Abstract
Purpose
There is uncertainty regarding the optimal strategy for identifying mutation carriers among those with hereditary nonpolyposis colorectal cancer (HNPCC).
Methods
We used decision analysis to compare the cost-effectiveness of 4 strategies among those with newly diagnosed colon cancer: (1) clinical and family history followed by microsatellite instability testing and germline testing (Bethesda guidelines); (2) universal microsatellite instability testing; (3) germline testing of those who meet clinical and family history criteria; and (4) universal germline testing.
Results
The added cost per year of life saved (YLS) for each strategy was as follows: (1) $11,865/YLS, (2) $35,617/YLS, (3) $49,702/YLS, and (4) $267,548/YLS.
Conclusions
The Bethesda guidelines are the most cost-effectiveness approach to screen persons for HNPCC.
Keywords: cost-effectivenessness, hereditary nonpolyposis colorectal cancer, screening, Bethesda guideline, microsatellite instability
Hereditary nonpolyposis colorectal cancer (HNPCC) is an autosomal-dominant inherited cancer susceptibility syndrome characterized by inherited mutations in one of the DNA mismatch repair (MMR) genes, and may account for approximately 2% of annual incident cases of colorectal cancer in the United States.1–6 A recent study suggests that regular colonoscopic screening of carriers, combined with early removal of benign and malignant tumors, reduces cancer morbidity and mortality.7 Although tumors from HNPCC mutation carriers have the characteristic phenotype of high levels of microsatellite instability (MSI), identifying those who should undergo HNPCC mutation testing is problematic because there are no overt symptoms or signs before the onset of colorectal cancer.
In this article, we estimate the clinical and economic tradeoffs associated with different strategies for detecting persons with HNPCC when a patient with recently diagnosed colorectal cancer is the starting point. Several approaches begin with newly diagnosed colorectal cancer patients who have pedigrees or other clinical characteristics that are suggestive of HNPCC.5,8,9 One of these, known as the Bethesda guidelines (Table 1), appears to be highly cost-effectiveness if it can be implemented as recommended.10 Nevertheless, patients’ reporting of cancer family histories is often inaccurate,11–13 and may be even more problematic in the future as wider use of colorectal cancer screening (along with excision of adenomatous polyps) reduces the rate of clinically detected cancers. As a result, others propose universal MSI or DNA analysis of persons with newly diagnosed colorectal cancers.14–16 These strategies would improve the sensitivity and specificity of screening, but universal tumor phenotype testing or germline testing would be initially costly compared to screening using personal and family cancer history. For example, routinely assaying all new colorectal tumors for MSI would cost the U.S. health care system more than $20 million annually; counseling and gene sequencing individuals for MMR mutations would cost more than $3.8 billion annually.
Table 1.
Bethesda guidelines for identifying potential HNPCC carriers among those with new colorectal cancers5
Test colorectal tumor for microsatellite instability if any of the below criteria are met:
|
Amsterdam criteria: (1) at least three relatives with histologically verified colorectal cancer. One family member a first degree relative of the other two (familial adenomatous polyposis excluded); (2) at least two successive generations affected with colorectal cancer; (3) in one of the individuals, diagnosis of colorectal cancer before the age of 50; and (4) diagnosis of familial adenomatous polyposis excluded.
Endometrial, ovarian, gastric, hepatobiliary, or small-bowel cancer or transitional cell carcinoma of the renal pelvis or ureter.
To address these issues, we designed a decision model to evaluate the clinical and economic implications of alternative strategies for identifying HNPCC carriers among individuals with newly diagnosed colorectal cancer.
METHODS
Overview
The decision model was developed to evaluate the cost-effectiveness of alternative case-finding methods for detecting HNPCC mutation carriers among persons with newly diagnosed colorectal cancer. The analysis includes direct medical care costs, nonmedical costs related to care (e.g., patient transportation costs), and health benefits (years of life saved) that accrue to individuals who are affected by testing.
Alternative screening approaches
We consider four potential strategies to identify MMR mutation carriers among those with newly diagnosed primary colorectal cancers (Fig. 1). Strategy 1 (Bethesda guidelines) involves MSI testing for those who meet clinical and family history, followed by mutation analysis for those with MSI high tumors. Strategy 2 (MSI all) involves MSI testing of all individuals, regardless of family or personal history. For Strategies 1 and 2, MSI-high individuals are then offered DNA testing for MMR mutations. Strategy 3 (DNA analysis for Bethesda clinical (+)) involves MMR mutation testing for all of those who meet the clinical and family history outlined for the Bethesda guidelines (i.e., MSI testing is bypassed). Strategy 4 (MMR all) involves MMR mutation testing of all individuals, regardless of personal or family history.
Fig. 1.
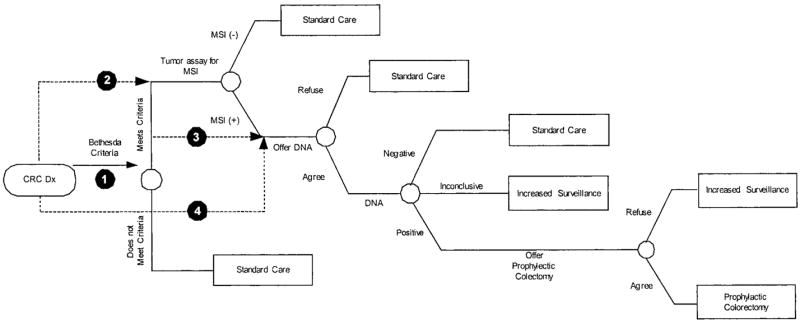
Decision tree representing HNPCC screening options for individuals with newly diagnosed colorectal cancers. CRC, colorectal cancer; HNPCC, hereditary nonpolyposis colorectal cancer; MSI, microsatellite instability; DNA, DNA analysis for mismatch repair mutation; Increased Surveillance, colonoscopy every 3 years; Standard Care, standard post colorectal cancer care.17 Shaded numbers represent the 4 alternative screening strategies. Outcomes (not shown) are either colorectal cancer followed by death or no colorectal cancer followed by death. This Figure is reprinted with permission from Ramsey et al. Cost-effectiveness of microsatellite instability screening as a method for detecting hereditary nonpolyposis colorectal cancer. Ann Intern Med 2001;135:577–588.
Clinical management
Those who do not meet personal, family history, or tumor criteria for MSI or MMR mutation testing (Strategies 1 or 3), are MSI-low (Strategy 1 or 2), or have DNA tests that are definitively negative (all 4 strategies) receive standard postcolorectal cancer care and surveillance.17 Those found to have an HNPCC mutation are offered subtotal colectomy.17 Those who refuse subtotal colectomy (25% in the base case) receive lifelong surveillance with regular colonoscopy.17 Those with indeterminate DNA evaluations also receive lifelong surveillance with colonoscopy.18
Siblings and children of those who are identified as mutation carriers and those with indeterminate results are contacted and offered testing. Those who are found to have mutations are offered lifelong colonoscopy surveillance. Carriers who develop colorectal cancer are offered subtotal colectomy.
Data sources and assumptions
The appendix contains a complete list of assumptions and data sources for the model. We highlight several assumptions here. The distribution of ages and stages of sporadic colorectal cancer cases at colorectal cancer diagnosis is based on data from the National Cancer Institute’s Surveillance, Epidemiology and End Results (SEER) cancer registry.19 We assume a 2% prevalence of HNPCC mutation carriers among newly diagnosed colorectal cancer patients.3,20 The age and stage distribution at diagnosis for those who are HNPCC carriers is based on the data provided by the Creighton International HNPCC Registry (Patrice Watson, personal communication on April 11, 2001).
Appendix.
List of parameters informing the decision model of MSI testing for hereditary colorectal cancer
| Input Variables and Sources | ||||
|---|---|---|---|---|
| Range
|
||||
| Variable | Base | Low | High | References |
| Disease | ||||
| Proband | ||||
| Prevelance of HNPCC in colorectal cancer (CRC) patients | 0.02 | 0.005 | 0.06 | 1,2 |
| Probability of second dx for HNPCC+, no treatment | 0.95 | 0.7 | 1 | 3–5 |
| Mean time to second dx in absence of intervention | 5 | 4 | 6 | 3–5 |
| Life expectancy (LE) under standard care (SC) for HNPCC− | a | a | a | 6 |
| LE under SC for HNPCC+ | a | a | a | 1,6–9 |
| LE under increased surveillance (IS) for HNPCC− | a | a | a | 6 |
| LE under IS for HNPCC+ | a | a | a | 1,6–9 |
| LE post-colectomy (PC) HNPCC− | a | a | a | 6 |
| LE PC HNPCC+ | a | a | a | 1,6–9 |
| First Degree Relatives | ||||
| P (1st dx | HNPCC+) | 0.8 | 0.7 | 1 | 10 |
| Mean number of years until first dx for HNPCC+ | 2 | 1 | 4 | |
| Relative mortality risk for IS in HNPCC+ | 0.348 | 0.122 | 0.999 | 11 |
| Operation and Acceptance of Tests | ||||
| Proband | ||||
| P (agree to guidelines assessment) | 1 | 0.5 | 1 | b |
| Sensitivity of guidelines | 0.7 | 0.5 | 0.8 | 12 |
| Specificity of guidelines | 0.85 | 0.75 | 0.95 | 12 |
| P (accept microsatellite instability (MSI) test) | 1 | 0.75 | 1 | 13,14 |
| Sensitivity of MSI | 0.91 | 0.75 | 0.95 | 15–17 |
| Specificity of MSI | 0.93 | 0.6 | 0.98 | 15–17 |
| P (accept mismatch repair mutation (MMR) test) | 0.6 | 0.3 | 1 | 13,14 |
| P (MMR Test + or incomplete (Inc) | HNPCC+) | 0.925 | 0.87 | 0.95 | 18 |
| Proportion of (+ or Inc) that are inconclusive | 0.06 | 0.02 | 0.1 | 18 |
| P (MMR Test −or Inc | HNPCC−) | 0.997 | 0.98 | 1 | 18 |
| Proportion of (− or Inc) that are inconclusive | 0.002 | 0.005 | 0 | 18 |
| P (adhere to colectomy) | 0.75 | 0.5 | 1 | 19 |
| P (death due to colectomy) | 0 | 0 | 0 | 20–22 |
| First Degree Relatives | ||||
| P (sibling complies with counseling) | 0.6 | 0.5 | 0.85 | 13,14 |
| P (sibling complies with MMR test) | 0.6 | 0.5 | 0.85 | 13,14 |
| P (child complies with counseling) | 0.7 | 0.65 | 0.8 | 13,14 |
| P (child complies with MMR test) | 0.7 | 0.65 | 0.8 | 13,14 |
| P (first degree relative (FDR) HNPCC+ | Proband is HNPCC+) | 0.5 | 0.5 | 0.5 | |
| P (FDR HNPCC+ | Proband is HNPCC−) | 0.02 | 0.01 | 0.04 | |
| Contact Information, First Degree Relatives | ||||
| Mean number of siblings per MMR+ | 0.75 | 0.5 | 1 | 23,24 |
| P (locate sibling) | 0.65 | 0.5 | 0.75 | b |
| Mean number of children per MMR+ | 1.5 | 1 | 2 | 23,24 |
| P (locate child) | 0.75 | 0.65 | 0.9 | b |
| Mean age of child bearing | 30 | 25 | 35 | 23,24 |
| Costs | ||||
| Discount rate | 0.03 | 0 | 0.05 | 25 |
| Cost of guidelines assessment | 33 | 25 | 100 | 26 |
| Cost of MSI | 120 | 40 | 285 | c |
| Cost of MMR | 2,030 | 1,378 | 3,600 | c |
| Cost of MMR counseling | 365 | 300 | 1,000 | 27 |
| Cost of colectomy | 30,673 | 25,000 | 45,000 | 28 |
| Cost to locate sibling | 150 | 100 | 200 | b |
| Cost to locate child | 50 | 25 | 100 | b |
| Cost of counseling FDR | 100 | 75 | 125 | 27 |
| Cost of FDR MMR (one primer) | 78 | 65 | 100 | c |
| Per person yearly cost SC HNPCC− | 100 | 50 | 150 | b |
| Per person yearly cost SC HNPCC+ | 100 | 50 | 150 | b |
| Per person yearly cost IS HNPCC− | 325 | 250 | 400 | 26 |
| Per person yearly cost IS HNPCC+ | 400 | 325 | 450 | 26 |
| Per person yearly cost PC HNPCC+ | 33 | 25 | 100 | b |
| Per person yearly cost PC HNPCC+ | 33 | 25 | 100 | b |
| Mean attributable cost for 2nd dx: Stage 1 | 27,794 | 22,235 | 33,353 | 29,30 |
| Mean attributable cost for 2nd dx: Stage 2 | 28,872 | 23,098 | 34,647 | 29,30 |
| Mean attributable cost for 2nd dx: Stage 3 | 33,658 | 26,927 | 40,390 | 29,30 |
| Mean attributable cost for 2nd dx: Stage 4 | 49,352 | 39,482 | 59,223 | 29,30 |
| Mean attributable cost for 1st dx: Stage 1 | 25,516 | 20,413 | 30,620 | 29,30 |
| Mean attributable cost for 1st dx: Stage 2 | 28,166 | 22,533 | 33,800 | 29,30 |
| Mean attributable cost for 1st dx: Stage 3 | 31,907 | 25,526 | 38,289 | 29,30 |
| Mean attributable cost for 1st dx: Stage 4 | 45,393 | 36,314 | 54,471 | 29,30 |
Columns list base case values, high and low values for the sensitivity analysis (range), and references.
SC, standard care (standard postcolorectal cancer care31); IS, increased surveillance; CRC, colorectal cancer; MMR, mismatch repair mutation; MSI, microsatellite instability test; FDR, first degree relative (siblings, children); PC, postcolectomy; LE, life expectancy; Inc, inconclusive; Mean attributable cost, due to colorectal cancer; Increased surveillance, colonoscopy every 3 years following cancer treatment; Increased surveillance FDR, colonoscopy every 3 years.
Values vary based on the subject’s age at the start of the intervention.
Expert opinion.
Facility survey.
Aaltonen LA, Sankila R, Mecklin JP, Jarvinen H, Pukkala E, Peltomaki P, et al. A novel approach to estimate the proportion of hereditary nonpolyposis colorectal cancer of total colorectal cancer burden. Cancer Detect Prev. 1994;18:57–63.
Houlston RS, Collins A, Slack J, Morton NE. Dominant genes for colorectal cancer are not rare. Ann Hum Genet. 1992;56:99–103. doi: 10.1111/j.1469-1809.1992.tb01136.x.
Ponz de Leon M. Descriptive epidemiology of hereditary non-polyposis colorectal cancer. Tumori. 1996;82:102–106. doi: 10.1177/030089169608200202.
Lynch HT, Smyrk TC. Hereditary colorectal cancer. Semin Oncol. 1999;26:478–484.
Vasen HF, Mecklin JP, Watson P, Utsunomiya J, Bertario L, Lynch P, et al. Surveillance in hereditary nonpolyposis colorectal cancer: an international cooperative study of 165 families. The International Collaborative Group on HNPCC. Dis Colon Rectum. 1993;36:1–4. doi: 10.1007/BF02050292.
Surveillance, Epidemiology, and End Results (SEER) Program Public-Use CD-ROM (1973–1997). 2000, National Cancer Institute, DCCPS, Cancer Surveillance Research Program, Cancer Statistics Branch.
Myrhoj T, Bisgaard ML, Bernstein I, Svendsen LB, Sondergaard JO, Bulow S. Hereditary non-polyposis colorectal cancer: clinical features and survival: Results from the Danish HNPCC register. Scand J Gastroenterol. 1997;32:572–576. doi: 10.3109/00365529709025102.
Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201.
Percesepe A, Benatti P, Roncucci L, Sassatelli R, Fante R, Ganazzi D, et al. Survival analysis in families affected by hereditary non-polyposis colorectal cancer. Int J Cancer. 1997;71:373–376. doi: 10.1002/(sici)1097-0215(19970502)71:3<373::aid-ijc12>3.0.co;2-h.
Boland CR. Hereditary nonpolyposis colorectal cancer. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. McGraw-Hill; New York: 1998.
Jarvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomaki P, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5.
Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758.
Kolata G. Genetic testing falls short of public embrace. New York Times(Print) 1998:A16.
Lerman C, Hughes C, Trock BJ, Myers RE, Main D, Bonney A, et al. Genetic testing in families with hereditary nonpolyposis colon cancer. JAMA. 1999;281:1618–1622. doi: 10.1001/jama.281.17.1618.
Bubb VJ, Curtis LJ, Cunningham C, Dunlop MG, Carothers AD, Morris RG, et al. Microsatellite instability and the role of hMSH2 in sporadic colorectal cancer. Oncogene. 1996;12:2641–2649.
Lothe RA, Peltomaki P, Meling GI, Aaltonen LA, Nystrom-Lahti M, Pylkkanen L, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852.
Gonzalez-Garcia I, Moreno V, Navarro M, Marti-Rague J, Marcuello E, Benasco C, et al. Standardized approach for microsatellite instability detection in colorectal carcinomas. J Natl Cancer Inst. 2000;92:544–549. doi: 10.1093/jnci/92.7.544.
Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, et al. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA. 1999;282:247–253. doi: 10.1001/jama.282.3.247.
Burke W, Petersen G, Lynch P, Botkin J, Daly M, Garber J, et al. Recommendations for follow-up care of individuals with an inherited predisposition to cancer, I: Hereditary nonpolyposis colon cancer. Cancer Genetics Studies Consortium. JAMA. 1997;277:915–919.
Lieberman DA, Weiss DG, Bond JH, Ahnen DJ, Garewal H, Chejfec G. Use of colonoscopy to screen asymptomatic adults for colorectal cancer. Veterans Affairs Cooperative Study Group 380. N Engl J Med. 2000;343:162–168. doi: 10.1056/NEJM200007203430301.
Jentschura D, Raute M, Winter J, Henkel T, Kraus M, Manegold BC. Complications in endoscopy of the lower gastrointestinal tract: Therapy and prognosis. Surg Endosc. 1994;8:672–676. doi: 10.1007/BF00678564.
Macrae FA, Tan KG, Williams CB. Towards safer colonoscopy: A report on the complications of 5000 diagnostic or therapeutic colonoscopies. Gut. 1983;24:376–383. doi: 10.1136/gut.24.5.376.
Bachu A. Current population reports: Population characteristics: Fertility of American women: June 1995 (Update) Census Bureau; 1997.
Statistical Abstract of the United States, 1995–2000. 2001, US Census Bureau.
Gold MR, Siegal JE, Russel LB, Weinstein MC, editors. Cost-effectiveness in health and medicine. New York: Oxford University Press; 1996.
Reimbursement: physician fee schedule. 1999–2003, Centers for Medicare and Medicaid Services.
Bernhardt BA, Peshkin BN, Kemel Y. Billing and record-keeping for familial cancer risk counseling: A national survey. J Genet Couns. 1998;7:317–330. doi: 10.1023/A:1022067916528.
Reimbursement: Medicare reimbursement for subtotal colectomy. 2001, Centers for Medicare and Medicaid Services.
Potosky AL, Riley GF, Lubitz JD, Mentnech RM, Kessler LG. Potential for cancer related health services research using a linked Medicare-tumor registry database. Med Care. 1993;31:732–748.
Etzioni R, Urban N, Baker M. Estimating the costs attributable to a disease with application to ovarian cancer. J Clin Epidemiol. 1996;49:95–103. doi: 10.1016/0895-4356(96)89259-6.
Benson AB, III, Desch CE, Flynn PJ, Krause C, Loprinzi CL, Minsky BD, et al. 2000 update of American Society of Clinical Oncology colorectal cancer surveillance guidelines. J Clin Oncol. 2000;18:3586–3588. doi: 10.1200/JCO.2000.18.20.3586.
The sensitivity and specificity of MSI testing (91% and 93%, respectively) is based on the current recommendation that a minimum of 4 markers are used, with instability defined as alterations in at least 2 of the 4 markers.21 The sensitivity and specificity for hMSH2 and hMLH1 assays are 87% and 99.5%, respectively (also accounting for the likelihood of inconclusive tests).22
Survival for individuals with colorectal cancer not due to HNPCC is based on the SEER registry,19 adjusted to account for the higher colon cancer–specific survival observed for HNPCC carriers compared with sporadic colorectal cancers.23–25 Mortality risks include those associated with endoscopic surveillance, total colectomy, and partial resection.26–33
We incorporate evidence that probands and first degree relatives are likely to have imperfect adherence to recommendations regarding genetic testing.34–36 Based on expert opinion, it is assumed that 75% of HNPCC mutation carriers with localized disease who are informed of their gene status will elect to have a total colectomy. Those who refuse and unaffected carriers receive regular surveillance with colonoscopy. Regular colonoscopy surveillance of mutation carriers has been shown to confer a survival benefit.7
MSI assays and genetic testing and counseling costs were based on national surveys.37 Lifetime costs related to colorectal cancer treatment and the cost of prophylactic colectomy are based on published estimates using the SEER-Medicare database.38–40 Future costs are discounted at 3% per annum in the base case.
Sensitivity analysis
To address uncertainty in the data that was used to inform our model, we performed 2 types of uncertainty analysis: (1) one-way sensitivity analysis, where each parameter is tested over a range of values while holding all other parameters at their base-case values; (2) multiway sensitivity analysis, where parameters are sampled from distributions over their ranges and all parameters are changed simultaneously. Ranges for each parameter were based on 95% confidence intervals if available from the source data, or by expert opinion if no range data were available.
The multiway probabilistic sensitivity analysis was performed by creating probability density functions for each of the top 15 most sensitive parameters identified in the one-way analysis then randomly drawing observations from those distributions and rerunning the simulation, resampling the parameters each time until the result space did not change significantly with additional simulations (typically 1000–5000 simulations). This allowed 90% confidence intervals to be tabulated for each cost-effectivenessness ratio.
The decision model used for the analysis was implemented using Microsoft Excel (Microsoft Corporation, Redmond, Washington) and @RISK 4.0 Pro (Palisade Corporation, Newfield, New York).
RESULTS
Clinical endpoints
Base case: Bethesda guidelines
Compared to no screening, the Bethesda guidelines strategy (Strategy 1) would identify 23,417 individuals out of 148,300 persons with newly diagnosed colorectal cancer who would be eligible for MSI tumor testing (Table 2). After MSI analysis, 2,810 probands would be eligible for DNA testing. Based on DNA results, 667 probands would be offered prophylactic colectomies. Based on the estimated benefits of colectomy and increased surveillance (for those refusing colectomy) in mutation carriers, implementing the Bethesda guidelines would add 833 years of life (undiscounted) to those with newly diagnosed colorectal cancers.
Table 2.
Clinical outcomes for alternative HNPCC screening strategies, starting with all persons with newly diagnosed colorectal cancer in the United Statesa
| Strategy 1
|
Strategy 2
|
Strategy 3
|
Strategy 4
|
|
|---|---|---|---|---|
| Outcome | Bethesda guidelines | MSI all | DNA analysis for Bethesda clinical (+) | DNA analysis for all |
| Number of colorectal cancer cases per year, United States | 148,300 | 148,300 | 148,300 | 148,300 |
| Number of tumors tested for microsatellite instability (MSI) | 23,417 | 148,300 | 0 | 0 |
| Number of false positive MSI tests | 1,542 | 10,277 | NA | NA |
| Number of false negative MSI tests | 125 | 133 | NA | NA |
| Number offered mismatch repair (MMR) mutation counseling and analysis | ||||
| Probands | 2,810 | 11,627 | 23,417 | 148,300 |
| Probands + relatives | 3,562 | 12,521 | 24,394 | 149,924 |
| Number of false positive MMR assays | ||||
| Probands | 5 | 31 | 66 | 440 |
| Probands + relatives | 5 | 32 | 67 | 442 |
| Number of false negative MMR assays | ||||
| Probands | 57 | 61 | 63 | 67 |
| Probands + relatives | 75 | 80 | 82 | 88 |
| Number of HNPCC carriers identified | ||||
| Probands | 662 | 704 | 727 | 774 |
| Probands + relatives | 869 | 925 | 956 | 1,023 |
| Years of life gained (no discount) | ||||
| Probands | 833 | 886 | 915 | 973 |
| Probands + relatives | 7,615 | 8,111 | 8,390 | 9,059 |
Strategy 1, Bethesda guidelines treatment algorithm; Strategy 2, Microsatellite instability testing of tumors of all persons with newly diagnosed colorectal cancer; Strategy 3, Mismatch repair gene mutation testing of persons meeting the clinical criteria for the Bethesda guidelines (no microsatellite instability testing); Strategy 4, Mismatch repair gene testing of all persons with newly diagnosed colorectal cancer.
When siblings and children are considered along with probands, this increases the number of mutation carriers identified and greatly improves the clinical impact of screening. A total of 752 surviving siblings and children of the probands would be eligible for genetic counseling and testing. Accounting for the likelihood of contacting these relatives and their expected rate of testing uptake, 207 siblings and children would be identified as mutation carriers. Regular surveillance of these individuals, followed by subtotal colectomy for those where malignancies are detected would add 6782 nondiscounted life years to this cohort.
Alternatives to the Bethesda guidelines
Table 2 lists the clinical outcomes for each strategy. The Bethesda guidelines approach (Strategy 1) identifies the fewest number of mutation carriers, whereas DNA testing of all persons with new colorectal cancer (Strategy 4) identifies the greatest number of carriers. Strategy 1 results in 85% fewer false positive MSI tests and 7% fewer false negative MSI tests than an alternative approach of testing all persons with new colorectal cancers for MSI (Strategy 2). Strategy 1 also results in far fewer false positive and negative DNA assays than the alternative strategies.
DNA analysis of all new colorectal cancer cases (Strategy 4) identifies the greatest number of mutation carriers and yields the greatest number of life years gained compared to no testing. The Bethesda guidelines (Strategy 1) identify the fewest number of HNPCC mutation carriers and yields the least number of life years gained. MSI testing of all new cancers (Strategy 2) and DNA analysis for MMR mutations of those with personal and family history meeting the Bethesda guidelines (Strategy 3) have similar outcomes and are between Strategies 1 and 4 in terms of mutation carriers identified and years of life gained.
Economic Endpoints
The average cost per carrier detected using the Bethesda guidelines testing strategy was $15,787 (Table 3). Expanding the proposed strategy to include costs and benefits for siblings and children greatly improves the cost-effectiveness of the program. Overall, screening siblings and children is cost saving, because the savings from intensive screening exceeds the costs of testing. The cost-effectiveness of Strategy 1 is $73,711 per life year gained when considering only costs and benefits for probands and $11,865 [90% CI: $8,005–$80,226] per life year gained when considering probands, siblings, and children. The cost-effectiveness of all alternative strategies was less favorable than Strategy 1 for identifying mutation carriers.
Table 3.
Economic outcomes for alternative HNPCC screening strategies
| Strategy 1
|
Strategy 2
|
Strategy 3
|
Strategy 4
|
|
|---|---|---|---|---|
| Economic Outcome | Bethesda guidelines | MSI all | DNA analysis for Bethesda clinical (+) | DNA analysis for all |
| Cost per carrier detected: probands | $20,313 | $ 57,027 | $ 63,980 | $ 336,745 |
| Cost per carrier detected: probands + relatives | $15,787 | $ 43,737 | $ 49,020 | $ 255,160 |
| Added cost per life year gained in 2002 dollars (3% discount, costs and benefits): | ||||
| Probands | $73,711 | $213,290 | $296,793 | $1,625,687 |
| Probands +relatives (90% CI) | $11,865 ($8,005–$80,226) | $ 35,617 ($15,091–$180,056) | $ 49,702 ($19,100–$252,151) | $ 267,548 ($68,328–$637,007) |
Table 4 compares the incremental cost-effectiveness of each testing strategy to the alternatives, to no screening, and to the other approaches, with benefits for probands and relatives as the measure of effectiveness. Compared to the Bethesda guidelines, the incremental cost-effectiveness of the other strategies was unfavorable, with ratios ranging from $394,067 per life year gained to $2.6 million per life year gained. In other words, when compared to the Bethesda guidelines approach, the gain in life years for the population (proband+relatives) was small relative to the incremental cost for the more technology-intensive programs.
Table 4.
Incremental cost-effectivenessness ratios of each screening strategy relative to the other strategies (cost per year of life saved), with years of life gained for probands and relatives (2002 dollars)a
| Strategy B
|
|||||
|---|---|---|---|---|---|
| Strategy 1
|
Strategy 2
|
Strategy 3
|
Strategy 4
|
||
| Strategy A | None | Bethesda guidelines | MSI all | DNA analysis for Bethesda clinical (+) | DNA analysis for all |
| None | 11,865 | 35,617 | 49,702 | 267,548 | |
| Strategy 1 | 0 | 394,067 | 411,339 | 1,485,640 | |
| Strategy 2 | 0 | 441,172 | 1,989,197 | ||
| Strategy 3 | 0 | 2,553,345 | |||
Sensitivity and uncertainty analysis
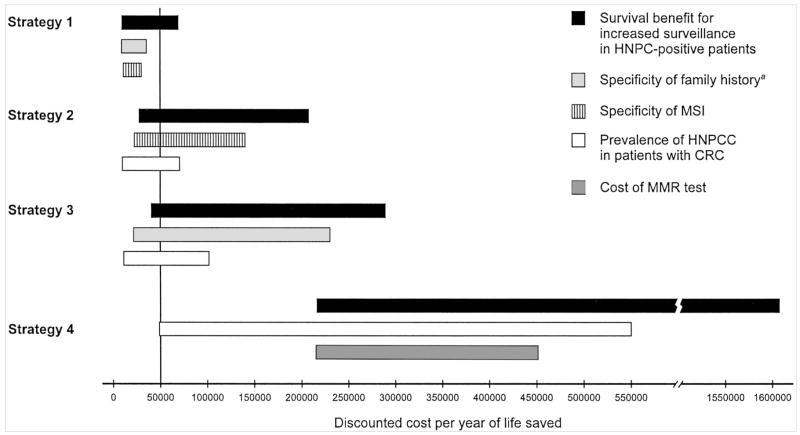
All programs were sensitive to the survival benefits from aggressive surveillance in mutation carriers without cancer and the prevalence of HNPCC in the population (Fig. 2). Although the ordering of the two most influential parameters does not change, the relative influence of each parameter varies substantially from strategy to strategy. For example, in a worst-case scenario for the prevalence of HNPCC among individuals with newly diagnosed colorectal cancer (0.5% of new cases), the cost-effectiveness of the Bethesda guidelines is $21,478 per life year gained, whereas the cost-effectivenessness of DNA analysis for MMR mutations of all persons with new cancers is $552,880 per life year gained. The influence of the cost of the DNA analysis on the cost-effectivenessness of each strategy is much lower for Strategy 1 than it is for the other strategies. This is due to the fact that far fewer individuals are offered DNA analysis for mutations under Strategy 1 compared to the other alternatives.
Fig. 2.
One-way sensitivity analysis demonstrating the most influential parameters for each screening strategy. For a description of each strategy, see Figure 1 and the text. Vertical line is set at $50,000 per life year saved, a commonly used threshold to judge whether medical interventions are considered cost-effectiveness.55 aIncludes personal and family history as outlined in the Bethesda guidelines (Table 1).
Figure 3 displays confidence ellipses for each strategy from the multivariate probabilistic sensitivity analysis. The expected outcomes for each strategy for the multivariate sensitivity analysis differ from the base-case results because the asymmetrical distributions of individual parameters lead to a slight shift in expected value between the confidence intervals and the ellipses. The ellipses overlap for Strategies 1 to 3, indicating that one cannot say with certainty that one strategy is clearly superior to another. Strategy 4 is clearly less cost effective than the other strategies.
Fig. 3.
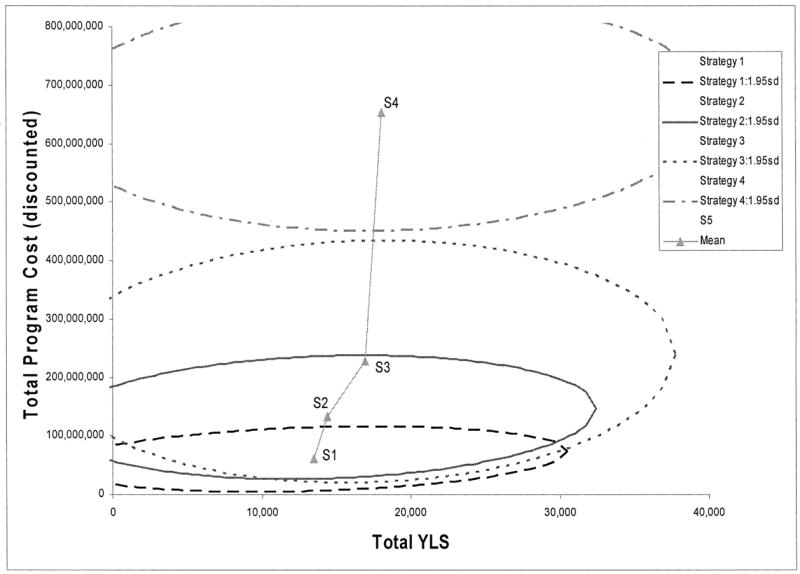
Cost-effectiveness confidence ellipses for alternative HNPCC screening strategies. Ellipses represent the degree of uncertainty across a cost-effectivenessness plane (95% confidence intervals). Plane displays the difference in costs (y-axis) and the difference in benefits (x-axis) for each strategy compared to standard care (no screening). Points in the middle of each ellipse represent the expected outcome for each strategy. Lines connecting two strategies represent the incremental cost-effectivenessness of one strategy versus another. Slopes closer to horizontal are more cost effective. For a description of each strategy, see Figure 1 and the text.
DISCUSSION
In today’s cost-conscious environment, it is important to consider both clinical and economic consequences of new screening programs. Because of their expense and social implications, this may be particularly true for screening programs that involve genetic testing. Screening for mutations linked to cancer susceptibility is not yet part of routine clinical practice. Because population-wide genetics-based screening is not yet feasible and may not be desirable due to privacy concerns,41–43 experts in screening policy have focused on those with colorectal cancer as the initial point for considering genetic testing. Previous analyses suggest that this strategy is preferable when the population prevalence of the mutation is likely to be low.10,44 The economic efficiency of the Bethesda strategy will still be within what many consider reasonable at lower prevalence rates of HNPCC. This is not true for the other strategies, particularly those that bypass MSI analysis. Thus, from an economic standpoint, this article supports the role of MSI testing in identifying persons with HNPCC.45
The sensitivity and specificity of MSI testing is variable, because the test exhibits a Receiver Operating Curve-type relationship, where sensitivity may be exchanged for specificity depending on the combination of microsatellite markers that are used to identify replication errors in the tumor DNA. Although the Bethesda criteria allow up to 10 markers if needed, we chose a 4-marker MSI test based on a balance of sensitivity and cost.5 Recent studies suggest that immunohistochemical assay may be superior to MSI testing for identifying hMLH1 and hMSH2 protein products in colorectal tumors.46 Our sensitivity analysis suggests that increasing the specificity of phenotype testing at a stable cost could substantially improve the cost-effectiveness of the Bethesda guidelines. Some laboratories now test for the MSH6 mutation in addition to MLH1 and MSH2, although we do not include the added costs and potential change in specificity in this analysis.
Other clinical risk-assessment algorithms for identifying persons with HNPCC have been proposed, including the Amsterdam criteria, which uses more restrictive family history criteria than does the Bethesda guidelines.8,9,47,48 Comparing other strategies, including the Amsterdam criteria, Reyes and colleagues found that a mixed strategy including MMR mutation evaluation of those who meet the Amsterdam criteria and those who meet less stringent modified criteria and show high MSI was the most cost effective option.49 From an economic perspective, the specificity of the initial clinical screening algorithm is critical, as it will reduce the number of false positive screenings, and thus the number of downstream follow-up tests for those who are HNPCC negative. To incorporate new knowledge about HNPCC, testing strategies, and technological advances, an update of the Bethesda criteria is underway.50
Efforts to promote genetics education for primary care providers emphasize the use of family history as a triage tool in routine practice, to identify patients who might be candidates for early screening and/or genetic testing.51 Our analysis does not address this approach to HNPCC detection. Some may question the choice to offer genetic counseling before mutation testing but not before MSI testing. We believe this approach accurately reflects current practice; however, we recognize a potential argument for some form of genetic counseling before MSI testing to alert the patient that the testing pathway leads to the potential outcome of a genetic diagnosis.
The cost-effectiveness of each strategy is highly dependent on our health system’s ability to identify and test unaffected relatives of those found to be mutation carriers. There is less to gain from an HNPCC diagnosis for the colorectal cancer patient than for a relative who is not yet affected with cancer. The level of physicians’ responsibility for ensuring that relatives of their patients know about their risk is not yet resolved. A New Jersey court case suggested that physicians might have an obligation to contact relatives directly to inform them of genetic risk; by contrast, a Florida ruling stated that a physician’s duty was discharged by informing the patient of a genetic risk to family members.52,53 This issue is notably absent from the American Gastroenterological Association’s recent position statement on hereditary colorectal cancer and genetic testing.54 More research is needed to determine the most effective and efficient method for disseminating risk information to family members after a diagnosis of HNPCC is made, and the feasibility of using family history as a screening tool in primary care practice.
Clinicians may be troubled by the fact that of the four approaches we consider in this study, the Bethesda strategy “misses” the greatest number of mutation carriers. This issue is at the crux of the controversy between the economic and clinical perspective when evaluating diagnostic tests. Table 4 highlights the implications of choosing a strategy with higher sensitivity, in terms of the cost of adding an additional year of life for those being screened. This issue is worthy of debate. Our purpose has been to make explicit the clinical and economic consequences of alternative strategies.
We note several important limitations of the analysis. Some of the input variables are based on expert opinion. Our uncertainty analysis suggests that changing the values of these variables will not influence the rank of cost-effectivenessness outcomes of each strategy, although the magnitude of difference may change. For the multivariate analysis, the large ranges and, more importantly, assumed independence of the individual parameters lead to rather large predictive regions in the cost/effectiveness space. If it were possible to specify the correlation between parameters, these regions (and thus the overall uncertainty) could be reduced. This analysis does not consider indirect costs associated with screening, such as the cost of job discrimination as a result of screening. Although important, including these costs are not likely to influence the ranking of the strategies.
Strategies that start with MSI testing of all persons regardless of history and those that bypass MSI testing altogether are also reasonably cost-effectiveness and not beyond a range of uncertainty at which options would be clearly inferior to the Bethesda guidelines strategy. The benefit of intensive surveillance and the population prevalence of HNPCC are important issues influencing the cost-effectiveness of all strategies. If the true prevalence of HNPCC mutations and the benefits of surveillance are lower than current estimates, then this will further favor the Bethesda guidelines from an economic perspective, as the cost-effectiveness of this strategy is less influenced by changes in the estimates than are the other approaches.
Acknowledgments
Financial support for this study was provided by Dr. Ramsey’s Howard Temin Career Development Award from the National Cancer Institute (K01 CA 76189) and by a grant from the National Human Genome Research (R01-HG02263).
References
- 1.Lynch HT, Smyrk TC. Hereditary colorectal cancer. Semin Oncol. 1999;26:478–484. [PubMed] [Google Scholar]
- 2.Ponz de Leon M. Descriptive epidemiology of hereditary non-polyposis colorectal cancer. Tumori. 1996;82:102–106. doi: 10.1177/030089169608200202. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen LA, Sankila R, Mecklin JP, Jarvinen H, Pukkala E, Peltomaki P, et al. A novel approach to estimate the proportion of hereditary nonpolyposis colorectal cancer of total colorectal cancer burden. Cancer Detect Prev. 1994;18:57–63. [PubMed] [Google Scholar]
- 4.Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, et al. A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: Meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 6.Ponz de Leon M. Prevalence of hereditary nonpolyposis colorectal carcinoma (HNPCC) Ann Med. 1994;26:209–214. doi: 10.3109/07853899409147892. [DOI] [PubMed] [Google Scholar]
- 7.Jarvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomaki P, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 8.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 9.Nakahara M, Yokozaki H, Yasui W, Dohi K, Tahara E. Identification of concurrent germ-line mutations in hMSH2 and/or hMLH1 in Japanese hereditary nonpolyposis colorectal cancer kindreds. Cancer Epidemiol Biomarkers Prev. 1997;6:1057–1064. [PubMed] [Google Scholar]
- 10.Ramsey SD, Clarke L, Etzioni R, Higashi M, Berry K, Urban N. Cost-effectiveness of microsatellite instability screening as a method for detecting hereditary nonpolyposis colorectal cancer. Ann Intern Med. 2001;135:577–588. doi: 10.7326/0003-4819-135-8_part_1-200110160-00008. [DOI] [PubMed] [Google Scholar]
- 11.Love RR, Evans AM, Josten DM. The accuracy of patient reports of a family history of cancer. J Chronic Dis. 1985;38:289–293. doi: 10.1016/0021-9681(85)90074-8. [DOI] [PubMed] [Google Scholar]
- 12.Aitken J, Bain C, Ward M, Siskind V, MacLennan R. How accurate is self-reported family history of colorectal cancer? Am J Epidemiol. 1995;141:863–871. doi: 10.1093/oxfordjournals.aje.a117522. [DOI] [PubMed] [Google Scholar]
- 13.Kerber RA, Slattery ML. Comparison of self-reported and database-linked family history of cancer data in a case-control study. Am J Epidemiol. 1997;146:244–248. doi: 10.1093/oxfordjournals.aje.a009259. [DOI] [PubMed] [Google Scholar]
- 14.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 15.Marx J. Gene defect identified in common hereditary colon cancer. Science. 1993;262:1645. doi: 10.1126/science.8259507. [DOI] [PubMed] [Google Scholar]
- 16.Danks DM. Colon cancer screening. Science. 1994;264:13–14. doi: 10.1126/science.8140413. [DOI] [PubMed] [Google Scholar]
- 17.Benson AB, III, Desch CE, Flynn PJ, Krause C, Loprinzi CL, Minsky BD, et al. Update of American Society of Clinical Oncology colorectal cancer surveillance guidelines. J Clin Oncol. 2000;18:3586–3588. doi: 10.1200/JCO.2000.18.20.3586. [DOI] [PubMed] [Google Scholar]
- 18.Burke W, Petersen G, Lynch P, Botkin J, Daly M, Garber J, et al. Recommendations for follow-up care of individuals with an inherited predisposition to cancer, I: Hereditary nonpolyposis colon cancer. Cancer Genetics Studies Consortium. JAMA. 1997;277:915–919. [PubMed] [Google Scholar]
- 19.Surveillance, Epidemiology, and End Results (SEER) Program Public-Use CD-ROM (1973–1997). National Cancer Institute, DCCPS, Cancer Surveillance Research Program, Cancer Statistics Branch, 2000.
- 20.Houlston RS, Collins A, Slack J, Morton NE. Dominant genes for colorectal cancer are not rare. Ann Hum Genet. 1992;56:99–103. doi: 10.1111/j.1469-1809.1992.tb01136.x. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Garcia I, Moreno V, Navarro M, Marti-Rague J, Marcuello E, Benasco C, et al. Standardized approach for microsatellite instability detection in colorectal carcinomas. J Natl Cancer Inst. 2000;92:544–549. doi: 10.1093/jnci/92.7.544. [DOI] [PubMed] [Google Scholar]
- 22.Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, et al. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. JAMA. 1999;282:247–253. doi: 10.1001/jama.282.3.247. [DOI] [PubMed] [Google Scholar]
- 23.Myrhoj T, Bisgaard ML, Bernstein I, Svendsen LB, Sondergaard JO, Bulow S. Hereditary non-polyposis colorectal cancer: clinical features and survival: Results from the Danish HNPCC register. Scand J Gastroenterol. 1997;32:572–576. doi: 10.3109/00365529709025102. [DOI] [PubMed] [Google Scholar]
- 24.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 25.Percesepe A, Benatti P, Roncucci L, Sassatelli R, Fante R, Ganazzi D, et al. Survival analysis in families affected by hereditary non-polyposis colorectal cancer. Int J Cancer. 1997;71:373–376. doi: 10.1002/(sici)1097-0215(19970502)71:3<373::aid-ijc12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 26.Lieberman DA, Weiss DG, Bond JH, Ahnen DJ, Garewal H, Chejfec G. Use of colonoscopy to screen asymptomatic adults for colorectal cancer: Veterans Affairs Cooperative Study Group 380. N Engl J Med. 2000;343:162–168. doi: 10.1056/NEJM200007203430301. [DOI] [PubMed] [Google Scholar]
- 27.Jentschura D, Raute M, Winter J, Henkel T, Kraus M, Manegold BC. Complications in endoscopy of the lower gastrointestinal tract: Therapy and prognosis. Surg Endosc. 1994;8:672–676. doi: 10.1007/BF00678564. [DOI] [PubMed] [Google Scholar]
- 28.Lo AY, Beaton HL. Selective management of colonoscopic perforations. J Am Coll Surg. 1994;179:333–337. [PubMed] [Google Scholar]
- 29.Macrae FA, Tan KG, Williams CB. Towards safer colonoscopy: a report on the complications of 5000 diagnostic or therapeutic colonoscopies. Gut. 1983;24:376–383. doi: 10.1136/gut.24.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouffet F, Hay JM, Vacher B, Fingerhut A, Elhadad A, Flamant Y, et al. Curative resection for left colonic carcinoma: hemicolectomy vs. segmental colectomy: A prospective, controlled, multicenter trial. French Association for Surgical Research. Dis Colon Rectum. 1994;37:651–659. doi: 10.1007/BF02054407. [DOI] [PubMed] [Google Scholar]
- 31.Stelzner M, Fonkalsrud EW. The endorectal ileal pullthrough procedure in patients with ulcerative colitis and familial polyposis with carcinoma. Surg Gynecol Obstet. 1989;169:187–194. [PubMed] [Google Scholar]
- 32.Coran AG. A personal experience with 100 consecutive total colectomies and straight ileoanal endorectal pull-throughs for benign disease of the colon and rectum in children and adults. Ann Surg. 1990;212:242–247. doi: 10.1097/00000658-199009000-00002. discussion 247–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen T, Eliasen K, Henriksen E. A prospective study of mortality associated with anaesthesia and surgery: risk indicators of mortality in hospital. Acta Anaesthesiol Scand. 1990;34:176–182. doi: 10.1111/j.1399-6576.1990.tb03066.x. [DOI] [PubMed] [Google Scholar]
- 34.Lerman C, Hughes C, Trock BJ, Myers RE, Main D, Bonney A, et al. Genetic testing in families with hereditary nonpolyposis colon cancer. JAMA. 1999;281:1618–1622. doi: 10.1001/jama.281.17.1618. [DOI] [PubMed] [Google Scholar]
- 35.Croyle RT, Lerman C. Interest in genetic testing for colon cancer susceptibility: Cognitive and emotional correlates. Prev Med. 1993;22:284–292. doi: 10.1006/pmed.1993.1023. [DOI] [PubMed] [Google Scholar]
- 36.Kinney AY, Choi YA, DeVellis B, Kobetz E, Millikan RC, Sandler RS. Interest in genetic testing among first-degree relatives of colorectal cancer patients. Am J Prev Med. 2000;18:249–252. doi: 10.1016/s0749-3797(99)00162-2. [DOI] [PubMed] [Google Scholar]
- 37.Bernhardt BA, Peshkin BN, Kemel Y. Billing and record-keeping for familial cancer risk counseling: A national survey. J Genet Couns. 1998;7:317–330. doi: 10.1023/A:1022067916528. [DOI] [PubMed] [Google Scholar]
- 38.Ramsey SD, Berry K, Etzioni R. Lifetime cancer-attributable cost of care for long term survivors of colorectal cancer. Am J Gastroenterol. 2002;97:440–445. doi: 10.1111/j.1572-0241.2002.05483.x. [DOI] [PubMed] [Google Scholar]
- 39.Etzioni R, Ramsey SD, Berry K, Brown M. The impact of including future medical care costs when estimating the costs attributable to a disease: A colorectal cancer case study. Health Econ. 2001;10:245–256. doi: 10.1002/hec.580. [DOI] [PubMed] [Google Scholar]
- 40.Potosky AL, Riley GF, Lubitz JD, Mentnech RM, Kessler LG. Potential for cancer related health services research using a linked Medicare-tumor registry database. Med Care. 1993;31:732–748. [PubMed] [Google Scholar]
- 41.Moore AD. Owning genetic information and gene enhancement techniques: Why privacy and property rights may undermine social control of the human genome. Bioethics. 2000;14:97–119. doi: 10.1111/1467-8519.00184. [DOI] [PubMed] [Google Scholar]
- 42.Anderlik MR, Rothstein MA. Privacy and confidentiality of genetic information: What rules for the new science? Annu Rev Genomics Hum Genet. 2001;2:401–433. doi: 10.1146/annurev.genom.2.1.401. [DOI] [PubMed] [Google Scholar]
- 43.ASHG statement. Professional disclosure of familial genetic information: The American Society of Hum Genet Social Issues Subcommittee on Familial Disclosure. Am J Hum Genet. 1998;62:474–483. [PMC free article] [PubMed] [Google Scholar]
- 44.Brown ML, Kessler LG. The use of gene tests to detect hereditary predisposition to cancer: Economic considerations. J Natl Cancer Inst. 1995;87:1131–1136. doi: 10.1093/jnci/87.15.1131. [DOI] [PubMed] [Google Scholar]
- 45.Terdiman JP. HNPCC: An uncommon but important diagnosis. Gastroenterology. 2001;121:1005–1008. doi: 10.1053/gast.2001.28634. [DOI] [PubMed] [Google Scholar]
- 46.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 47.Bapat BV, Madlensky L, Temple LK, Hiruki T, Redston M, Baron DL, et al. Family history characteristics, tumor microsatellite instability and germline MSH2 and MLH1 mutations in hereditary colorectal cancer. Hum Genet. 1999;104:167–176. doi: 10.1007/s004390050931. [DOI] [PubMed] [Google Scholar]
- 48.Fornasarig M, Viel A, Bidoli E, Campagnutta E, Minisini AM, Cannizzaro R, et al. Amsterdam criteria II, and endometrial cancer index cases for an accurate selection of HNPCC families. Tumori. 2002;88:18–20. [PubMed] [Google Scholar]
- 49.Reyes CM, Allen BA, Terdiman JP, Wilson LS. Comparison of selection strategies for genetic testing of patients with hereditary nonpolyposis colorectal carcinoma: Effectiveness and cost-effectivenessness. Cancer. 2002;95:1848–1856. doi: 10.1002/cncr.10910. [DOI] [PubMed] [Google Scholar]
- 50.Diagnostic Criteria for Hereditary Non-Polyposis Colorectal Cancer, and Microsatellite Instability. National Cancer Institute, Division of Cancer Prevention, 2002.
- 51.Burke W, Emery J. Genetics education for primary-care providers. Nat Rev Genet. 2002;3:561–566. doi: 10.1038/nrg845. [DOI] [PubMed] [Google Scholar]
- 52.Safer v. Estate of Pack. Atl Report. 1996;677:1188–1193. [PubMed] [Google Scholar]
- 53.Pate v. Threlkel. Wests South Report. 1995;661:278–282. [PubMed] [Google Scholar]
- 54.American Gastroenterological Association medical position statement: hereditary colorectal cancer and genetic testing. Gastroenterology. 2001;121:195–197. doi: 10.1053/gast.2001.25580. [DOI] [PubMed] [Google Scholar]
- 55.Winkelmayer WC, Weinstein MC, Mittleman MA, Glynn RJ, Pliskin JS. Health economic evaluations: The special case of end-stage renal disease treatment. Med Decis Making. 2002;22:417–430. doi: 10.1177/027298902236927. [DOI] [PubMed] [Google Scholar]