Abstract
In most organisms 10-formyl-tetrahydrofolate (10-CHO-THF) participates in the synthesis of purines in the cytosol and formylation of mitochondrial initiator methionyl-tRNAMet. Here we studied 10-CHO-THF biosynthesis in the protozoan parasite Leishmania major, a purine auxotroph. Two distinct synthetic enzymes are known, a bifunctional methylene-tetrahydrofolate dehydrogenase/cyclohydrolase (DHCH) or formyl-tetrahydrofolate ligase (FTL), and phylogenomic profiling revealed considerable diversity for these in trypanosomatids. All species surveyed contain a DHCH1, which was shown recently to be essential in L. major. A second DHCH2 occurred only in L. infantum, L. mexicana and T. cruzi, and as a pseudogene in L. major. DHCH2s bear N-terminal extensions and we showed a LiDHCH2-GFP fusion was targeted to the mitochondrion. FTLs were found in all species except Trypanosoma brucei. L. major ftl− null mutants were phenotypically normal in growth, differentiation, animal infectivity and sensitivity to a panel of pteridine analogs, but grew more slowly when starved for serine or glycine, as expected for amino acids that are substrates in C1-folate metabolism. Cell fractionation and western blotting showed that both L. major DHCH1 and FTL were localized to the cytosol and not the mitochondrion. These localization data predict that in L. major cytosolic 10-formyl-tetrahydrofolate must be transported into the mitochondrion to support methionyl-tRNAMet formylation. The retention in all the trypanosomatids of at least one enzyme involved in 10-formyl-tetrahydrofolate biosynthesis, and the essentiality of this metabolite in L. major, suggests that this pathway represents a promising new area for chemotherapeutic attack in these parasites.
1. Introduction
Leishmania are group of parasitic protozoa that pose substantial threats to health across a large part of the world. Different species of Leishmania cause a variety of pathologies that range from disfiguring skin lesions to fatal visceral infections. No vaccine is available for leishmaniasis and current drugs suffer from drawbacks such as toxicity, high cost or emerging resistance. Due to such problems, new drugs and new targets for drug development are needed. Historically, folate metabolism in parasitic protozoa has been a rich source of targets for chemotherapy, with folate biosynthesis and dihydrofolate reductase (DHFR) inhibitors showing good efficacy against malaria [1].
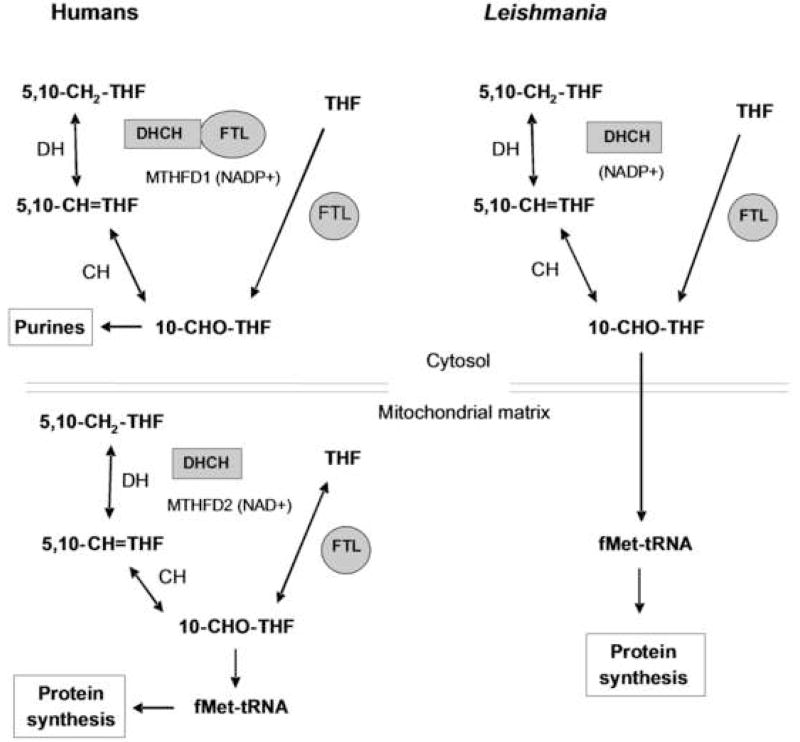
In this work we focus on the biosynthesis of 10-formyl-tetrahydrofolate (10-CHO-THF), which is used in cytosolic purine biosynthesis, or the formylation of the mitochondrial initiator methionyl-tRNAMet to make fMet-tRNAMet [2] (Fig. 1). There are two 10-CHO-THF biosynthetic pathways known (Fig. 1). One begins with 5,10-methylenetetrahydrofolate (5,10-CH2-THF), which arises from serine via serine hydroxymethyltransferase (SHMT), or from glycine by the mitochondrial glycine cleavage complex (GCC). 5,10-CH2-THF can be oxidized to 5,10-methenyltetrahydrofolate (5,10-CH=THF) by 5,10-CH2-THF dehydrogenase (DH), and subsequently hydrolyzed by 5,10-CH=THF cyclohydrolase (CH) to yield 10-CHO-THF. These two sequential reactions are mediated by a single enzymatic site, so this bifunctional enzyme is termed DHCH [3]. In a second pathway, formate is added onto THF by an ATP-dependent formate-tetrahydrofolate ligase (FTL). In plants and prokaryotes, these enzymes are usually found as bifunctional DHCH and monofunctional FTL enzymes [2, 4–7]. However, in humans and yeast the DHCH and FTL activities can be joined in a multienzyme polypeptide called the C1-synthase, which in yeast exists in two isoforms, one located in the cytosol and one in the mitochondrion [8, 9]. In humans the trifunctional enzyme is cytosolic [10] while a bifunctional DHCH [11] and a monofunctional FTL are found in the mitochondria [12, 13]. The coordination of 10-CHO-THF metabolism between these two compartments is thought to depend primarily on the shuttling of metabolites such as glycine, serine and formate [2]. However the situation may be more complex, as while initial studies suggested that reduced and polyglutamylated forms of tetrahydrofolate did not cross the inner mitochondrial membrane [14–16], recent studies suggest that trafficking of various C1-THF derivatives may occur in several species [17–19].
Figure 1. 10-formyl-THF metabolism in humans and L. major.
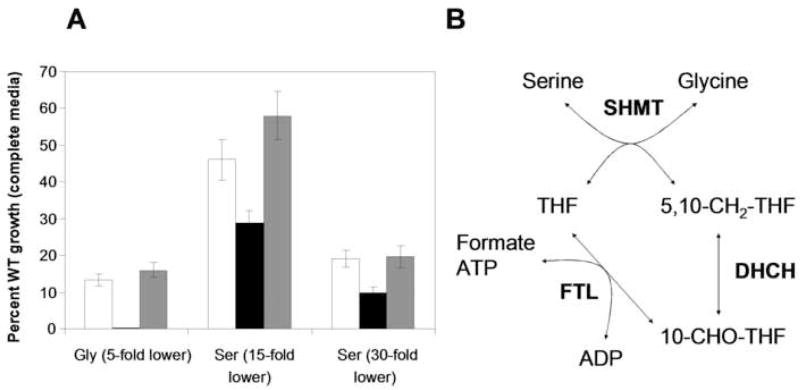
Two pathways of 10-CHO-THF synthesis are known; in the first pathway, formate is added onto THF by formate-tetrahydrofolate ligase (FTL). Alternatively, 10-CHO-THF is generated in two steps from 5,10-methylene-tetrahydrofolate (5,10-CH2-THF): through oxidation to 5,10-methenyl-tetrahydrofolate (5,10-CH=THF) by a methylene-tetrahydrofolate dehydrogenase (DH), and then hydrolysis to 10-CHO-THF by methenyl-tetrahydrofolate cyclohydrolase (CH). In humans 10-CHO-THF is generated in the cytosol through either pathway by the trifunctional DHCH/FTL protein encoded by MTHFD1, often referred to as C1-THF synthase. In contrast the human mitochondrion contains a bifunctional DHCH and a monofunctional FTL. In L. major, 10-CHO-THF is generated in the cytosol by a bifunctional DHCH and a monofunctional FTL.
In the trypanosomatids the metabolic pathways using and producing 10-CHO-THF differ in several respects from those of their mammalian hosts. These parasites lack de novo purine biosynthesis and instead obtain purines through salvage pathways [20]. Therefore the major function for 10-CHO-THF that remains in these organisms is the production of fMet-tRNAMet [21]. While tRNA formylation has not been studied in Leishmania, it has been characterized in Trypanosoma brucei, where it shows a number of unusual properties. Firstly, the trypanosomatid mitochondrial genomes do not encode tRNAs, all of which must be imported from the cytosol [22]. Secondly, following import into the mitochondrion, the cytosolic elongator tRNAMet-e acts as either an elongator tRNAMet, or is formylated and used as an initiator tRNAMet; remarkably, the tRNAMet-i was not used in the trypanosome mitochondrion [23]. Lastly, methionyl-tRNA formylation is required for binding of the fMet-tRNAMet to initiation factor 2, and appears to be essential in trypanosomes [24]. In contrast, the requirement for initiator tRNAMet formylation appears highly variable in evolution; while initially considered essential in bacteria, later data suggested otherwise [25, 26]. In some eukaryotic species it may be essential, in others it is dispensable, but aids growth in some circumstances [27–30].
Previous genomic predictions of trypanosomatid folate metabolism suggested that 10-CHO-THF is synthesized by a bifunctional DHCH (DHCH1) in Leishmania, T. brucei and T. cruzi, as well as by a monofunctional FTL in Leishmania and T. cruzi [21]. We showed previously that the L. major DHCH1 is essential for parasite survival, although its loss could be rescued by FTL overexpression [31]. This identified 10-CHO-THF as an essential metabolite and established that its metabolism is a validated drug target. In this study we used database mining to fully catalog the DHCH and FTL gene families in the Trypanosomatidae, revealing the existence of a second family of DHCH homologues (DHCH2) in some of the Leishmania and T. cruzi, but absent in L. major and T. brucei. Several of these DHCH2 proteins bear N-terminal extensions that resemble mitochondrial targeting sequences and the L. infantum DHCH2 was indeed shown to be a mitochondrial protein. We then studied the function, role in sensitivity to antifolates and compartmentalization of the DHCH1 and FTL in Leishmania major. We found that the L. major FTL exhibits the expected enzymatic activity in vitro, but this enzyme was not inhibited by any pteridine analogues tested. A null ftl− mutant L. major was produced by homologous replacement, with no detectable effect on growth, drug sensitivity, differentiation or infectivity in a susceptible mouse model, eliminating FTL as a unique drug target. However, nutritional studies suggest that this enzyme may play a role in the ability of Leishmania to withstand serine or glycine starvation. Interestingly, our data establish that L. major FTL and DHCH1 are localized to the cytosol and cannot be detected in the mitochondrion. As the only known essential use of 10-CHO-THF is in the mitochondrion, this suggests that L. major differs from mammals in that its survival may depend on mitochondrial 10-CHO-THF uptake.
2. Materials and methods
2.1. Reagents
(6-R,S)-Tetrahydrofolate (THF) monoglutamate was obtained from Schircks Laboratories (Jona, Switzerland). Blasticidin (BSD) and G418 were from Invitrogen (San Diego, CA) and nourseothricin (SAT) was from Werner BioAgents (Jena, Germany). The DHCH inhibitor 5,10-CO-THF was synthesized as described [31]. All other chemicals and reagents were of analytical grade.
2.2. Parasite growth and virulence
L. major Friedlin (MHOM/IL/81/Friedlin) clone V1 or the L. donovani Bob clonal derivative of L. donovani strain 1S2D (MHOM/SD/62/1S-CL2D) [32] were routinely grown as promastigotes at 27°C in M199 medium (US Biologicals) supplemented with 40 mM 4-(2-hydroxyethyl)-1- piperazine-ethanesulfonic acid (HEPES) pH 7.4, 50 μM adenosine, 1 μg ml−1 biotin, 5 μg ml−1 hemin, 2 μg ml−1 biopterin and 10 % (v/v) heat-inactivated fetal calf serum. Growth under semi-defined conditions used RPMI medium (Gibco-BRL) supplemented with 30 mM HEPES pH 7.4, 62.5 μM adenosine, 2 μg ml-1 biopterin, 5 μg ml-1 hemin and 1% (v/v) heat-inactivated fetal calf serum. The infectivity of wild-type (WT) and ftl− cell lines was determined by infection of 1×105 metacyclic parasites (purified by negative selection with peanut agglutinin [33] into the foot-pads of susceptible BALB/c mice, followed by measurement of lesion growth, [34].
2.3. Molecular cloning of FTL and DHCHs
The FTL coding sequence was amplified with recombinant Pfu DNA polymerase (Strata-gene) from L. major genomic DNA, which was prepared by the LiCl mini-prep method,[35] using the primers SMB3537 and SMB3538, which added 5′ NdeI and 3′ BamHI sites, respectively (oligonucleotides are described in Table S1). The amplified product was inserted by blunt-ended cloning into pCR.blunt (Invitrogen), according to the manufacturer’s protocol, to create pCR.blunt.FTL. This plasmid was digested with NdeI and BamHI and the fragment released subcloned between the NdeI and BamHI sites of pET15b (Novagen) to create pET15b.LmjFTL (B5885). pXG-FTL (B5917) was created by excising the FTL ORF from pCR.blunt.FTL by digestion with BamHI and insertion into the BamHI site of the pXG vector,[36]. The C-terminal GFP L. infantum DHCH1 and DHCH2 fusions were created by amplification of the respective ORFs from genomic DNA (primers SMB3601/SMB3597 and SMB3602/SMB3599 respectively). These primers added a 5′ BamHI site and replaced the stop codon with an EcoRV site, allowing cloning as an in-frame fusion with GFP in the vector pXG-/GFP+ (B2863), yielding constructs pXG-LiDHCH2-GFP (B6326) and pXG-LiDHCH2-GFP (B6327). The L. donovani DHCH1 and 2 genes were amplified with the same primers as the L. infantum genes, cloned into pCRblunt as before and then sequenced using the M13 primer sites located in the vector. The L. donovani genes were amplified a second time and the PCR product sequenced directly, to confirm that the clones sequenced were not mutants. These sequences were deposited in GenBank, with the accession numbers FJ487909 and FJ487910. Similarly, the pIR1.PHLEO-LiDHCH1 and pIR1.PHLEO-LiDHCH2 constructs (B6328 and B6329 respectively) were created by amplification of the respective ORFs from genomic DNA using the primers SMB3601/SMB3598 and SMB3602/SMB3600, respectively. These added flanking BamHI sites to the genes, allowing subcloning into the BglII site in the pIR1-PHLEO vector.
2.4. Expression and purification of recombinant LmjFTL
pET15b-LmjFTL DNA was transformed into E. coli strain Rosetta2(DE3)pLysS. Cells were grown at 37°C in 800 ml LB media containing 100 μg ml−1 carbenicillin and 12.5 μg ml−1 chloramphenicol to an OD of 0.6. Expression was induced for 4 h with 1 mM isopropyl-β-D-galactopyranoside and the cells then harvested and frozen. Cells were thawed on ice and resuspended in 30 ml of lysis buffer (75 mM Na-phosphate, pH 7.5, 500 mM NaCl, 1 mM benzamidine, 3 μg ml−1 leupeptin, 250 μM 4-(2-aminoethyl)benzenesulphonyl fluoride, 10 U ml−1 DNAase I (Sigma), 10,000 U ml−1 lysozyme (Sigma), 1 mM 2-mercaptoethanol). The cells were then lysed by sonication, (4 × 30 s bursts), with cooling to <4°C between pulses. After centrifugation (30,000 × g, 1 h, 4°C), clarified extract was applied at 2 ml min−1 at 4°C to a nickel-charged 20 ml (1.2 × 11 cm) Ni-NTA chelating agarose column (Qiagen) equilibrated with buffer A (50 mM Na-phosphate, pH 7.5, 200 mM NaCl). Unbound protein was removed by washing with 50 ml of this buffer. Retained proteins were eluted with a linear gradient at 1 ml min−1 of 0–100% of buffer B (50 mM Na-phosphate, pH 7.5, 200 mM NaCl, and 500 mM imidazole).
2.5 Enzyme assays
FTL activity was measured using a temperature-controlled Beckman DU-640 spectrophotometer, essentially as described [37]. Assays were carried out in a 0.5 ml volume at 30°C and contained 25 mM K+-HEPES, pH 7.5, 1 mM THF, 200 mM sodium formate, 100 mM KCl, 10 mM MgSO4 and 5 mM ATP. Reactions initiated by the addition of enzyme and stopped by the addition of 500 μl 0.5 M HCL. The 5,10-CH=THF produced was quantified at 350 nm, using a extinction coefficient of 24.9 mM−1 cm−1. Background rates were subtracted from all data and at maximal substrate concentrations were less than 2% of the enzymatic rate. THF was dissolved just before use in 250 mM triethanolamine-Cl, pH 7, 40 mM 2-mercaptoethanol. Enzyme inhibition was measured using the same assay, with stocks of hydrophobic diaminoquinazolines and diaminopteridines dissolved in DMSO. Inhibition was expressed as a percentage of a no-drug DMSO control. DHCH activity was assayed as described previously, using either 1 mM NAD+ or 1 mM NADP+ in the reaction mixture [31].
2.6. Generation of ftl− null mutants
Fusion PCR was used to generate the two targeting constructs, containing either Blasticidin (BSD) or nourseothricin (SAT) drug-resistance cassettes inserted between the 5′ and 3′ LmjFTL flanking sequences [38]. First, the flanking sequences and drug resistance cassettes were amplified separately by PCR using primers that produce overlapping ends. The 5′ and 3′ LmjFTL flanking sequences (842 and 781 bp), were amplified using the primers SMB2651/2652 and SMB2653/2654 that added an XbaI and NdeI site, respectively. BSD and SAT drug-resistance cassettes (399 and 526 bp) were amplified using the primers SMB2555/2556 and SMB2630/2631. The final targeting construct was then made by mixing the purified PCR products for the flanking sequences and a drug-resistance cassette and using these products as templates in a second round of PCR with the external primers 2651 and 2654. These targeting fragments were then inserted by TA cloning into pGEM-T (Promega), yielding pGEM-T.5′.BSD.3′.LmjFTL (B5897) and pGEM-T.5′.SAT.3′.LmjFTL (B5896). The sequences of these constructs were confirmed. These DNAs were digested with XbaI and NdeI, and the BSD or SAT targeting fragments (1.9 or 2.0 kb respectively) were gel-purified.
Parasite transfection was performed as described [39]. First, the BSD or SAT targeting fragments were electroporated separately into WT L. major FV1 promastigotes, and colonies were obtained following plating on semisolid M199 medium containing 10 μg ml−1 Blasticidin or 100 μg ml−1 nourseothricin, after 1–2 weeks. Clonal lines were generated and PCR tests confirmed the presence of heterozygous +/ΔFTL::BSD or +/ΔFTL::SATlines respectively, using a forward primer located 5′ of the FTL ORF targeting fragment (2729) and a reverse primer located within the BSD or SAT genes (2556/2631), or a reverse primer located 3′ of the FTL ORF targeting fragment (2730) with a forward primer located within the BSD or SAT genes (2555/2630).
For deletion of the second allele, heterozygous +/ΔFTL::BSD or +/ΔFTL::SATclonal lines were transfected with targeting constructs containing SAT or BSD, respectively, and parasites were plated as before on media containing both antibiotics. Recovery of the planned replacements was confirmed by PCR using primer pairs specific for either the integrated drug resistance cassette (described above), and loss of the FTL confirmed using primers specific for the chromosomal FTL allele (3123/3124). Fourteen homozygous ftl− null mutants were characterized (ΔFTL::SAT/ΔFTL::BSD) nine from the SAT/BSD and five from the BSD/SAT construct transfections. One clone of each line, clone 1 from SAT/BSD and clone 4 BSD/SAT, which were termed ftl− (SB) and ftl− (BS) respectively, were studied further.
To restore FTL expression, ftl− (SB) L. major were electroporated with pXG-FTL (B5917) and plated on semisolid media containing 10 or 12.5 μg ml−1 G418; clonal lines (ΔFTL::SAT/ΔFTL::BSD [pXG-FTL], referred to ftl−/+FTL) were recovered and the presence of the plasmid was confirmed by PCR from genomic DNA with primers specific for the NEO marker (2727/2728) and FTL open reading frame (3123/3124). Similarly, L. donovani pXG-DHCH1-GFP or pXG-DHCH2-GFP transfectants were isolated by electroporation of WT L. donovani and isolation of transformants on plates containing 12 μg ml−1 G418.
To generate lines to test the ability of LiDHCH1 and LiDHCH2 to rescue loss of DHCH1 in L. major, pIR1-PHLEO- LiDHCH1 and LiDHCH2 were linearized by SwaI digestion and electroporated into either WT or dhch1−/+pXNG-LmjDHCH1 L. major [31]. This allows integration of the vector into the ribosomal small subunit locus. Transfectants were recovered by plating on semisolid media containing either 20 μg ml−1 phleomycin for WT, or 20 μg ml−1 phleomycin and 100 μg ml−1 nourseothricin for the dhch1−/+pXNG-LmjDHCH1 line. Single-cell sorting into standard M199 media was performed as described previously [31].
2.8. Subcellular fractionation
Extracts from L. major WT, ftl− (SB), dhch1−/+pXNG-FTL and FV1 pXG-GCVP-GFP parasites were prepared and fractionated as described [40], yielding fractions enriched for mitochondrial and cytoplasmic proteins. Briefly, parasites in the log phase (108 cells) were resuspended in 500 μl lysis buffer containing 125 mM sucrose, 50 mM KCl, 4 mM MgCl2, 0.5 mM EDTA, 20 mM K+-Hepes pH 7.2 and protease inhibitor cocktail (Sigma). The cells were lysed by sonication, and centrifuged at 12,000 × g for 20 min to obtain supernatant and pellet fractions. The pellet fraction was washed 3 times with lysis buffer. Protein concentrations were determined using the Bradford dye-binding assay.
2.9. Immunization and antibody purification
Polyclonal antisera were raised against purified FTL and LmjDHCH1 antigens by the commercial company Proteintech. Recombinant LmjDHCH1 was purified as described elsewhere [31]. Two rabbits were used for each antigen, with the primary injection being followed by four boosts, each two weeks apart. Prior to use, the anti-FTL antiserum was affinity-purified by binding to FTL protein that had been linked to cyanogen bromide-activated Sepharose 4B (Sigma), using a modified version of a published protocol [41]. All steps were carried at room temperature, using a rotating tube holder to keep the resin in solution. A 2 ml sample of the activated resin was washed in 10 ml 1 mM HCl and 10 ml water, and then incubated with 5.8 mg FTL protein in 3 ml 50 mM Na-phosphate, pH 7.5, 200 mM NaCl. This procedure gave a resin with a ligand density of 6.5 mg ml−1. Unreacted cyanogen bromide groups were then blocked by incubation for 1 h with 10 ml 1 M ethanolamine pH 8, and unreacted protein removed by washing twice with 10 ml PBS and twice with 10 ml PBS containing 1 M NaCl. A 750 μl sample of the FTL-resin was then incubated for 1 h with 12 ml anti-FTL antisera, washed again with 2 × 10 ml in PBS and 2 × 10 ml PBS containing 1 M NaCl, and bound antibodies eluted in 2 ml 100 mM glycine pH 2.7. The eluate was then rapidly neutralized by the addition of 100 μl 3 M Tris, pH 8.4.
2.10. Western blot analysis
Proteins from sub-cellular fractions (5 μg) were separated by electrophoresis on a 10% SDS polyacrylamide gel and electrotransferred onto nitrocellulose membranes (Hybond-ECL; Amersham Biosciences). A rabbit polyclonal antibody anti-GFP purchased from AbCam (Cambridge, MA) was used at a 1:10,000 dilution; antisera raised against LmFTL and LmjDHCH1 were used at a 1:1,000 and 1:3,000 dilution, respectively. Secondary antibodies conjugated to peroxidase were from Amersham Biosciences, and bound antibodies detected using the PerkinElmer Life Sciences chemiluminescence kit, following the manufacturer’s instructions. The specificities of the antisera were assessed by comparisons between WT and mutant lines lacking either DHCH1 or FTL genes: ftl− (SB) above and also dhch1− [31]. The relative molecular masses of the DHCH1 and FTL bands were calculated using a standard curve produced by a linear fit to the migration of the standards plotted against the reciprocal of their molecular mass [42].
2.11. Immunofluorescence
Log-phase L. donovani expressing GFP-tagged LiDHCH2 were collected by centrifugation and resuspended in RPMI media containing Mitotracker Red CMXRos (Invitrogen; 125 nM) for 15 min. Parasites were then washed in media and fixed in 2% paraformaldehyde (in PBS) for 2 min, washed in PBS, and deposited on coverslips by centrifugation. The coverslips were then incubated in blocking buffer (PBS containing 0.1% (v/v) Triton-X-100 and 5% (v/v) normal goat sera) for 30 min prior to staining with a rabbit anti-GFP polyclonal antibody (Abcam, diluted 1:500 in blocking buffer) for 1 h. Following extensive washing in PBS, the coverslips were then stained with an Alexafluor488 goat anti-rabbit antibody (Invitrogen, diluted 1:1000 in blocking buffer) and Topro-3 (Invitrogen, 2 μM) for 45 min. Coverslips were then washed again in PBS and mounted on ProLong Gold reagent (Invitrogen). Images were acquired on a Zeiss 510 META confocal laser scanning microscope and levels were adjusted with Photoshop software (Adobe).
3. Results
3.1. Phylogenomic profiling of Trypanosomatid genes implicated in 10-CHO-THF synthesis
We used dehydrogenase/cyclohydrolase (DHCH) and formyl-THF ligase (FTL) sequences from both eukaryotes and prokaryotes to probe the trypanosomatid sequence database GeneDB for related genes (http://www.genedb.org/, searches were performed in October 2008). These searches yielded three groups, with two showing similarity to DHCHs and one related to FTLs. The predicted genes, key properties and their distribution within trypanosomatids are summarized in Table 1. As the genomes for several of these trypanosomatids are incomplete, ‘absent’ assignments must be considered provisional. No genes related to DHCH or FTL were found in the predicted products of the trypanosomatid mitochondrial genomes.
Table 1.
10-CHO-THF synthetic enzymes in Trypanosomatid protozoans
| Organism | FTL | DHCH1 | DHCH2 |
|---|---|---|---|
| Leishmania major | FTL (c) | DHCH1 (c) | pseudo |
| L. infantum (L. donovani*) | FTL (c) | DHCH1 (c) | DHCH2 (m) |
| L. mexicana | FTL (c) | DHCH1 (c) | DHCH2 (m) |
| L. braziliensis | FTL (c) | DHCH1 (c) | — |
| Crithidia fasciculata | FTL (c) | DHCH1 (c) | DHCH2 (m) |
| Trypanosoma cruzi | FTL (c) | DHCH1 (c) | DHCH2 (?) |
| T. brucei | — | DHCH1 (c) | — |
Proteins whose cellular localization has been experimentally established are shown in bold. Assignment of other proteins to cytosol (c) or mitochondrion (m) is based upon the absence of N-terminal amino acid extensions, and/or PSORT II predictions. Dashes signify that related sequences were not found in blastp and tblastn searches of the genomes available (www.genedb.org).
The closely related species L. donovani was shown in Section 2.3 to contain DHCH1 and DHCH2; the presence of FTL was not surveyed.
A single FTL homolog was found in Leishmania species, Crithidia fasciculata and T. cruzi, while no FTL-related sequences were detected in the African trypanosomes T. brucei, T. vivax and T. congolense datasets (Table 1). Comparison of the sequence of L. major FTL with that of FTL from the bacterium Moorella thermoacetica, where the structure of the enzyme is known [43], showed an overall identity of 54%, and conservation of key amino acid motifs (Fig. S1). Of the eleven residues predicted to be involved in the active site of the M. thermoacetica enzyme, 9 were identical in L. major with two conservative substitutions (E9D and T75S) (Fig. S1). Phylogenetic analysis suggests that the trypanosomatid FTLs are related to those of eukaryotes, with the trees having a topology similar to that expected from organismal relationships (data not shown).
Two DHCH-related gene clusters were identified. The first, termed DHCH1, occurs in all trypanosomatid species and shows high levels of similarity to other eukaryotic DHCH enzymes (Fig S2). The L. major DHCH1 was shown previously to have the expected enzymatic properties [31]. The second, termed DHCH2, occurs as a pseudogene in L. major (LmjF22.0340, termed pseDHCH2) encoding a truncated product lacking amino acid residues that form part of the active site in the human MTHFD1 (Fig. S2 and [3]). DHCH2-related sequences were not detected in L. braziliensis, T. congolense, T. vivax and T. brucei, but DHCH2 occurred in a seemingly intact form in L. infantum, L. mexicana and T. cruzi (Fig S2 and Table 1). Unlike DHCH1s, most of the predicted DHCH2 proteins have short leader sequences resembling mitochondrial targeting peptides (for example, the PSORT II algorithm assigns L. infantum DHCH2 as mitochondrial with 57% confidence; [44] Fig. S2.). However, the T. cruzi DHCH2 lacks such an extension (Fig. S2).
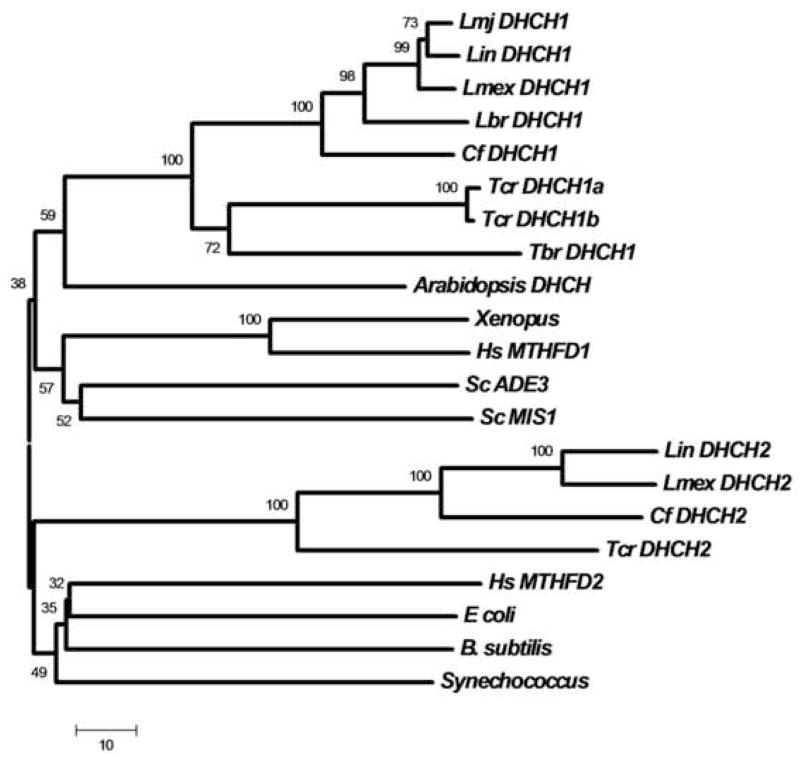
Alignments and phylogenetic trees of the predicted trypanosomatid DHCH1 and DHCH2 proteins with DHCH domains of prokaryotes and eukaryotes showed strong evolutionary conservation (Fig. 2, S2). These studies clearly grouped trypanosomatid DHCH1s with tri or bifunctional DHCH enzymes from other eukaryotes. Trypanosomatid DHCH2s in contrast constituted a distinct group, associated with the human mitochondrial DHCH encoded by MTHFD2 and prokaryotic DHCHs in most evolutionary trees (Fig. 2 or data not shown). In contrast, compelling studies have established that the human MTHFD2 most likely arose from the trifunctional enzyme, as evidenced by homology of the MTHFD2 3′ UTR with the FTL domain of the trifunctional cytosolic protein [45].
Figure 2. Evolutionary relationships amongst DHCHs (or DHCH domains).
A phylogenetic tree was constructed from the indicated DHCHs (or DHCH domains) by the Neighbor Joining method as implemented in the MEGA 4 software package [59]. A consensus bootstrap tree (50,000 replicates) is shown; the small numbers above each branch represent the boostrap confidence percentages and the bar corresponds to 10 amino acid substitutions. Trypanosomatid sequences were taken from the GeneDB database unless otherwise indicated (www.genedb.org). Abbreviations are Lmj, L. major; Lin, L. infantum; Lmex, L. mexicana, Lbr, L. braziliensis; Cf, Crithidia fasciculata, Tcr, Trypanosoma cruzi; Tbr, T. brucei, Hs, Homo sapiens, and Sc, Saccharomyces cerevisae. Sequences considered were Lmj DHCH1 (LmjF26.0320), Lin DHCH1 (XP_001470379), Lmex DHCH1 (Contig_0001325), Lbr DHCH1 (XP_001562267), Cf DHCH1 (SMB, in preparation), Tcr DHCH1a and b (XP_806444, XP_818429), Tbr DHCH1 (XP_845784), Lin DHCH2 (LinJ22_V3.0220), Lmex DHCH2 (Contig_0001368), Cf DHCH2 (SMB, in preparation), Tcr DHCH2 (XP_813647), Arabidopsis DHCH (NP_191971), Xenopus (NP_001080574), Hs MTHFD1 (P11586), Sc ADE3 (NP_011720), Sc MIS1 (EDN64699), Hs MTHFD2 (P13995), E. coli FolD (P24186), B. subtilis (NP_390311), and Synechococcus (YP_399796).
Sequence alignments (Fig. S2) showed the presence of conserved residues implicated in substrate binding predicted from the human DHCH domain structure, including the active site tyrosine and lysine as well as neighboring amino acids, in both trypanosomatid DHCH1s and DHCH2s [3]. Importantly, trypanosomatid DHCH1s share the conserved residues that confer specificity for NADP+ in MTHFD1, most notably Arg173, which interacts with the coenzyme 2′ phosphate group in the human enzyme [46]. This corresponds to the observation that the L. major DHCH1 (LmjF26.0320) encodes a NADP+-dependent DHCH activity [31]. However, the low level of conservation in the residues implicated in determining the specificity of cofactor binding in the human MTHFD2 or bacterial enzymes hinders the prediction of the specificity of the DHCH2 enzymes [47, 48]. While the predicted DHCH2 proteins contain insertions of approximately 100 residues, relative to bacterial or human DHCH domains, these differences would be predicted, by modeling against the human DHCH domain structure, to fall in an exposed ‘loop’. However, despite the conservation of key catalytic residues in DHCH2s, we have as yet been unable to demonstrate enzymatic activity associated with the L. infantum DHCH2 protein (Section 3.8).
3.2. FTL activity and inhibition
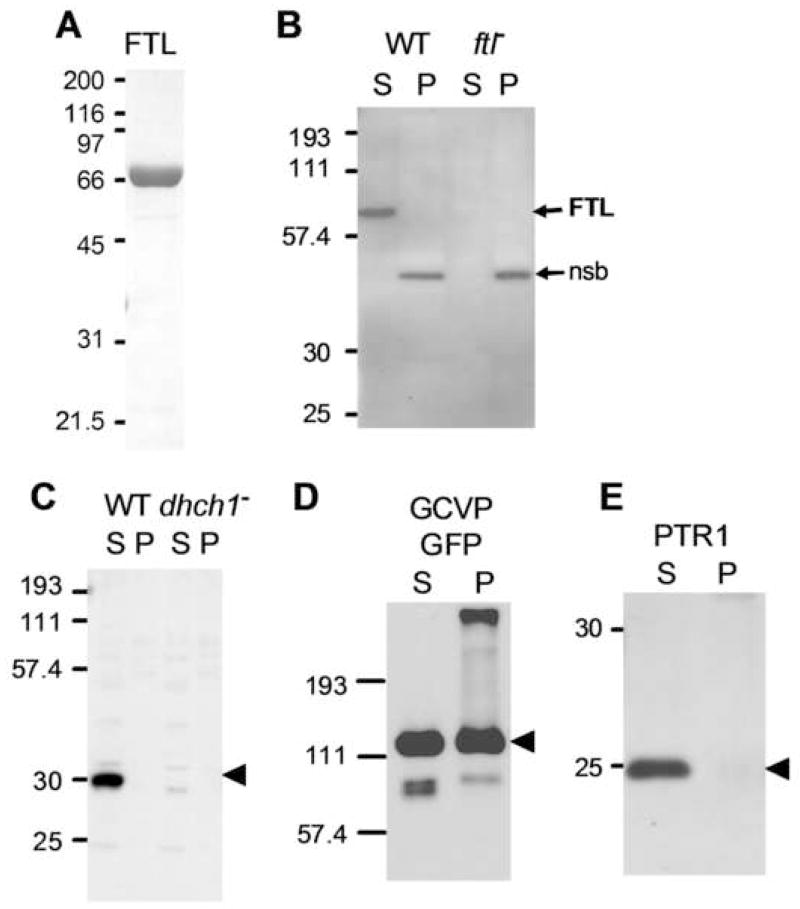
L. major FTL was expressed in E. coli as a hexahistidine-tagged fusion protein and purified by metal-affinity chromatography, yielding preparations more than 95% pure by SDS-PAGE (Fig. 5A). As expected, the recombinant FTL catalyzed the ATP-dependent production of 10-CHO-THF from formate and THF (Table 2). The specific activity of the purified L. major FTL was 3.3 μmol min−1 mg−1, comparable to that reported for the FTL encoded by the cytosolic human C1-synthase (12.5 μmol min−1 mg−1) [49]. The lack of saturation observed with THF-monoglutamate is also seen with human mitochondrial FTL and the rabbit liver C1 synthase, where this results from a preference by these mammalian enzymes for the polyglutamylated form of THF [13, 50].
Figure 5. Localization of L. major FTL and DHCH1 to the cytosol.
A. Purification of L. major FTL. 5 μg of purified recombinant protein was separated by SDS-PAGE and stained with Coomassie blue. B. Western blot with anti-FTL antiserum of soluble (S) and pellet (P) fractions of WT and ftl− lines. The positions of FTL and a non-specific band (nsb) are shown to the right of the blot. C. Western blot with an anti-DHCH1 antiserum of soluble (S) and pellet (P) fractions of WT and dhch1−/+FTL lines [31]. This line lacks the DHCH1 gene, but is viable due to provision of 10-CHO-FTL from FTL overexpression. The arrow marks the position of DHCH1. D. Western blot with an anti-GFP antiserum of soluble (S) and pellet (P) fractions from a WT transfectant expressing a GCVP-GFP fusion protein (pXG-GCVP-GFP) targeted to the parasite mitochondrion [40]. The arrow marks the position of GCVP-GFP. E. Western blot with an anti-PTR1 antiserum of soluble (S) and pellet (P) fractions from WT. The arrow marks the position of PTR1.
Table 2.
Kinetic constants of Leishmania major FTL
| Substrate | Km (app) (μM) | kcatd (s−1) | kcat/Km(M−1 s−1) × 104 |
|---|---|---|---|
| THF a | > 1,500 | - | - |
| ATP b | 360 ± 50 | 3.7 ± 0.1 | 1 |
| Formate c | 21 ± 2 | 4.4 ± 0.1 | 21 |
Data are from a single determination and are given with the standard error of a non-linear fit to the Michaelis-Menten equation.
Measured with 5 mM ATP and 200 mM formate as the fixed substrates.
Measured with 1 mM THF and 200 mM formate as the fixed substrates.
Measured with 1 mM THF and 5 mM ATP as the fixed substrates.
Calculated assuming one active site per polypeptide.
A selection of pteridine analogues were tested as inhibitors at a concentration of 100 μM. No inhibition was observed with methotrexate or with two hydrophobic pteridine analogues, 2,4-diamino-6-(3,4-dichlorophenoxy)-quinazoline or 2,4-diamino-6-benzyl-5-(3-phenylpropyl)-pyrimidine (numbered 34 or 70, respectively in reference [51]). These pteridine analogues were previously characterized as potent inhibitors of both the L. major dihydrofolate reductase and pteridine reductase 1, but have additional uncharacterized targets in Leishmania [51]. We also tested N5-N10-carbonyl tetrahydrofolate, a DHCH inhibitor active against mammalian and L. major DHCH1 [31, 52], which failed to inhibit FTL activity when tested at 100 μM (data not shown).
3.3. Generation of ftl− null mutants and FTL overexpressors
Leishmania are predominantly diploid and therefore require two rounds of gene targeting to generate null mutants [53]. The ORFs of the two FTL alleles were deleted sequentially by homologous gene replacement, and replaced by BSD and SAT drug-resistance ORFs [53]. The null mutants were viable and control PCR tests confirmed the absence of the FTL and the generation of the planned SAT and BSD replacements (Fig. S3). Restoration of FTL was achieved by introduction of an episomal FTL expression construct, yielding add-back lines termed ftl−/+FTL. The null mutants and the add-back promastigotes grew at rates similar to WT during in vitro culture (data not shown).
3.4. Differentiation and virulence of ftl− null mutants
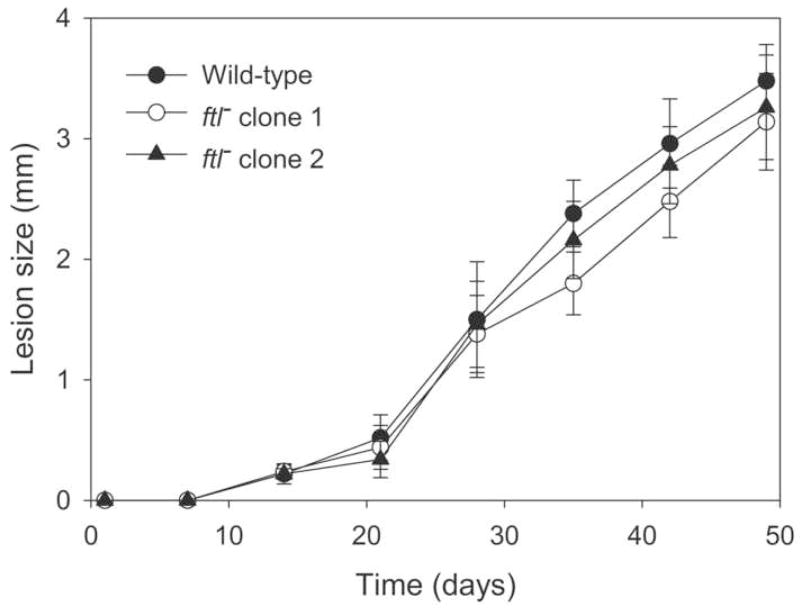
The ability of two independent ftl− lines to differentiate into the infective metacyclic promastigote form in vitro was assessed by the peanut agglutinin assay [33]. Metacyclic promastigotes do not bind to this lectin and can thus be scored as peanut agglutinin negative (PNA−) cells. Maximal numbers of metacyclics were observed 6 days after entry into stationary phase; at that time PNA− metacyclics comprised 18% of the WT cells, compared with 21% and 12% for two independent ftl− clonal lines. PNA− metacyclics were purified from WT and the two ftl− lines and 105 were injected into the footpads of BALB/c mice, which are highly susceptible to L. major infection. All infections produced lesions which appeared at similar times and progressed similarly thereafter (Fig. 3). FTL activity is therefore neither required for differentiation into the infective form of the parasite, the establishment of infection by metacyclics, nor for replication as amastigotes within the host.
Figure 3. Pathology induced in BALB/c mice by WT and ftl− L. major.
Mice were injected with 1×105 metacyclic promastigotes in the footpad, and the increase in footpad thickness caused by lesion progression was measured; the mean and standard deviation is shown. (●) WT, (○) and (▲) two independent ftl− lines.
3.5. Drug sensitivities of the ftl− null mutant and FTL overexpressor
We were interested in whether 10-CHO-THF metabolism might represent the uncharacterized target that was proposed as an explanation for the lack of effect produced by varying PTR1 and dihydrofolate reductase levels on the sensitivity of L. major to cytotoxic pteridine analogues [51]. Therefore we measured the sensitivity of WT, ftl− and ftl−/+ FTL lines towards methotrexate and to the two hydrophobic pteridine analogues that were tested above for FTL enzymatic inhibition (Table 3). As expected, western blots confirmed that the episomal ftl−/+ FTL line over expresses the FTL enzyme (Fig. S4). Despite these manipulations leading to either loss or over expression of FTL-dependent activities, no differences in drug sensitivity were seen amongst any of the lines tested (Table 3).
Table 3.
Drug sensitivities of ftl− mutant and FTL overexpressing lines
| EC50 (nM) | |||
|---|---|---|---|
| Line | Methotrexate | Compound 70 | Compound 34 |
| WT | 60 ± 10 | 170 ± 10 | 310 ± 30 |
| ftl− (BS) | 60 ± 10 | 200 ± 40 | 400 ± 150 |
| ftl− (SB) | 50 ± 40 | 130 ± 60 | 340 ± 50 |
| ftl−/+pXG-FTL | 50 ± 20 | 150 ± 50 | 400 ± 200 |
The numbers shown are the mean ± standard deviation for two determinations. Compound 34 is 2,4-diamino-6-(3,4-dichlorophenoxy)-quinazoline) and compound 70 is 2,4-diamino-6-benzyl-5-(3-phenylpropyl)-pyrimidine) [51]. ftl− lines obtained by sequential BSD then SAT replacements (BS), or by SAT and then BSD (SB).
3.6. Serine and glycine requirements of the ftl− null mutant and FTL overexpressor
Serine and glycine are interconverted in both the cytosol and mitochondrion by SHMT, which either generates or consumes 5,10-CH2-THF (Fig. 4B). The 5,10-CH2-THF pool is influenced by the combined activities of FTL, DHCH, and the mitochondrial glycine cleavage complex (GCC), with these enzymes either incorporating formate into the C1-folate pool to supply 5,10-CH2-THF for serine biosynthesis, or converting the 5,10-CH2-THF produced during glycine biosynthesis back into THF. Therefore the effects of FTL ablation or over expression on the ability of L. major to grow at low levels of these amino acids were examined. The lines were first adapted to growth in a semidefined media containing 1% fetal calf serum. When placed in media containing reduced serine (1 or 2 mg ml−1, 15 or 30-fold lower) or glycine (2 mg ml−1, 5 fold lower) WT parasites showed reduced growth, with the lowest concentrations of serine and glycine inhibiting the growth to 13% and 16% of that seen with complete media (Fig. 4A). The growth of the ftl− null mutant was strongly inhibited under these conditions, with glycine limitation preventing growth entirely (<1%) and serine limitation reducing growth to 10% of that of WT in complete media (Fig. 4A). These effects were reversed in the control ftl−/+FTL line, which closely resembled WT (Fig. 4A).
Figure 4. Growth of WT, ftl− and ftl−/+FTL L. major in low concentrations of glycine and serine.
A. Parasites were adapted and grown in semidefined RPMI media as described in the methods. Growth is shown relative to that of WT in complete RPMI media (= 100%). Amino acid levels were altered as follows: 5-fold reduced levels of glycine (Gly 5-fold lower), or serine at 15 or 30-fold lower levels, (Ser 5-fold lower or Ser 30-fold lower, respectively). White bars, WT; black bars, ftl−; and gray bars, ftl−/+FTL. The data are the mean of three independent experiments, with the standard deviation shown. B. Pathway for FTL and DHCH-dependent interconversion of serine and glycine.
3.7. Localization of L. major FTL and DHCH1 to the cytosol
Antisera were raised in rabbits against purified L. major FTL and DHCH1 proteins, and their specificity confirmed by tests with WT and null mutants (Fig. 5B, 5C; below; [31]). Sub-cellular fractionation of L. major extracts by low-speed centrifugation yields a large particulate fraction (designated P), which is enriched for mitochondrial proteins, and a soluble fraction enriched for cytosolic proteins (designated S) [40]. Anti-FTL antisera recognized a band with a relative molecular mass (Mr) of 68 kDa in WT parasites, in good agreement with the predicted mass of 67 kDa, which was absent in extracts prepared from the ftl− mutant (Fig. 5B). Importantly, the FTL band was present exclusively in the cytosolic fraction (S) with no reactivity in the particulate (P) fraction (Fig. 5B). In contrast, a 42 kDa nonspecific cross reacting band (nsb) was localized exclusively to the particulate fraction, fortuitously providing a useful internal fractionation control. Anti-DHCH1 antisera recognized a protein with a Mr of 30 kDa in WT parasites, in good agreement with the predicted mass of 32 kDa, and this band was absent in a dhch1−/+FTL line (dhch1− lines are not viable, but can be rescued by FTL overexpression; [31]). As with FTL, the DHCH1 protein was only detectable in the cytosolic fraction (Fig. 5C).
As a mitochondrial protein control we examined the localization of a GFP-tagged glycine cleavage complex P-protein (GCVP) [40]. Western blotting with anti-GFP antisera revealed high levels of reactivity in the particulate fraction, as well as strong reactivity in the soluble fraction (Fig. 5D). The GCVP-GFP band was not detected in WT parasites (not shown). The soluble GCVP-GFP signal presumably arises from breakage of organelles during cell lysis and extraction, as previous studies on this line using immunofluorescence had observed an exclusively mitochondrial localization [40]. Interestingly, the ‘nsb’ band in the FTL blot did not show this distribution, being localized entirely to the particulate fraction. This may reflect association of this unknown immunoreactive protein with other organelles, or perhaps associations with cell membranes or the cytoskeleton. Nevertheless, this does not detract from the use of the GCVP-GFP fusion protein to establish that this extraction method does localize a known mitochondrial protein to the particulate fraction. As a cytosolic protein control, an antibody against L. major pteridine reductase 1 localized this protein exclusively to the cytosolic fraction, as expected from this protein’s lack of a predicted mitochondrial targeting sequence (Fig. 5E). These data establish that in L. major, both DHCH1 and FTL are localized exclusively to the cytosol and are not detectable in the mitochondrion or other vesicular compartments.
3.8. L. infantum DHCH2 is localized to the mitochondrion
We were curious about the localization of DHCH2, given its potentially overlapping metabolic role with DHCH1, and possible mitochondrial localization. For these studies we turned to L. donovani, a closely related parasite used routinely in our laboratory. We sequenced the L. donovani DHCH1 and DHCH2, and found that the LdDHCH1 differed by a single conservative amino acid substitution from LiDHCH1 (Ala232 versus Thr232), while the DHCH2 proteins were identical. We then generated GFP fusions to the C-termini of L. infantum DHCH1 and DHCH2, expressed these in L. donovani, and visualized their cellular localization.
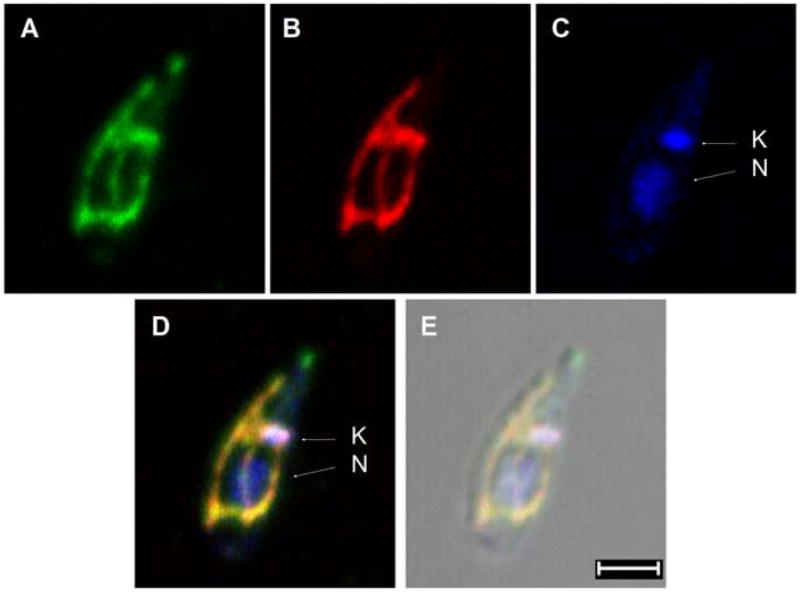
As expected from the studies above on L. major DHCH1, L. infantum DHCH1-GFP was detected throughout the parasite cytosol (data not shown). In contrast, the L. infantum DHCH2-GFP fusion protein showed a branched tubular pattern running the length of the cell, which is typical for proteins localized to the single large mitochondrion present in Leishmania. Co-localization was also seen with the parasite kinetoplast, but not the nucleus (Figs. 6A and 6C, merge 6D). To confirm this localization, cells were incubated with a fixable mito-tracker dye, which showed extensive co-localization with the LiDHCH2-GFP protein and the kinetoplast (Fig. 6A and 6B, merge 6D). Thus we conclude that while DHCH1 is cytosolic, L. infantum DHCH2 is targeted to the mitochondrion.
Figure 6. Immunolocalization of L. infantum DHCH2.
L. donovani transfected with pXG-LiDHCH2-GFP were stained with an anti-GFP antibody (green, panel A), Mitotracker Red CMXRos (red, panel B) and Hoechest 33342 (blue, panel C). The kinetoplast (K) and nucleus (N) labeled with arrows. Merged fluorescence images are shown in panel D and the corresponding differential interference contrast image is shown in panel E, scale bar is 2 μm. No GFP staining was seen in negative controls where the primary anti-GFP antibody was omitted, or in untransfected wild-type cells (not shown).
Thus far we have been unable to demonstrate enzymatic activity with LiDHCH2 expressed heterologously in L. major. We introduced LiDHCH1 and LiDHCH2 into WT L. major using integrating expression vectors (pIR1SAT), which yield high level expression of passenger genes due to insertion into the highly expressed rRNA locus. Using a spectrophotometric assay [31], the NADP-dependent activity of the endogenous LmjDHCH1 in WT cells was barely detectable, with a specific activity of 0.07 ±0.03 nmol/min/mg, while overexpression of LiDHCH1 increased this NADP+-dependent activity approximately 50-fold, to a specific activity of 3.7 ±0.07 nmol/min/mg. As expected from the coenzyme specificity of LmjDHCH1 [31], no NAD+-dependent activity was detected in WT or LiDHCH1-overexpressing extracts. In contrast, no increase in activity over WT was seen in extracts from the LiDHCH2-overexpressors, using either NADP+ or NAD+ as the coenzyme. Consistent with these data, preliminary tests using selection for the loss of an episomal rescue vector [31] suggested that the loss of LmDHCH1 from a L. major dhch1− chromosomal null mutant was not rescued by LiDHCH2, but was rescued by LiDHCH1 (data not shown).
4. Discussion
Leishmania major synthesizes 10-CHO-THF through a monofunctional FTL and a bifunctional DHCH1. Unlike their human host, the biosynthesis of 10-CHO-THF in Leishmania major appears to be restricted to the cytosol. Such compartmentalization of 10-CHO-THF biosynthesis is curious, since methionyl-tRNAMet formylation is the only reaction thought to require this metabolite in trypanosomatids, and this modification only occurs in mitochondrial methionyl-tRNAMet [23]. As FTL is not essential for the growth or virulence of L. major, this could either indicate that 10-CHO-THF is not an essential metabolite, or that the contribution of FTL to this pool is not significant. However, studies on the L. major DHCH1 have demonstrated that this gene is required for viability, and that dhch1− null mutants could be rescued by overexpression of FTL [31]. These results argue strongly that 10-CHO-THF is indeed essential in Leishmania, but that FTL plays a secondary role under normal conditions. Consistent with this, we could find little difference in the distribution of C1-THF species in the WT, ftl− and ftl−/+ FTL lines 1.
In humans, mitochondrial and cytosolic 10-CHO-THF metabolism has distinct functions, which arise from the different coenzyme specificities and localizations of the enzymes involved [2]. In the cytosol, the NADP+-dependent DHCH and FTL activities of the C1 synthase provide 10-CHO-THF for purine biosynthesis and supply 5,10-CH2-THF for serine biosynthesis. In the cytosolic compartment the high NADPH/NADP+ and ATP/ADP ratios will favor flux from formate to 5,10-CH2-THF. In contrast, in the mitochondrion the NAD+-dependence of the DHCH activity and the low ATP/ADP ratio instead favors conversion of 5,10-CH2-THF to formate, which is thought to make the mitochondrion a source of formate for cytosolic purine or serine biosynthesis under conditions when serine is limiting [2]. The second function of mitochondrial 10-CHO-THF is as a formyl donor in methionyl-tRNA modification, although the importance of this reaction in eukaryotic cells has been the focus of some debate [27, 28].
In Leishmania the loss of purine biosynthesis has removed a major role for 10-CHO-THF metabolism [20], with the provision of 5,10-CH2-THF for serine biosynthesis by SHMT and mitochondrial tRNA formylation the two remaining known functions. In serine biosynthesis the 10-CHO-THF pathway is reversed, with DHCH reducing 10-CHO-THF produced by FTL into 5,10-CH2-THF, which is then used by SHMT to convert glycine into serine [54]. Previous studies have shown that L. major requires serine for growth, and that growth at low serine concentrations could be restored by the provision of high concentrations of glycine and formate [40]. However, since serine is essential for the growth of WT L. major, this pathway appears insufficient under normal conditions [40]. Moreover, the lack of growth or virulence phenotypes in the ftl− null mutant indicates that this pathway is not required for the growth of promastigotes in vitro, or of amastigotes in vivo. Although the ability of Leishmania to use the cytosolic SHMT, FTL and DHCH1 to synthesize serine is not essential, this function could be useful under conditions of high C1-THF pathway flux, perhaps induced by serine limitation.
The remaining known metabolic role known for 10-CHO-THF is as a substrate for the formylation of the mitochondrial initiator methionyl-tRNAMet [21]. While not studied in Leishmania, methionyl-tRNAMet-e formylation appears essential for mitochondrial protein biosynthesis in the related trypanosomatid T. brucei [24]. Since both DHCH1 and FTL are located in the cytosol of L. major, and previous studies in T. brucei showed that the initiator methionyl-tRNAMet-e is formylated in the mitochondrial matrix [23], this implies that 10-CHO-THF can be transported across the mitochondrial membrane in trypanosomatids. While there is some debate about the nature and extent of organellar C1-THF transport in other organisms [15], recent studies in several species with some studies reveal evidence of organellar transport of C1-THFs including 5-CH3-THF and 5-CHO-THF [17–19]. Although the mammalian mitochondrial reduced folate transporter has been identified [55, 56], its specificity is controversial and no other transporters specific for C1-folates have been identified [57]. If a 10-CHO-THF transporter does exist in Leishmania, it might present a target for inhibition, but such inhibitors would have to selective and not affect mammalian folate transporters. We emphasize that our data do not exclude the possibility that instead of 10-CHO-THF trafficking into the mitochondrion, this metabolite arises in situ by an entirely novel route. However, we favor the simpler transport hypothesis because of its agreement with data from other organisms, as well as the fact that the existence of a novel, hypothetical pathway for mitochondrial 10-CHO-THF biosynthesis in L. major would also require the existence of a novel, hypothetical, and essential function for cytosolic 10-CHO-THF, to explain why the cytosolic DHCH1 is essential [31]. Future studies will be required to address this possibility.
Our phylogenomic analysis further develops the initial genome-based view of the 10-CHO-THF synthesis presented by Opperdoes and Coombs [21], extending to additional species, and identified a second family of DHCH homologues in the trypanosomatids. These DHCH2 genes are absent in L. major, L. braziliensis and Trypanosoma brucei, but present in several other species of Leishmania, as well as in Crithidia and Trypanosoma cruzi. The predicted products of these DHCH2 genes retain residues that form the active site in the human MTHFD1 protein (Fig. S2). Howver, thus far we have been unable to find any evidence for enzymatic activity of LiDHCH2 (Section 3.8). One unlikely possibility is that despite their sequence similarity to NAD(P)-dependent dehydrogenases, the DHCH2 enzymes might use non-nucleotide cofactors, such as direct donation of electrons to flavoenzymes in the respiratory chain or to ubiquinone in the inner mitochondrial membrane. This would not account for the inability of LiDHCH2 to rescue loss of LmDHCH1 in L. major in vivo however. Interestingly, the Leishmania and Crithidia DHCH2s have short N-terminal extensions that resemble mitochondrial targeting sequences, and our data show that an L. infantum DHCH2-GFP fusion protein was localized to the mitochondrion (Fig. 6). One model consistent with these data would be that the evolutionary ancestor of Leishmania possessed a mitochondrial DHCH activity, which was lost independently in L. braziliensis and L. major. The forces leading to and the consequences of such a transition are not obvious based on current knowledge of Leishmania metabolism or of the roles of 10-CHO-THF in other organisms. This question could be addressed by future studies of DHCH2-containing species such as L. donovani. Intriguingly, the presence of a pseudogene in L. major that, if expressed, would produce a truncated protein containing only the N-terminus of DHCH2 is reflected in humans by the production of a short splice variant of the mitochondrial DHCH that encodes a similar truncated open reading frame [58]. However, the protein encoded by this alternative transcript has no detectable enzymatic activity and its function in humans is unknown.
The pathways of 10-CHO-THF biosynthesis seem much reduced in T. brucei, as neither DHCH2 nor FTL homologues are evident in this parasite’s genome (Table 1). In addition, T. brucei cannot carry out 5,10-CH2-THF-dependent serine biosynthesis, since this parasite lacks SHMT [21]. However, consistent with our previous finding that 10-CHO-THF biosynthesis is essential in L. major, all the trypanosomatid genomes examined encode least one enzyme that can produce this metabolite. The retention of this biosynthetic pathway, combined with the probability that mitochondrial methionyl-tRNA formylation is essential in these organisms, indicates that 10-CHO-THF synthesis and mitochondrial C1-folate transport may occur in all these protozoa. This pathway may therefore represent a promising new area for chemotherapy in this family of human pathogens.
Acknowledgments
We thank D. Scott, Andrew Hanson and Jesse Gregory III for discussions, Valeria Naponelli, J. Gregory and A. Hanson for preliminary studies of C1-THF levels. This work was supported by National Institutes of Health Grant AI21903 (to S.M.B.); Brazilian agencies: CNPq and FAPEMIG (to S.M.F.M.) and European Molecular Biology Organization long term fellowship ALTF 106–2005 (to T. V.).
The abbreviations used are
- 10-CHO-THF
10-formyl-tetrahydrofolate
- 5,10-CH2-THF
5,10-methylenetetrahydrofolate
- 5,10-CH=THF
5,10-methenyltetrahydrofolate
- FTL
formyl tetrahydrofolate ligase
- DHCH
methylene tetrahydrofolate dehydrogenase/cyclohydrolase
- PTR1
pteridine reductase 1
- SHMT
serine hydroxymethyltransferase
- GCC
glycine cleavage complex
- THF
tetrahydrofolate
- GFP
green fluorescent protein
- WT
wild-type
Footnotes
Valeria Naponelli, Jesse F. Gregory III, Andrew D. Hanson, T. Vickers, S. Murta, and S.M. Beverley, preliminary data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nzila A. The past, present and future of antifolates in the treatment of Plasmodium falciparum infection. J Antimicrob Chemother. 2006;57:1043–54. doi: 10.1093/jac/dkl104. [DOI] [PubMed] [Google Scholar]
- 2.Christensen KE, MacKenzie RE. Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. Bioessays. 2006;28:595–605. doi: 10.1002/bies.20420. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt A, Wu H, MacKenzie RE, Chen VJ, Bewly JR, Ray JE, Toth JE, Cygler M. Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Biochemistry. 2000;39:6325–35. doi: 10.1021/bi992734y. [DOI] [PubMed] [Google Scholar]
- 4.Nour JM, Rabinowitz JC. Isolation, characterization, and structural organization of 10-formyltetrahydrofolate synthetase from spinach leaves. J Biol Chem. 1991;266:18363–9. [PubMed] [Google Scholar]
- 5.D’Ari L, Rabinowitz JC. Purification, characterization, cloning, and amino acid sequence of the bifunctional enzyme 5,10-methylenetetrahydrofolate dehydrogenase/5,10-methenyltetrahydrofolate cyclohydrolase from Escherichia coli. J Biol Chem. 1991;266:23953–8. [PubMed] [Google Scholar]
- 6.Ragsdale SW, Ljungdahl LG. Purification and properties of NAD-dependent 5,10-methylenetetrahydrofolate dehydrogenase from Acetobacterium woodii. J Biol Chem. 1984;259:3499–503. [PubMed] [Google Scholar]
- 7.Hanson AD, Roje S. One-Carbon Metabolism in Higher Plants. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:119–137. doi: 10.1146/annurev.arplant.52.1.119. [DOI] [PubMed] [Google Scholar]
- 8.Shannon KW, Rabinowitz JC. Purification and characterization of a mitochondrial isozyme of C1-tetrahydrofolate synthase from Saccharomyces cerevisiae. J Biol Chem. 1986;261:12266–71. [PubMed] [Google Scholar]
- 9.Staben C, Rabinowitz JC. Nucleotide sequence of the Saccharomyces cerevisiae ADE3 gene encoding C1-tetrahydrofolate synthase. J Biol Chem. 1986;261:4629–37. [PubMed] [Google Scholar]
- 10.Hum DW, Bell AW, Rozen R, MacKenzie RE. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988;263:15946–50. [PubMed] [Google Scholar]
- 11.Yang XM, MacKenzie RE. NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian homolog of the mitochondrial enzyme encoded by the yeast MIS1 gene. Biochemistry. 1993;32:11118–23. doi: 10.1021/bi00092a022. [DOI] [PubMed] [Google Scholar]
- 12.Christensen KE, Patel H, Kuzmanov U, Mejia NR, MacKenzie RE. Disruption of the mthfd1 gene reveals a monofunctional 10-formyltetrahydrofolate synthetase in mammalian mitochondria. J Biol Chem. 2005;280:7597–602. doi: 10.1074/jbc.M409380200. [DOI] [PubMed] [Google Scholar]
- 13.Walkup AS, Appling DR. Enzymatic characterization of human mitochondrial C1-tetrahydrofolate synthase. Arch Biochem Biophys. 2005;442:196–205. doi: 10.1016/j.abb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Cybulski RL, Fisher RR. Uptake of oxidized folates by rat liver mitochondria. Biochim Biophys Acta. 1981;646:329–33. doi: 10.1016/0005-2736(81)90339-4. [DOI] [PubMed] [Google Scholar]
- 15.Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. Faseb J. 1991;5:2645–51. doi: 10.1096/fasebj.5.12.1916088. [DOI] [PubMed] [Google Scholar]
- 16.Lin BF, Huang RF, Shane B. Regulation of folate and one-carbon metabolism in mammalian cells. III. Role of mitochondrial folylpoly-gamma-glutamate synthetase. J Biol Chem. 1993;268:21674–9. [PubMed] [Google Scholar]
- 17.Horne DW, Holloway RS, Said HM. Uptake of 5-formyltetrahydrofolate in isolated rat liver mitochondria is carrier-mediated. J Nutr. 1992;122:2204–9. doi: 10.1093/jn/122.11.2204. [DOI] [PubMed] [Google Scholar]
- 18.Prabhu V, Chatson KB, Lui H, Abrams GD, King J. Effects of sulfanilamide and methotrexate on 13C fluxes through the glycine decarboxylase/serine hydroxymethyltransferase enzyme system in arabidopsis. Plant Physiol. 1998;116:137–44. doi: 10.1104/pp.116.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson AD, Gage DA, Shachar-Hill Y. Plant one-carbon metabolism and its engineering. Trends Plant Sci. 2000;5:206–13. doi: 10.1016/s1360-1385(00)01599-5. [DOI] [PubMed] [Google Scholar]
- 20.Carter NS, Yates P, Arendt CS, Boitz JM, Ullman B. Purine and pyrimidine metabolism in Leishmania. Adv Exp Med Biol. 2008;625:141–54. doi: 10.1007/978-0-387-77570-8_12. [DOI] [PubMed] [Google Scholar]
- 21.Opperdoes FR, Coombs GH. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 2007;23:149–58. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Simpson AM, Suyama Y, Dewes H, Campbell DA, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–45. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan TH, Bochud-Allemann N, Horn EK, Schneider A. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc Natl Acad Sci U S A. 2002;99:1152–7. doi: 10.1073/pnas.022522299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charriere F, Tan TH, Schneider A. Mitochondrial initiation factor 2 of Trypanosoma brucei binds imported formylated elongator-type tRNA(Met) J Biol Chem. 2005;280:15659–65. doi: 10.1074/jbc.M411581200. [DOI] [PubMed] [Google Scholar]
- 25.Newton DT, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem. 1999;274:22143–6. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 26.RajBhandary UL. More surprises in translation: initiation without the initiator tRNA. Proc Natl Acad Sci U S A. 2000;97:1325–7. doi: 10.1073/pnas.040579197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Holmes WB, Appling DR, RajBhandary UL. Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J Bacteriol. 2000;182:2886–92. doi: 10.1128/jb.182.10.2886-2892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibbetts AS, Oesterlin L, Chan SY, Kramer G, Hardesty B, Appling DR. Mammalian mitochondrial initiation factor 2 supports yeast mitochondrial translation without formylated initiator tRNA. J Biol Chem. 2003;278:31774–80. doi: 10.1074/jbc.M304962200. [DOI] [PubMed] [Google Scholar]
- 29.Spencer AC, Spremulli LL. Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res. 2004;32:5464–70. doi: 10.1093/nar/gkh886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vial L, Gomez P, Panvert M, Schmitt E, Blanquet S, Mechulam Y. Mitochondrial methionyl-tRNAfMet formyltransferase from Saccharomyces cerevisiae: gene disruption and tRNA substrate specificity. Biochemistry. 2003;42:932–9. doi: 10.1021/bi026901x. [DOI] [PubMed] [Google Scholar]
- 31.Murta SM, Vickers TJ, Scott DA, Beverley SM. Methylene tetrahydrofolate dehydrogenase/cyclohydrolase and the synthesis of 10-CHO-THF are essential in Leishmania major. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyard S, Segawa H, Gordon J, Showalter M, Duncan R, Turco SJ, Beverley SM. An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol. 2003;130:31–42. doi: 10.1016/s0166-6851(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 33.Sacks DL, Perkins PV. Development of infective stage Leishmania promastigotes within phlebotomine sand flies. Am J Trop Med Hyg. 1985;34:456–9. doi: 10.4269/ajtmh.1985.34.456. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham ML, Titus RG, Turco SJ, Beverley SM. Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science. 2001;292:285–7. doi: 10.1126/science.1057740. [DOI] [PubMed] [Google Scholar]
- 35.Medina-Acosta E, Cross GA. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol. 1993;59:327–9. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 36.Ha DS, Schwarz JK, Turco SJ, Beverley SM. Use of the Green Fluorescent Protein as a marker in transfected Leishmania. Molec. Biochem. Parasitol. 1996;77:57–64. doi: 10.1016/0166-6851(96)02580-7. [DOI] [PubMed] [Google Scholar]
- 37.Appling DR, Rabinowitz JC. Regulation of expression of the ADE3 gene for yeast C1-tetrahydrofolate synthase, a trifunctional enzyme involved in one-carbon metabolism. J Biol Chem. 1985;260:1248–56. [PubMed] [Google Scholar]
- 38.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–20. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 39.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–28. doi: 10.1016/s0166-6851(03)00079-3. [DOI] [PubMed] [Google Scholar]
- 40.Scott DA, Hickerson SM, Vickers TJ, Beverley SM. The role of the mitochondrial glycine cleavage complex in the metabolism and virulence of the protozoan parasite Leishmania major. J Biol Chem. 2008;283:155–65. doi: 10.1074/jbc.M708014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukas TJ, Watterson DM. Purification of calmodulin and preparation of immobilized calmodulin. Methods Enzymol. 1988;157:328–39. doi: 10.1016/0076-6879(88)57088-x. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Brush JS, Wang T. New method for analyzing the molecular weights of proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochem Biophys Res Commun. 2001;281:976–8. doi: 10.1006/bbrc.2001.4447. [DOI] [PubMed] [Google Scholar]
- 43.Radfar R, Shin R, Sheldrick GM, Minor W, Lovell CR, Odom JD, Dunlap RB, Lebioda L. The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica. Biochemistry. 2000;39:3920–6. doi: 10.1021/bi992790z. [DOI] [PubMed] [Google Scholar]
- 44.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–6. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 45.Patel H, Christensen KE, Mejia N, MacKenzie RE. Mammalian mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase derived from a trifunctional methylenetetrahydrofolate dehydrogenase-cyclohydrolase-synthetase. Arch Biochem Biophys. 2002;403:145–8. doi: 10.1016/S0003-9861(02)00203-5. [DOI] [PubMed] [Google Scholar]
- 46.Allaire M, Li Y, MacKenzie RE, Cygler M. The 3-D structure of a folate-dependent dehydrogenase/cyclohydrolase bifunctional enzyme at 1.5 A resolution. Structure. 1998;6:173–82. doi: 10.1016/s0969-2126(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 47.Christensen KE, Mirza IA, Berghuis AM, Mackenzie RE. Magnesium and phosphate ions enable NAD binding to methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase. J Biol Chem. 2005;280:34316–23. doi: 10.1074/jbc.M505210200. [DOI] [PubMed] [Google Scholar]
- 48.Shen BW, Dyer DH, Huang JY, D’Ari L, Rabinowitz J, Stoddard BL. The crystal structure of a bacterial, bifunctional 5,10 methylene-tetrahydrofolate dehydrogenase/cyclohydrolase. Protein Sci. 1999;8:1342–9. doi: 10.1110/ps.8.6.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan LU, Drury EJ, MacKenzie RE. Methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. A multifunctional protein from porcine liver. J Biol Chem. 1977;252:1117–22. [PubMed] [Google Scholar]
- 50.Strong W, Joshi G, Lura R, Muthukumaraswamy N, Schirch V. 10-Formyltetrahydrofolate synthetase. Evidence for a conformational change in the enzyme upon binding of tetrahydropteroylpolyglutamates. J Biol Chem. 1987;262:12519–25. [PubMed] [Google Scholar]
- 51.Hardy LW, Matthews W, Nare B, Beverley SM. Biochemical and genetic tests for inhibitors of Leishmania pteridine pathways. Exp Parasitol. 1997;87:157–69. [PubMed] [Google Scholar]
- 52.Tonkinson JL, Habeck LL, Toth JE, Mendelsohn LG, Bewley J, Shackelford KA, Gates SB, Ray J, Chen VJ. The antiproliferative and cell cycle effects of 5,6,7, 8-tetrahydro-N5, N10-carbonylfolic acid, an inhibitor of methylenetetrahydrofolate dehydrogenase, are potentiated by hypoxanthine. J Pharmacol Exp Ther. 1998;287:315–21. [PubMed] [Google Scholar]
- 53.Cruz A, Coburn CM, Beverley SM. Double targeted gene replacement for creating null mutants. Proc Natl Acad Sci USA. 1991;88:7170–7174. doi: 10.1073/pnas.88.16.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenzie KQ, Jones EW. Mutants of formyltetrahydrofolate interconversion pathway of Saccharomyces cerevisiae. Genetics. 1977;86:85–102. doi: 10.1093/genetics/86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Titus SA, Moran RG. Retrovirally mediated complementation of the glyB phenotype. Cloning of a human gene encoding the carrier for entry of folates into mitochondria. J Biol Chem. 2000;275:36811–7. doi: 10.1074/jbc.M005163200. [DOI] [PubMed] [Google Scholar]
- 56.McCarthy EA, Titus SA, Taylor SM, Jackson-Cook C, Moran RG. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of chinese hamster cells. J Biol Chem. 2004;279:33829–36. doi: 10.1074/jbc.M403677200. [DOI] [PubMed] [Google Scholar]
- 57.Bedhomme M, Hoffmann M, McCarthy EA, Gambonnet B, Moran RG, Rebeille F, Ravanel S. Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J Biol Chem. 2005;280:34823–31. doi: 10.1074/jbc.M506045200. [DOI] [PubMed] [Google Scholar]
- 58.Prasannan P, Appling DR. Human mitochondrial C1-tetrahydrofolate synthase: submitochondrial localization of the full-length enzyme and characterization of a short isoform. Arch Biochem Biophys. 2009;481:86–93. doi: 10.1016/j.abb.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]