Abstract
Understanding the basic neuronal building blocks of the neocortex is a necessary first step toward comprehending the composition of cortical circuits. Neocortical layer VI is the most morphologically diverse layer and plays a pivotal role in gating information to the cortex via its feedback connection to the thalamus and other ipsilateral and callosal corticocortical connections. The heterogeneity of function within this layer is presumably linked to its varied morphological composition. However, so far, very few studies have attempted to define cell classes in this layer using unbiased quantitative methodologies. Utilizing the Golgi staining technique along with the Neurolucida software, we recontructed 222 cortical neurons from layer VI of mouse barrel cortex. Morphological analyses were performed by quantifying somatic and dendritic parameters, and, by using principal component and cluster analyses, we quantitatively categorized neurons into six distinct morphological groups. Additional systematic replication on a separate population of neurons yielded similar results, demonstrating the consistency and reliability of our categorization methodology. Subsequent post hoc analyses of dendritic parameters supported our neuronal classification scheme. Characterizing neuronal elements with unbiased quantitative techniques provides a framework for better understanding structure-function relationships within neocortical circuits in general.
Indexing terms: neocortex layer VI, Golgi, neuronal morphology, barrel cortex
The neocortex is composed of six distinct layers, with each layer containing different morphological phenotypes that presumably subserve different processing roles (Mountcastle, 1997). For example, the excitatory spiny stellate cells and small pyramidal neurons in layer IV receive strong thalamo-cortical input (see Davis and Sterling, 1979; White, 1989). By contrast, the excitatory neurons of the infragranular layers (V and VI) are composed predominantly of larger pyramidal cells (Jones, 1984; White, 1989; Rocco and Brumberg, 2007). Although layer IV is the principal input layer, layers V and VI originate most of the cortical output (Jones, 1984; White, 1989). Within the infragranular layers, layer VI is of particular interest, primarily because of the diverse population of neurons and its complicated microcircuitry. Layer VI receives thalamic and cortical inputs and gives rise to corticothalamic feedback projections as well as corticocortical, corticoclaustral, and commissural fibers (Tombol, 1984; Katz, 1987; Brumberg et al., 2003; Chakrabarti and Alloway, 2006). Therefore, this layer is at the pivotal point to affect thalamocortical/corticothalamic loops, which may gate the information that ultimately ascends to the cerebral cortex.
As a first step toward understanding the structural and functional relationships within layer VI, studies have linked some characteristic neuronal morphologies with specific functional classes (Katz, 1987; Van Brederode and Snyder, 1992; Yang et al., 1996). However, because of the morphological diversity of the neuronal population in layer VI, the full spectrum of phenotypic classes remains unclear. Understanding the cytoarchitecture of layer VI neurons provides a basis upon which models of cortical circuits can be built. Prior studies have used qualitative descriptions to identify the cell types (Lorente de Nό, 1949; Simons and Woolsey, 1984; Prieto and Winer, 1999). One approach to remove possible selection bias is to perform multidimensional quantitative analysis of the morphological phenotypes (Cauli et al., 2000; Tamas et al., 2000; Tsiola et al., 2003), which utilizes statistical procedures to categorize neuronal groups objectively.
In the present study, we focused on layer VI of the mouse barrel field in the somatosensory cortex (S1). The barrel cortex has been shown to be a model system for the study of local cortical circuits and their relation to behavior (see Petersen, 2007). Furthermore, bercause of layer VI’s pivotal role in the modulation of sensory activity (Sherman and Guillery, 2002) and in many different cortical circuits (see Mendizabal-Zabiaga et al., 2007), it is of importance to clarify and identify the cytoarchitectural components of this morphologically and functionally diverse layer. In both the cat and the mouse visual systems, it was found that the apical dendrites of corticothalamic neurons do not extend superficially to layer IV, whereas the neighboring corticoclaustral neurons have different morphologies (Katz, 1987; Brumberg et al., 2003). These data highlight the correlations between the morphology and the functional role that neurons play in cortical circuits. We extend these studies by morphologically characterizing the neurons of layer VI in mouse barrel cortex using unbiased and quantitative methods as a means of determining the basic building blocks of local cortical circuits.
MATERIALS AND METHODS
Golgi staining protocol
The FD Rapid Golgi Stain Kit (FD Neurotechnologies, Inc.), a simplified and a reliable kit for Golgi impregnation, was used to label neurons. It is based on the staining principles previously described by Ramό n-Moliner (1970) and Glaser and Van der Loos (1981), which have demonstrated complete labeling of neuronal somata and dendrites. The procedure stains approximately 10% of the total neurons in the neocortex, thereby minimizing background noise to allow accurate neuronal reconstruction. Numerous studies (Ramanan et al., 2005; Dang et al., 2006; Elia et al., 2006; Furtak et al., 2007; Sadakata et al., 2007) as well as the present report have used this procedure to produce high-quality Golgi-impregnated tissue (see Fig. 1) for studying the morphological properties of neurons.
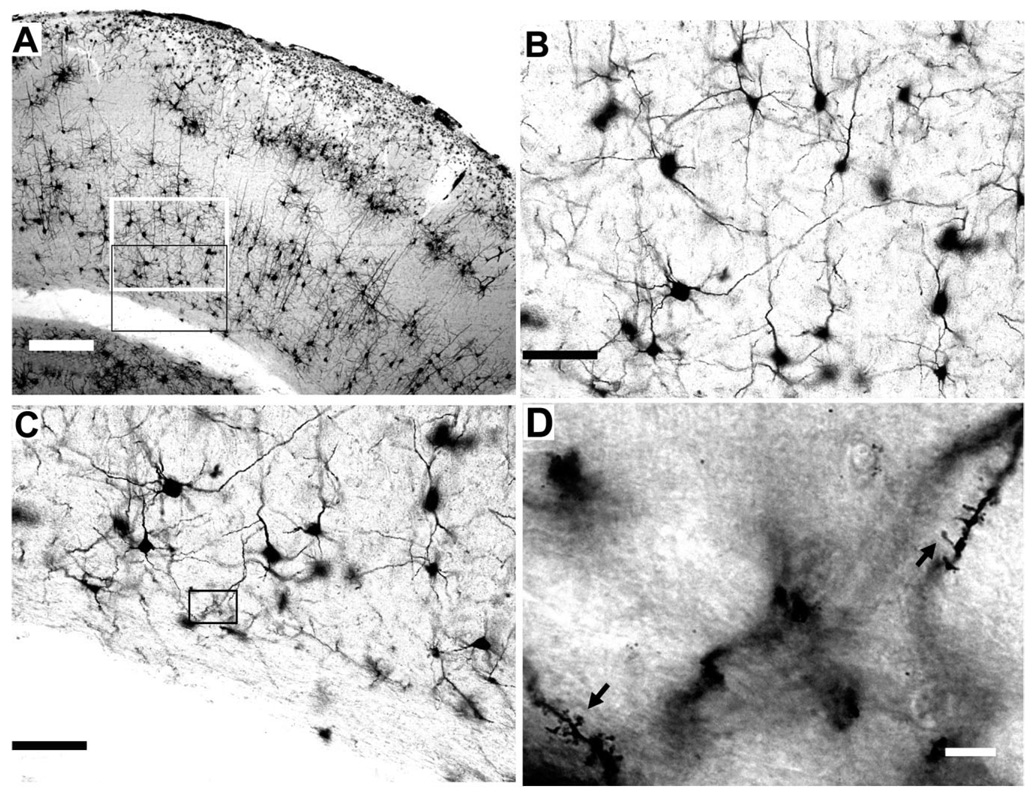
Figure 1. Golgi-stained section of the barrel cortex.
A: Low-magnification image (×4) illustrating barrel cortex and its six distinct layers bounded by pia and the white matter. Inset: The regions that are magnified in B (white boundary) and C (black boundary). B,C: Higher magnification (×20) highlighting the morphologies of layer VIa (B) and VIb (C) neurons. Note the diversity of neuronal morphologies. Inset: The region that is magnified in D. D: High magnification (×100) of dendrites that exhibit spines. Scale bars = 150 µm in A; 50 µm in B,C; 3 µm in D.
Preparation of animals and tissue processing
Animals (CD1 mice, N = 7) of either sex at 80–90 postnatal days (PND) of age were randomly selected and anesthetized with an intraperitoneal injection of Euthasol (Virbac AH, Inc). All procedures were in accordance with the Queens College, CUNY Institutional Animal Care and Use Committee (protocol No. 100) and National Insitutes of Health guidelines concerning the responsible use of animals in research. The brains were immediately removed and rinsed in 0.1 M phosphate buffer (PB; pH 7.13) for 3 minutes. After the PB rinse, retrieved brains were immersed in a Golgi-Cox solution comprising potassium dichromate, mercuric chloride, and potassium chromate. This mixture of solutions was replaced once after 12 hours of initial immersion, with storage at room temperature in darkness for 2–3 weeks.
After the immersion period in the Golgi-Cox solution, the embedded brains were transferred to a cryoprotectant solution (FD Rapid Golgi Stain Kit) and stored at 4°C for at least 1 week in the dark before cutting. The brain slices were sectioned in the coronal plane at approximately 200–250 µm thickness on a freezing cryostat (approximately −25°C). To prevent ice crystal damage, tissues were rapidly frozen with dry ice and quickly embedded in optimal cutting temperature (OCT) medium. Sliced tissues were transferred onto triple-dipped gelatin slides and were coated with additional cryoprotectant solution. Cut sections were air dried at room temperature in the dark for at least 2–3 weeks before further processing. After drying, sections were rinsed with distilled water and were subsequently stained in a developing solution (FD Rapid Golgi Stain Kit) and dehydrated with 50%, 75%, 95%, and 100% ethanol. Finally, the sections were defatted in xylene substitute and coverslipped with either Permount (Fisher Scientific, Fair Lawn, NJ) or SHUR/Mount (Triangle Biomedical Sciences, Inc.).
Identification of barrel field and cortical layer VI
Barrel cortex location was identified by observing the characteristic clusters of cells that are typically found in granular and supragranular layers (see Fig. 1A) and by matching with a previously published atlas of Golgi-stained mice brain (Valverde, 1998). In the rostral-caudal axis, the barrel cortex was defined by the initial appearance of the anterior commissure. Golgi-impregnated neurons were located with respect to the layer VI boundaries. Subdivision of layer VI can also be observed, in which VIa borders the large pyramidal neurons of layer V (Fig. 1B) whereas VIb is immediately lateral and superior to the white matter (Fig. 1C). Layer VI neurons were obtained at all depths, providing an unbiased sampling of morphologies. Images were acquired from prepared slides using a ×10 air objective lens on a Nikon E4 microscope. These images were used to keep track of which neurons had been reconstructed.
Neuronal selection and reconstruction
Neurons were viewed with Neurolucida 7.5 software (MBF Bioscience, Inc.) and an Olympus B×51 microscope equipped with a high-resolution digital camera (Optronics Microfire), a mechanical stage (Ludl, Thornwood, NY), and an x-y-z axis encoder connected to a Windows Pentium 4 PC. Each neuron was scanned under high (×20, oil immersion, NA = 0.8) magnification by varying the depth of the Z plane, to ensure that all parts of the cell (especially dendrites) were intact. Dendrites that tapered to a point were assumed to be complete and uncut. Typically, the reconstructed neuronal somata were located at ~100 µm to the section surface, although a few exceptions exist. Only the neurons that exhibited complete Golgi impregnation with a limited amount of staining artifacts were traced.
Two hundred twenty-two neurons were selected from layer VI of the neocortex (animal 1: n = 50 neurons, animal 2: n = 52 neurons, animal 3: n = 48 neurons, animal 4: n = 21 neurons, animal 5: n = 17 neurons, animal 6: n = 21 neurons, animal 7: n = 13 neurons). 3D neuronal reconstruction took place under a ×60 objective (oil immersion, 1.4 NA) of an Olympus B×51 microscope.
Statistical methods of classifying neuronal groups
The software NeuroExplorer (MBF Bioscience, Inc.) was used to conduct various morphological measurements. Specific morphological characteristics including somatic shape and size, dendritic structure, and branching patterns were analyzed. For each reconstructed neuron, we measured the following 10 somatic and 25 dendritic components: 1) Somatic perimeter and 2) somatic area and all somatic measurements, referring to an XY projection because all somata were reconstructed as a 2D image at the plane of sharpest focus; the plane of the sharpest focus is typically calculated, for each neuron, at the midpoint of where the Z-plane first and last focuses for that particular neuron; 3) somatic feret maximum, referring to the longest diameter of the soma, and 4) somatic feret minimum, the longest diameter perpendicular to the feret maximum; 5) aspect ratio = (feret max)/(feret min); as aspect ratio approaches 1, it is indicative that the soma is closer to a symmetric shape (e.g., circle or square); 6) somatic compactness = [√(4/π) × area]/(feret max); numerical values of somatic compactness closer to 1 represent a more compact soma; 7) convexity = (convex contour)/(perimeter); this parameter is indicative of the somatic morphological profile; higher somatic convexity yields to more indentations, which translates to higher estimated surface area to cellular volume ratio; 8) somatic form factor = (4π × area)/(perimeter2); this value directly reflects the complexity of somatic perimeter; a higher numerical value represents a more complex somatic perimeter; 9) somatic roundness = (4 × area)/(π × feret max2), i.e., similar to compactness but making it easier to differentiate objects with small compactness values; 10) somatic solidity, the ratio of somata area as a whole over convex area, where values closer to 1 represent more solid (i.e. smooth, uniform) somata; 11) quantity of apical dendrites; 12) quantity of apical dendritic nodes (including bifurcating as well as trifurcating nodes); 13) quantity of apical dendritic endings; 14) total length of apical dendrite in micrometers; 15) mean length of apical dendrites in micrometers; this number is derived from taking total length of apical dendrites and divided by the number of apical dendrites; 16) total surface area of apical dendrites; 17) mean surface area of apical dendrites; 18) total volume; 19) mean volume of apical dendrites; 20) quantity of basilar dendrites; 21) quantity of basilar dendritic nodes; 22) quantity of basilar dendritic ends; 23) total length of basilar dendrites; 24) mean length of basilar dendrites; 25) total surface area of basilar dendrites; 26) mean surface area of basilar dendrites; 27) total volume of basilar dendrites; 28) mean volume of basilar dendrites; 29) quantity of total dendrites; this includes the summed number of apical dendrite and the basilar dendrites in a particular neuron; 30) quantity of total dendritic nodes; 31) quantity of total dendritic ends; 32) total dendritic length, i.e., regardless of basilar or apical, the summation of the total dendritic length; 33) mean dendritic length, i.e., regardless of basilar or apical, the mean of the overall dendritic length; 34) total dendritic surface area; and 35) mean of total dendritic surface area.
After the compilation of descriptive variables, a principal component analysis (PCA) was conducted to determine which morphological variables accounted for the greatest degree of variance within the traced cell sample. One concern is that the PCA is influenced more by variables with larger absolute numbers (e.g., the total dendritic length in micrometers) than smaller numbers (e.g., number of dendritic nodes). To address this concern, we normalized each variable across the reconstructed neuronal sample using Z-scores transformation. Another concern is the high correlations between two variables; hence, we would eliminate one of the two variables to avoid double-weighing on the cluster analysis. After the selection of significant principal components (with absolute loading values = 0.7), we conducted a cluster analysis using Ward’s method (Ward, 1963) and Euclidean distance, which, based on several Monte Carlo tests, is a highly reliable and valid method for population clustering (for review see Scheibler and Schneider, 1985). The neuronal groups and subgroups were identified in Statistica (StatSoft, Inc.) based on the factor loadings of specific measurements of somata and dendrites.
Postclustering validation
A number of methods were performed “post hoc” in order to support the validity and the reliability of our clustering scheme. Each methodology provides added validity from several different aspects such as location of soma with respect to cortical plate, dendritic fanning pattern as a function of distance away from the soma, and dendritic branching polarity across the clustered neuronal groups.
Systematic replication
In addition to our original N = 150 population of neurons, we reconstructed another separate population (N = 72) of neurons from an independent group of animals comprising four mice. The statistical methods and the morphological variables that were employed for this separate population of neurons were identical to what was described above. This systematic replication was conducted to support the reliability, validity, and consistency of our initial morphological classification scheme.
Laminar location of somata
Location of neuronal somata in relation to the cortical plate thickness supports the notion that the reconstructed neurons are indeed within layer VI. For each reconstructed neuron, laminar data are exemplified by the following measurements: the shortest distance from pia mater to soma (PIA-SOMA), the shortest distance from pia mater to the most distal dendritic process (PIA-DISTAL), and the shortest distance from the white matter to pia (WM-PIA), which bisects the soma of that particular neuron. From the obtained measurements, we calculated two metrics: the ratio of somatic location over cortical plate thickness (PIA-SOMA/WM-PIA) and the ratio of distal process over cortical plate thickness (PIA-DISTAL/WM-PIA).
Sholl analyses
Based on Sholl’s (1956) method, concentric spheres were placed around the center of each cell’s soma. These concentric spheres increased in preselected radii of 10 µm. Such analyses provide useful information regarding the number of dendritic intersections, nodes, endings, spines, and length of dendrites passing through the shells within each incrementing sphere. An ANOVA was performed for the number of dendritic intersections, spines, and nodes. These variables were withheld from the original PCA list and did not participate as the loading factors that determined the neuronal clustering.
Dendritic fanning polarity
From the center of each cell’s soma as the pivot point, 12 pie-shaped regions were created, with each region occupying a 30° angle. Arbitrarily setting pia mater as 0° and white matter as 180°, we analyzed the total dendritic length that falls under the particular jurisdictions of the wedged regions. Furthermore, we corrected the cerebral hemispheric (left vs. right) orientation by flipping the left hemispheric images 180°. We arbitrarily set the lateral/inferior direction as 90° and the proximal/superior direction as 270° (in terms of gross anatomy of the brain). We flattened the Z-axis and investigated only the XY plane. This was accomplished by selecting the “3D wedge analysis” function that is included in the Neurolucida 7.5 software package (NeuroExplorer version 4.0). In this way, elucidation of polarity of dendritic patterning was provided in the coronal plane.
Statistical analysis
We did not perform between-groups ANOVA across the 11 dimensions that were responsible for splitting the sampled neurons into morphological groups. This is due to the fact that Ward’s method using Euclidean distance is already an ANOVA-type approach in itself (see also Results). However, because the postclustering data were withheld from the original PCA matrix, it was appropriate to conduct between-groups ANOVA on these “post hoc” data. Similarly, it would also be statistically appropriate to conduct ANOVA-type comparison for the other branch structure parameters that were not selected by the PCA (and therefore were not responsible for splitting the neuronal sample into groups). All of the postclustering pair-wise testings are two-tailed independent t-tests, unless otherwise indicated. Statistical significance was set at P < 0.05.
Photomicrograph software and imaging protocol
All photomicrographs were imaged with Picture Frame 2.3 software (Optronics) that comes bundled with Neurolucida 7.5 (MBF Bioscience, Inc.). Photomicrographs were taken under a Microfire (Optronics) camera and processed digitally (color: RBG autowhite-balanced, exposure: 32 msec, contrast: 60, brightness: 50, gamma: 1.0). Acquired color images were imported into Photoshop 7.0 and transformed into a black-and-white photomicrographs.
RESULTS
Determination of principal components
Our Golgi material produced excellent resolution of dendritic arborization, dendritic spines, and somatic labeling, with minimal background staining artifacts (Fig. 1A–D). The somatic and dendritic data were derived from 150 reconstructed neurons from layer VI of the adult mouse barrel cortex. In total, 10 somatic and 25 dendritic variables (Table 1) were chosen as candidates for a PCA matrix that determined the parameter( s) that accounted for the greatest degree of variance within the reconstructed cell sample. Among these 35 parameters, total apical dendritic length and mean apical dendritic length significantly overlapped one another (r > 0.9). Other significant overlapping pairs included total apical dendritic surface area and mean apical dendritic surface area (r > 0.9) along with total apical dendritic volume and mean apical dendritic volume (r > 0.9). Mean apical dendritic length, mean apical dendritic surface area, and mean apical dendritic volume, therefore, were removed from the variable list that participated in the PCA. Eleven of the thirty-two variables (italicized in Table 1) had absolute loading values greater than or equal to 0.7, and thus were used to separate neurons into specific morphological groups with a subsequent cluster analysis.
TABLE 1.
Morphological Parameters of Neurons That Were Analyzed With Principal Component Analysis
| Somatic variables | Apical dendrite | Dendrites | Total dendrites |
|---|---|---|---|
| Perimeter (µm) | Quantity | Quantity | Quantity |
| Area (µm2) | Nodes | Nodes | Nodes |
| Feret max (µm) | Ends | Ends | Ends |
| Feret min (µm) | Length (µm) | Length (µm) | Length (µm) |
| Aspect ratio | Surface area (µm2) | Mean length (µm) | Mean length(µm) |
| Compactness | Accumulative volume (µm3) | Surface area (µm2) | Surface area (µm2) |
| Convexity | Mean surface area (µm2) | Mean surface area (µm2) | |
| Form factor | Accumulative volume (µm3) | ||
| Roundness | Mean volume (µm3) | ||
| Solidity |
Cluster analysis of morphological neuronal classifications
The selected 11 principal components were loaded into a cluster analysis matrix that maximizes between-cluster differences and minimizes within-cluster differences (Ward’s method, Euclidean distance). Figure 2 shows the results of a dendrogram, indicating that six distinct groups were categorized based on our objective criteria. Whereas each point on the X-axis is arbitrarily preselected and signifies a neuron, the Y-axis depicts the linkage distance across the 11 dimensions selected; i.e., neurons clustered within the same group share more similarity than neurons in other groups. Two of the strongest splitting factors, total dendritic length and quantity of apical dendrites, were primarily responsible for dividing the reconstructed neurons into six groups. Because of the sum of squares (SS) calculations that were utilized in the cluster analysis, it is inappropriate to use between-groups ANOVA or pairwise testing to contrast the groups for the 11 selected morphological factors (which were responsible for splitting the neurons into groups). Insofar as the cluster analysis itself is already a reversed ANOVA-type approach, performing an ANOVA and/or pairwise testing would be statistically circular.
Figure 2. The results of cluster analysis (Ward’s method, Euclidean distances) of 150 digitally reconstructed neurons.
Each point on the X-axis is arbitrary and represents one cell. The Y-axis depicts linkage distance, the degree of variance among neurons based on surveyed parameters. Six distinct groups and several subgroups are defined.
Group 1: pyramidal neurons with elaborate dendrites
Common features of group 1 (n = 16 in this group, 10.6% of the total reconstructed population)
Group 1 (Fig. 3) is composed of pyramidal neurons with elaborate dendrites. The most distinctive characteristics are the extremely long apical dendrites (679.5 µm ± 56.9; all data are reported as mean ± SEM), the numerous apical dendritic bi-/trifurcations (14.7 ± 2.6), and the extensive overall dendritic length (1,347.9 µm ± 81.9). Other common features of group 1 include extremely large total and apical dendritic surface area (5,000.9 µm2 ± 305.4, 2,827.2 µm2 ± 231.8, respectively). The apical dendrites tend to have a large number of branching points as well as secondary oblique dendrites that originate from the apical trunk. These neurons also exhibited the longest overall basilar dendritic processes among all pyramidal neuronal groups (group 1 = 668.4 µm ± 60.4, group 3 = 431.8 µm ± 22.9, group 4 = 232.6 ± 18.3). Group 1 could be further divided into two subgroups based on our objective morphological splitting criteria.
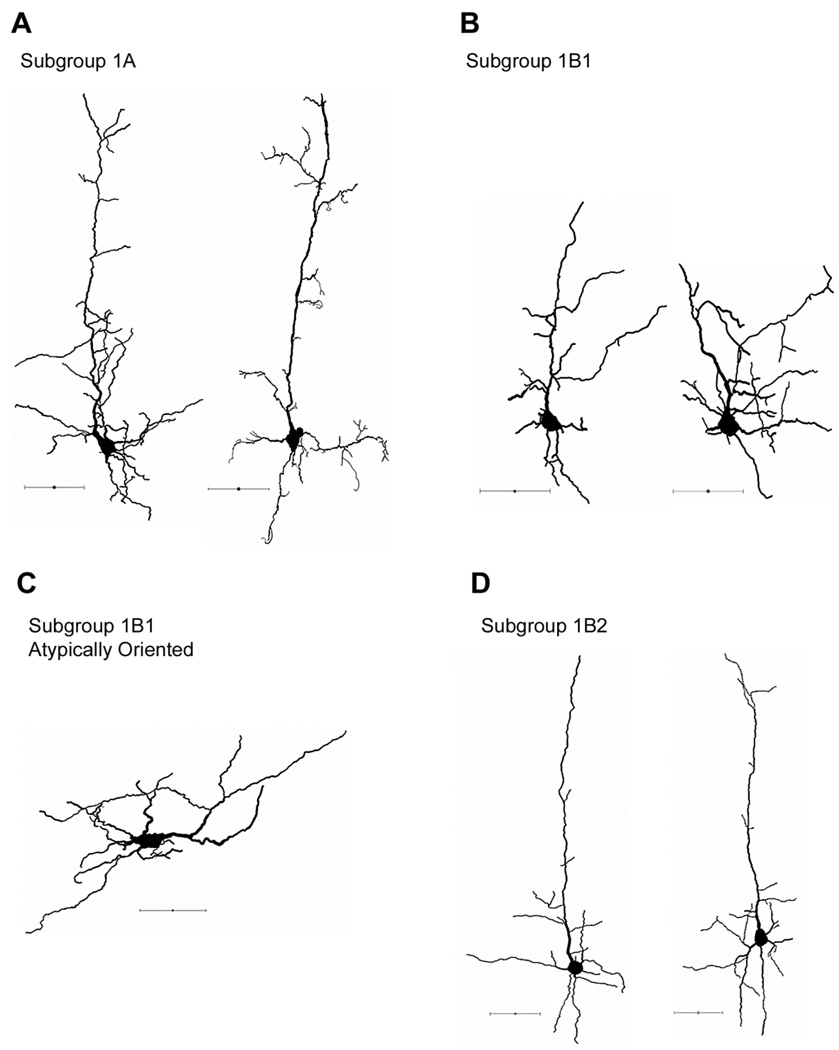
Figure 3. Group 1 representative cells. Pyramidal neurons with elaborate dendrites.
A: Examples of cells in subgroup 1A, characterized by an extensive and elaborate pattern of apical dendrites. B: Examples of cells in subgroup 1B1, characterized by slightly shorter but thicker apical tufts. C: Example of an atypically oriented pyramidal neuron. Note that the apical tuft is pointing horizontally. D: Examples of subgroup 1B2, characterized by an elaborate basilar skirt and a long and relatively simple apical dendrite. Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 1
Subgroups of group 1 include 1A (n = 4, representative neurons shown in Fig. 3A) and 1B (n = 12, Fig. 3B–D). Although morphological differences are minimal, cells in subgroup 1A possess relatively more apical dendritic nodes (26.3 ± 7.2 vs. 10.8 ± 1.4), basilar dendritic nodes (30.25 ± 7.2 vs. 8.3 ± 1.2), and total dendritic nodes (56.5 ± 6.1 vs. 19.2 ± 1.9) than cells in subgroup 1B. In addition, neurons in subgroup 1A also exhibit a higher number of apical (25.5 ± 6.9 vs. 11.9 ± 1.4), basilar (36.3 ± 6.8 vs. 15.3 ± 1.1), and total (61.8 ± 7.2 vs. 27.2 ± 2) dendritic endings than subgroup 1B.
Subgroup 1B can be further subdivided into 1B1 (n = 3, Fig. 3B) and 1B2 (n = 8, Fig. 3D). In terms of somatic components, subgroup 1B1 displays significantly fewer convexities and form factors than subgroup 1B2 (convexity 0.85 ± 0.03 vs. 0.96 ± 0.01, P < 0.05; form factor 0.53 ± 0.08 vs. 0.73 ± 0.05, P < 0.05), indicating that cells in 1B2 have smoother somatic edges yet more complex perimeters compared with cells in 1B1. With regard to dendritic components, neurons in subgroup 1B2 demonstrate notably higher numbers in total dendritic length, total dendritic mean length, and total dendritic mean surface area (1,367.3 µm ± 70.3 vs. 1,036.8 µm ± 87.1, 201.7 µm ± 17.7 vs. 111.7 µm ± 18.1, and 5,155 µm2 ± 345.1 vs. 4,278.5 µm2 ± 300.6, respectively). Among the 16 pyramidal neurons clustered in group 1, two neurons in 1B1 were observed to have an atypically oriented dendritic pattern (example in Fig. 3C).
Group 2: complex interneurons
Common features of group 2 (n = 8, 5.3%)
Figure 4 represents examples of neurons clustered in group 2. These neurons tend to have large circular/oval cell bodies (somatic area = 275.9 µm2 ± 48.5). Their most noticeable features are the high numbers of dendritic nodes and dendritic ends compared with the other five groups (dendritic nodes = 27.6 ± 4.2, dendritic ends = 33.3 ± 7.6). Among six distinct neuronal groups, group 2 neurons exhibited the highest dendritic surface area (4,357.2 µm2 ± 520.2) and dendritic volume (1,631.9 µm3 ± 309.7), suggesting the important roles that these neurons play in integrating signals within deep cortical layers. Our data indicated that this particular group of neurons displays highly complicated dendritic fanning patterns (see below).
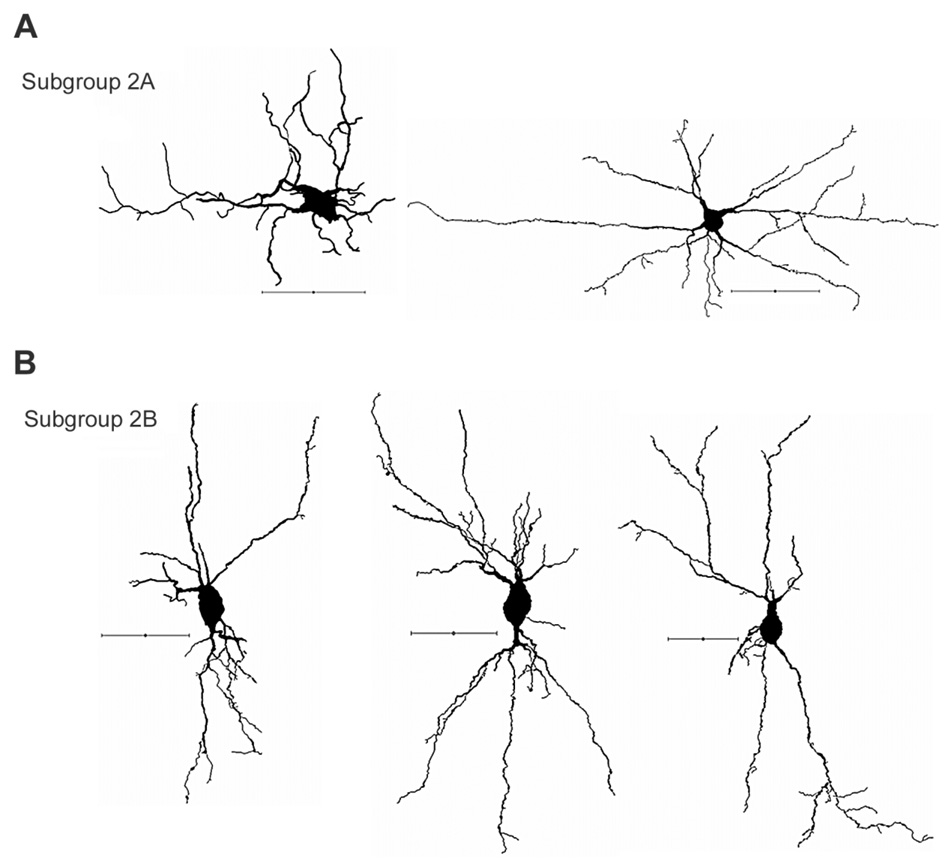
Figure 4. Group 2 representative cells. Nonpyramidal neurons with elaborate dendrites.
A: Examples of cells in subgroup 2A, characterized by elaborate and relatively spherical dendritic fanning patterns. B: Examples of cells in subgroup 2B, characterized by bipolar fanning patterns particularly toward the pia mater and the white matter. Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 2
Two subgroups can be further divided within this group: subgroups 2A (n = 3, Fig. 4A) and 2B (n = 5, Fig. 4B). Neurons in subgroup 2A are exemplified by their relatively circular/spherical dendritic fanning pattern, whereas neurons in subgroup 2B have more bitufted dendritic patterns throughout. It is therefore postulated that neurons in subgroup 2B may play crucial roles in integrating information between layer VIa and VIb. In comparing subgroups 2A and 2B, although there are negligible differences in somatic parameters (P > 0.15 for all), considerable disparities are observed in dendritic parameters. Neurons in subgroup 2A possess significantly higher dendritic surface area as well as dendritic volume compared with the neurons in subgroup 2B (dendritic surface area 6,007.3 µm2 ± 171.7 vs. 3,367.1 µm2 ± 308.8; dendritic volume 2,564.3 µm2 ± 368 vs. 1,072.5 µm2 ± 139.2).
Group 3: pyramidal neurons with moderate apical and basilar dendritic pattern
Common features of group 3 (n = 46, 30.6%)
Neurons in group 3 (Fig. 5) are characterized by relatively small somata (somatic perimeter = 48.83 µm ± 1.14, somatic area = 164 µm2 ± 7.7, feret max = 17.76 µm ± 0.52, feret min = 12.48 µm ± 0.27). Despite their small cell bodies, it was observed that several somatic parameters such as compactness (0.812 ± 0.01), convexity (0.98 ± 0.002), form factor (0.85 ± 0.01), and solidity (0.97 ± 0.003) are particularly high for neurons clustered in this group. The commonly shared dendritic features include similar range of apical dendritic parameters (apical dendritic nodes = 6.86 ± 0.4, apical dendritic length = 461.8 µm ± 19.5, apical dendritic surface area = 1,835.2 µm2 ± 93.3, apical dendritic volume = 750.9 µm3 ± 54.2). Similarity in ranges of basilar dendritic parameters was also demonstrated (basilar dendritic nodes = 7.77 ± 0.86, basilar dendritic total length = 431.8 µm ± 22.9, basilar dendritic surface area = 1,119.8 µm2 ± 61.3, basilar dendritic volume = 305.3 µm3 ± 22.8). Compared with group 1, neurons in group 3 possess considerably shorter and noticeably less complicated basilar dendrites. It was also observed that the apical dendritic complexities of group 3 cells are not as elaborate as neurons in group 1. However, the neurons in group 3 are still distinctly more elaborate than neurons clustered in group 4, which also possess apical dendrites. Furthermore, the overall morphological structures of group 3 neurons suggest that a subset of corticothalamic neurons may belong in this particular neuronal cluster when compared with previously conducted studies (Brumberg et al., 2003).
Figure 5. Group 3 representative cells. Neurons with moderate apical dendritic length and elaborate basilar dendritic fanning pattern.
A: Examples of cells in subgroup 3A1−α, characterized by long apical dendrites that typically terminate in upper layer V. B: Examples of cells in subgroup 3A1−β, characterized by medium-length apical dendrites that typically terminate in lower layer V. C: Examples of cells in subgroup 3A2; note the spherical soma compared with those of 3A1. D–F: Examples of cells in subgroup 3B, all with long apical dendrites that typically terminate in upper layer V (D) or lower layer IV (E,F). Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 3
Two main subgroups of neurons were observed: subgroups 3A (n = 24, Fig. 5A–C) and 3B (n = 22, Fig. 5D–F). Overall, neurons clustered in subgroup 3A have significantly larger somata than those in 3B, as indicated by several somatic variables (somatic perimeter = 58.7 µm ± 5.44 vs. 48.83 µm ± 3.13, P < 0.05; somatic area = 200.31 µm2 ± 24.57 vs. 164.04 µm2 ± 21.13, P < 0.05, feret max = 20.30 µm ± 1.78 vs. 17.76 µm ± 1.43, P < 0.05; feret min = 14.00 µm ± 1.10 vs. 12.48 µm ± 0.74, P < 0.05; two-tailed independent t-tests for all). In various indicators of somatic shape/contour, neurons in subgroup 3B displayed significantly larger convexity (0.98 ± 0.01 vs. 0.94 ± 0.02, P < 0.05), form factor (0.84 ± 0.01 vs. 0.75 ± 0.06, P < 0.05), and solidity (0.97 ± 0.01 vs. 0.94 ± 0.02, P < 0.05) than neurons in subgroup 3A. This type of pattern in somatic variants reflects considerable somatic disparities in both size and shape between subgroup 3A and 3B. For dendritic parameters, our data suggest that cells in subgroup 3A possess significantly larger basilar dendritic surface area as well as larger basilar dendritic volume than subgroup 3B (basilar dendritic surface area 1,812.61 µm2 ± 144.88 vs. 1,119.80 µm2 ± 168.36, basilar dendritic volume 612.22 µm3 ± 71.03 vs. 305.30 µm3 ± 62.49). It is therefore postulated that neurons in subgroup 3A may process more intralaminar information than 3B. However, neurons clustered in subgroup 3B exhibited more extensively patterned and longer apical dendrites than subgroup 3A (apical dendritic nodes = 6.86 ± 1.08 vs. 3.58 ± 0.87, apical dendritic ends = 7.82 ± 1.14 vs. 4.58 ± 0.83, apical dendritic length = 461.80 µm ± 53.48 vs. 264.95 µm ± 39.83, all significant at P < 0.05). Therefore, the results strongly indicated that the neurons in subgroup 3B may be important for interlaminar communication, whereas neurons in subgroup 3A are possibly important for intralaminar communication in the barrel cortex.
Subgroup 3A can be further subdivided into 3A1 (n = 15, Fig. 5A,B) and 3A2 (n = 9, Fig. 5C). Neurons in subgroup 3A1 have considerably more elongated cell bodies compared with those in 3A2, as indicated by elevated values in somatic components such as perimeter (63.89 µm ± 3.48 vs. 50.05 µm ± 3.34, P < 0.05), feret max (22.10 ± 1.12 vs. 17.28 ± 1.03, P < 0.05), and aspect ratio (1.59 ± 0.07 vs. 1.26 ± 0.05, P < 0.05). This elongation of somata in subgroup 3A1 is further validated by significantly decreased values in compactness (0.74 ± 0.01 vs. 0.86 ± 0.02), somatic roundness (0.55 ± 0.02 vs. 0.75 ± 0.04), and solidity (0.92 ± 0.01 vs. 0.98 µm ± 0.01) compared side by side with subgroup 3A2. Consequently, neurons in subgroup 3A2 possess relatively more spherical somata than those in 3A1. In general, subgroup 3A1 demonstrated significantly increased apical dendritic components than 3A2 when it comes to overall length and area (apical dendritic length = 314.96 µm ± 22.23 vs. 181.59 µm ± 21.58, apical dendritic surface area = 1,779.93 µm2 ± 103.54 vs. 1058.05 µm2 ± 128.07, apical dendritic volume = 947.51 µm3 ± 86.49 vs. 597.61 µm3 ± 93.50). With regard to basilar dendritic components, subgroup 3A2 exhibited far more dendritic nodes and longer processes than subgroup 3A1 extending into other cortical laminae (basilar dendritic node = 8.44 ± 1.75 vs. 4.53 ± 0.71, P < 0.05; basilar dendritic mean length = 111.3 µm ± 10.76 vs. 76.29 µm ± 8.39), hinting that the neurons in 3A2 may be more involved than 3A1 in roles such as intralaminar communication. Within subgroup 3A1, two subdivisions can be defined: 3A1−α (Fig. 5A) and 3A1−β (Fig. 5B). In comparing 3A1−α and 3A1−β in somatic parameters, 3A1−β exhibited higher values than 3A1−α in convexity (0.95 ± 0.02 vs. 0.88 ± 0.03, P < 0.05) and form factor (0.75 ± 0.04 vs. 0.59 ± 0.04, P < 0.05). Upon visual inspection, there seems to be a dramatic difference in apical dendritic parameters between 3A1−α and 3A1−β. Numerical calculations and analyses, however, revealed that there were no significant disparities in terms of apical dendritic parameters (P > 0.05 for all apical dendritic parameters). Some basilar dendritic variables of 3A1−β were significantly greater than in 3A1−α, specifically, basilar dendritic quantity (9.67 ± 0.99 vs. 5.00 ± 0.50, P < 0.05), basilar dendritic ends (13.67 ± 1.43 vs. 10.13 ± 0.72, P < 0.05), basilar dendritic length (591.82 µm ± 40.92 vs. 401.18 µm ± 27.47, P < 0.05), and basilar dendritic surface area (2,045.69 µm2 ± 99.32 vs. 1,556.20 µm2 ± 111.26).
Subgroup 3B gives rise to three smaller subclusters of neurons: 3B0 (n = 2, not shown), 3B1 (n = 13, Fig. 5D), and 3B2 (n = 7, Fig. 5E,F). Subgroup 3B0, comprising only two neurons, shared limited morphological characteristics with the rest of subgroup 3B. Therefore, this independent subgroup was separated from the rest of 3B, according to our objective clustering criteria. In comparing the somatic parameters, 3B1 displayed several significantly higher variables over 3B2, including somatic area (178.30 µm2 ± 15.56 vs. 130.89 µm2 ± 16.21, P < 0.05), feret maximum (19.18 µm ± 1.15 vs. 15.16 µm ± 1.01, P < 0.05), convexity (0.99 ± 0.002 vs. 0.96 ± 0.008, P < 0.05), and solidity (0.98 ± 0.003 vs. 0.95 ± 0.01, P < 0.05). It is therefore inferred that 3B1’s somata are generally larger and more elongated, with more complicated somatic perimeter shapes. In terms of apical dendritic variables, despite the fact that 3B1 exhibited significantly less branching than 3B2 (apical dendritic nodes = 5.83 ± 0.58 vs. 8.86 ± 1.37, P < 0.05), neurons in 3B1 still displayed significantly larger dendritic surface area and dendritic volume than 3B2 (apical dendritic surface area 1,911.35 µm2 ± 133.08 vs. 1,307.56 µm2 ± 167.02, P < 0.05; apical dendritic volume 839.50 µm3 ± 94.77 vs. 392.92 µm3 ± 50.33, P < 0.05). This suggests the possibility that neurons in 3B1 process significantly more interlaminar information than those in 3B2. Although there were no significant disparities between 3B1 and 3B2 in the quantity of basilar dendrites (P > 0.05), neurons of 3B1 showed several significant decreases in the number of basilar dendritic branching (4.91 ± 1.00 vs. 14.57 ± 2.14, P < 0.05), basilar dendritic endings (9.91 ± 1.19 vs. 19.71 ± 2.42, P < 0.05), total basilar dendritic length (372.42 µm ± 43.75 vs. 554.50 µm ± 51.40, P < 0.05), and average basilar dendritic length (72.75 µm ± 5.25 vs. 120.04 µm ± 14.24, P < 0.05). This suggests that neurons in 3B2 may play a significant role in integration of intralaminar information within neocortical layer VI.
Group 4: pyramidal neurons with short overall dendrites
Common features of group 4 (n = 40, 26.6%)
Similarly to group 3, neurons clustered in group 4 (Fig. 6) also exhibited small somata (somatic perimeter = 51.56 µm ± 2.45, somatic area ± 166.51 µm2 ± 12.90, feret max = 18.01 µm ± 0.83, feret min = 12.87 µm ± 0.65). Every neuron clustered in group 4 has an apical dendrite. A subset of neurons in group 4 has anatomical features that highly resemble the morphology of star-pyramidal neurons in layer IV. Despite the similarities in somatic variables, there are distinct differences in dendritic parameters between group 4 neurons and neurons in other groups. Among the neuronal groups that possess apical dendrites (groups 1, 3, and 4), group 4 neurons have the shortest and least complicated apical as well as basilar dendritic processes. This morphological uniqueness is exemplified by low values obtained in apical dendritic bi/trifurcations (3.33 ± 0.30), apical dendritic endings (4.40 ± 0.31), apical dendritic length (225.51 µm ± 12.79), apical dendritic surface area (903.61 µm2 ± 63.69), and apical dendritic volume (399.58 µm3 ± 45.38). Consequently, it is rare that the apical dendrites of group 4 neurons extend beyond layer VI; the few exceptions are described below. Neurons of group 4 also displayed a limited fanning pattern of basilar dendrites, as reflected by low numbers in basilar dendritic nodes (3.21 ± 0.33), basilar dendritic endings (8.19 ± 0.57), total basilar dendritic length (232.58 µm ± 18.25), mean basilar dendritic length (50.18 µm ± 4.48), total basilar dendritic surface area (652.56 µm2 ± 68.26), mean basilar dendritic surface area (131.39 µm2 ± 11.73), total basilar dendritic volume (193.90 µm3 ± 27.64), and mean basilar dendritic volume (37.52 µm3 ± 4.45). Because of the restricted branching pattern of dendritic processes, we postulate that group 4 pyramidal neurons manage information in a more spatially localized fashion compared with neurons in group 1 and/or group 3 and perhaps are the layer VI homolog of the layer IV spiny stellate cell.
Figure 6. Group 4 representative cells. Small pyramidal neurons with short apical and basilar dendrites.
A,B: Examples of cells in subgroup 4A, typically with apical dendrites terminating within layer VI. Note that subgroup 4A1 (A) exhibits more elaborative fanning pattern than subgroup 4A2 (B). C–E: Examples of cells in subgroup 4B, characterized by circular/oval somata and short total length of dendrites. Note that, although E has longer apical dendrites, the averaged total lengths of dendrites are comparable to C,D. F: Observed cells with atypical orientation. Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 4
Two main subgroups were identified within group 4, subgroups 4A (n = 17, Fig. 6A,B) and 4B (n = 23, Fig. 6C–E). Within somatic parameters, cells in subgroup 4A demonstrated considerably longer perimeter than cells in subgroup 4B (59.74 µm ± 3.18 vs. 45.42 µm ± 1.98, P < 0.05), with no significant disparity in somatic area (193.26 µm2 ± 21.24 vs. 146.44 µm2 ± 14.63, P > 0.05). Subgroup 4A neurons exhibited significantly longer feret maximum than subgroup 4B (20.67 µm ± 1.06 vs. 16.02 µm ± 0.66, P < 0.05). This discrepancy between the two subgroups was, however, not observed in feret minimum (14.06 µm ± 0.99 vs. 11.98 µm ± 0.64, P > 0.07). This type of data pattern suggests that the neurons in subgroup 4A have more elongated cell bodies than the neurons in subgroup 4B. A statistical significance between subgroup 4A and 4B in their aspect ratios (1.54 ± 0.07 vs. 1.36 ± 0.04, P < 0.05) further suggests that notable differences exist in their overall somatic shape, in terms of how elongated their cell bodies are. Various indicators revealed that the neurons in subgroup 4B have significantly more intricate cell bodies than those in subgroup 4A (compactness 0.833 ± 0.012 vs. 0.738 ± 0.019, P < 0.05; convexity 0.978 ± 0.005 vs. 0.924 ± 0.013, P < 0.05; form factor 0.854 ± 0.015 vs. 0.650 vs. 0.024, P < 0.05; solidity 0.971 ± 0.005 vs. 0.902 ± 0.009, P < 0.05). In comparing subgroups 4A and 4B with respect to apical dendritic parameters, no significant differences were found, with the exception of apical dendritic length differences, in which neurons clustered in 4B revealed considerably longer apical dendrites than neurons in 4A (249.75 µm ± 13.31 vs. 193.20 µm ± 18.92). Although there was no significant difference between 4A and 4B in their basilar dendritic length (240.76 µm ± 29.36 vs. 226.45 µm ± 23.10, P ± 0.65), there was a significant difference between 4A and 4B in their basilar dendritic quantity (6.61 ± 0.59 vs. 3.79 ± 0.30, P < 0.05). Consequently, neurons in subgroup 4B possess considerably longer mean basilar dendritic length than subgroup 4A (59.73 µm ± 6.27 vs. 37.44 µm ± 4.74, P < 0.05). Although there is no significant difference in mean basilar dendritic surface area or mean basilar dendritic volume (P ± 0.84 and 0.38, respectively), subgroup 4A exhibited dramatically larger basilar dendritic surface area (868.35 µm2 ± 129.64 vs. 490.71 µm2 ± 53.54) and total basilar dendritic volume (291.57 µm3 ± 53.70 vs. 120.64 µm3 ± 17.90) than subgroup 4B. It is inferred that neurons in subgroup 4B play crucial roles in interlaminar communication and that neurons in subgroup 4A are important for intralaminar information processing within layer VI.
Subgroup 4A is composed of two smaller clusters of neurons: 4A1 (Fig. 6A) and 4A2 (Fig. 6B). Surprisingly, when comparing the somata between 4A1 and 4A2, no significant differences were found across all 10 somatic variables or in any apical dendritic variables. The only exception was apical dendritic volume, in which neurons in 4A1 have significantly larger volume than 4A2 (590.37 µm3 ± 89.35 vs. 316.88 µm3 ± 65.12). Therefore, it is inferred that the neurons in 4A1 possess thicker apical dendrites than neurons in 4A2. Despite the similarities between 4A1 and 4A2 in both somatic and apical dendritic variables, basilar dendritic variables between 4A1 and 4A2 were distinctively different. With the exception of basilar dendritic quantity, which yielded no significant difference (P > 0.05), cortical neurons in 4A1 exhibited greater values in all basilar dendritic parameters comparated with 4A2 (P < 0.05 for all). The data suggest that, although basilar dendritic quantity may be similar, subgroup 4A1 has significantly broader spatial processing capacity than 4A2 when it comes to intralaminar cortical communication.
Subgroup 4B gives rise to three smaller clusters: 4B1 (Fig. 6C), 4B2 (Fig. 6D), and 4B3 (Fig. 6E). Subgroup 4B is populated primarily by neurons in subgroup 4B1 and 4B2. In comparing subgroup 4B1 and 4B2, there were no distinct differences in somatic morphology (P > 0.05 for all), except for somatic solidity (indicator of unitary, smooth somata), in which subgroup 4B2 displayed significantly smaller average values than 4B1 (0.960 ± 0.007 vs. 0.981 ± 0.002, P < 0.01). Although there were no significant morphological differences in apical dendritic quantity, apical dendritic nodes, apical dendritic endings, or apical dendritic length (P < 0.05 for all), there were considerable morphological differences in apical dendritic surface area (727.67 µm2 ± 68.87 vs. 1,120.38 µm2 ± 154.44, P < 0.05) and apical dendritic volume (252.67 µm3 ± 36.88 vs. 532.81 µm3 ± 141.68, P < 0.05), in which 4B1 displayed lower numbers than 4B2 for both variables. This pattern of data indicates that neurons in subgroup 4B2 possess significantly thicker apical dendrites than neurons in 4B1. Among the basilar dendritic parameters, the data suggest that neurons in subgroup 4B2 have significantly broader spatial processing capacity than those in subgroup 4B1, exemplified by significantly higher numbers in variables such as total basilar dendritic surface area (642.55 µm2 ± 81.82 vs. 362.55 µm2 ± 48.77, P < 0.05), mean basilar dendritic surface area (156.58 µm2 ± 18.51 vs. 90.87 µm2 ± 13.16, P < 0.05), total basilar dendritic volume (175.93 µm3 ± 22.90 vs. 66.82 µm3 ± 12.01, P < 0.05), and mean basilar dendritic volume (42.63 µm3 ± 4.95 vs. 17.02 µm3 ± 3.35, P < 0.05). Because subgroup 4B3 has only two neurons, quantitative analyses were not performed in comparison with 4B1 and/or 4B2.
Atypically oriented neurons in group 4
Compared with group 1 and group 3 neurons, a high proportion of group 4 neocortical neurons demonstrated an atypical orientation (Fig. 6F), in which the apical dendrites are pointing horizontally or toward the white matter. Among the 40 observed neurons in group 4, 25% (n =10) exhibited atypical orientation, whereas only 12.5% of group 1 (2 of 16) and 2.2% of group 3 (1 of 46) neurons exhibited atypical orientation.
Group 5: large nonpyramidal neurons
Common features of group 5 (n =28, 18.6%)
Figure 7 depicts representative cells from group 5. Among the six neuronal groups defined by the present cluster analysis, these neurons have the largest cell bodies, indicated by the highest value obtained in somatic variables such as somatic perimeter (76.43 µm ± 2.94), somatic area (317.48 µm2 ± 19.56), feret maximum (28.27 µm ± 1.35), and feret minimum (16.35 µm ± 0.54). These neurons do not possess apical dendrites and have limited dendritic node to quantity ratio (0.76), which indicates that the amount of bifurcations is rather restricted. However, despite the paucity of dendritic nodes, group 5 neurons still exhibited the second highest dendritic surface area (2,661.01 µm2 ± 155.30) as well as dendritic volume (1,031.63 µm3 ± 70.90), trailing only group 2. These data suggest that the dendrites of group 5 neurons are quite thick in diameter.
Figure 7. Group 5 representative cells. Large interneurons.
A: Examples of cells in subgroup 5A1, characterized by circular/oval somata and moderate dendritic fanning pattern. B: Representative cells in subgroup 5A2; note that the dendritic pattern is simpler compared with A. C: Examples of cells in subgroup 5B, characterized by a dendritic fanning pattern typically oriented along the contours of white matter. Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 5
Group 5 can be further divided into two subgroups, 5A (n = 17, Fig. 7A,B) and 5B (n = 11, Fig. 7C). On visual inspection, the morphology of subgroup 5A resembles that of neurons that have been previously described as stellate neurons, whereas subgroup 5B strongly resembles horizontal (bipolar) neurons (Furtak, 2007). Therefore, it was no surprise that somatic comparison revealed many significant differences, in which subgroup 5A exhibited significantly smaller and less elongated somata than subgroup 5B (somatic perimeter = 67.86 µm ± 2.42 vs. 89.45 µm ± 4.45, P < 0.05; feret max = 33.75 µm ± 2.38 vs. 24.64 µm ± 0.96 vs. 33.75 µm ± 2.38). Somatic data further suggested that these two subclusters of group 5 exhibit differences in somatic shape and somatic surface complexity, reflected by significant differences in compactness (0.800 ± 0.019 vs. 0.614 ± 0.023), convexity (0.969 ± 0.009 vs. 0.900 ± 0.025), form factor (0.810 ± 0.029 vs. 0.521 ± 0.039), roundness (0.629 ± 0.029 vs. 0.382 ± 0.029), and solidity (0.960 ± 0.013 vs. 0.865 ± 0.030). In terms of dendrites, data indicated that, although bipolar neurons on average possess significantly more dendrites than stellate neurons (dendritic quantity 9.18 ± 0.736 vs. 6.44 ± 0.483, P < 0.05), stellate neurons have significantly longer as well as thicker dendrites (dendritic mean length 100.99 µm ± 5.19 vs. 73.75 ±m ± 9.79; P < 0.05; dendritic mean surface area 425.88 µm2 ± 24.64 vs. 318.11 µm2 ± 35.40, P < 0.05).
Subgroup 5A gives rise to two smaller subclusters: subgroups 5A1 (Fig. 7A) and 5A2 (Fig. 7B). As previously mentioned, visual examination suggests that both of these two subclusters are stellate neurons compared with the results of prior studies. Quantitative comparison further supported the idea that there are no significant somatic differences between subgroups 5A1 and 5A2 (P > 0.25 for all somatic variables). Dendritic comparisons, however, revealed that there are some subtle differences between these subclusters. Specifically, neurons in subgroup 5A1 displayed a higher number of dendrites (7.75 ± 0.49 vs. 5.13 ± 0.52, P < 0.05), longer dendrites (832.88 µm ± 35.62 vs. 461.15 µm ± 47.06, P < 0.05), and more dendritic surface area (3,334.03 µm2 ± 185.86 vs. 1989.72 µm2 ± 142.34) and dendritic volume (1,204.45 µm3 ± 144.86 vs. 814.53 µm3 ± 68.80) 5A2. Neurons in 5A1 have more dendritic processes as well as longer dendrites than those in 5A2, suggesting that neurons in 5A1 have larger spatial processing capacity in cortical layer VI than neurons in 5A2.
Group 6: simple neurons without apical dendrites
Common characteristics of group 6 (n = 12, 8%)
Representative cells from group 6 are presented in Figure 8. This particular group contained neurons with relatively simple dendritic organization. Collectively, neurons clustered in this group have somata that are medium in size (perimeter = 63.73 µm ± 3.61, somatic area = 247.19 µm2 ± 25.45), symmetrical (aspect ratio = 1.50 ± 0.074), and very uniform (0.938 ± 0.017). Although overall dendritic quantity is comparable to that of other neuronal groups (6.08 ± 0.81), neurons in group 6 possess surprisingly few dendritic nodes (2.58 ± 0.379) as well as the shortest dendrites on average (mean dendritic length = 42.08 µm ± 4.70). Despite exhibiting the least complex dendritic fanning pattern and the shortest dendrites, group 6 neurons still showed relatively high mean dendritic surface area (183.72 µm2 ± 16.93) as well as mean dendritic volume (76.56 µm3 ± 8.52), suggesting that the dendritic diameters of these neurons are quite thick on average.
Figure 8. Group 6 representative cells. Small interneurons. Cells in this group are characterized by small somata and absence of an apical dendrite.
A,B: Examples of cells in subgroups 6A and 6B. Note that cells in A have more circular somata compared with cells in B. C: Cells in subgroup 6C are suspected to be incompletely stained and/or incompletely reconstructed neurons. Only a small (three of 150) portion of neurons belongs in this category. Cells are oriented so that the pia mater is at the top and medial is to the left. Scale bars = 50 µm.
Subgroups of group 6
Several subgroups are observed: 6A (n = 3, Fig. 8A), 6B (n = 6, Fig. 8B), and 6C (n = 3, Fig. 8C). Quantitatively, the three subgroups exhibited very few significant differences in somatic variables except for somatic complexity [between-groups ANOVA: F(2,9) = 4.66, P < 0.05]. A post hoc test using Fisher LSD criterion revealed that the average somatic complexity was significantly lower in subgroup 6B than in subgroup 6C (P < 0.05). Statistical significance was also found for form factor [F(2,9) ± 15.32, P < 0.05], in which Fisher LSD post hoc suggested that subgroup 6B exhibited lower values than both subgroup 6A and subgroup 6C (P < 0.05 for both pairs). Subgroups 6A and 6C, however, did not significantly differ from one another in form factor. In addition, subgroup 6B exhibited lower solidity than subgroups 6A and 6C [F(2,9) ± 6.70, P < 0.05; Fisher LSD post hoc, P < 0.05]. With regard to dendritic components, there were no statistical significances across all three subgroups. To summarize, we have defined six neuronal groups using quantitative and objective data in barrel cortex layer VI.
Results of postclustering analyses
In an attempt to provide additional support for our results, we held back a number of variables from the PCA/cluster analyses. We then performed conventional analyses of variance on these variables using the groups determined by the cluster analysis as the categorical predictor. Our intent was to use these analyses as “post hoc” confirmations of our classification scheme, because it is not possible to assign significance to the results of a cluster analysis.
Systematic replication
To determine the reliability and repeatability of our methodology, we reconstructed an additional 72 neurons and quantified their morphologies exactly as described above. The clustering results that we obtained from our systematic replication were similar to the results of our original clustering. This is especially evident in the similarities in the classification dendrogram (compare Supp. Info. Fig. 1A with Fig. 2). Our clustering algorithm once again was able to discriminate six morphological classes, and, when we analyzed the distribution of the neurons within these six groups, there was no difference from our original data set [χ2 test of independence, χ2 (5, N = 222) = 0.805, P = 0.977]. In addition, the morphological characteristics of this replicated population were also strikingly similar, if not almost identical, to those of our original clustering (compare Supp. Info. Fig. 1B–G with Fig. 3–Fig.8; see also Supp. Info. Table 1). Taken together, the similarities of results between our original and replicated neuronal populations strongly suggested that our clustering scheme is a reliable and valid way of objectively categorizing neurons based on morphological quantifications.
Laminar location
It has been previously demonstrated that neurons in layer VIa differ from those in the VIb (Ferrer et al. 1986), which suggested the possibility of different roles in cortical processing. Furthermore, converging data have demonstrated that layers VIa and VIb developed with different embryological timelines, in which VIb develops at approximately the same time as the subplate and is earlier than VIa (Valverde et al., 1989; Del Río et al., 2000; Arimatsu et al., 2003). This diversity between VIa and VIb has been further supported by recent studies showing that neocortex VIa and VIb express genetically different mRNA (Heuer et al., 2003; Watakabe et al., 2007). By measuring the ratio of somatic location over the cortical width, we intend to demonstrate that the overall morphological characteristics of cortical neurons are strongly associated with their roles in information processing.
To determine whether traced neurons are located in layer VI, the width of the cortical plate was measured at the midpoint of the reconstructed soma from the pia to the white matter (Fig. 9A). For each data set, the distance is expressed as a percentage, in which the total distance (from pia to white matter) for that particular data set is 100%. Therefore, we calculated the relative location of the reconstructed neuron. This was done to account for the shrinkage resulting from tissue processing. Neuronal group 6 as well as group 4 displayed the closest relative distance to white matter, followed by group 5 and 2. Groups 1 and 3, composed predominantly of excitatory pyramidal neurons, had the closest relative distance to the pia mater. Figure 9B shows that the different neuronal groups, although overlapping, indeed have their somata located at different cortical depths [Kruskal-Wallis, H(5, N = 150) ± 14.09, P < 0.05]. Additionally, the most proximal dendritic process in relation to the pia mater was also measured. These data sets are also expressed in percentage form, in which relative location of the most distal dendritic process was obtained and analyzed. Figure 9C depicts the trends of the relative distance from distal dendritic process to the white matter. The present data indicate that group 3 has the closest distal processes to pia, whereas groups 5 and 6 have the farthest. Statistical analyses revealed that each neuronal group indeed demonstrated significant differences in terms of distal dendritic location [Kruskal-Wallis: H(5, N = 150) ± 38.18, P < 0.001] relative to cortical plate width. In summary, these analyses provided additional support for our classification methodology, insofar as these relative distance data were withheld from the initial PCA/cluster analysis, and statistical significance was still found when postclustering analyses were performed between the neuronal groups.
Figure 9. Laminar location.
A: Schematics of laminar location. Each component (cortical plate distance, distance from distal process to white matter, and distance from cell body to pia) was obtained for all neurons. B: Relative distances (%) from soma to pia. C: Relative distances (%) from distal process to white matter. Center white lines represent numerical means, whereas center black lines represent medians.
Sholl analyses
Sholl analyses were utilized to determine the complexity of the traced dendrites. The number of intersections, dendritic spines, and dendritic nodes in 10-µm increments away from the soma were calculated as described in Materials and Methods. Figure 10A illustrates how the measurements were made.
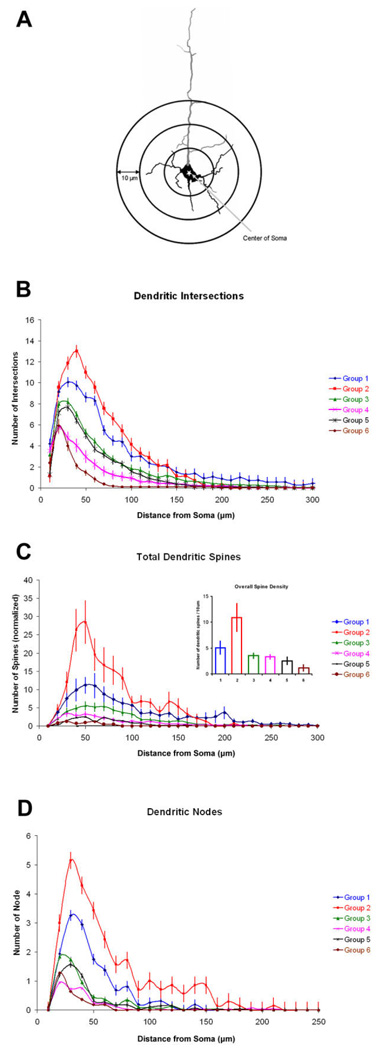
Figure 10. Sholl analyses.
A: Schematics of Sholl analysis. Each increment of concentric sphere radii is 10 µm. Total dendritic intersections, total dendritic spines, and total dendritic nodes were measured and analyzed. B: Number of dendritic intersections as a function of distance away from the soma (in micrometers). C: The number of spines as a function of distance from the soma (in micrometers) for each neuronal group. Inset: Overall density of dendritic spines per 10 µm of total dendritic length. D: Number of dendritic nodes as a function of distance from the soma (in micrometers) for each neuronal group. For all panels, error bars show SEM.
Intersections
Figure 10B depicts the number of intersections with the Sholl annuli for each group of their basilar dendrites. Not all groups have definable apical dendrites, so we confined our between-group analyses to nonapical dendrites. The peak number of intersections for all groups is approximately at 30–50 µm away from the soma. In general, group 2 neurons contained significantly more intersections at every distance point, followed by groups 1, 3, 5, 4, and 6. Statistical results (Kruskal-Wallis) indicated that the number of dendritic intersections significantly varied across neuronal groups (P < 0.001), and there is also a significant effect of the distance from the soma (one-way repeated ANOVA, P < 0.001). These differences across neuronal groups were no longer significant at distance points greater than 180 µm from the soma, most likely because of the small numbers of basilar dendrites exceeding this length.
Spines
Dendritic spines are the principal sites of excitatory inputs onto cortical pyramidal cells (White, 1989). During reconstruction of the neurons, the location of the spines was noted, and Figure 10C shows the number of total dendritic (i.e., apical + basilar) spines for each group. Overall, the number of observable dendritic spines peaked at approximately 30–50 µm away from the soma, with group 2 neurons exhibiting the highest number of spines at every distance compared with other groups. Statistical results (Kruskal-Wallis) indicated that the number of dendritic spines significantly varied across neuronal groups (P < 0.001), and a main effect of distance away from the soma was also found (one-way repeated ANOVA, P < 0.001). These differences between groups are no longer significant past the 250 µm mark from the soma, mainly because of the small number of dendrites present. In addition, there is a significant difference in the overall, averaged dendritic spine density across the clustered six neuronal groups [Kruskal Wallis, H(5, N = 120) = 26.13, P < 0.01]. We corrected the density of spines by taking the accumulated dendritic length into account. Figure 10C (inset) shows that neuronal group 2 exhibited the highest average density of dendritic spines, followed by groups 1, 3, and 4. Group 5, and particularly group 6, displayed very few spines overall.
Nodes
Dendritic nodes are indicators of the complexity of neuronal morphologies (Uyling et al., 1986; Libersat and Duch, 2004). To investigate the variability of dendritic morphology across neuronal groups, we examined the number of nodes as a function of distance from the center of somata. A significant statistical finding suggested that distinct variability of dendritic nodes indeed exists across groups (Kruskal-Wallis, P < 0.01). Although there is a significant effect of distance on the number of dendritic nodes displayed (one-way repeated ANOVA, P > 0.01), very much like dendritic intersections and spines, the differences between groups are no longer significant past the 170 µm mark from the soma.
In summary, our Sholl analyses sampled neurons in parameters of dendritic intersections, spines, and nodes. The obtained data were consistent, in that group 2 neurons exhibited the highest values in all surveyed dimensions, followed by groups 1 and 3. The vast number of statistical significances found across multiple zones of concentric spheres as a function of distance away from the somata have further supported the idea that our clustering methodology is an effective method for objectively determining neuronal classes.
Dendritic fanning polarity
With the center of soma as the pivot point, 12 30° pie-shaped regions were created for each neuron. We arbitrarily selected the direction of pia to be 0°, and the direction of inferior/lateral to be 90°. The directions toward the white matter and the superior/medial path would be 180° and 270°, respectively (Fig. 11A). Results of polarity analyses revealed that each neuronal group has a unique dendritic fanning pattern (Fig. 11B–G). For this part of the analysis, atypically oriented pyramidal neurons were excluded in order to preserve the internal consistency and the reliability of the data, especially with regard to the projecting orientation of the apical dendrites.
Figure 11. Polar plots of dendritic fanning pattern.
A: Schematics of polarity; 0° represents the direction toward the pial surface, whereas 90° and 270° represent the directions toward lateral/inferior side and medial/superior side, respectively. B–G: Polar plots of dendritic pattern for each neuronal group. Theta axes depict length of dendrites (in micrometers), whereas the radian depicts degree in angles (°) of the fanning pattern. Solid lines represent length of basilar dendrites, whereas dotted lines represent length of apical dendrites. Atypically oriented neurons were excluded.
Group 1 (Fig. 11B)
It is clear that this particular group of neurons exhibited extremely long apical dendrites (shown by dotted line in Fig. 11). With the atypically oriented pyramidal neurons (n = 2) excluded, results of repeated-measure ANOVA followed by Tukey’s post hoc criterion revealed that the apical dendrites of this neuronal group have a strong tendency to point toward the pia (0°) [F(11,143) = 39.50, P < 0.001; post hoc P < 0.01]. Surprisingly, results of the basilar dendritic analysis (shown by solid line) also reached statistical significance [F(11,143) = 2.10, P < 0.05]. The findings with Tukey’s post hoc criterion suggested that these basilar dendrites tend to avoid the 30° and 330° wedges, which are the regions immediately adjacent to 0°, and have a strong tendency to spread in the 180° direction (P < 0.05).
Group 2 (Fig. 11C)
This group of neurons displayed a relatively spherical dendritic fanning pattern, as suggested by nonsignificant results of ANOVA [F(11,77) = 1.19, P = 0.31]. However, because of the small sample of neurons clustered in this particular group (n = 8), a more powerful pairwise test was utilized to compensate for the lack of power. Indeed, results of the pairwise Hayter-Fisher LSD suggest that the dendrites of this neuronal group tend to point toward the white matter (180°) and avoid horizontal projection (evident by small values obtained at 60° and 300°).
Group 3 (Fig. 11D)
Morphologically, this group of cells exhibited morphometric characteristics that are consistent with classical pyramidal neurons. With the atypically oriented pyramidal neuron (n = 1) excluded, statistical analyses revealed that there is a tendency for the apical dendrites to project toward the pia (0°) as well as a slight predilection for the 30° lateral/inferior direction [F(11,473) = 133.49, P < 0.001; Tukey post hoc criteria P < 0.05]. The differences in basilar dendritic fanning as a function of geographic polarity are also significant [F(11,473 = 8.65), P < 0.01]. Results of post hoc analyses using Tukey criterion showed that group 3 neurons exhibit high basilar dendritic fanning in 60° to 240° from pia mater and tend to avoid the 0°, 30°, and 270° regions that the apical dendrites usually occupy.
Group 4 (Fig. 11E)
As previously mentioned, this group exhibited a high percentage (25%) of atypically oriented neurons. By excluding the atypically oriented pyramidal neurons (n = 10), it is clear that the apical dendrites of this group have a strong predilection for the pia mater (0°) [F(11,275) = 104.73, P < 0.01; Tukey post hoc P < 0.05], with no fanning preference for other polar-wedge regions. Their basilar dendrites are inclined to spread 210° and 240° from pia mater and shun the direction of the pia [F(11,27)5 = 4.93, P < 0.01; Tukey post hoc P < 0.05]. In converse, statistical analyses of the atypically oriented pyramidal neurons revealed that the apical dendrites of these neurons have a strong predilection to project toward the 270° direction (medial and superior), with no preference for any other direction [repeated-measure ANOVA, F(11,99) = 2.96, P < 0.01; post hoc criterion Fisher LSD P < 0.01]. However, there was no clear indication of the preferred projecting direction for these atypically oriented neurons when it comes to the basilar dendritic fanning pattern [F(11,99) = 0.88, P > 0.55].
Group 5 (Fig. 11F)
For this group, there is a significant correlation between dendritic fanning pattern and polarity region [F(11,286) = 2.87, P < 0.01]. Post hoc criterion using Fisher LSD indicated that, overall, dendrites in this group tend to project in the directions toward 0°, 150°, and 330° (P < 0.05) and to avoid directions toward 30°, 90°, 120°, 180°, 210°, 240°, and 270° (P < 0.05).
Group 6 (Fig. 11G)
Neurons clustered in this particular group exhibited a weak predilection for dendritic projection toward the pia at 0° [F(11,121) = 3.64, P < 0.01; Fisher LSD post hoc P < 0.05]. Furthermore, there are no predilections for other polarity regions displayed by group 6 neurons.
The results of this postclustering analysis suggest that each distinct neuronal group displayed its own unique dendritic fanning polarity, indicating that perhaps each neuronal group is sampling different inputs. Moreover, they reflect the difference in spatial capacity across classified neuronal group. For example, it can be inferred that group 1 neurons have much better interlaminar processing capacity than group 4 neurons, because of the dramatically higher dendritic length in the 0° direction. The results also support the validity of our initial morphological classification.
DISCUSSION
The goal of the current research is to understand the cellular components of layer VI of the mouse barrel field. We reconstructed 150 Golgi-impregnated neurons and analyzed 35 morphological variables that included somatic and dendritic factors. By using a PCA followed by a cluster analysis (Ward’s method, Euclidean distance), we were able to employ strict and objective morphological criteria to cluster the neurons effectively into six distinct groups.
The reconstructed neurons in the present study fall into two major categories: neurons with and without apical dendrites. Specifically, the neurons clustered in groups 1, 3, and 4 exhibited apical dendritic features, whereas groups 2, 5, and 6 did not. Group 1 neurons are pyramidal neurons with elaborate dendrites, characterized by extensively long overall dendritic length. Group 2 includes complex multipolar and bipolar neurons, which are presumed to be interneurons. These neurons possessed an intricate dendritic fanning pattern and thick dendrites. Group 3 consists of pyramidal neurons with apical dendrites that typically terminate in lower layer IV or upper layer V. Group 4 is composed of simple, small pyramidal neurons that have limited apical and basilar dendritic processes. In addition, a high proportion of group 4 neurons displayed atypical fanning orientation. Group 5 nonpyramidal neurons, based on their somatic properties and lack of apical dendrites, included stellate, bipolar, and horizontal cells. Group 6 contained small neurons with limited dendritic processes. A subgroup of group 6, despite possessing triangular somata, did not present with apical dendritic features. Collectively, the results indicate that a variety of neuronal morphological classes can be quantitatively defined within this heterogeneous layer of the mouse barrel field.
The postclustering analyses strongly support the validity of our initial choice of statistical methods (PCA, followed by a cluster analysis) and therefore suggest that neuronal populations can be quantitatively analyzed and classified with this methodology. Nonparametric statistical procedures (e.g., Kruskal-Wallis) were the tests of choice because of the highly unequal number of cells clustered across neuronal groups. Homogeneity of the relative somata locations indicates that most likely all of the reconstructed neurons were within layer VI. Moreover, cortical plate analyses reveal that the distal dendritic processes of each neuronal group varied significantly, which further supported the validity of our clustering method. The data derived from our Sholl analyses indicate that the neuronal groups differ considerably in all surveyed parameters of dendritic intersections, spines, and nodes. These Sholl results further support the trends of significance across the categorized neuronal groups, suggesting that each neuronal group exhibits its own unique dendritic pattern as the distance from the somata varies. Additionally, analyses of the dendritic polarity also reveal remarkable differences in dendritic fanning pattern across neuronal groups. In summary, these postclustering data provide support for the validity of our initial classifying methodology. Previous studies exploring the neuronal morphologies have also used similar classification techniques (Sultan and Bower, 1998; Tsiola et al., 2003). The finding that each group had distinctly different dendritic architectures and spine densities suggests that they might have different roles in local cortical circuits.
Methodological considerations
In the present study, we treated each case (neuron) independently, and we opted not to use the general linear model that is classically used in regression analyses, where the predictor is “known,” which does not apply to our data set, where we were seeking to determine the number of anatomical groups that existed within layer VI of the mouse barrel cortex. By contrast, cluster analysis is designed to assist in the exploratory determination of what groupings exist within a data set without making any prior assumptions. We utilized a PCA as both a data reduction tool and a mechanism for determining what anatomical variables are the most salient in discriminating morphological classes (Cattell, 1966; Jolliffee, 1972). These statistical methods are not novel and have been previously used (Scheibler and Schneider, 1985; Sultan and Bower, 1998; Tsiola et al., 2003). We have extended this technique by using post hoc validation of the resultant groupings. In the present study, we conducted post hoc validation of our clustering results with variables (location within the cortical plate, Sholl analysis, dendritic fanning pattern) that were not put into either the PCA or clustering algorithms, followed by an ANOVA with the groups as the categorical predictor.
Recent studies have demonstrated that the intensity of Golgi impregnation varies as a function of the developmental age of the animals (Furtak et al., 2007). Specifically, the intensity of impregnation increases as the age of animal increases, such that, by PND15, silver chromate impregnation appears to be complete. Since we chose to use adult mice with ages of ~PND90, the Golgi impregnation method should completely fill all, or at least nearly all, of the neurons that were reconstructed. Moreover, although they were rare during our neuronal reconstruction phase, when we encountered a neuron that was suspected of being incompletely filled, we discarded that particular neuron from the reconstruction. Therefore, it is very likely that our reconstructed neurons are an extremely accurate reflection of the cellular components in layer VI.
To avoid tracing error, during the reconstruction phase we avoided neurons that clumped together with a high proportion of staining artifacts. It is possible that these neurons exhibited yet another morphological type compared with the neurons presented in the current study. However, technical problems arise from tracing neurons with adjoined cell bodies, because their dendrites are located very closely together, making it difficult to distinguish which dendrite belongs to which neuron. In terms of validity of the sampling, prior studies have suggested that the Golgi staining method has a slight predilection for labeling neurons with larger somata (Pasternak and Woolsey, 1975). We reconstructed every possible neuron that exhibited complete staining, with the exception of neurons with cut dendrites and neurons that were clumped together. It is important to note that we observed a wide range of somata diameters (small to large) and that, because we did not attempt to predict the relative frequency of the different groups, the Golgi method is well suited for our study. Therefore, our “combing” method of neuronal sampling is most likely an extremely effective and accurate presentation of all Golgi-labeled neurons in mouse barrel field layer VI. In conclusion, our accurate tracing of the completely filled neurons and our relatively unbiased sample are very likely a correct reflection of the morphologically diverse layer VI.
Many prior studies interested in neuronal classification have relied on qualitative and subjective criteria for categorizing neuronal groups (Lorente de Nό, 1922; Ferrer et al., 1986; Prieto and Winer, 1999; Furtak et al., 2007). By contrast, our study uses quantitative measurements as the criteria for categorizing neurons based on their morphologies. Other prior studies, although limited in number, have also used PCA and cluster analyses to classify neurons quantitatively (Sultan and Bower, 1998; Tsiola et al., 2003). We followed the progression of these studies’ objective and unbiased methodology of characterizing neuronal groups and at the same time performed additional confirmatory and postclustering analyses to support the validity of our neuronal categorization results. It is important to note that our quantitative method still allowed the neurons with similar phenotype to be clustered in the same group or subgroup, as if they were qualitatively grouped together. This further supports the validity of our methodology, in that the human element of bias is removed, yet we were still able to classify the neurons based on their morphology efficiently. Additionally, our data indicate that the density of spines significantly differs across the classified neuronal groups. Insofar as the spine data were withheld from the initial PCA and cluster analysis, our significant findings in the post-clustering analysis strongly support the validity of our methodological choice. Layer VI of the mouse cortical system may be different from that of other mammals, such as felines (Prieto and Winer, 1999) and primates (Brigs and Callaway, 2001). The overall results of the current study indeed had some discrepancies compared with these prior studies. For instance, Briggs and Callaway (2001) found several distinct subgroups of neurons with apical dendrites extending well beyond layer IV in the primary visual cortex. Prieto and Winer (1999) observed horizontal cells with very elaborate dendritic patterning in the primary auditory cortex, whereas we saw the same neurons with less elaborate dendritic arborizations. It is likely that the mouse barrel system required adaptations different from those for these other mammals. It is also possible that the observed discrepancies are correlated with the differences in perceptual modality between studies.
Functional implications
Neocortical layer VI provides important feedback to the thalamus and layer IV and, therefore, plays a crucial role in gating/modulating sensory information. This in turn influences sensational and perceptual experiences in general. It is highly probable that the differences in cellular morphology are correlated with different cortical roles, as well as different micro-circuits. For instance, the larger dendritic fields of group 2 suggest that they may integrate information from a greater part of its associated cortical column than neurons from groups 4 and 6, which have more confined dendritic trees. Our spine density analyses can be used as a possible correlate of the number of synaptic inputs that a specific neuronal class generally receives, and, based on that analysis, group 2 neurons receive the greatest density of neuronal inputs. By applying the size principal (Fromm and Evarts, 1981), it may be inferred that the larger pyramidal neurons (e.g., group 1) are more difficult to recruit to fire than pyramidal neurons with smaller somata (e.g., group 4). It is also possible that the diversity in layer VI is reflective of the neurons participating in the many different networks of afferent and efferent projections, such as corticothalamic, ipsilateral and contralateral corticocortical, and corticostriatal pathways. In sum, characterizing the neuronal elements within layer VI is an important first step toward understanding the building blocks of cortical circuits and their role(s) in cortical computations.
Following this approach, we can start to elucidate this extremely diverse cortical layer by understanding the physiology of each type of neuron. It is also essential to investigate the functional anatomical properties of the categorized neurons based on afferent/efferent locations. By implementing recently developed genetic labeling techniques, future studies may aim to investigate the nature of monosynaptic restriction of transynaptic tracing in the barrel field (Wickersham et al., 2007). In summary, layer VI is a poorly understood layer in terms of the microcircuitry of the neocortex, mainly because of its complexity. Furthermore, it is also due to this complexity, along with the role of this layer as a gateway for feedback to subcortical structures such as the thalamus (Tombol, 1984; White, 1989), that this layer is of great interest. Therefore, it becomes important to analyze layer VI quantitatively, because this analytical approach provides a more accurate and unbiased way of studying neuronal cytoarchitectures. Identifying specific types of neurons within layer VI is the first step in understanding the role that this complicated cortical layer plays in sensation and perception.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Asaf Keller, Carolyn Pytte, Raddy Ramos, Areti Tsiola, and Philip Ramsey for insightful discussions.
Grant sponsor: Doctoral Students Council (DSC), CUNY (to C.-C.C.); Grant sponsor: CUNY Equipment Grant (to J.C.B.); Grant sponsor: SOMAS (to J.C.B.); Grant sponsor: National Institutes of Health; Grant number: NS058758-01A1 (to J.C.B.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Arimatsu Y, Ishida M, Kaneko T, Ichinose S, Omori A. Organization and development of corticocortical associative neurons expressing the orphan nuclear receptor Nurr1. J Comp Neurol. 2003;466:180–196. doi: 10.1002/cne.10875. [DOI] [PubMed] [Google Scholar]
- Briggs F, Callaway EM. Layer-specific input to distinct cell types in layer 6 of monkey primary visual cortex. J Neurosci. 2001;21:3600–3608. doi: 10.1523/JNEUROSCI.21-10-03600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumberg JC, Hamzei-Sichani F, Yuste R. Morphological and physiological characterization of layer VI corticofugal neurons of mouse primary visual cortex. J Neurophysiol. 2003;89:2854–2867. doi: 10.1152/jn.01051.2002. [DOI] [PubMed] [Google Scholar]
- Cattell RB. The scree test for the number of factors. Multivariate Behav Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc Natl Acad Sci U S A. 2000;97:6144–61449. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Alloway KD. Differential origin of projections from SI barrel cortex to the whisker representations in SII and MI. J Comp Neurol. 2006;498:624–636. doi: 10.1002/cne.21052. [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, Yin HH, Loving DM, Wang Y, Li Y. Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc Natl Acad Sci U S A. 2006;103:15254–15259. doi: 10.1073/pnas.0601758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Sterling P. Microcircuitry of cat visual cortex: classification of neurons in layer IV of area 17, and identification of the patterns of lateral geniculate input. J Comp Neurol. 1979;188:599–628. doi: 10.1002/cne.901880407. [DOI] [PubMed] [Google Scholar]
- Del Río JA, Martinez A, Anladell C, Soriano E. Developmental history of the subplate and developing white matter in the murine neocortex. Neuronal organization and relationship with the main afferent systems at embryonic and perinatal stages. Cereb Cortex. 2000;10:784–801. doi: 10.1093/cercor/10.8.784. [DOI] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 Catenin regulates dendritic spine and synapse development through rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Fabregues I, Condom EA. Golgi study of the sixth layer of the cerebral cortex. I. The lisencephalic brain of Rodentia, Lagomorpha, Insectivora, and Chiroptera. J Anat. 1986;145:217–234. [PMC free article] [PubMed] [Google Scholar]
- Fromm C, Evarts EV. Relation of size and activity of motor cortex pyramidal neurons during skills movements in the monkey. J Neurosci. 1981;1:453–460. doi: 10.1523/JNEUROSCI.01-05-00453.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak SC, Moyer JR, Brown TH. Morphology and ontogeny of rat perirhinal cortical neurons. J Comp Neurol. 2007;505:493–510. doi: 10.1002/cne.21516. [DOI] [PubMed] [Google Scholar]
- Glaser ME, Van der Loos H. Analysis of thick brain sections by obverse-reverse computer microscopy: application of a new, high clarity Golgi-Nissl stain. J Neurosci Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- Heuer H, Christ S, Friedrichsen S, Brauer D, Winckler M, Bauer K, Raivich G. Connective tissue growth factor: a novel marker of layer VII neurons in the rat cerebral cortex. Neuroscience. 2003;119:43–52. doi: 10.1016/s0306-4522(03)00100-3. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Discarding variables in a principal component analysis. I: artificial data. Appl Statist. 1972;21:160–173. [Google Scholar]
- Jones EG. Laminar distribution of cortical efferent cells. In: Peters A, Jones EG, editors. Cerebral cortex, cellular componenets of the cerebral cortex. vol 1. New York: Plenum; 1984. pp. 521–553. [Google Scholar]
- Katz LC. Local circuitry of identified projection neurons in cat visual cortex brain slices. J Neurosci. 1987;7:1223–1249. doi: 10.1523/JNEUROSCI.07-04-01223.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libersat F, Duch C. Mechanisms of dendritic maturation. Mol Neurobiol. 2004;29:303–320. doi: 10.1385/MN:29:3:303. [DOI] [PubMed] [Google Scholar]
- Lorente de Nó R. Physiology of the nervous system. New York: Oxford University Press; 1949. Cerebral cortex: architecture, intracortical connections, motor projections; pp. 288–330. [Google Scholar]
- Mendizabal-Zubiaga JL, Reblet C, Bueno-Lopez JL. The Underside of the cerebral cortex: layer V/VI spiny inverted neurons. J Anat. 2007;211:223–236. doi: 10.1111/j.1469-7580.2007.00779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle VB. The cerebral cortex. Cambridge, MA: Harvard University Press; 1998. Perceptual neuroscience. [Google Scholar]
- Pasternak JF, Woolsey TA. On the “selectivity” of the Golgi-Cox method. J Comp Neurol. 1975;160:307–312. doi: 10.1002/cne.901600304. [DOI] [PubMed] [Google Scholar]
- Petersen CCH. The functional organization of the barrel cortex. Neuron. 2007;56:339–354. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Prieto JJ, Winer JA. Layer VI in cat primary auditory cortex: Golgi study and sublaminar origins of projection neurons. J Comp Neurol. 1999;404:332–358. doi: 10.1002/(sici)1096-9861(19990215)404:3<332::aid-cne5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schütz G, Linden DJ, Ginty D. SRF mediates activity-induced gene expression and synaptic plasticity but not neuron viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Ramón-Moliner E. The Golgi-Cox technique. In: Nauta WJH, Ebbesson SOE, editors. Contemporary methods in neuroanatomy. New York: Springer; 1970. [Google Scholar]
- Rocco MM, Brumberg JC. The sensorimotor slice. J Neurosci Methods. 2007;162:139–147. doi: 10.1016/j.jneumeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Sadakata T, Kakegawa W, Mizoguchi A, Washida M, Katoh-Semba R, Shutoh F, Okamoto T, Nakashima H, Kimura K, Tanaka M, Sekine Y, Itohara S, Yuzaki M, Nagao S, Furuichi T. Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J Neurosci. 2007;27:2472–2482. doi: 10.1523/JNEUROSCI.2279-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibler D, Schneider W. Monte Carlo tests of the accuracy of cluster analysis algorithms: a comparison of hierarchical and nonhier-archical methods. Multivariate Behav Res. 1985;20:283–304. doi: 10.1207/s15327906mbr2003_4. [DOI] [PubMed] [Google Scholar]
- Sherman SM, Guillery RW. The role of the thalamus in the flow of information to the cortex. Philos Trans R Soc Lond B Biol Sci. 2002;357:1695–1708. doi: 10.1098/rstb.2002.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. The organization of the cerebral cortex. New York: John Wiley; 1956. [Google Scholar]
- Simons DJ, Woolsey TA. Morphology of Golgi-Cox impregnated barrel neurons in rat SmI cortex. J Comp Neurol. 1984;230:119–132. doi: 10.1002/cne.902300111. [DOI] [PubMed] [Google Scholar]
- Sultan F, Bower JM. Quantitative Golgi study of the rat cerebellar molecular layer interneurons using principal component analysis. J Comp Neurol. 1998;393:353–373. [PubMed] [Google Scholar]
- Tamas G, Buhl EH, Lorincz A, Somogyi P. Proximally targeted GABAergic synapses and gap junctions synchronize cortical interneurons. Nat Neurosci. 2000;3:366–371. doi: 10.1038/73936. [DOI] [PubMed] [Google Scholar]
- Tombol T. Cellular components of the cerebral cortex. New York: Plenum Press; 1984. Layer VI cells. [Google Scholar]
- Tsiola A, Hamzei-Sichani F, Paterlin Z, Yuste R. Quantitative morphologic classification of layer 5 neurons from mouse primary visual cortex. J Comp Neurol. 2003;461:415–438. doi: 10.1002/cne.10628. [DOI] [PubMed] [Google Scholar]
- Uylings HBM, Ruiz-Marcos A, van Pelt J. The metric analysis of thee-dimensional dendritic patterns: a methodological review. J Neurosci Methods. 1986;18:129–151. doi: 10.1016/0165-0270(86)90116-0. [DOI] [PubMed] [Google Scholar]
- Valverde F. Golgi atlas of the postnatal mouse. Vienna: Springer-Verlag; 1998. [Google Scholar]
- Valverde F, Facal-Valverde MV, Satacana ML, Heredia M. Development and differentiation of early generated cells of sublayer VIb in the somatosensory cortex of the rat: a correlated Golgi and autoradio-graphic study. J Comp Neurol. 1989;290:118–140. doi: 10.1002/cne.902900108. [DOI] [PubMed] [Google Scholar]
- Van Brederode JF, Snyder GL. A comparison of the electrophysiological properties of morphologically identified cells in layer 5B and 6 of the rat neocortex. Neuroscience. 1992;50:315–337. doi: 10.1016/0306-4522(92)90426-3. [DOI] [PubMed] [Google Scholar]
- Ward JH., Jr Hierarchical grouping to optimize an objective function. J Am Statist Assoc. 1963;58:236–244. [Google Scholar]
- Watakabe A, Ichinohe N, Ohsawa S, Hashikawa T, Komatsu Y, Rockland K, Yamamori T. Comparative analysis of layer-specific genes in mammalian neocortex. Cereb Cortex. 2007;17:1918–1933. doi: 10.1093/cercor/bhl102. [DOI] [PubMed] [Google Scholar]
- White EL. Cortical circuits, synaptic organization of the cerebral cortex, structure, function, and theory. Boston: Birkhauser; 1989. [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young JAT, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CR, Seanmens JK, Gorelova N. Electrophysiological and morphological properties of layer V-VI principal pyramidal cells in rat prefrontal cortex in vivo. J Neurosci. 1996;16:1904–1921. doi: 10.1523/JNEUROSCI.16-05-01904.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.