Abstract
Friedreich’s Ataxia (FA) is an inherited neurodegenerative disease caused by reduction in levels of the mitochondrial protein frataxin. Currently there are no simple, reliable methods to accurately measure the concentrations of frataxin protein. We designed a lateral-flow immunoassay that quantifies frataxin protein levels in a variety of sample materials. Using recombinant frataxin we evaluated the accuracy and reproducibility of the assay. The assay measured recombinant human frataxin concentrations between 40 and 4000 pg/test or approximately 0.1 – 10 nM of sample. The intra and inter-assay error was < 10% throughout the working range. To evaluate clinical utility of the assay we used genetically defined lymphoblastoid cells derived from FA patients, FA carriers and controls. Mean frataxin concentrations in FA patients and carriers were significantly different from controls and from one another (p = 0.0001, p = 0.003, p = 0.005, respectively) with levels, on average, 29% (patients) and 64% (carriers) of the control group. As predicted, we observed an inverse relationship between GAA repeat number and frataxin protein concentrations within the FA patient cohort. The lateral flow immunoassay provides a simple, accurate and reproducible method to quantify frataxin protein in whole cell and tissue extracts, including primary samples obtained by non-invasive means, such as cheek swabs and whole blood. The assay is a novel tool for FA research that may facilitate improved diagnostic and prognostic evaluation of FA patients and could also be used to evaluate efficacy of therapies designed to cure FA by increasing frataxin protein levels.
Keywords: Frataxin, Friedreich’s Ataxia, diagnostic, prognostic, theranostic, lateral flow immunoassay, mitochondria
Introduction
Friedreich’s Ataxia (FA) is an inherited recessive neurodegenerative disorder caused by the partial reduction in levels of the mitochondrial protein frataxin [1, 2]. FA is the most common inherited cause of ataxia, with an incidence estimated between 1:30,000 to 1:50,000 in both the US and Europe [3–5]. The disease is characterized by a progressive, unrelenting sensory neuropathy due to death of primary sensory neurons of the dorsal root ganglia while other neurological symptoms present variably [4]. FA is also often accompanied by cardiomyopathy and an increased incidence of diabetes. Typically, FA patients are normal at birth and in early childhood and the majority of affected individual's exhibit onset of symptoms by age 20. However, FA penetrance is highly variable and can be incomplete and/or delayed for reasons unknown at present [6].
The genetic basis of FA is now well-established [4]. It is a triplet-nucleotide disease with > 95% of cases attributable to expanded GAA repeats in intron 1 of both alleles of the frataxin gene, FXN [2, 7]. The remaining cases are all compound heterozygotes in which one FXN allele contains an expanded GAA repeat while the second allele carries a deleterious point mutation [8, 9]. Most individuals carry only small numbers of GAA repeats in the frataxin gene, with most (> 80%), carrying “small normal” (6–12) repeats and the remainder carrying “large normal” repeats (14–34) [10]. To date, the functional role and pathological consequences, if any, of these small normal and large normal repeats is still unknown. However, individuals with large normal repeats are at increased risk of undergoing rapid germ-line expansions resulting in offspring with expanded repeats (66 to more than 1500 repeats) which result in disease [10]. An approximate inverse correlation exists between the age of onset (and disease severity) and the size of the expanded GAA repeats, especially the smaller allele, accounting for approximately 50%–70% of the variance in age of onset of FA [7, 10–12]. However some cases present much later than expected, with a milder form of the disease in spite of large GAA repeats, and this idiosyncratic penetration cannot be explained by repeat number alone [10, 13].
At the molecular level the large expanded GAA repeats (>66) in intron 1 of the FXN gene interfere with FXN transcription, resulting in reduced frataxin mRNA and protein levels in FA patients [14–17]. The decrease in frataxin protein has broad, far-reaching effects because the protein is an essential iron chaperone required for the biogenesis of iron-sulfur clusters, aconitase activation, and heme biosynthesis [18–21], and further performs a critical role in iron detoxification and anti-oxidant protection [22–24]. Thus a loss of frataxin leads to widespread impairment of energy metabolism, increased oxidative stress, and a generally dysregulated iron metabolism, including accumulation of iron in the heart and nervous system [25].
Currently much focus on FA research is on providing early diagnosis of the disease and in developing therapies to ameliorate symptoms and even cure the disease. One key for potential curative therapies is that the genetic defect is in an intron and not in a coding sequence of the frataxin gene. This opens the possibility of using small molecule drugs to boost frataxin concentrations by increasing transcription of the unaltered, normal coding sequence and recent experimental advances have demonstrated the feasibility of this approach in vitro [26–29]. In addition, other compounds, such as recombinant human erythropoietin, have been shown to increase frataxin protein levels in vitro by a presently unknown mechanism [30].
While a genetic diagnosis of FA is now possible and widely used, measurement of frataxin protein concentrations by simple lateral-flow immunoassay would have the advantages of speed, reduced costs and may have greater diagnostic and prognostic value as frataxin protein levels, not GAA repeat number per se, likely define disease severity. Moreover, as frataxin upregulation therapies enter clinical trials and eventual application, it will be necessary to measure the concentrations of this protein accurately, routinely and in a minimally invasive way to monitor the molecular efficacy of the drug candidates. To address this need we report here a lateral-flow immunoassay to quantify frataxin protein levels.
Materials and Methods
Monoclonal Antibodies (mAbs)
Anti-frataxin mAbs were generated by immunizing mice (F1 BALB/cJ x SLJ/J) with soluble, native recombinant human frataxin; amino acids 56–210 prepared as previously described [31]. This construct corresponds to the 155 amino acid form of frataxin shown to be present inside mitochondria immediately after proteolytic removal of the mitochondrial targeting sequence from the precursor protein [32]. Splenocytes were harvested from mice with strong anti-frataxin antibody titers, fused with null mouse myeloma cells (X63-Ag8.653) and the resulting hybridomas cultured and subcloned as previously described [33]. The mAbs were then screened with ELISA, western blot, immunocytochemistry and frataxin immunocapture lateral flow assays (dipsticks). The two mAbs selected as dipstick immunocapture (clone ID# 17A11AC7) and detector (clone ID# 18A5DB1) mAbs recognize frataxin in each of these assay formats.
Lateral Flow Immunoassay
The frataxin-specific dipsticks were prepared as follows. First, a 3.5 cm × 30 cm nitrocellulose membrane (#SA3J44IH7, Millipore) was laminated to the lower part of an adhesive backing support card (GL-187, 0.010" white matte vinyl; G&L Precision). A cellulose wicking pad was then laminated to the top of the card, slightly overlapping the nitrocellulose membrane along the full length of the card (Whatman #17CHR). An Imagene Isoflow reagent dispense was then used to apply the anti-frataxin capture mAb (clone ID# 17A11AC7) in a narrow zone the length of the membrane (18 µL/card at 2 g/L in PBS, or approximately 0.5 µg capture mAb per device). A parallel zone of goat anti-mouse (GAM) antibody (#115-005-164, GAM-IgG reactive with all mouse sub-classes, Jackson ImmunoResearch) was also applied in a zone between the anti-frataxin line and the wicking pad. The GAM line serves as an assay procedural control to verify that the entire sample has passed through the anti-frataxin capture zone. Antibody-stripped cards were then incubated in a dry 37 °C incubator for 1 hour, followed by incubation in a desiccator chamber at room temperature for at least 24 hours. Each card was cut into 4 mm wide strips using a Kinematic Matrix 2360 guillotine cutter. The devices were stored desiccated.
Gold/mAb Conjugations
The anti-frataxin gold-conjugated detector mAb (clone ID# 18A5DB1) was prepared by conjugating the mAb to colloidal gold particles (approximately 40 nm in diameter) prepared by the controlled reduction of gold chloride with trisodium citrate. In short, 20 mL of colloidal gold (524 nmol/L, absorbance = 1), pH 8, was mixed with detector mAb (10 mg/L colloid) and incubated for 10 minutes at room temperature. Bovine serum albumin (BSA) was then added to a final concentration of 10 g/L to block non-specific binding sites, the gold-mAb suspension centrifuged at 5000g for 20 minutes and the pellet of gold conjugated mAb resuspended in 10 g/L BSA, 100 mmol/L phosphate buffer, pH 7.4 for a final absorbance of 10. The conjugate was stored at 4 ° C until use.
Lymphoblastoid Cell Culture
Lymphoblast cells derived from FA patients, carriers and unaffected individuals were obtained from the Coriell Institute. A list of these cell lines, along with other relevant information from Coriell, is provided in the Results section. The cell lines were selected to provide a wide range of GAA repeat values to allow a comparison of GAA repeat #s with frataxin levels measured by the dipsticks. Cells were grown in suspension culture and maintained in log phase using HGDMEM supplemented with 100 mL/L fetal calf serum, 1 mmol/L pyruvate, 4 mmol/L glutamine, 50 mg/L uridine,1 mg/L insulin, 50 µmol/L 2-mercaptoethanol, 20 µmol/L ethanolamine and 10 mmol/L HEPES. Dense cultures of actively growing cells were harvested by centrifugation (6 minutes at 300g), washed three times with serum-free Dulbecco’s calcium and magnesium free PBS (6 minutes at 300g), and then quickly frozen at −85 °C as wet whole cell pellets in aliquots of approximately 1 × 107 cells.
Sample Preparation
Frozen pellets of cultured cells were thawed and immediately dissolved in 10 volumes of ice cold extraction buffer (1.5% Lauryl Maltoside (Anatrace), 100 mmol/L NaCl, 25mmol/L Hepes, pH 7.4) with protease inhibitors (Sigma # P8340), with gentle repetitive micropipetting. After 15 minute incubation on ice the samples were cleared of insoluble material by centrifugation at 16,000g for 15 min at 4 °C. Cheek cells (i.e. buccal cells) were harvested using Epicentre Buccal Swab Brushes # MB030BR. With firm pressure, a brush is applied to the inner check and swabbed for 30 seconds on each cheek. The brush is then immersed in 600 µL of extraction buffer and twirled gently back and forth to dislodge the collected cells. After 15 min. incubation on ice the samples were cleared of insoluble material by centrifugation at 16,000g for 15 min at 4 °C. For preparation of muscle tissue extracts samples were mixed with 10 volumes of ice cold extraction buffer with protease inhibitors/weight of sample, homogenized with an Ultra-turrax (IKA Works) hand held homogenizer, kept on ice for 15 minutes and then cleared of insoluble material by centrifugation at 16,000g for 15 min. at 4°C. In all of the above preparations, the supernatant was saved and total soluble protein concentration was determined by the BCA method (Pierce). Whole blood (either fresh finger-prick samples or samples collected in EDTA tubes and stored frozen until use) protein were extracted by mixing 1 volume of whole blood to 3 volumes of extraction buffer (see above). After 15 minute incubation on ice, the samples were cleared as above and the supernatant saved. Extracted blood samples were diluted further as desired in extraction buffer and loaded with reference to whole blood volume.
Assay Protocol
First, 25 µL of sample (in extraction buffer) was mixed with 25 µL of 2X blocking buffer (Sigma Block) and 5 µL of gold-mAb conjugate in one well of a microtiter plate. A lateral-flow device was then added to the well and the sample was allowed to wick up through the membrane (~15 min). 30 µL of wash buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.4) was then added to each well and allowed to wick laterally through the membrane, clearing background from around the capture zones. The developed dipsticks were dried before immunocapture zone signals were quantified with a Hamamatsu ICA-1000 immuno-chromato reader. The results were expressed as Area units per capture zone.
Western Blotting
Human frataxin56–210 and frataxin78–210 were expressed in E. coli and purified as described previously [34]. Whole lymphoblast cell extracts were prepared from frozen cell pellets as described above. For optimal separation of frataxin bands, samples were analyzed by SDS/PAGE using T = 12.5% for the separating gel (total length, 12.5 cm) and T = 4% for the stacking gels (T denotes the total concentration of acrylamide and bisacrylamide), from a stock solution of 40:1.7 acrylamide:bisacrylamide. Electrophoresis was started at 180 V, shifted to 240 V after the samples had completely entered the separating gel, and continued for an additional 90 min after the samples had reached the bottom of the separating gel. Frataxin bands were detected by western blotting and chemiluminescence with ECL Plus™ reagents (GE Healthcare).
Results
Assay specificity, sensitivity, and reproducibility
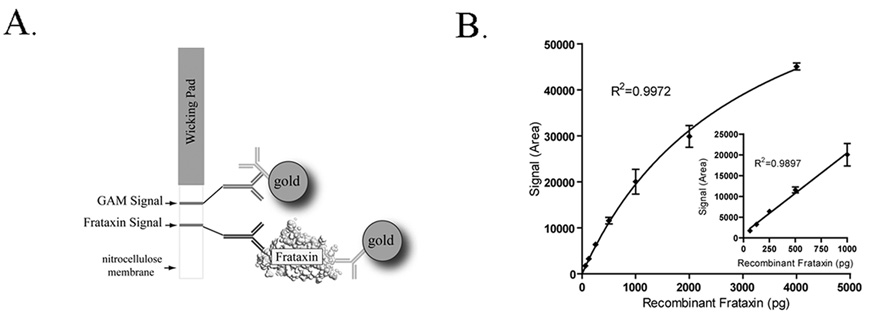
Lateral flow immunoassay devices (dipsticks) are simple, self-developing 2-site sandwich immunoassays that allow rapid quantitation of specific target antigens (Fig. 1A). As shown in Figure 1B, the frataxin dipstick assay is sensitive and saturable, consistent with the use of specific, high-affinity mAbs as both capture and detector reagents. Recombinant frataxin56–210 (the immunogen used to generate the paired capture and detector mAbs used in the assay) can be measured over a wide range (40 – 4000 pico-grams (pg)/test or approximately 0.1 – 10 nM sample concentrations) and the assay is linear up to approximately 1000 pg/test (Fig. 1B).
Fig. 1. Frataxin Lateral flow device and binding profile.
(a) Schematic of the Frataxin Immunossay. (b) The assay is sensitive and accurate over a wide range of frataxin concentrations (>60-fold) and begins to approach saturation at 4000 picograms (pg) of recombinant frataxin. Error bars show standard deviations at each dilution (n=3). Dipstick signal is expressed as Hamamatsu ICA-1000 Area units. One-site hyperbola non-linear and linear regressions (inset) were generated using Graph Pad Prism software.
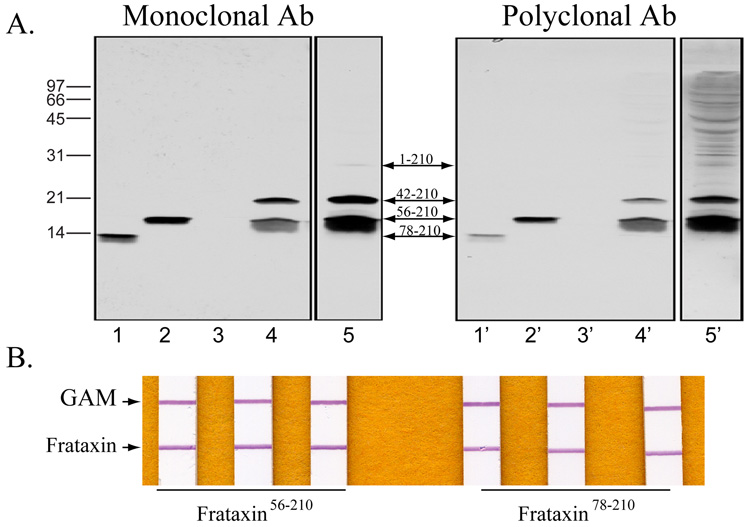
Frataxin is encoded in the nucleus and synthesized in the cytoplasm as a precursor polypeptide (frataxin1–210) that is transported to the mitochondrial matrix and proteolytically cleaved to the mature form (frataxin56–210) via a processing intermediate (frataxin42–210) [32, 35]. Independent groups have reported that frataxin56–210 is susceptible to either proteolytic or iron-mediated cleavage to ~14 kDa products ranging from residues 77–210 to 81–210 [25, 36, 37], which can bind and deliver iron in vitro [34, 36] and can also rescue aconitase defects when overexpressed in frataxin deficient cells [37]. Western blot examination of lymphoblast whole cell extracts showed significant amounts of the intermediate and mature forms of frataxin (frataxin42–210 and frataxin56–210), while only trace amounts of the precursor were revealed (Fig. 2A). Although a smear of products shorter than frataxin56–210 was detected, no products equal or smaller than frataxin78–210 were observed (Fig. 2A). Similarly, frataxin56–210 was the predominant form detected in human fibroblast or heart extracts (not shown). In all tissues analyzed, the monoclonal antibody detected the same frataxin protein bands recognized by a previously described anti-frataxin polyclonal antibody [31, 32], except for several bands larger than the frataxin precursor that were only observed with the polyclonal antibody and most likely resulted from non specific cross reacting proteins (Fig. 2A). Both antibodies reacted well with purified frataxin78–210 and frataxin56–210 in western blots, although each showed slightly less reactivity to frataxin78–210. Similarly, frataxin56–210 was slightly more immunoreactive (+30%) than frataxin78–210 in the quantitative 2-mAb-based dipstick assay (Fig. 2B).
Fig. 2. Binding affinities of mAbs to different sized forms of frataxin.
(a) Lymphoblast cell extracts were prepared from five clones (GM07521, GM14907, GM14406, GM05398, and GM03798) of the control cohort and equal total protein amounts from each extract were pooled. An aliquot (100 µg total protein) was analyzed by western blotting (lanes 4 and 4’) next to 1.5 ng purified frataxin78–210 (lanes 1 and 1’) or frataxin56–210(lanes 2 and 2’). The membrane was first incubated with monoclonal antibody 18A5DB1 (lanes 1–5), stripped, and reprobed with a polyclonal anti-frataxin antibody (lanes 1’-5’). Autoradiographies were performed for 1 min (lanes 1–4) or 5 min (lane 5) for the monoclonal antibody, and for 5 min (lanes 1’–4’) or 1 h (lane 5’) for the polyclonal antibody. After stripping, absence of residual signal was verified by treating the membrane with ECL Plus reagents followed by a 60 min autoradiography. Lanes 3 and 3’ were empty. (b) Equal amounts of recombinant ftrataxin 56–210 and frataxin 78–210 were loaded per dipstick in triplicate. Signals was quantified, averaged, and interpolated off of a standard curve using recombinant frataxin 56–210.
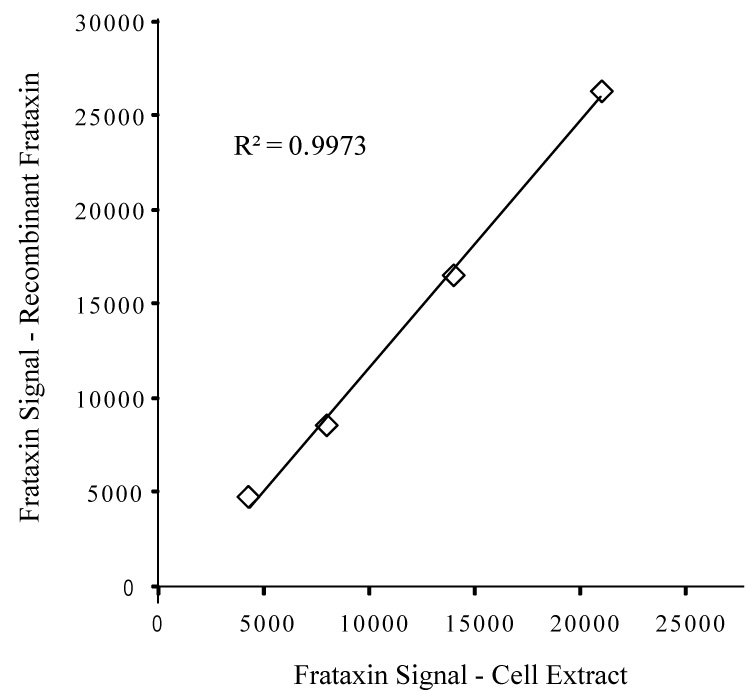
To determine the ability of the frataxin dipsticks to accurately measure endogenous frataxin levels in complex cell extracts, and to further establish utility of recombinant frataxin56–210 as a reference protein, we compared the binding profiles of recombinant frataxin56–210 and endogenous frataxin in lymphoblast whole cell extracts. As shown in Figure 3, the immunoreactivities of these two samples show a 1:1 relationship, indicating that the assay measures recombinant frataxin56–210 and endogenous frataxin with equal facility and that the assay is unaffected by variable levels of non-frataxin proteins present in complex mixtures of whole cell extracts. Finally, repetitive assays run using recombinant frataxin or whole cell extracts demonstrated that the dipstick assay is highly reproducible, with low intra-assay (<5%) and inter-assay (<10%) coefficient of variation (CV) throughout the working range (Table 1 and Table 2). Therefore, frataxin levels in lymphoblast whole cell extracts can be measured accurately with reference to a standard curve prepared from recombinant frataxin56–210.
Fig. 3. Recombinant human frataxin56–210 and endogenous human frataxin in whole cell extracts exhibit similar binding profiles.
Samples of recombinant frataxin56–210 and cellular frataxin (from lymphoblast cell extract) were prepared at concentrations empirically determined to generate approximately equal dipstick signals (~25,000 units). The two samples were then serially diluted in 2-fold steps, frataxin levels measured, and the results plotted by pairing the starting high end samples and each pair of subsequent 2-fold diluted samples. The strong linear relationship between the two sample sets indicates immunological equivalence of the two forms of frataxin as measured in the dipstick assay.
Table 1.
Working ranges for various human sample materials.
| Sample type* | Working range |
|---|---|
| Lymphoblasts | 0.2 – 5 µg |
| Fibroblasts | 0.5 – 10 µg |
| Cheek cell (Buccal) | 2 – 30 µg |
| Whole blood | 0.2 – 3 µL |
| Muscle tissue | 0.3 – 5 µg |
Whole cell and tissue extracts were prepared as described in the Materials and Methods section.
Table 2.
Frataxin Lateral Flow Assay Intra- and Inter-assay Imprecision.
| Intra-assay Imprecision | ||||||
|---|---|---|---|---|---|---|
| Experiment | Dipstick 1 | Dipstick 2 | Dipstick 3 | Mean | SD | CV (%) |
| Day 1 | 19,380 | 17,696 | 19,168 | 18,748 | 917 | 4.9 |
| Day 2 | 19,867 | 19,535 | 18,260 | 19,221 | 848 | 4.4 |
| Day 3 | 19,107 | 18,531 | 18,343 | 18,660 | 398 | 2.1 |
| Day 4 | 21,502 | 23,164 | 23,717 | 22,794 | 1,152 | 5.1 |
| Inter-assay Imprecision (Day 1- Day 4) | ||||||
|---|---|---|---|---|---|---|
| Mean Signal | SD | CV (%) | ||||
| 19,856 | 1,974 | 9.9 | ||||
A single sample of normal cultured fibroblast whole cell extract (within working range of assay) was prepared as described in the Materials and Methods. Total protein concentration of the extract was determined by the BCA method, aliquots frozen at −80° C and then retrieved each day for analysis. Dipstick signal was determined using a Hamamatsu ICA immune-chormato reader and is expressed as Area units. Intra-assay imprecision was determined from n=3 dipsticks/ day. Inter-assay imprecision data was compiled from 4 separate days. Coefficient of variation (CV) was determined by the ratio of standard deviation (SD)/Mean × 100.
Characterization of FA Patients and Carriers
To evaluate clinical utility, we measured frataxin levels in cultured lymphoblastoid cells derived from FA patients and FA carriers and compared these values to those in a control group (Table 3 and Supplemental Table 1).
Table 3.
Human Lymphoblastoid cell lines used in this study.
| # | Coriell # | GAA repeat # | Gender | Onset age/Age |
|---|---|---|---|---|
| Controls | ||||
| 1 | GM07521 | n.a. | F | −/19 |
| 2 | GM14907 | n.a. | M | −/28 |
| 3 | GM14406 | n.a. | F | −/41 |
| 4 | GM05398 | n.a. | M | −/44 |
| 5 | GM03798 | n.a. | M | −/10 |
| FA Carriers | ||||
| 6 | GM15847a | 760 | F | −/45 |
| 7 | GM15848b | 830 | M | −/46 |
| 8 | GM16219 | 570 | F | −/43 |
| 9 | GM15849c | 920 | M | −/10 |
| FA Patients | ||||
| 10 | GM16216 | 200/500 | F | n.a./45 |
| 11 | GM16210 | 580/580 | M | 17/39 |
| 12 | GM16223 | 400/630 | M | 19/41 |
| 13 | GM15850 | 650/1030 | M | n.a./13 |
| 14 | GM16244 | 550/920 | F | 9/18 |
| 15 | GM16243 | 670/1170 | M | 14/23 |
| 16 | GM14518 | 925/1122 | F | n.a |
Lymphoblast cell lines derived from clinically unaffected controls, clinically unaffected FA carriers and clinically - affected FA patients were obtained from the Coriell Institute for Medical Research. Coriell provided the numbers of expanded GAA repeats. Within each group, samples are in order from highest to lowest based on the dipstick signal strength. Precise values for GAA repeats were not available (n.a.) for controls or for the normal allele of FA carriers, but these alleles were described as lacking expanded GAA repeats. Gender is expressed as male (M) or female (F). Age of onset is reported if the information was provided by Coriell. Age corresponds to the age when the cellular material was isolated for cell culture.
Mother, father and brother of affected child, GM15850.
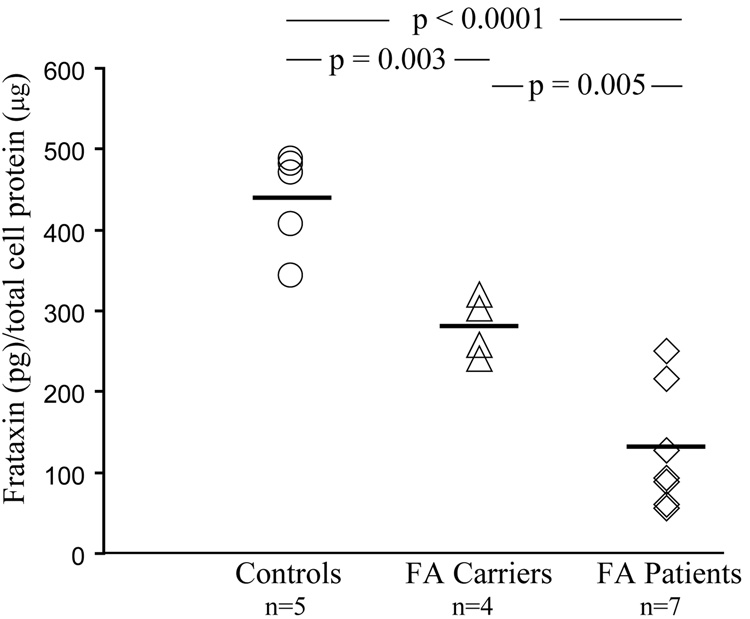
The FA patient cell lines were chosen to provide a sample set with a wide range of expanded GAA repeats on both alleles. The mean expansion size was 708 GAA repeats (SD = 283) with a range of 200 – 1170 repeats (Table 3). As shown in Figure 4, these FA patient-derived cells had significantly lower frataxin levels than the control group (p < 0.001). The control mean was 438 pg frataxin per µg of total cell protein (range: 343–488 pg frataxin per µg of total cell protein; SD = 62) while the FA patient mean was 127 pg frataxin per µg of total cell protein (range: 56–251 pg frataxin per µg of total cell protein; SD = 77; 29% of Control mean) (Fig. 4). Frataxin levels in FA carrier-derived cells were intermediate between the controls and FA patient-derived cells (p=0.003). The FA carrier group mean was 281 pg frataxin per µg of total cell protein (range: 241–320 pg frataxin per µg of total cell protein; SD = 37; 64% of Control mean). Furthermore, the carrier range does not overlap with the control group and is significantly different from the FA patient group (p = 0.005) (Fig.4).
Fig. 4. Frataxin dipsticks identify FA patients and FA carriers from Controls.
Frataxin protein levels were measured in whole cell extracts from lymphoblastoid cells derived from controls (n=5), FA carriers (n=4) and FA patients (n=7). Two measurements from each sample (2 µg total cell protein per dipstick) were averaged and extrapolated from a standard curve to pico-grams (pg) of recombinant frataxin before analysis. The p values shown (Student’s t-test) were calculated for pair-wise comparisons as indicated. Solid lines indicate the mean value for each group; 100% for Controls, 64% for FA carriers and 29% for FA patients.
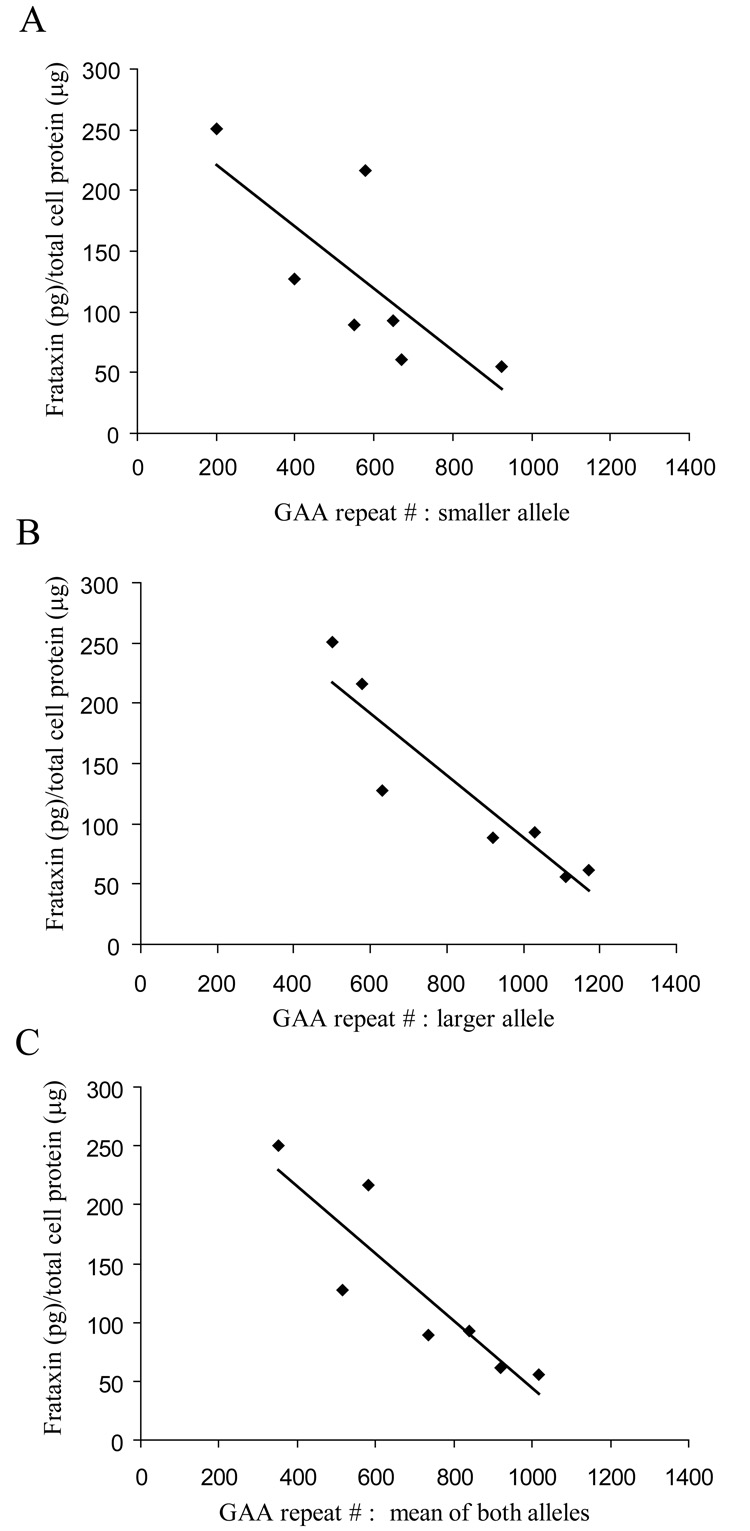
The correlation between frataxin levels and length of homozygous GAA repeats in the frataxin gene is shown in Figure 5. The inverse relationship is strong whether the frataxin levels are plotted relative to the length of GAA repeats on the longer allele (r = − 0.918), the shorter allele (r = − 0.755), or the combined alleles (r = − 0.887). Consistent with this relationship, the single FA carrier/FA patient overlap involves the carrier, GM15849, with the largest GAA repeat number (920) and the FA patient, GM16216, with the least number of total GAA repeats in the FA patient group (200/500).
Fig. 5. Frataxin dipsticks confirm a strong correlation between frataxin protein level and GAA triplet repeat size in FA patients.
Analysis of the relationship between GAA repeat number and dipstick detected frataxin in FA patients. The repeat number for the smaller allele (A), the larger allele (B), and the mean of both alleles (C) were compared to the frataxin dipstick signal extrapolated from the standard curve. All comparisons show linear relationships with the strongest association by the larger allele (B). r-values are A = −0.76, B = −0.92, C = −0.89. p values are A = 0.05, B = 0.0035 and C = 0.0077.
Measurement of Frataxin in Tissues of Clinical Relevance
The frataxin dipstick can be used to quantify frataxin from a variety of cells and tissues, including cells collected by non-invasive (cheek cell) or minimally invasive (whole blood) sampling. The sample range was found to vary depending on the tissue of origin (0.2 to 30 µg total cell protein, intra-assay error <8%) which presumably is proportional to the relative concentrations of frataxin per mg protein in the different tissues (Table 1.). Importantly, in each case, only a very small amount of whole cell extract is needed for testing. For example, a single buccal swab provides approximately 150–300 µg total cell protein extract, which is enough material to perform 15–30 tests at the assay midpoint. Similarly, only ~ 1.5 µL of whole blood is needed for a measurement at the assay midpoint, an amount easily obtained by a finger prick sample (Table 1.).
Discussion
Friedreich’s Ataxia is a devastating neurodegenerative disease caused by reduced levels of the nuclear encoded, mitochondrial protein frataxin. The basic genetics of this disorder are understood and clinical researchers have developed a set of diagnostic molecular genomic tests to detect the expanded GAA repeats (and associated rare point mutations) responsible for FA [4]. These genomic tests are currently used to confirm or deny a clinically based diagnosis of FA and to identify carriers related to FA patients. The diagnostic utility of objective molecular tests is important as FA typically has a gradual onset that can make it difficult to distinguish clinically from other neurological conditions [4]. The genomic tests also have prognostic value, as the number of GAA repeats in homozygous affected individuals is correlated roughly with disease severity and age of onset [7]. However, the prognostic value of the current genomic diagnostic tests is limited, as they do not take into account the effects of background genetics and/or environmental modifiers of frataxin expression that result in atypical cases. Therefore, the correlation between GAA repeat number and disease is estimated to account for only approximately 50–70% of the variance in age of onset of FA [7, 10–12]. An additional significant shortcoming of genomic testing is that the tests cannot be used to monitor molecular efficacy of new therapies designed to treat or prevent onset of FA by boosting levels of the frataxin protein [26, 27, 29, 30].
Therefore, there is a very strong need for a test that measures the functional entity altered in FA, namely the protein frataxin. It is clear that such tests could have diagnostic, prognostic and theranostic utility, as FA is caused by a reduction in frataxin levels and therapeutic interventions are being designed to increase levels of frataxin. Most data to data has shown that this can be achieved by boosting mRNA levels, although other mechanisms may exist to alleviate the reduced levels of frataxin seen in FA patients, as seems to be the case with recombinant human erythropoietin [30].
The diagnostic and prognostic utility of frataxin protein levels has not been exploited to date due to technical limitations in existing frataxin assays. Previously, frataxin protein levels could only be estimated by Western blot, which is cumbersome and semi-quantitative at best [15]. In contrast, the lateral flow immunoassay described here is quantitative, accurate and reproducible, with low intra and inter-assay error throughout a wide working range. Importantly, there is no need to purify mitochondria, allowing simple 1-step sample preparation and rapid assay performance (less than 45 minutes). Measurements can therefore be made using a variety of easily obtained cell and tissue samples such as finger prick blood or cheek swabs, making the assay suitable for both routine diagnostic and theranostic applications. For some clinical samples, where loading by protein concentration is not feasible or practical (i.e. whole blood), incorporating appropriate mitochondrial loading controls may be necessary. This could be achieved by simply adding a second detector zone specific for another mitochondrial protein to normalize mitochondrial load and serve as an internal control. It would of course be critical to determine empirically that FA does not affect levels of the chosen mitochondrial marker since levels of many nuclear-encoded mitochondrial proteins and mRNAs are altered in FA [15, 38–42].
The frataxin dipsticks should therefore facilitate a wide range of new studies to determine the diagnostic and prognostic utility of measuring frataxin levels in FA and to better understand the natural history of the disease and the mechanics of disease progression. For example, frataxin protein expression patterns have not been carefully examined in late-onset FA individuals and other currently unexplained “atypical” cases of FA. Such work may identify patients with mutations in regulatory elements that affect frataxin expression. These putative modifiers might not only contribute to disease progression, but might also be manipulated to up regulate frataxin expression in FA patients.
In the current work we have documented the diagnostic utility of the frataxin lateral flow assay with FA patient-derived lymphoblast cells and show that the assay successfully distinguishes FA patients from controls. Importantly, the assay also identifies FA carriers by their intermediate levels of frataxin and a range that does not overlap with the controls. In addition, we show that FA carrier group is significantly different then FA patient group with only one patient overlapping with the FA carriers. It should be noted that this individual (GM 16216) had the smallest amount of total expanded GAA triplet repeats within the FA patient cohort and it only overlapped a single FA carrier –the one that has the most GAA repeats (GM 15849) (Table 2). Finally, we demonstrate that there is an inverse correlation between the number of triplet repeats in the frataxin gene and the levels of the protein, which is consistent with current understanding of the pathology of FA and GAA repeats [4, 15]. Previous studies which quantified either frataxin protein levels by Western blot analysis or measured frataxin mRNA levels by real time PCR have found that frataxin levels in FA patients are between 6% and 30% of controls for protein levels and between 13% and 30% for mRNA levels [2, 17]. Our data using the frataxin lateral flow assay gives a similar range; 12–30% of control cohort for five of the seven FA patients analyzed.
In summary, the lateral flow test for frataxin described here is a rapid and simple method to quantify frataxin protein levels from a variety of cell material. The assay should find widespread use in basic research into frataxin biology and in clinical practice involving FA.
Supplementary Material
Acknowledgements
We would like to thank Beth Prescott of the University of Oregon Monoclonal Antibody Facility for expert technical assistance with hybridoma cell culture, and Mayen Obette for lymphoblast cell culture work. We would also like to sincerely thank Dr. Michael Makler and Ian Buchanan for introducing MM and JW to lateral flow technology and Diagnostic Consulting Network (DCN) for further immunoassay development. We would also like to thank Dr. Heather O'Neill for performing the initial screening of the antibodies by western blotting.
Grant/funding Support: Dr. G. Isaya acknowledges FARA and NIH support (AG015709 from NIH/NIA) and Dr. M. Marusich acknowledges NIH support (5R42GM71052-3).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures: JW, RC and MM are employees of MitoSciences and, therefore, have financial interests. None declared for G.I or O.G.
References
- 1.Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- 2.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 3.Delatycki MB, Williamson R, Forrest SM. Friedreich ataxia: an overview. J Med Genet. 2000;37:1–8. doi: 10.1136/jmg.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandolfo M. Friedreich ataxia: Detection of GAA repeat expansions and frataxin point mutations. Methods Mol Med. 2006;126:197–216. doi: 10.1385/1-59745-088-X:197. [DOI] [PubMed] [Google Scholar]
- 5.Patel PI, Isaya G. Friedreich ataxia: from GAA triplet-repeat expansion to frataxin deficiency. Am J Hum Genet. 2001;69:15–24. doi: 10.1086/321283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Michele G, Filla A, Cavalcanti F, Di Maio L, Pianese L, Castaldo I, Calabrese O, Monticelli A, Varrone S, Campanella G, et al. Late onset Friedreich's disease: clinical features and mapping of mutation to the FRDA locus. J Neurol Neurosurg Psychiatry. 1994;57:977–979. doi: 10.1136/jnnp.57.8.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, Campanella G, Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 8.Bidichandani SI, Ashizawa T, Patel PI. Atypical Friedreich ataxia caused by compound heterozygosity for a novel missense mutation and the GAA triplet-repeat expansion. Am J Hum Genet. 1997;60:1251–1256. [PMC free article] [PubMed] [Google Scholar]
- 9.Cossee M, Durr A, Schmitt M, Dahl N, Trouillas P, Allinson P, Kostrzewa M, Nivelon-Chevallier A, Gustavson KH, Kohlschutter A, Muller U, Mandel JL, Brice A, Koenig M, Cavalcanti F, Tammaro A, De Michele G, Filla A, Cocozza S, Labuda M, Montermini L, Poirier J, Pandolfo M. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200–206. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 10.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, Turano M, Filla A, De Michele G, Cocozza S. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 11.Durr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A, Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N Engl J Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 12.Mateo I, Llorca J, Volpini V, Corral J, Berciano J, Combarros O. GAA expansion size and age at onset of Friedreich's ataxia. Neurology. 2003;61:274–275. doi: 10.1212/01.wnl.0000073537.08141.77. [DOI] [PubMed] [Google Scholar]
- 13.Bidichandani SI, Garcia CA, Patel PI, Dimachkie MM. Very late-onset Friedreich ataxia despite large GAA triplet repeat expansions. Arch Neurol. 2000;57:246–251. doi: 10.1001/archneur.57.2.246. [DOI] [PubMed] [Google Scholar]
- 14.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, Authier FJ, Durr A, Mandel JL, Vescovi A, Pandolfo M, Koenig M. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 16.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 17.Pianese L, Turano M, Lo Casale MS, De Biase I, Giacchetti M, Monticelli A, Criscuolo C, Filla A, Cocozza S. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of Friedreich ataxia patients and carriers. J Neurol Neurosurg Psychiatry. 2004;75:1061–1063. doi: 10.1136/jnnp.2003.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Monomeric yeast frataxin is an iron-binding protein. Biochemistry. 2006;45:7767–7777. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C, Cortopassi G. Frataxin knockdown causes loss of cytoplasmic iron-sulfur cluster functions, redox alterations and induction of heme transcripts. Arch Biochem Biophys. 2007;457:111–122. doi: 10.1016/j.abb.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulteau AL, O'Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 22.Gakh O, Park S, Liu G, Macomber L, Imlay JA, Ferreira GC, Isaya G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum Mol Genet. 2006;15:467–479. doi: 10.1093/hmg/ddi461. [DOI] [PubMed] [Google Scholar]
- 23.Anderson PR, Kirby K, Orr WC, Hilliker AJ, Phillips JP. Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich's ataxia. Proc Natl Acad Sci U S A. 2008;105:611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runko AP, Griswold AJ, Min KT. Overexpression of frataxin in the mitochondria increases resistance to oxidative stress and extends lifespan in Drosophila. FEBS Lett. 2008;582:715–719. doi: 10.1016/j.febslet.2008.01.046. [DOI] [PubMed] [Google Scholar]
- 25.Babady NE, Carelle N, Wells RD, Rouault TA, Hirano M, Lynch DR, Delatycki MB, Wilson RB, Isaya G, Puccio H. Advancements in the pathophysiology of Friedreich's Ataxia and new prospects for treatments. Mol Genet Metab. 2007 doi: 10.1016/j.ymgme.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant L, Sun J, Xu H, Subramony SH, Chaires JB, Hebert MD. Rational selection of small molecules that increase transcription through the GAA repeats found in Friedreich's ataxia. FEBS Lett. 2006;580:5399–5405. doi: 10.1016/j.febslet.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 28.Sarsero JP, Li L, Wardan H, Sitte K, Williamson R, Ioannou PA. Upregulation of expression from the FRDA genomic locus for the therapy of Friedreich ataxia. J Gene Med. 2003;5:72–81. doi: 10.1002/jgm.320. [DOI] [PubMed] [Google Scholar]
- 29.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB, Gottesfeld JM. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich's ataxia. Proc Natl Acad Sci U S A. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturm B, Stupphann D, Kaun C, Boesch S, Schranzhofer M, Wojta J, Goldenberg H, Scheiber-Mojdehkar B. Recombinant human erythropoietin: effects on frataxin expression in vitro. Eur J Clin Invest. 2005;35:711–717. doi: 10.1111/j.1365-2362.2005.01568.x. [DOI] [PubMed] [Google Scholar]
- 31.Cavadini P, O'Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Genet. 2002;11:217–227. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- 32.Cavadini P, Adamec J, Taroni F, Gakh O, Isaya G. Two-step processing of human frataxin by mitochondrial processing peptidase. Precursor and intermediate forms are cleaved at different rates. J Biol Chem. 2000;275:41469–41475. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- 33.Marusich MF. Efficient hybridoma production using previously frozen splenocytes. J Immunol Methods. 1988;114:155–159. doi: 10.1016/0022-1759(88)90167-6. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill HA, Gakh O, Isaya G. Supramolecular assemblies of human frataxin are formed via subunit-subunit interactions mediated by a non-conserved amino-terminal region. J Mol Biol. 2005;345:433–439. doi: 10.1016/j.jmb.2004.10.074. [DOI] [PubMed] [Google Scholar]
- 35.Koutnikova H, Campuzano V, Koenig M. Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Hum Mol Genet. 1998;7:1485–1489. doi: 10.1093/hmg/7.9.1485. [DOI] [PubMed] [Google Scholar]
- 36.Yoon T, Dizin E, Cowan JA. N-terminal iron-mediated self-cleavage of human frataxin: regulation of iron binding and complex formation with target proteins. J Biol Inorg Chem. 2007;12:535–542. doi: 10.1007/s00775-007-0205-2. [DOI] [PubMed] [Google Scholar]
- 37.Condo I, Ventura N, Malisan F, Rufini A, Tomassini B, Testi R. In vivo maturation of human frataxin. Hum Mol Genet. 2007;16:1534–1540. doi: 10.1093/hmg/ddm102. [DOI] [PubMed] [Google Scholar]
- 38.Foury F. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- 39.Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 40.Shan Y, Napoli E, Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the Nfs1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- 41.Stehling O, Elsasser HP, Bruckel B, Muhlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 42.Coppola G, Choi SH, Santos MM, Miranda CJ, Tentler D, Wexler EM, Pandolfo M, Geschwind DH. Gene expression profiling in frataxin deficient mice: microarray evidence for significant expression changes without detectable neurodegeneration. Neurobiol Dis. 2006;22:302–311. doi: 10.1016/j.nbd.2005.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.