNA binding specificity and hence allow ATF4 to mediate multiple signalling pathways (Hai and Curran, 1991).
In osteoblasts, one of the target genes of ATF4 is the differentiation marker, osteocalcin (Yang et al., 2004). The potent activation of the osteocalcin gene is crucial for osteoblast differentiation and requires cooperative interactions between RUNX2, SATB2, and ATF4 at the osteocalcin promoter (Dobreva et al., 2006; Xiao et al., 2005).
Factor Inhibiting ATF4-mediated Transcription (FIAT) is a protein whose name was coined for its interaction with ATF4 and subsequent blockage of ATF4-directed osteocalcin gene transcription (Yu et al., 2005). FIAT is a ~66-kDa protein that lacks a basic DNA-binding domain but contains three identifiable leucine zipper domains. FIAT heterodimerizes with ATF4 through its second leucine zipper (Yu et al., 2008) and thereby prohibits ATF4 from binding to its cognate DNA sequence (St-Arnaud and Yu, 2005; Yu et al., 2005; Yu et al., 2006). Transgenic mice overexpressing FIAT under the control of the osteoblast-specific α1(I) collagen promoter fragment are osteopenic, with reduced bone mineral density, trabecular volume, and bone rigidity (Yu et al., 2005). This low bone mass phenotype was shown to be caused by impaired osteoblast activity, without changes in osteoblast proliferation or apoptosis (Yu et al., 2005), thus mimicking several aspects of the ATF4-deficient mice phenotype(Yang et al., 2004). We interpret these data to mean that FIAT controls osteoblast activity by regulating ATF4-dependent functions. To achieve a better understanding of the expression of FIAT in osteoblasts, we measured its mRNA and protein levels relative to its dimerization target, ATF4, during osteoblastogenesis.
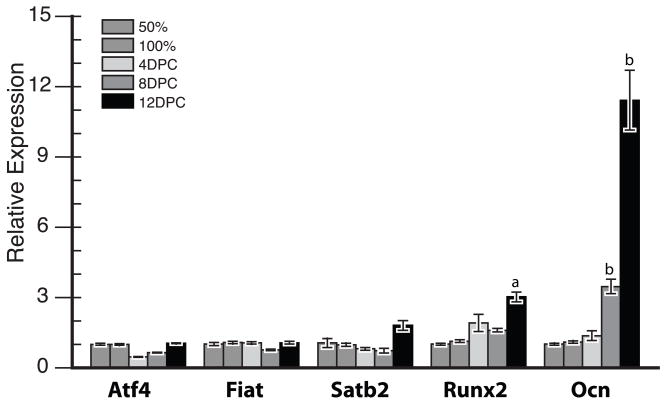
We measured the RNA expression levels for Atf4, Fiat, Satb2, Runx2, and osteocalcin(Ocn) in primary osteoblastic cultures obtained from calvaria of 5–8 days old C57BL/6 mice. Osteoblasts were cultured in the presence of 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10−8 M dexamethasone, and were harvested at 50% sub-confluence, 100% confluence, and 4, 8, and 12 days post-confluence for RT-qPCR analysis. RNA expression of Atf4, Fiat, and Satb2 was generally low and did not vary throughout the differentiation sequence (Fig. 1). At 12 days post-confluence, Runx2 expression increased significantly. The transcript levels of Ocn increased significantly at 8 days post-confluence and higher levels were also measured at 12 days post-confluence (Fig. 1).
Figure 1. RNA expression of Atf4, Fiat, Satb2, Runx2, and osteocalcin during osteoblast differentiation in culture.
Transcript expression was measured in primary osteoblast cell cultures at 50% cell confluence, 100% cell confluence, and 4, 8, and 12 days post-confluence (DPC) by reverse-transcription quantitative PCR and normalized to Gapdh transcript levels. Expression at 50% confluence was ascribed a relative value of 1. Ocn, osteocalcin. a, p<0.001 vs. Runx2 at 50% confluence; b, p<0.001 vs. Ocn at 50% confluence.
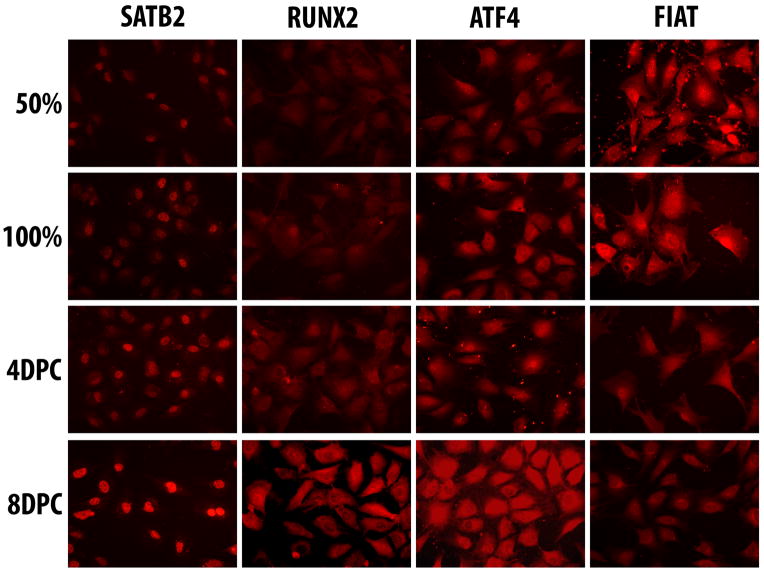
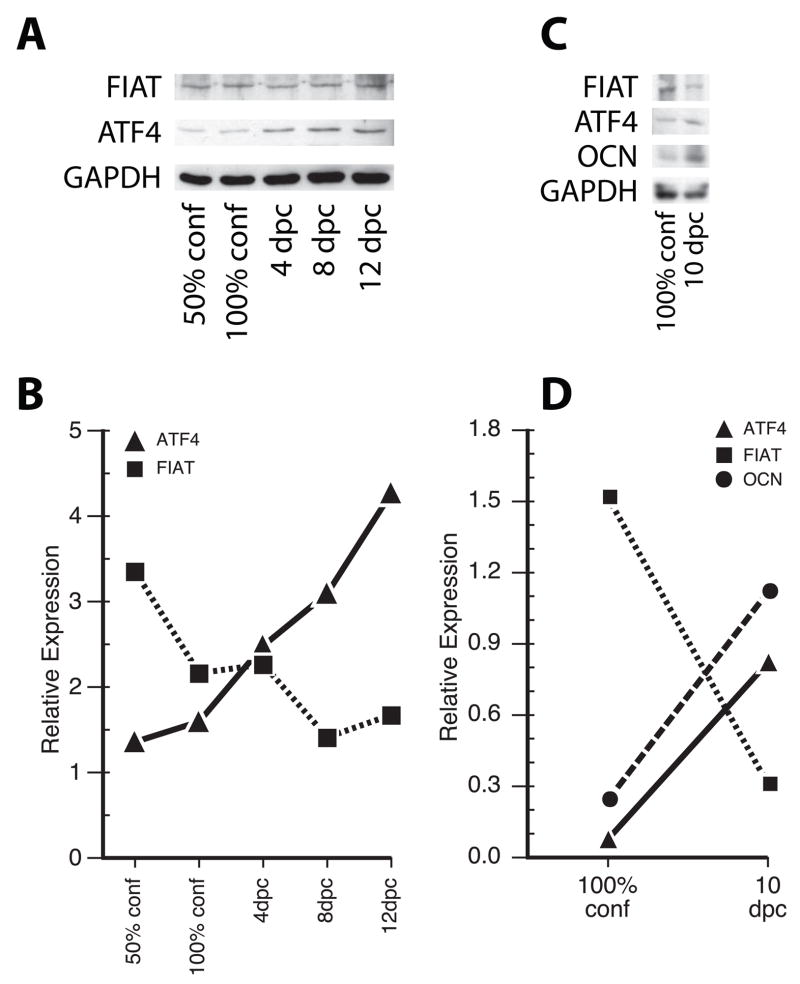
Next, we assessed SATB2, RUNX2, ATF4, and FIAT protein expression using immunocytofluorescence in primary osteoblast cultures. SATB2 was detected exclusively in the nucleus, while RUNX2, ATF4, and FIAT were detected in both the cytoplasmic and nuclear compartments (Fig. 2). SATB2, RUNX2, and ATF4 protein expression appeared to increase as a function of time in culture and the highest signal intensity was observed in differentiated, 8 days post-confluence cultures (Fig. 2). FIAT protein expression followed a different pattern. The protein was readily detected in early cultures (50% and 100% confluence), but signal intensity decreased thereafter (Fig. 2). When immunoblotting was used to quantify the relative amounts of both proteins (Fig. 3A), the expression levels of the two proteins were found to be inversely correlated (Fig. 3B). In subconfluent cultures, the expression of FIAT relative to the housekeeping marker GAPDH exceeded the ATF4/GAPDH ratio. As the cultures differentiated, FIAT relative expression decreased whereas ATF4 relative expression increased (Fig. 3B), leading to an excess of ATF4 molecules relative to FIAT molecules in differentiated osteoblasts. This pattern of expression was observed in five independent experiments. When the relative expression of the FIAT, ATF4, and OCN proteins was evaluated in parallel by immunoblotting in confluent and late cultures, the stoichiometric excess of ATF4 protein over FIAT coincided with increased OCN protein levels (Fig. 3C, D).
Figure 2. SATB2, RUNX2, ATF4, and FIAT protein expression during osteoblastogenesis.
Primary cultures of osteoblasts were trypsinized at the indicated times and plated onto coverslips 16 h prior to fixation, permeabilization, and incubation with the corresponding primary antibody and a fluorescent secondary antibody. 50%, half-confluence; 100%, confluence; DPC, days post-confluence.
Figure 3. FIAT, ATF4, and OCN protein expression during osteoblastogenesis.
(A) Protein expression levels of FIAT, ATF4, and the ubiquitous marker GAPDH were detected in primary osteoblast cell cultures at 50% cell confluence, 100% cell confluence, and 4, 8, and 12 days post-confluence (dpc) by Western blotting. A representative example of five experiments that yielded similar results is shown. (C) Protein expression levels of FIAT, ATF4, OCN, and GAPDH were detected in primary osteoblast cell cultures at 100% cell confluence and 10 days post-confluence (dpc) by Western blotting. (B, D) Band intensity of FIAT, ATF4, and OCN in immunoblots were quantified, normalized against GAPDH protein level, and plotted. Conf, confluence; dpc, days post-confluence.
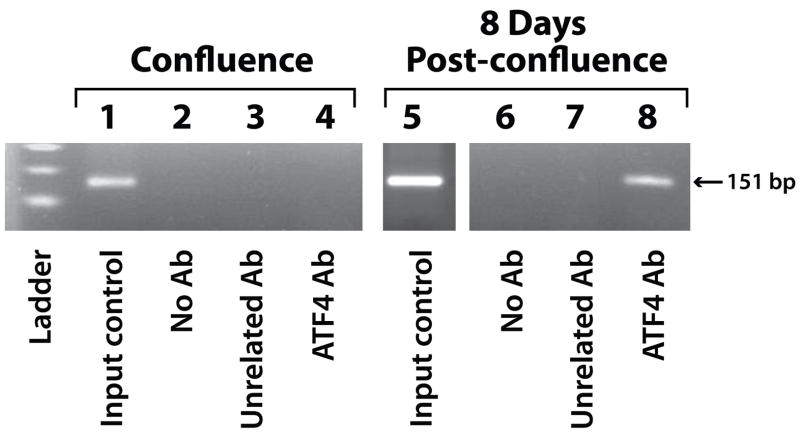
To correlate the expression level of ATF4 and its activity in regulating gene transcription crucial for osteoblast differentiation, we investigated ATF4-binding to the osteocalcin promoter during early as well as late osteoblast differentiation. Primary osteoblast cultures were harvested at cell confluence and 8 days post-confluence for chromatin immunoprecipitation using an anti-ATF4 antibody, followed by PCR probing for the osteocalcin promoter DNA fragment from position −152 to −2, which includes the ATF4 binding site (Yang et al., 2004). No osteocalcin promoter DNA fragments were precipitated in the absence of antibody or with an unrelated antibody (directed against the yeast protein GAL4) at either time points (Fig. 4, lanes 2, 3, 6, 7). However, osteocalcin promoter chromatin was precipitated by the anti-ATF4 antibodies in primary osteoblasts harvested at 8 days post-confluence (Fig. 4, lane 8), but not at cell confluence (Fig. 4, lane 4). These data support the hypothesis that when ATF4 protein expression exceeds the levels of FIAT protein at 8 days post-confluence, it is freed from FIAT inhibition and can bind the osteocalcin gene promoter to activate its transcription.
Figure 4. ATF4 binds to the proximal osteocalcin promoter in primary osteoblasts at 8 days post-cell confluence.
Binding of ATF4 to the osteocalcin promoter was assayed by chromatin immunoprecipitation using an anti-ATF4 antibody, followed by PCR amplification of the ATF4 binding site at the proximal osteocalcin promoter (position −152 to −2 relative to the transcription start site). Immunoprecipitation by the anti-ATF4 antibody was specific as no DNA were detected when no antibody (No Ab, lanes 2 and 6) or an unrelated antibody (directed against the yeast transcription factor GAL4) were used (lanes 3 and 7). ATF4 did not bind to the osteocalcin promoter during early osteoblast differentiation (lane 4) but binding occurred in mature osteoblasts (lane 8).
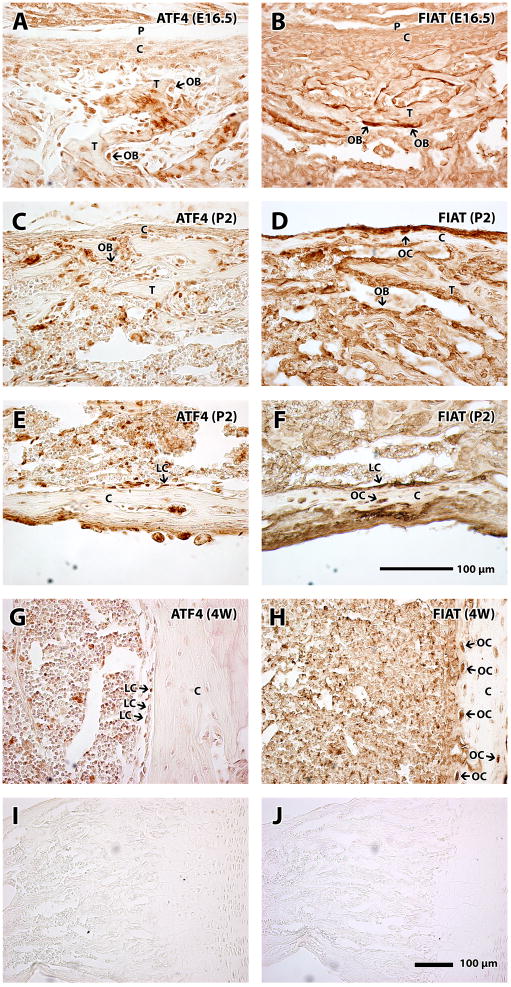
The in vivo expression patterns of FIAT and ATF4 were also monitored in the long bones of C57BL/6 mice harvested at E16.5, post-natal day 2, and 4 weeks. Antibodies raised against FIAT or ATF4 were used to probe cognate protein expression on paraffin-embedded femur or tibia sections and signals were revealed by an enzyme-substrate detection method. As trabecular bone began to form and mineralize at the mid-shaft of the diaphysis at E16.5, anti-FIAT and anti-ATF4 antibodies-stained osteoblasts were obvious surrounding the newly formed trabecular bone (Fig. 5A, B). Osteoblasts stained with the anti-ATF4 antibody appeared to be more cuboidal, whereas anti-FIAT antibody-stained osteoblasts seemed more flattened. As observed in primary cultures of osteoblasts (Fig. 2), FIAT protein expression did not appear restricted to the cell’s nucleus (Fig. 5B). At post-natal day 2, ATF4-positive staining was observed in most of the osteoblasts (Fig. 5C) and lining cells (Fig. 5E). On the other hand, FIAT stained positive in osteoblasts (Fig. 5D), lining cells (Fig. 5F), and was particularly strongly expressed in osteocytes (Fig. 5D, F). At post-natal four weeks, we observed less osteoblasts that stained positive for ATF4 compared to bones obtained from a younger age; however, ATF4 was still present in some osteoblasts and lining cells (Fig. 5G). In contrast, strong FIAT protein staining was mostly detected in osteocytes (Fig. 5H). Of note, ATF4 was not detected in osteocytes of 4-week-old bones (Fig. 5G). No staining was detected when the primary antibody was omitted from the reaction (Fig. 5I, J).
Figure 5. Expression of ATF4 and FIAT protein during skeletal development.
Long bones (tibia or femur) were harvested at the indicated times, fixed, demineralized, embedded in paraffin, and sectioned for immunodetection using specific anti-ATF4 or anti-FIAT antibodies. At E16.5, localization of ATF4 (A) and FIAT (B) in early osteoblasts was detected. At post-natal day 2, both ATF4 (C, E) and FIAT (D, F) were detected in osteoblasts and lining cells, while FIAT also stained strongly in osteocytes. In bones from 4 week-old skeletons, ATF4 stained the nucleus of lining cells (G), whereas FIAT was only expressed strongly in osteocytes (H). No staining was detected when the primary antibody was omitted from the reaction (I, J). E16.5, embryonic day 16.5; P2, post-natal day 2; 4W, 4 weeks; P, periosteum; C, cortex; T, trabeculae; OB, osteoblast; LC, lining cell; OC, osteocyte. Magnification: 40X.
We have shown that FIAT and ATF4 RNA levels remained stable at all stages of osteoblast differentiation. The relative excess of ATF4 over FIAT protein at 8 days post-cell confluence coincided with the binding of ATF4 to the osteocalcin promoter and with increased osteocalcin transcription. These data suggest that a stoichiometric excess of ATF4 over FIAT in mature osteoblasts releases the inhibitory effect of FIAT on ATF4, allowing ATF4 to homodimerize and subsequently transactivate the osteocalcin gene and possibly other downstream signalling targets important for bone formation.
This model relies on the assumption that the antibodies against ATF4 and against FIAT recognize their target antigen with the same affinity, which may of course not be the case. Nevertheless, protein expression was quantified relative to the same ubiquitous internal reference, GAPDH, and the changes over the differentiation sequence were measured using the same antibodies at each time point. Similar results were obtained when expression levels were normalized against a different ubiquitous protein marker, TBP (TATA binding protein; data not shown). Therefore, while the stoichiometric relationship of the two proteins may not be numerically precise, the expression of the ATF4 protein at 8, 10, and 12 days post-confluence is higher than the FIAT protein expression, supporting our model that in differentiated osteoblasts, ATF4 escapes FIAT inhibition.
Osteocalcin gene transcription depends on the complex interplay between several transcriptional regulators, including cooperative interactions between RUNX2, SATB2, and ATF4 (Dobreva et al., 2006; Xiao et al., 2005). The immunofluorescence detection signal for SATB2 and RUNX2 appeared to increase during differentiation in culture. No interaction has ever been reported between FIAT and SATB2 or RUNX2, and it is not known whether FIAT can influence their activity.
Both ATF4 and FIAT were detected in the cytosol and nucleus of differentiating osteoblasts. The regulation of ATF4 activity by FIAT could thus happen in both compartments. Detection of FIAT in the cytosol is in accord with the identification of FIAT as a protein that also interacts with the cytoplasmic syntaxin family members (Nogami et al., 2004). FIAT was also shown to contact the transcriptional coactivator αNAC (Yoshida et al., 2005; Yu et al., 2006), a protein that shuffles between the cytoplasm and the nucleus (Quelo et al., 2004a; Quelo et al., 2004b; Quelo et al., 2005). It will prove interesting to determine if the subcellular localization of FIAT depends solely on its interaction with partner targets or whether it is controlled by more elaborate post-translational mechanisms.
In bone sections, we detected ATF4 protein in cuboidal osteoblasts and in lining cells, but not in osteocytes. On the other hand, strong FIAT protein expression was detected in osteocytes as early as postnatal day 2. The exclusive localisation of FIAT to osteocytes suggests that FIAT interacts with other targets in these cells. Whether this putative target represents a previously identified partner such as αNAC, which is expressed in bone (Akhouayri et al., 2005; Akhouayri and St-Arnaud, 2007; Moreau et al., 1998), or a different bZIP factor that is involved in the regulation of osteocyte f unction will be important to elucidate.
2. Experimental procedures
2.1 Primary Cell Culture
Primary cultures of osteoblasts were obtained from 5–8-day-old calvaria of C57BL/6 mice, and were prepared as described in Yu et al. (Yu et al., 2005). Cells were cultured in αMEM (Invitrogen, Burlington, ON) supplied with 10% FBS (Invitrogen), 50 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10−8 M dexamethasone (Sigma-Aldrich, Oakville, ON). Media was changed every 2–3 days, and cells were trypsinized with 0.05% trypsin-EDTA (Invitrogen) when required.
2.2 Reverse Transcription Quantitative PCR (RT-qPCR)
Primary osteoblasts were plated at 5 × 104 cells per well in a 12-well plate, and were harvested for RNA at 50% sub-confluence, 100% confluence, and 4, 8, or 12 days post-confluence using the RNAqueous-4PCR kit, adhering to the manufacturer’s instructions (Ambion Inc., Austin, TX). Ten μg of RNA were reverse-transcribed into cDNA using the High Capacity cDNA Archive kit as per the manufacturer’s recommendations (Applied Biosystems, Foster City, CA). RT-qPCR amplification was performed on an Applied Biosystems 7500 instrument using specific TaqMan probes for the Atf4, Fiat, Satb2, Runx2, osteocalcin (Ocn), and Gapdh genes, and the Taqman Universal PCR Master Mix (Applied Biosystems). Expression level of each mRNA was quantified by the ΔCt method and normalized to Gapdh levels.
2.3 Immunocytofluorescence
Primary cultures of osteoblasts were seeded at 106cells per well in a 6 -well plate. Cells were trypsinized with 0.05% trypsin-EDTA 16 h prior to harvesting at 50% confluence, 100% confluence, and 4 and 8 days post-confluence, and plated on gelatin-coated coverslips. The primary osteoblasts were fixed the following morning in 4% paraformaldehyde and permeabilized with 1.0% Triton X-100 for 15 min. Following blocking with 1% Blocking Reagent (Roche Molecular Diagnostics, Indianapolis, IN) supplemented with 0.2% Tween-20, the cells were incubated with the primary antibodies at a 1:200 dilution (anti-SATB2, catalog No. ab34735, Abcam Inc., Cambridge, MA; anti-RUNX2, catalog No. R9403, Sigma-Aldrich; anti-ATF4, catalog No. sc-22800, SantaCruz Biotechnologies, SantaCruz, CA; anti-FIAT raised against residues 111–125 (Yu et al., 2005)). The cells were then incubated for 1 hour at room temperature with a 1:2000 dilution of an AlexaFluor 594-conjugated goat anti-rabbit IgG secondary antibody (Molecular Probes-Invitrogen). Coverslips were mounted in Vectashield (with DAPI) mounting medium (Vector Laboratories, Burlingame, CA). Images were acquired using an Olympus DP70 digital camera with the DP Controller image management software.
2.4 Western Blotting
Primary osteoblasts were plated at 7.5 × 106 cells per 100 mm Petri dish and were harvested for nuclear proteins at 50% sub-confluence, 100% confluence, and 4, 8, 10, or 12 days post-confluence. The purified anti-FIAT antibody (180 μg/mL, 1:200 dilution), anti-ATF4 antibody (1:200 dilution) (Santa Cruz), anti-OCN antibody (1:400 dilution) (Santa Cruz), or anti-GAPDH antibody (1:1000 dilution) (Ambion) were used to probe immunoblots of nuclear extracts from primary osteoblasts. Anti-rabbit or anti-mouse antibodies conjugated with horseradish peroxidase (1:10000 dilution) were used as secondary antibodies (Amersham Biosciences, Piscataway, NJ) and detected by ECL Western Blotting Detection Reagents (GE Healthcare Bio-Sciences, Baie d’Urfé, QC). Quantification of the signal intensity was performed using the software Scion Image Alpha 4.0.3.2 (Scion Corporation, Frederick, MD) following the manufacturer’s instructions. Signal intensity of FIAT, ATF4, or OCN normalized to GAPDH intensity was graphed.
2.5 Chromatin Immunoprecipitation
Primary osteoblasts were plated at 7.5 × 106 cells per 100 mm Petri dish and cells were harvested at confluency as well as 8 days post-confluence. Chromatin Immunoprecipitation was performed as described in detail elsewhere (Akhouayri et al., 2005). Five μl of purified DNA was used as template for PCR with the following primers which amplified the promoter region of the osteocalcin gene: Forward, 5′-AGGCAGCTGCAATCACCA-3′ and Reverse, 5′-GCACCCTGCAGCATCCA-3′.
2.6 Histological Sample Preparation
Femurs and tibiae of C57BL/6 mice were collected at E16.5, post-natal day 2, and post-natal day 28. Samples were fixed in 4% paraformaldehyde, demineralized with formic acid-based Immunocal™ (Decal Chemical Co., Tallman, NY), and dehydrated in increasing concentrations of ethanol before embedding in paraffin blocks. Embedded samples were sectioned at 6 μm using a Leica microtome (Leica Microsystems, Richmond Hill, ON).
2.7 Immunohistochemistry
Demineralized paraffin-embedded tibia and femur sections (6 μm) were used for immunohistochemical experiments. Sections were deparaffinized in xylene and rehydrated in 100%, 75%, 50% ethanol and deionized H2O. Antigen unmasking was achieved in 0.1% trypsin (Sigma-Aldrich) for 20 min at room temperature, followed by 1% H2O2 peroxidase blockage for 10 min. After blocking with PBS-T supplemented with 5% normal serum, 1% BSA, and/or 0.1% FIAT peptide from the C-terminus, sections were incubated with either anti-FIAT antibody (1:150 dilution) or anti-ATF4 antibody (1:200 dilution) (Santa Cruz). Biotinylated anti-rabbit IgG provided by the VECTASTAIN Elite ABC Kit (Vector Laboratories Inc, Burlington, ON) was used as secondary antibody (1:200 dilution) and signals were detected with the DAB Substrate Kit (Vector Laboratories).
Acknowledgments
We thank Mark Lepik for preparation of the figures. This work was supported by a grant from the U.S. National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR53287) to R. St-A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhouayri O, Quelo I, St-Arnaud R. Sequence-Specific DNA Binding by the αNAC Coactivator Is Required for Potentiation of c-Jun-Dependent Transcription of the Osteocalcin Gene. Mol Cell Biol. 2005;25:3452–3460. doi: 10.1128/MCB.25.9.3452-3460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhouayri O, St-Arnaud R. Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by jun family members. Calcif Tissue Int. 2007;80:123–131. doi: 10.1007/s00223-006-0102-7. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, Clement K, Vaisse C, Karsenty G. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514–520. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau A, Yotov WV, Glorieux FH, St-Arnaud R. Bone-specific expression of the alpha chain of the nascent polypeptide- associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol Cell Biol. 1998;18:1312–1321. doi: 10.1128/mcb.18.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S, Satoh S, Tanaka-Nakadate S, Yoshida K, Nakano M, Terano A, Shirataki H. Identification and characterization of taxilin isoforms. Biochem Biophys Res Commun. 2004;319:936–943. doi: 10.1016/j.bbrc.2004.05.073. [DOI] [PubMed] [Google Scholar]
- Quelo I, Akhouayri O, Prud’homme J, St-Arnaud R. GSK3beta-dependent phosphorylation of the alphaNAC coactivator regulates its nuclear translocation and proteasome-mediated degradation. Biochemistry. 2004a;43:2906–2914. doi: 10.1021/bi036256+. [DOI] [PubMed] [Google Scholar]
- Quelo I, Gauthier C, Hannigan GE, Dedhar S, St-Arnaud R. Integrin-linked kinase regulates the nuclear entry of the c-Jun coactivator alpha-NAC and its coactivation potency. J Biol Chem. 2004b;279:43893–43899. doi: 10.1074/jbc.M406310200. [DOI] [PubMed] [Google Scholar]
- Quelo I, Gauthier C, St-Arnaud R. Casein kinase II phosphorylation regulates alphaNAC subcellular localization and transcriptional coactivating activity. Gene Expr. 2005;12:151–163. doi: 10.3727/000000005783992070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud R, Yu VW. [Inhibition of bone mass accrual by the FIAT transcriptional repressor] Med Sci (Paris) 2005;21:1020–1021. doi: 10.1051/medsci/200521121020. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- Yang X, Karsenty G. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004;279:47109–47114. doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 Is a Substrate of RSK2 and an Essential Regulator of Osteoblast Biology; Implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Nogami S, Satoh S, Tanaka-Nakadate S, Hiraishi H, Terano A, Shirataki H. Interaction of the taxilin family with the nascent polypeptide-associated complex that is involved in the transcriptional and translational processes. Genes Cells. 2005;10:465–476. doi: 10.1111/j.1365-2443.2005.00848.x. [DOI] [PubMed] [Google Scholar]
- Yu VW, Ambartsoumian G, Verlinden L, Moir JM, Prud’homme J, Gauthier C, Roughley PJ, St-Arnaud R. FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J Cell Biol. 2005;169:591–601. doi: 10.1083/jcb.200412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VW, Gauthier C, St-Arnaud R. FIAT represses bone matrix mineralization by interacting with ATF4 through its second leucine zipper. J Cell Biochem. 2008 doi: 10.1002/jcb.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VWC, Gauthier C, St-Arnaud R. Inhibition of ATF4 transcriptional activity by FIAT/γ-taxilin modulates bone mass accrual. Ann N Y Acad Sci. 2006;1068:131–142. doi: 10.1196/annals.1346.027. [DOI] [PubMed] [Google Scholar]