Summary
Menin displays the unique ability to either promote oncogenic function in the hematopoietic lineage or suppress tumorigenesis in the endocrine lineage, however its molecular mechanism of action has not been defined. We demonstrate here that these discordant functions are unified by menin's ability to serve as a molecular adaptor that physically links the MLL histone methyltransferase with LEDGF, a chromatin-associated protein previously implicated in leukemia, auto-immunity and HIV-1 pathogenesis. LEDGF is required for both MLL-dependent transcription and leukemic transformation. Conversely, a subset of menin mutations in MEN1 patients abrogates interaction with LEDGF while preserving MLL interaction, but nevertheless compromises MLL/menin-dependent functions. Thus, LEDGF critically associates with MLL and menin at the nexus of transcriptional pathways that are recurrently targeted in diverse diseases.
Keywords: LEDGF, MLL, menin, leukemia, MEN1
Significance
Menin is a tumor suppressor whose loss of function causes the human cancer syndrome known as multiple endocrine neoplasia type 1. Conversely, menin serves as an essential oncogenic cofactor for MLL oncoproteins in leukemic transformation. These discordant functions are attributed in part to tissue-specific differences in critical MLL target genes, however the molecular role of menin in these contrasting oncogenic settings remains undefined. We demonstrate here that menin serves as a molecular adaptor that tethers the MLL histone methlytransferase and its oncogenic counterparts to LEDGF, a transcriptional co-activator previously implicated in cancer, auto-immunity and HIV proviral integration. A requirement for LEDGF in MLL/menin transcriptional and oncogenic actions places it at the center of a key epigenetic pathway in cancer pathogenesis.
Introduction
Cancer results from genetic and epigenetic perturbations that cause the unbalanced actions of oncoproteins and tumor suppressors. The menin tumor suppressor is implicated in cancer pathogenesis and transcriptional regulation as an integral component of the MLL (Mixed Lineage Leukemia) histone methyltransferase (HMT) complex, which promotes specific trimethylation of lysine 4 on histone H3, an epigenetic mark associated with transcriptionally active chromatin (Hughes et al., 2004; Milne et al., 2002; Nakamura et al., 2002; Yokoyama et al., 2004). Menin is a product of the MEN1 gene whose loss of function causes the human cancer syndrome known as multiple endocrine neoplasia type 1 (MEN1) (Chandrasekharappa et al., 1997). It is also implicated in the dynamic regulation of pancreatic beta cell proliferation in response to normal physiologic demands during pregnancy whose failure may promote gestational diabetes (Karnik et al., 2007). These normal and pathologic roles in endocrine cells reflect specific requirements for the MLL/menin HMT complex to maintain expression of the CDKN1B and CDKN2C genes, which encode cyclin-dependent kinase inhibitors (CDKIs) p27KIP1 and p18Ink4c, respectively (Karnik et al., 2005; Milne et al. 2005). Their compromised expression following mutation or deletion of MEN1 leads to hyper-proliferation of endocrine lineage cells (Bertolino et al., 2003; Franklin et al., 1998; Crabtree et al., 2001). Many mutations of menin in endocrine tumors abrogate its ability to associate with the MLL HMT complex (Hughes et al., 2004), however the specific molecular function of menin is currently unknown.
Transcriptional regulation of HOX genes is also exquisitely dependent on MLL, which is required to establish the embryonic body plan during development (Yu et al., 1995) and to promote progenitor expansion and stem cell self-renewal in the hematopoietic lineage (Jude et al., 2007; McMahon et al., 2007). In a subset of acute leukemias, chromosomal aberrations generate chimeric MLL oncoproteins that cause constitutive expression of HOX genes, a key feature of MLL leukemia pathogenesis (Daser and Rabbitts, 2004; Hess, 2004). In contrast to its tumor suppressor role in MEN1 tumorigenesis, menin serves as an essential cofactor for MLL oncoproteins to sustain HOX gene mis-expression and maintain leukemic transformation (Caslini et al., 2007; Chen et al., 2006; Yokoyama et al., 2005). Thus, menin has the unusual ability to either promote oncogenic function in the hematopoietic lineage or suppress tumorigenesis in the endocrine lineage. These discordant functions are attributed to tissue-specific differences in critical target genes that encode oncogenic versus tumor suppressor proteins in hematopoietic versus endocrine cells, respectively. However a unifying molecular role for menin underlying transcriptional regulation by normal and oncogenic MLL proteins has not been defined.
Here we demonstrate that menin tethers MLL with the p75 isoform of LEDGF (Lens Epithelium-Derived Growth Factor; also called DFS70/p75/PSIP1), a chromatin-associated protein that is also targeted in various diseases including cancer (Ahuja et al., 2000; Daugaard et al., 2007; Huang et al., 2007), auto-immunity (Ganapathy and Casiano, 2004), and AIDS (Ciuffi and Bushman, 2006), thereby providing a crucial molecular link with chromatin for an epigenetic complex at the center of multiple pathologic processes.
Results
Wild type and oncogenic MLL/menin protein complexes specifically associate with LEDGF
Menin is essential for MLL-dependent transcriptional regulation and leukemic transformation (Caslini et al., 2007; Chen et al., 2006; Yokoyama et al., 2005), however its molecular mechanism of action has not been defined. We hypothesized that menin might further recruit an unknown factor to the MLL/menin complex. Since stably associated factors of MLL have been extensively characterized (Dou et al., 2005; Yokoyama et al., 2004), we inferred that potential novel factors might associate more weakly. Thus, the MLL-ENL/menin complex was biochemically purified by one-step affinity purification from cells transiently expressing high levels of the respective proteins to minimize the loss of weakly associated proteins during the purification procedure. In the purified materials, we identified a 75 kDa nuclear protein termed LEDGF (Figure 1A) previously implicated in transcriptional co-activation (Ge et al., 1998) and lentiviral integration (Ciuffi and Bushman, 2006).
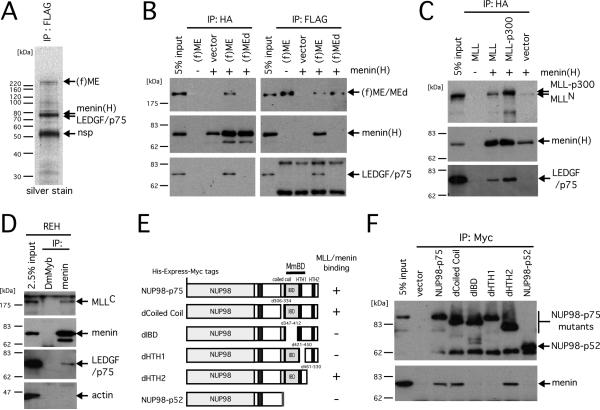
Figure 1. LEDGF and NUP98-LEDGF associate with the MLL/menin histone methyltransferase complex.
A. Silver stained image of an SDS-PAGE analysis shows presence of LEDGF in the purified MLL-ENL/menin immunoprecipitate. FLAG-tagged MLL-ENL [(f)ME] and HA-tagged menin [menin(H)] were transiently expressed in large scale culture of 293T cells. A nuclear extract was prepared from the cells and subjected to immunopurification using anti-FLAG antibody. Protein bands from SDS-PAGE analysis were analyzed by mass spectrometry and identified as indicated by arrows. The band at 52 kDa is a non-specific (nsp) product that was also observed in the control IP of a nuclear extract from non-transduced cells (not shown). Protein standards are shown on the left.
B. Reciprocal immunoprecipitations of the MLL-ENL/menin complex. (f)ME, or a mutant [(f)MEd] lacking the high affinity menin binding motif, were transiently expressed in 293T cells with (+) or without (-) HA-tagged menin. Nuclear extracts from the transfected cells were subjected to IP with anti-FLAG or anti-HA antibodies followed by immunoblotting with anti-FLAG, anti-HA, and anti-LEDGF (p75) antibodies.
C. MLL or MLL-p300 was transiently expressed with (+) or without (-) HA-tagged menin in 293T cells and subjected to IP with anti-HA antibody, followed by immunoblotting with anti-MLLN, anti-HA, and anti-LEDGF antibodies.
D. To detect endogenous MLL/menin/LEDGF association, nuclear extract of REH cells was subjected to IP with anti-menin antibody followed by immunoblotting with anti-MLLC, anti-menin, anti-LEDGF, and anti-ACTIN antibodies. Anti-Drosophila Myb (DmMyb) antibody was used as a negative control.
E. Schematic representations of the NUP98-LEDGF mutants are shown with a summary of their binding properties with MLL/menin on the right. The identified minimum MLL/menin binding domain (MmBD, residues 335-460) is indicated.
F. His-Express-Myc-tagged NUP98-LEDGF proteins were expressed in 293T cells and subjected to IP with anti-MYC antibody followed by immunoblotting with anti-Express epitope and anti-menin antibodies.
Specific association of LEDGF with the MLL-ENL/menin complex was confirmed by reciprocal immunoprecipitation (IP) experiments using anti-FLAG or anti-HA antibodies to pull down MLL-ENL or menin, respectively, which showed that endogenous LEDGF consistently co-precipitated with transiently expressed menin and MLL-ENL (Figure 1B). However, expression of menin or MLL-ENL alone did not result in co-IP of LEDGF at a detectable level. Consistent with these observations, a mutant MLL-ENL (MEd) that lacks the high affinity menin-binding motif (hMBM) and therefore does not interact with menin efficiently (Yokoyama et al., 2005), failed to co-precipitate endogenous LEDGF despite the presence of abundant menin (Figure 1B). These data demonstrate that LEDGF associates conjointly with MLL-ENL and menin but not with either protein separately.
LEDGF also co-precipitated with menin in association with transiently expressed wild type MLL and other MLL fusion proteins (Figures 1C and S1A). At a very low level, LEDGF was detected in the endogenous MLL/menin complex in REH cells as well (Figure 1D). Furthermore, the NUP98-LEDGF fusion protein created by chromosomal translocation in acute leukemia (Ahuja et al., 2000) also co-precipitated with the MLL/menin complex suggesting a possible pathogenic association. Interaction was dependent on the HIV-1 integrase-binding domain (IBD) (Cherepanov et al., 2004) as revealed by deletion mutagenesis (Figures 1E and 1F), consistent with the inability of a naturally occurring isoform of LEDGF (p52) that lacks the IBD to co-IP with MLL/menin (Figure S1B). Thus, the p75 isoform of LEDGF is a specific cofactor for both wild type and oncogenic MLL/menin protein complexes.
LEDGF co-localizes with MLL proteins and menin on crucial target genes
Chromatin immunoprecipitation (ChIP) analyses demonstrated that LEDGF co-localizes with MLL/menin complexes at target sites within HOXA, MEIS1 and CDKI genes (Figures 2 and S2), all previously linked with MLL/menin pathologies (Ayton and Cleary, 2003; Karnik et al., 2005; Milne et al., 2005; Wong et al., 2007). This was the case in U937 cells, which lack MLL gene rearrangement and express only wild type MLL (Dreying et al., 1996; Guenther et al., 2005), and in ML-2 cells, which exclusively express MLL-AF6 in the absence of wild type MLL (Tanabe et al., 1996; Yokoyama et al., 2005). The occupancy of MLL-AF6 or wild type MLL throughout the HOXA locus and other target loci extensively overlapped with menin and LEDGF (Figure 2B). Thus, LEDGF forms stable complexes with MLL (or MLL oncoproteins) in vivo on genes implicated to critically mediate MLL/menin-associated biology and disease.
Figure 2. MLL proteins co-localize with LEDGF on chromatin of cancer-associated genes.
A. ChIP was performed on ML-2 cells using anti-MLLN, anti-menin and anti-LEDGF antibodies. MLL-AF6, menin and LEDGF occupancy was specifically observed at the HOXA7 and HOXA9 loci, but not on the MEIS1 and GAPDH loci. Negative and positive controls consisted of no antibody and anti-histone H3 antibody, respectively.
B. ChIP followed by quantitative PCR was performed on ML-2 and U937 cells using anti-MLLN, anti-menin and anti-LEDGF antibodies. Values are expressed relative to the maximum value (arbitrarily set at 100%) in each group with error bars representing standard deviations for triplicate PCR analyses.
LEDGF is required for initiation of leukemic transformation by MLL oncoproteins
A structure/function analysis of MLL-ENL revealed that deletions spanning MLL residues 112-153 completely abolished LEDGF binding, while preserving menin association (Figures 3A, 3B and S3). This defined an evolutionally conserved region within the amino-terminal portion of MLL as a specific LEDGF binding domain (LBD) distinct from the hMBM.
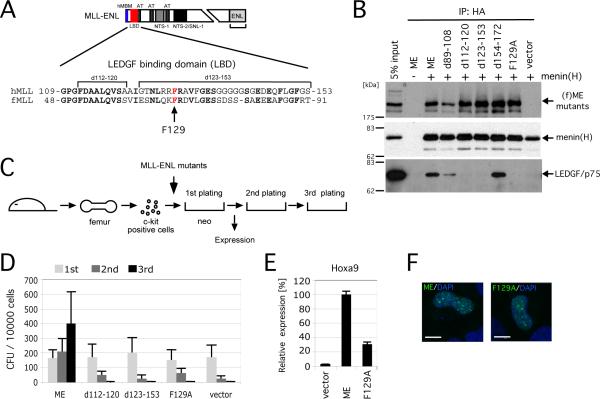
Figure 3. MLL oncoproteins associate with LEDGF to initiate myeloid transformation.
A. Schematic structure of the MLL-ENL oncoprotein. Amino acid sequence of MLL encompassing the LEDGF binding domain (LBD, residues 109-153 denoted by red box) is aligned with that of fugu MLL. Arrow indicates the phenylalanine residue substituted to alanine.
B. Various mutants of (f)ME were transiently expressed with (+) or without (-) HA-tagged menin in 293T cells and subjected to IP with anti-HA antibody followed by immunoblotting with anti-FLAG, anti-HA, and anti-LEDGF antibodies.
C. The experimental scheme for myeloid progenitor transformation assay. The time point at which Hoxa9 expression was measured (end of 1st plating) is indicated.
D. The colony forming units (CFU) per 104 plated cells are shown for each round of plating. Error bars represent standard deviations of three independent experiments.
E. Relative expression levels of Hoxa9 are shown for first round colonies. Expression levels are normalized to β-actin and expressed relative to the ME value (arbitrarily set at 100%). Error bars represent standard deviations of triplicate PCR analyses.
F. Subnuclear localizations of MLL fusion proteins in HeLa cells are shown as a merged image of signals for FITC (MLL) and DAPI (DNA). Scale bar, 10 μm.
The MLL-ENL deletion mutants were evaluated for oncogenic activity using a transformation assay that reads out the ability of MLL fusion genes to induce enhanced self-renewal of myeloid progenitors in vitro compared with control cells, which rapidly exhaust their clonogenic potentials. All deletion mutants unable to associate with LEDGF were also unable to transform myeloid progenitors (Figures 3C and 3D). Moreover, a single amino acid substitution within the LBD that converted an evolutionally conserved phenylalanine to alanine (F129A) completely abolished LEDGF binding and myeloid transformation. The F129A mutant was also transcriptionally incapable of maintaining Hoxa9 expression in transduced myeloid progenitors (Figure 3E) despite efficient expression and localization in nuclear bodies (Figure 3F). Thus, specific association with LEDGF is essential for mis-regulation of Hoxa9 expression and transformation of myeloid progenitors by the MLL oncoprotein.
Menin is an adapter that tethers MLL oncoproteins with LEDGF in transformed myeloid progenitors
LEDGF contains a highly conserved motif (PWWP) that is structurally related to the so-called “royal family” of domains present in a wide variety of chromatin-associated proteins and implicated in recognition of modified nucleosomes (Maurer-Stroh et al., 2003). The PWWP domain of LEDGF is necessary to associate with chromatinized DNA and target HIV-1 genome integration to transcriptionally active sites (Botbol et al., 2008; Shun et al. 2007). We hypothesized, therefore, that menin's role is to tether MLL proteins with LEDGF as an adaptor, which in turn promotes association of the complex with transcriptionally active chromatin through its PWWP domain.
To test this hypothesis, MLL oncoproteins were engineered to circumvent menin by replacing the hMBM with the PWWP domain of LEDGF. The resultant modified MLL-ENL (pME) was no longer capable of stably associating with menin and LEDGF (Figures 4A and 4B), but nevertheless strongly up-regulated Hoxa9 expression and efficiently transformed myeloid progenitors (Figures 4A and 4C). Consistent with these observations, pME function was not compromised by mutation (F129A) of the LBD, which otherwise abrogates non-covalent LEDGF interaction. Fusion with PWWP, however, did not bypass the requirement for the MLL fusion partner moiety in myeloid transformation by pME or a similarly engineered MLL-AF10 (Figures 4A and 4B). ChIP analysis of pME-transformed cells showed that pME occupied the Hoxa9 locus in the absence of menin and LEDGF (Figure 4E). Mutation of an evolutionally conserved tryptophan to alanine (W21A) in the PWWP motif abolished both Hoxa9 up-regulation and myeloid transformation, confirming a critical role of the PWWP domain in pME function (Figures 4A, 4C and 4D).
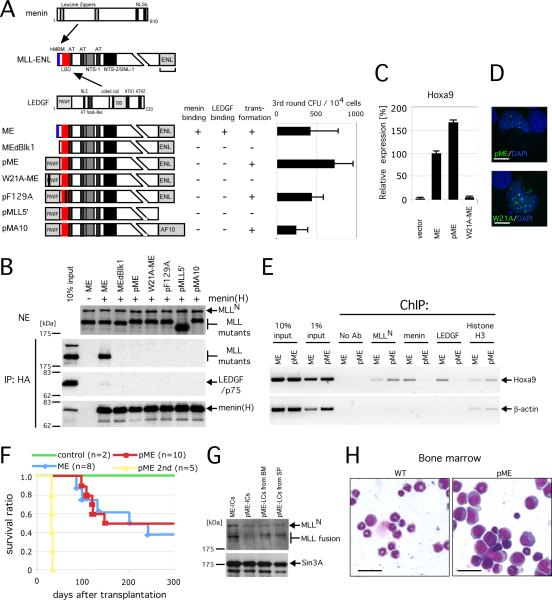
Figure 4. Menin tethers LEDGF with MLL oncoproteins.
A. Schematic structures of menin, MLL-ENL and LEDGF are shown at the top with sites of intermolecular interactions indicated by arrows. Binding properties and transforming abilities are summarized to the right of the respective MLL-ENL constructs. The numbers of 3rd round CFU in myeloid progenitor assays are shown with error bars representing the standard deviations of three independent experiments.
B. Various mutants of pME were transiently expressed with (+) or without (-) HA-tagged menin in 293T cells and subjected to IP with anti-HA antibody followed by immunoblotting with anti-MLLN, anti-HA, and anti-LEDGF antibodies. Upper panel shows immunoblot of the nuclear extracts (NE) to determine protein inputs. Mutants lacking the hMBM failed to co-IP with menin and LEDGF.
C. The relative expression levels are shown for Hoxa9 in first round colonies. Expression levels are normalized to β-Actin, and expressed relative to the ME value (arbitrarily set at 100%). Error bars represent standard deviations of triplicate PCR analyses.
D. Subnuclear localization of the pME mutants in HeLa cells is shown as a merged image of signals for FITC (MLL) and DAPI (DNA). Scale bar, 10 μm.
E. ChIP analysis of ME- or pME-transformed myeloid progenitors was performed using antibodies indicated at the top. Amplicons upstream of Hoxa9 and β-Actin genes were analyzed.
F. Survival curves are shown for mice transplanted with the indicated transduced cells. Numbers of mice analyzed (n) are shown.
G. Expression of ME and pME proteins in immortalized cells (IC) and leukemic cells (LC) from bone marrow (BM) and spleen (SP) were analyzed by immunoblotting with anti-MLLN and anti-SIN3A (control) antibodies.
H. Morphology is shown for pME leukemic blasts (pME: right panel) and normal BM (wt: left) following May-Grunwald/Giemsa staining of cytospin preparations. Scale bar, 15 μm.
Transplantation of pME-transformed cells into congenic recipients induced acute leukemias with latencies and penetrance similar to those of ME-transplanted mice (Figure 4F and 4G). In both cohorts, blast cells with morphologic features characteristic of acute monocytic leukemia massively infiltrated the spleen, liver and bone marrow (Figures 4H and S4), and primary leukemia cells expressing pME were transplantable to secondary recipients, which developed leukemia with short latencies (Figures 4F and 4G). Thus, pME induces leukemia in vivo despite its inability to associate with menin.
A conditional knockout approach was employed to further confirm that menin is dispensable for leukemic transformation by pME. Myeloid progenitors harvested from Men1-floxed mice were transformed by ME or pME, and subsequently transduced with Cre-ER (Cre recombinase fused with estrogen receptor), which was conditionally activated in the presence of 4-OHT (4-hydroxytamoxifen) to inactivate the Men1 gene (Figure 5A). While ME-transformed cells lost their clonogenicity and displayed predominantly differentiated colony morphologies after Men1 inactivation as previously reported (Yokoyama et al., 2005), pME-transformed cells retained their transformed phenotype (Figures 5B and 5C), and maintained high-level Hoxa9 expression in the absence of menin (Figures 5D and 5E). Thus, covalent fusion of the PWWP domain of LEDGF to MLL oncoproteins fully bypasses the requirement for menin in MLL-mediated leukemic transformation indicating that the sole molecular requirement for menin as an oncogenic cofactor in MLL-associated leukemogenesis is to tether the PWWP domain of LEDGF to the MLL oncoprotein.
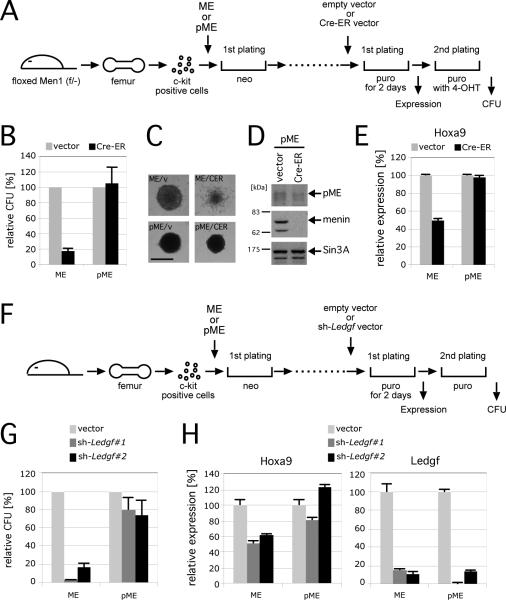
Figure 5. LEDGF is necessary for maintenance of MLL leukemic transformation.
A. The experimental scheme for conditional inactivation of Men1 in MLL transformed cells. The time points at which CFU or gene expression were measured are indicated.
B. The relative CFUs are shown for cells transformed by ME or pME in the absence (vector) or presence (Cre-ER) of Men1 inactivation (vector controls are arbitrarily set at 100%). Error bars represent the standard deviations of three independent analyses.
C. Morphologies are shown for representative colonies from experiment in panel B. Scale bar, 150 μm.
D. Western blot shows expression of pME, menin and SIN3A proteins after Men1 inactivation.
E. Relative expression levels of Hoxa9 are shown for first round colonies after Cre-ER transduction. Expression levels were normalized to β-Actin, and expressed relative to the vector control values (arbitrarily set at 100%). Error bars represent standard deviations of triplicate PCR analyses.
F. Experimental scheme for conditional inactivation of Ledgf by shRNA-mediated knockdown. The time points at which CFUs or gene expression were measured are indicated.
G. The relative CFU activity of ME- or pME-transformed cells is shown with (sh-Ledgf#1 or sh-Ledgf#2) or without (vector) Ledgf knockdown (the vector controls are arbitrarily set at 100%). Error bars represent the standard deviations of three independent analyses.
H. Relative expression levels of Hoxa9 and Ledgf are shown for first round colonies after shRNA vector transduction. Expression levels are normalized to β-actin, and expressed relative to the vector control values (arbitrarily set at 100%). Error bars represent the standard deviations of triplicate PCR analyses.
Sustained LEDGF expression is required for maintenance of leukemic transformation by MLL oncoproteins
To assess whether LEDGF must be continuously present for maintenance of MLL-dependent transformation, its expression was knocked down in ME- and pME-transformed progenitors using shRNA techniques (Figure 5F). Efficient (>80%) knockdown of Ledgf expression obtained by two different shRNAs (Figure 5H) markedly reduced the clonogenic potential of ME-transformed cells, whereas pME-transformed cells were unaffected (Figure 5G). Furthermore, Ledgf-knockdown impaired Hoxa9 expression in ME-transformed cells, but not in pME-transformed cells (Figure 5H). Thus, LEDGF is required to maintain aberrant Hox gene expression and transformation induced by MLL oncoproteins.
Implication of LEDGF in menin tumor suppression
To investigate the role of LEDGF in MEN1 tumorigenesis, menin proteins harboring point mutations found in MEN1 patients were analyzed for their abilities to associate with MLL proteins and LEDGF. Several menin mutants (H138D, A242V, T344R) displayed no or markedly reduced interactions with MLL (Figures 6A and 6B), consistent with their previously demonstrated inabilities to co-IP with HMT activity (Hughes et al., 2004). They also failed to associate with LEDGF, compatible with our foregoing results (Figure 1B) that LEDGF interacts conjointly with MLL/menin. Other menin mutants (P12L and L22R), however, retained competence to co-IP with MLL, but were nevertheless unable to associate with LEDGF (Figures 6A and 6B).
Figure 6. Menin functionally interacts with LEDGF.
A. Schematic structures of menin and its mutants. The abilities of each protein to associate with MLL or LEDGF and to rescue MLL-dependent transcription are summarized on the right. *, indicates impaired association as opposed to complete loss of association. ND, not determined.
B. Various HA-tagged menin proteins were transiently expressed with (+) or without (-) (f)ME in 293T cells and subjected to IP with anti-HA antibody, followed by immunoblotting with anti-FLAG, anti-HA, and anti-LEDGF antibodies.
C. Experimental scheme for rescue of Men1 knockdown by wild type or mutant menin proteins. The time points at which CFUs or gene expression were measured are indicated.
D. Western blot analysis shows expression of menin mutants in ME-transformed cells detected by anti-HA and anti-MLLN antibodies, respectively.
E. Relative CFU are shown for ME-transformed cells transduced with various menin mutants with (+) or without (-) Men1 knock down (vector control was arbitrarily set at 100%). Error bars represent the standard deviations of three independent analyses.
F. Relative expression levels of Hoxa9 and Men1 are shown for first round colonies after Men1 knockdown. Expression levels are normalized to Gapdh, and expressed relative to the ME/vector value (arbitrarily set at 100%). Error bars represent the standard deviations of triplicate PCR analyses. P values for differences compared to wild type menin rescue were determined by unpaired t test (*, P<0.0005; **, P<0.005).
The latter mutants were assessed for their potential to rescue loss of menin function following shRNA knockdown of endogenous murine Men1 expression in ME-transformed cells (Figure 6C). In contrast to transduced wild type menin, which prevented loss of clonogenicity caused by Men1 knockdown, the P12L and L22R mutants failed to maintain myeloid transformation (Figures 6D and 6E). They were also unable to optimally maintain Hoxa9 expression following inactivation of Men1 in ME-transformed cells compared with wild type menin-transduced cells (Figure 6F) consistent with a compromised ability of L22R to maintain CDKI transcription in menin-deficient MEFs (Milne et al., 2005). Therefore, a subset of menin mutations in MEN1 tumors specifically abrogates LEDGF interaction while preserving MLL interaction, but nevertheless compromises MLL-dependent functions. These data support a role for LEDGF in MEN1 tumor suppression as well as MLL-associated leukemogenesis.
Discussion
Our studies establish LEDGF as a crucial cofactor required for both the oncogenic and tumor suppressor functions of MLL/menin complexes. LEDGF interacts conjointly with MLL and menin on the chromatin of cancer-associated genes to mediate MLL-dependent transcription pathways (Figure 7). In this context, menin serves as an adaptor to link MLL with LEDGF. A subset of menin mutations in MEN1 tumors is particularly informative as they abrogate interactions with LEDGF, but not MLL, yet compromise MLL/menin activity. Genetic evidence that MLL functions with LEDGF in its normal developmental role to regulate HOX gene expression during establishment of the embryonic body plan is suggested by the phenotypic overlaps of Mll- and Ledgf-deficient mice, which both display skeletal malformations representative of anterior and posterior homeotic transformations (Sutherland et al., 2006; Yu et al., 1995). Thus, LEDGF is an essential component of the MLL/menin HMT complex in the setting of its normal and pathologic activities.
Figure 7. Model for the role of LEDGF in the normal and neoplastic functions of the MLL/menin HMT complex.
Upper: Specific association with LEDGF is required for transcriptional contributions by the MLL/menin HMT complex at its chromatin sites of action.
Lower: Similarly, the constitutive transcriptional properties of MLL chimeric oncoproteins in complex with menin are also dependent on association with LEDGF, which may provide a molecular target for therapeutic intervention.
LEDGF has previously been implicated in various transcriptional processes and cellular functions. Originally discovered based on its co-fractionation with the general transcriptional co-activator PC4, LEDGF reportedly associates with transcriptional activators and components of the basal transcriptional machinery including RNA pol II subunits (Ge et al., 1998), and contributes to the transcriptional response following environmental stress (Shinohara et al., 2002). Coincidently, menin is also a regulator of stress-induced response in fruit flies, which transcriptionally up-regulate expression of various heat shock proteins following stressful stimuli (Papaconstantinou et al., 2005). Menin has been reported to co-IP RNA pol II (Hughes et al., 2004), raising the possibility that it links MLL with LEDGF/p75 in higher-order complexes of dynamic composition to regulate specific stages of transcription.
In addition to its transcriptional role, LEDGF is important for lentiviral integration (Ciuffi et al., 2005; Llano et al., 2006; Shun et al., 2007). LEDGF is the dominant binding partner for HIV-1 integrase in human cells (Cherepanov et al., 2003), and tethers it to host chromosomes (Maertens et al., 2003), thereby serving a major role in determining the highly distinctive pattern of lentiviral genome integration within active transcription units (Ciuffi and Bushman, 2006). Thus, physical association of LEDGF with MLL/menin on chromatin may provide a molecular basis for the selective integration of HIV-1 into actively transcribed regions since the epigenetic mark placed by MLL is involved in maintaining chromatin in a state conducive for transcription (Li et al., 2007). A tethering role for LEDGF likely extends to other host proteins as well since the chromosomal association of JPO2, a MYC-interacting protein with transforming activity, is also strictly dependent on LEDGF (Maertens et al., 2006). It remains to be determined if these various interactions, which target the IBD of LEDGF, are mutually exclusive with MLL/menin and what their implications may be for anti-lentiviral therapy.
The association of LEDGF with chromatinized DNA is critically dependent on its PWWP domain (Botbol et al., 2008). This highly conserved motif is present in a variety of chromatin-associated proteins involved in transcriptional regulation, DNA repair and methylation (Stec et al., 2000). It has structural similarities with Tudor, chromo and MBT domains, all of which are implicated in binding methylated Lys residues on histones (Maurer-Stroh et al., 2003; Li et al. 2007). Structural similarities with the ligand binding cavities of these evolutionally related domains strongly suggest that the PWWP domain binds to a currently undefined component of chromatin, although a possible role in non-specific DNA binding has been suggested as well (Lukasik et al., 2006; Nameki et al., 2005; Sue et al., 2004; Qiu et al., 2002). Compelling evidence for the role of LEDGF in targeting the MLL/menin HMT complex to chromatin is provided by grafting of its PWWP onto the MLL oncoprotein, which was fully capable of bypassing the requirement for menin in oncogenesis, Hox gene mis-regulation and chromatin association. This artificial construct is structurally similar to the A. thaliana homologs of MLL (ATX1 and ATX2) (Alvarez-Venegas and Avramova, 2001), which contain PWWP domains in their amino-terminal portions, providing evolutionary support for the functional link between MLL and LEDGF.
Our study indicates significant roles for LEDGF in menin-dependent growth control. Menin has been reported to potentially interact with DNA (La et al., 2004) and several other proteins (Balogh et al., 2006), however our data suggest that menin's sole role in MLL leukemia is to recruit LEDGF. Our observation that some MEN1-associated mutations specifically disrupt LEDGF binding and compromise MLL-dependent transcription also implicates LEDGF in MEN1 tumorigenesis. Accumulating evidence indicates that the physiologic growth responses of endocrine lineage cells are heavily dependent on the MLL/menin pathway through regulated expression of CDKIs (Franklin et al., 1998; Milne et al., 2005; Karnik et al., 2005; 2007). In contrast to ATX proteins, the interaction of MLL with LEDGF critically mediated by menin is non-covalent, which provides a potential mechanism for regulating their conditional association. LEDGF is induced upon cellular stress stimuli including serum starvation (Huang et al., 2007) and, interestingly, is secreted from and re-enters lens epithelial cells by penetrating the plasma membrane (Singh et al., 1999). Thus, it is tempting to speculate that cell autonomous or non-autonomous induction of LEDGF may be implicated in the growth control of endocrine and other lineages, perhaps as part of a molecular switch for targeting of the MLL/menin HMT complex to chromatin (Figure 7).
Our results provide a broader context for conceptualizing the various pathologies associated with LEDGF and its binding partners, which appear to be frequently targeted in diverse diseases. In addition to making essential contributions to MLL-mediated leukemias and endocrine tumorigenesis, LEDGF itself is targeted by chromosomal translocations in leukemia that result in its fusion with the nucleoporin NUP98 (Ahuja et al., 2000). The molecular mechanism by which NUP98-LEDGF causes leukemia is unknown, however our data show that it is capable of associating with MLL/menin suggesting that it may also perturb the HOX pathway, compatible with the more frequent fusion of NUP98 with HOX proteins themselves in leukemias. Increased LEDGF expression is also a feature of some cancers including acute myeloid leukemia and tumors of breast and bladder origins (Daugaad et al., 2007; Huang et al., 2007), whereas auto-antibodies to LEDGF are frequently present in patients with atopic dermatitis and other auto-immune disorders (Ganapathy and Casiano, 2004). The specific roles of MLL/menin in these various diseases merit further investigation as they may have important implications for therapeutic interventions in cancer, auto-immunity and AIDS.
Experimental procedures
Cell culture and animal use
HB1119, ML-2, REH and U937 cells were cultured in RPMI 1640 medium supplemented with 15% fetal calf serum and non-essential amino acids. 293T, HeLa and plat-E cells (Morita et al., 2000) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal calf serum and non-essential amino acids. All experiments on mice in this study were performed with the approval and in accordance with Stanford's Administrative Panel on Laboratory Animal Care.
Vector construction
The pCMV5 FLAG-MLL-ENL vector (Ayton et al. 2004), and pMSCV-neo constructs encoding MLL-ENL and MLL-AF10 (Ayton and Cleary, 2003; DiMartino et al., 2002) were described previously. Expression vectors for various MLL mutants were generated by restriction enzyme digestion and PCR-based mutagenesis. The cDNA for menin was provided by Dr. F. Hosoda. A series of menin mutants tagged with HA were generated by PCR-mediated mutagenesis and cloned into pcDNA3.1/hygro (Invitrogen) or pMSCV hygro (Clontech). The expression vector for LEDGF was purchased from OriGene Technologies, Inc. The cDNA fragments for p75 and p52 were generated by PCR and cloned into pCMV6. The cDNA of myc tagged NUP98-LEDGF was generated by PCR using the pMSCV NUP98-HOX9 (Iwasaki et al., 2005) and pCMV6 LEDGF vectors as templates, and subsequently cloned into the pcDNA4 HisMax vector (Invitrogen). The sh-RNA expression vectors were purchased from Open Biosystems (TRCN0000012113 for Ledgf#1; TRCN0000012114 for Ledgf#2; TRCN000034394 for Men1).
One step immunopurification and identification of MLL/menin associated proteins
Preparation of nuclear extracts and large-scale immunoprecipitations were performed as described elsewhere (Yokoyama et al., 2004). 293T cells were cultured in ten 175 cm2 dishes and transfected with pCMV FLAG-MLL-ENL and pcDNA3.1 menin-HA vectors using Lipofection 2000 (Invitrogen). Nuclear extracts were prepared from the cells 48 hrs after transfection, cleared by ultracentrifugation, and then immunoprecipitated with anti-FLAG (M2) agarose beads for 4 h. After extensive washing, the purified material was subjected to SDS-PAGE analysis and visualized by Comassie Brilliant Blue staining. Gel bands containing proteins of interest were subjected to mass spectrometry by the Stanford Proteomics & Integrative Reseach Facility.
Immunoprecipitation and immunoblotting
Preparation of nuclear extracts, immunoprecipitation and immunoblotting were performed as described elsewhere (Yokoyama et al., 2004; 2005). Primary antibodies included mouse monoclonal anti-MLLN (mmN4.4) and anti-MLLC (mmC2.1), and rabbit polyclonal anti-MLLN (rpN1) as described previously (Yokoyama et al., 2002). Rabbit anti-DmMyb was provided by J. Lipsick. Goat anti-menin (C19), mouse anti-Express epitope (Omni probe; D-8) and rabbit anti-SIN3A (K-20) antibodies were purchased from Santa Cruz Biotechnology, Inc. Additional primary antibodies included rabbit anti-menin (BL342) and anti-LEDGF (BL3656) (Bethyl Laboratories, Inc.), mouse anti-LEDGF (BD Transduction Laboratories), and mouse anti-ACTIN (MAB 1501R) (Chemicon). Rat anti-HA antibody (3F10) conjugated with HRP or immobilized to matrix was purchased from Roche. Rabbit anti-FLAG (F-7425) antibody and agarose affinity beads coupled to mouse anti-Flag (M2) or anti-MYC (9E10) monoclonal antibody were purchased from Sigma.
Virus production
Ecotropic retrovirus was produced using plat-E packaging cells (Morita et al., 2000). Lentivirus was produced by co-transfection of viral vectors with pCMV gag-pol and pVSVG env packaging constructs into 293T cells (Dull et al., 1998). Medium containing virus was collected 48 h post transfection and used for transductions.
Myeloid progenitor transformation assay
Myeloid progenitor transformation assays were performed as described elsewhere (Lavau et al., 1997; Yokoyama et al., 2005). Cells (CD45.1) were harvested from the femurs of C57BL/6 or Men1 floxed (Men1floxed/-) mice. Progenitors (c-kit+) were enriched by immuno-magnetic selection (Miltenyi Biotech), transduced with recombinant retroviruses by spinoculation, and plated in methylcellulose media (M3231, Stemcell Technologies) containing SCF, IL-3, IL-6 and GM-CSF. In vitro Cre-dependent gene inactivation was performed as previously described (Yokoyama et al., 2005) using 0.1 nM 4-OHT to activate the Cre-ERtam protein. For secondary transductions, 105 cells were transduced with retrovirus by spinoculation, cultured in methylcellulose media overnight, and selected for drug resistance (hygromycin 750 μg/ml or puromycin 4 μg/ml) for at least 2 days.
In vivo leukemogenesis assay
Transformed cells (2×105) from methylcellulose cultures were transplanted intravenously into lethally irradiated C57BL/6 mice (900 rads) with 2×105 syngeneic bone marrow cells. Moribund mice were sacrificed, and tissues were fixed in 10% formalin and processed for H&E staining. Cells from bone marrow and spleen were subjected to cytospin preparation followed by May-Grunwald/Giemsa staining or cultured in methylcellulose media for secondary transplantation.
Quantitative RT-PCR
Reverse transcription and quantitative PCR were performed as described previously using Taqman probes for Hoxa9 (Mm00439364_m1), Men1 (Mm00484963_m1), Gapdh (Mm99999915_g1), Ledgf (Mm01259222_g1) and β-Actin (Mm00607939_m1) purchased from Applied Biosystems. Expression levels normalized to that of β-Actin or Gapdh were calculated using a standard curve and the relative quantitation method as described in ABI User Bulletin #2.
Chromatin immunoprecipitation
Chromatin immunoprecpitation was performed as previously described (Weinmann and Farnham 2002; Yokoyama et al., 2005) using primary antibodies specific for MLLN (rpN1), menin (BL342), LEDGF (BL3656) or histone H3 (ab1791 purchased from Abcam). Semi quantitative or quantitative real-time PCR was performed on the precipitated DNAs using primers and qPCR probes described in the supplemental data. The relative values to input were determined using a standard curve and the relative quantitation method as described in ABI User Bulletin #2.
Indirect immunofluorescence
Indirect immunofluorescence was performed as described elsewhere (Yokoyama et al., 2001) on HeLa cells tranfected with expression vectors encoding various MLL mutant proteins. The cells were fixed, incubated with rabbit anti-MLLN antibody (rpN1), and probed with a FITC-conjugated goat anti-rabbit IgG secondary antibody (Santa Cruz Biotechnology). Cells were stained with DAPI (Vector Laboratories) and analyzed by confocal immunofluoroscopy at the Stanford Cell Sciences Imaging Facility.
Supplementary Material
Acknowledgments
We thank Dr. J. Lipsick for providing anti-DmMyb antibody, Dr. T. Kitamura for providing the plat-E cell line, Dr. M. Myerson for Men1 floxed mice, and Dr. F. Hosoda for MEN1 cDNA. We acknowledge C. Nicolas, M. Ambrus, and B. T. Rouse for technical assistance, A. James for graphics support, and Drs. M. Murphy and M. Iwasaki for technical instruction. A.Y. was supported by a Special Fellow Award of the Leukemia and Lymphoma Society. These studies were supported by the Children's Health Initiative of the Packard Foundation and grants from the National Institutes of Health (CA55029 and CA116601).
References
- Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z. Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene. 2001;271:215–221. doi: 10.1016/s0378-1119(01)00524-8. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh K, Racz K, Patocs A, Hunyady L. Menin and its interacting proteins: elucidation of menin function. Trends Endocrinol Metab. 2006;17:357–364. doi: 10.1016/j.tem.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Bertolino P, Tong WM, Galendo D, Wang ZQ, Zhang CX. Heterozygous Men1 mutant mice develop a range of endocrine tumors mimicking multiple endocrine neoplasia type 1. Mol Endocrinol. 2003;17:1880–1892. doi: 10.1210/me.2003-0154. [DOI] [PubMed] [Google Scholar]
- Botbol Y, Raghavendra NK, Rahman S, Engelman A, Lavigne M. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008;36:1237–1246. doi: 10.1093/nar/gkm1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, Silva A, Brown EJ, Hess JL, Pear WS, Hua X. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci U S A. 2006;103:1018–1023. doi: 10.1073/pnas.0510347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Crabtree JS, Scacheri PC, Ward JM, Garrett-Beal L, Emmert-Buck MR, Edgemon KA, Lorang D, Libutti SK, Chandrasekharappa SC, Marx SJ, et al. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc Natl Acad Sci U S A. 2001;98:1118–1123. doi: 10.1073/pnas.98.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- Daugaard M, Kirkegaard-Sorensen T, Ostenfeld MS, Aaboe M, Hoyer-Hansen M, Orntoft TF, Rohde M, Jaattela M. Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res. 2007;67:2559–2567. doi: 10.1158/0008-5472.CAN-06-4121. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin DS, Godfrey VL, Lee H, Kovalev GI, Schoonhoven R, Chen-Kiang S, Su L, Xiong Y. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 1998;12:2899–2911. doi: 10.1101/gad.12.18.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy V, Casiano CA. Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us? Arthritis Rheum. 2004;50:684–688. doi: 10.1002/art.20095. [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. Embo J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Huang TS, Myklebust LM, Kjarland E, Gjertsen BT, Pendino F, Bruserud O, Doskeland SO, Lillehaug JR. LEDGF/p75 has increased expression in blasts from chemotherapy-resistant human acute myelogenic leukemia patients and protects leukemia cells from apoptosis in vitro. Mol Cancer. 2007;6:31. doi: 10.1186/1476-4598-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Kuwata T, Yamazaki Y, Jenkins NA, Copeland NG, Osato M, Ito Y, Kroon E, Sauvageau G, Nakamura T. Identification of cooperative genes for NUP98-HOXA9 in myeloid leukemogenesis using a mouse model. Blood. 2005;105:784–793. doi: 10.1182/blood-2004-04-1508. [DOI] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik SK, Chen H, McLean GW, Heit JJ, Gu X, Zhang AY, Fontaine M, Yen MH, Kim SK. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–809. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- Karnik SK, Hughes CM, Gu X, Rozenblatt-Rosen O, McLean GW, Xiong Y, Meyerson M, Kim SK. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proc Natl Acad Sci U S A. 2005;102:14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La P, Silva AC, Hou Z, Wang H, Schnepp RW, Yan N, Shi Y, Hua X. Direct binding of DNA by tumor suppressor menin. J Biol Chem. 2004;279:49045–49054. doi: 10.1074/jbc.M409358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Lukasik SM, Cierpicki T, Borloz M, Grembecka J, Everett A, Bushweller JH. High resolution structure of the HDGF PWWP domain: a potential DNA binding domain. Protein Sci. 2006;15:314–323. doi: 10.1110/ps.051751706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Hiew SYL, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady JM. MLL has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Nameki N, Tochio N, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Matsuda T, Fujikura Y, Saito M, et al. Solution structure of the PWWP domain of the hepatoma-derived growth factor family. Protein Sci. 2005;14:756–764. doi: 10.1110/ps.04975305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaconstantinou M, Wu Y, Pretorius HN, Singh N, Gianfelice G, Tanguay RM, Campos AR, Bedard PA. Menin is a regulator of the stress response in Drosophila melanogaster. Mol Cell Biol. 2005;25:9960–9972. doi: 10.1128/MCB.25.22.9960-9972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T, Singh DP, Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog Retin Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Ohguro N, Chylack LT, Jr., Shinohara T. Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Invest Ophthalmol Vis Sci. 1999;40:1444–1451. [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- Sue SC, Chen JY, Lee SC, Wu WG, Huang TH. Solution structure and heparin interaction of human hepatoma-derived growth factor. J Mol Biol. 2004;343:1365–1377. doi: 10.1016/j.jmb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Sutherland HG, Newton K, Brownstein DG, Holmes MC, Kress C, Semple CA, Bickmore WA. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol Cell Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Zeleznik-Le NJ, Kobayashi H, Vignon C, Espinosa R, 3rd, LeBeau MM, Thirman MJ, Rowley JD. Analysis of the t(6;11)(q27;q23) in leukemia shows a consistent breakpoint in AF6 in three patients and in the ML-2 cell line. Genes Chromosomes Cancer. 1996;15:206–216. doi: 10.1002/(SICI)1098-2264(199604)15:4<206::AID-GCC2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Kawaguchi Y, Kitabayashi I, Ohki M, Hirai K. The conserved domain CR2 of Epstein-Barr virus nuclear antigen leader protein is responsible not only for nuclear matrix association but also for nuclear localization. Virology. 2001;279:401–413. doi: 10.1006/viro.2000.0715. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.