Abstract
Objective
Protein citrullination is an important posttranslational modification recognized by rheumatoid arthritis (RA)–specific autoantibodies. One of the citrullinating enzymes, peptidyl arginine deiminase type 4 (PAD-4), is genetically associated with development of RA in some populations, although the mechanism(s) mediating this effect are not yet clear. There have been descriptions of anti–PAD-4 autoantibodies in different rheumatic diseases. This study was undertaken to investigate whether anti–PAD-4 antibodies are specific to RA, are associated with disease phenotype or severity, and whether PAD-4 polymorphisms influence the anti–PAD-4 autoantibody response.
Methods
Sera from patients with established RA, patients with other rheumatic diseases, and healthy adults were assayed for anti–PAD-4 autoantibodies by immunoprecipitation of in vitro–translated PAD-4. The epitope(s) recognized by PAD-4 autoantibodies were mapped using various PAD-4 truncations. PAD-4 genotyping was performed on RA patients with the TaqMan assay. Joint erosions were scored from hand and foot radiographs using the Sharp/van der Heijde method.
Results
PAD-4 autoantibodies were found in 36–42% of RA patients, and were very infrequent in controls. Recognition by anti–PAD-4 autoantibodies required the 119 N-terminal amino acids, which encompass the 3 nonsynonymous polymorphisms associated with disease susceptibility. Strikingly, the anti–PAD-4 immune response was associated with the RA susceptibility haplotype of PADI4. Anti–PAD-4 antibodies were associated with more severe joint destruction in RA.
Conclusion
Our findings indicate that anti–PAD-4 antibodies are specific markers of RA, independently associated with more severe disease, suggesting that an anti–PAD-4 immune response may be involved in pathways of joint damage in this disease. Polymorphisms in the PADI4 gene influence the immune response to the PAD-4 protein, potentially contributing to disease propagation.
Rheumatoid arthritis (RA), a systemic auto-immune disease affecting 0.5–1% of the population worldwide, is characterized by chronic joint inflammation and, in severe cases, joint erosions (1). Although the mechanisms of initiation and propagation of RA remain incompletely defined, autoimmunity and inflammatory effector pathways appear to play important pathogenetic roles. The notable efficacy of tumor necrosis factor (TNF) inhibitors has established that TNFα plays a central role in RA, and the therapeutic effects of rituximab and abatacept strongly indicate roles for B cells and T cells, respectively (2,3).
Although the specific autoantigens that drive B cells and T cells in RA remained elusive for decades, recent advances have identified protein citrullination as a primary focus of the RA-specific autoantibody response (4). Citrulline is generated posttranslationally by the deimination of arginine, and autoantibodies in RA recognize various naturally citrullinated proteins (including fibrin, vimentin, and filaggrin), as well as cyclic citrullinated peptides (CCPs) derived from them (5-7). Together with the extraordinary specificity (90–99%) of anti-CCP antibodies in RA (8,9), the observation that anti-CCP antibodies are frequently present early in the disease process and often precede development of the diagnostic phenotype (10-13) strongly suggests that these antibodies are markers of the specific events that initiate autoimmunity in RA.
The citrullination reaction is catalyzed by a family of enzymes known as peptidyl arginine deiminases (PADs). There are 5 isoforms (14), differentially expressed in various cells. PAD type 4 (PAD-4) has received particular attention in RA, since it is expressed in myelomonocytes, can be detected in inflamed RA synovium (14,15), and has recently been genetically associated with RA. The first group to describe the genetic association of PADI4 variants with RA defined 2 common haplotypes of the PADI4 gene segregated by 4 exonic single-nucleotide polymorphisms (SNPs) in linkage disequilibrium. These 2 haplotypes were designated “susceptible (haplotype 2)” or “nonsusceptible (haplo-type 1)” based on their relative frequency in a group of Japanese patients with RA versus controls (16). The odds ratio (OR) for association of the susceptibility haplotype with RA was 1.4.
In several other populations, similar associations of PADI4 susceptibility haplotypes with RA were observed, although the magnitude of the effect was lower (17-20). In some studies, no association of PADI4 genotype with RA was observed (21-23). Suzuki et al showed a modest increase in RNA stability for the susceptibility haplotype, and proposed that the genetic effect of PADI4 is mediated through increased PAD-4 levels and activity, with enhanced citrullination and increased levels of anti-CCP antibodies (16). Significant direct support for this model is still lacking, prompting us to explore whether additional mechanisms might mediate some of the genetic effect of PADI4.
We demonstrate here that autoantibodies against PAD-4 protein are a highly specific marker of RA. In a cross-sectional cohort of RA patients, these antibodies were independently associated with a more severe RA phenotype, characterized by worse joint damage and erosions. Notably, anti–PAD-4 autoantibodies were associated with the PADI4 susceptibility haplotype (OR 2.59), particularly with the heterozygous diplotype (OR 4.02). Interestingly, the epitopes recognized by anti–PAD-4 antibodies include the N-terminal region of PAD-4 containing the polymorphisms associated with RA susceptibility. Taken together, the specificity of the antibody response for the polymorphic N-terminal region of PAD-4, the magnitude of the association of autoantibody with susceptibility genotype, and the association of the strongest anti–PAD-4 autoantibody responses with RA severity are striking. They implicate unique PAD-4 structure and/or function in the generation of a PAD-4–specific immune response and, potentially, the downstream augmentation of joint damage in RA.
PATIENTS AND METHODS
Complementary DNA (cDNA) constructs and in vitro transcription and translation (IVTT).
Messenger RNA (mRNA) was extracted from differentiated HL-60 cells and reverse-transcribed to generate cDNA. PAD-4 cDNA was amplified by polymerase chain reaction (PCR) and cloned into the Gateway expression vector pEF-DEST51 (Invitrogen, San Diego, CA). Truncated PAD-4 constructs were generated by PCR amplification of full-length PAD-4 cDNA, and cloned into pDEST15 for expression with an N-terminal glutathione S-transferase (GST) tag, or into pEF-DEST51. 35Smethionine–labeled proteins were generated from cDNA samples by coupled IVTT (Promega, Madison, WI).
Patients
All patients diagnosed as having RA met the American College of Rheumatology (formerly, the American Rheumatism Association) classification criteria (24). Initially, sera from a convenience sample (pilot study) of patients with established RA (n = 38) followed up at the Johns Hopkins Arthritis Center were analyzed. Subsequently, we expanded our studies using sera from 129 patients with established RA enrolled in an ongoing study at Johns Hopkins, called the ESCAPE RA (Evaluation of Subclinical Cardiovascular Disease and its Predictors of Events in Rheumatoid Arthritis) trial. Inclusion criteria for this prospective, observational study of patients with RA for subclinical cardiovascular disease are RA of any duration, age 45–84 years, and absence of a prior clinical cardiovascular event. Sociodemographic and disease-related characteristics of these ESCAPE RA patients are shown in Table 1. Single-view, anteroposterior radiographs of the hands and feet were obtained on all ESCAPE RA patients, and scored using the Sharp/van der Heijde method (25) by a single, trained radiologist (WWS) who was blinded with regard to patient characteristics.
Table 1.
Selected demographic and disease-related characteristics of the patients in the ESCAPE RA cohort*
| All patients (n = 129) |
PAD-4– negative patients (n = 83) |
PAD-4– positive patients (n = 46) |
|
|---|---|---|---|
| Age, mean ± SD years | 59.4 ± 8.1 | 59.2 ± 8.0 | 59.7 ± 8.4 |
| Sex, % female | 63.6 | 62.7 | 65.2 |
| Ethnicity, % | |||
| White | 86.0 | 84.3 | 89.1 |
| African American | 8.6 | 12.0 | 2.2 |
| Asian | 3.1 | 3.6 | 2.2 |
| Other | 2.3 | 6.5 | |
| Body mass index, mean ± SD kg/m2 | 28.5 ± 5.8 | 28.9 ± 5.7 | 27.9 ± 6.0 |
| Disease duration, mean ± SD years | 12.5 ± 10.5 | 10.3 ± 9.7 | 16.3 ± 10.9† |
| Shared epitope status, % | |||
| None | 30.2 | 37.3 | 17.4 |
| Heterozygous | 50.4 | 45.8 | 58.7 |
| Homozygous | 19.4 | 16.9 | 23.9 |
| CRP level, mg/liter | |||
| Mean ± SD | 6.8 ± 14.2 | 5.9 ± 13.0 | 8.4 ± 16.2 |
| Median | 2.7 | 2.1 | 3.9 |
ESCAPE RA = Evaluation of Subclinical Cardiovascular Disease and its Predictors of Events in Rheumatoid Arthritis; PAD-4 = peptidyl arginine deiminase type 4; CRP = C-reactive protein.
P = 0.002 versus PAD-4–negative patients.
Sera from a study of early synovitis at the National Institutes of Health (26) (protocol 94-AR-194) were also tested. Patients in this study had persistent arthritis for <6 weeks but >1 year, and were followed up for development of RA or other arthritides. Only sera from the first visit were available for use in the present study, and only samples from patients who subsequently developed RA were analyzed. Sera from healthy adults and patients with other rheumatic diseases (myositis, scleroderma, Sjögren's syndrome, or systemic lupus erythematosus [SLE]) were used as comparison groups.
All patient samples were de-identified, with clinical and laboratory features linked only to the patient code. All subjects provided informed consent as approved by the Johns Hopkins Institutional Review Board.
Anti-CCP assays
Anti-CCP levels in patient sera were determined using the QUANTA Lite CCP IgG enzyme-linked immunosorbent assay (ELISA) kit (Inova Diagnostics, San Diego, CA).
Immunoprecipitation
Immunoprecipitation using 35Smethionine–labeled IVTT products was performed as previously described (27); products were electrophoresed on 7.5% or 10% sodium dodecyl sulfate–polyacrylamide gels, and visualized by fluorography. To normalize between experiments, one replicate each of the same negative and strong positive RA sera were included as reference sera in each experiment. After densitometric scanning of the autoradiograms, the positive reference serum was assigned a value of 1.0, thus enabling all sera to be scored on a scale of 0–1. Sera with values of >0.3 were designated as 3+. All immunoprecipitation studies were repeated on 2–4 separate occasions, with similar results.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using GenoPrep Cartridge B on a GenoM-6 Robotic Workstation (GenoVision, Exton, PA). Three SNPs from the gene encoding PADI4 were genotyped with the TaqMan assay using primers and probes from Applied Biosystems (Foster City, CA) (including rs11203366, rs11203367, and rs874881). The reactions were set up in 5 μl on 96-well plates in TaqMan Universal Master Mix (Applied Biosystems) with 5 ng DNA, 1 μM of each primer, and 0.2 μM of probe. The thermal cycling reactions (50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute) were run and analyzed on a 7900HT Sequence Detection System (Applied Biosystems) with Applied Biosystems Genotyper software (SDS system, version 2.2). As controls, each plate contained 8 randomly selected, duplicated samples and 4 blank wells without DNA.
Statistical analysis
Descriptive statistics (means and proportions) were calculated for sociodemographic and disease-related characteristics. Associations were evaluated using t-tests, chi-square tests, and calculations of relative odds. P values less than 0.05 were considered significant. For geno-typing studies, departures from Hardy-Weinberg equilibrium at each locus were tested separately among cases and controls, and minor allele frequencies were calculated. Distribution of the genotypes was consistent with Hardy-Weinberg equilibrium, with a P value cutoff of 0.01. To avoid the possibility of missing associations or finding spurious associations due to population substructure, we performed analyses on only the 111 white subjects from the ESCAPE RA cohort, excluding 18 subjects with other self-reported race and/or ethnicity. We used Phase, version 2.1 (28) to estimate haplotypes in the study population, and took best estimates of each subject's diplo-types for further analyses in Stata, version 8.2. Logistic regression models were constructed to obtain ORs and 95% confidence intervals (95% CIs) to estimate genotype and haplotype risks for detectable antibodies.
RESULTS
Anti–PAD-4 autoantibodies are highly specific markers of RA
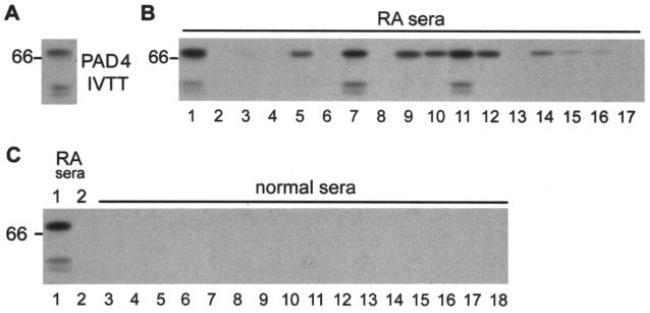
To screen for anti–PAD-4 autoantibodies, 35S-methionine–labeled PAD-4 was generated by IVTT of human PAD-4 cDNA, and was used to screen a convenience set of 38 RA sera for anti–PAD-4 antibodies by immunoprecipitation (pilot study) (Figure 1B). We also tested sera from 32 healthy controls, and from 126 patients with other systemic autoimmune diseases (31 patients with scleroderma, 31 patients with myositis, 32 patients with primary Sjögren's syndrome, and 32 patients with SLE). PAD-4 was frequently targeted in patients with RA (Figure 1B). Antibodies to PAD-4 were demonstrated in 16 (42%) of 38 RA patients, compared with 0 of 32 healthy controls and 1 (0.8%) of 126 control patients (P ≤ 0.0001 for both healthy controls and patients with other rheumatic diseases) (Figure 1C). Anti–PAD-4 antibodies therefore had a sensitivity of 42% and a specificity of 99% for RA.
Figure 1.
Peptidyl arginine deiminase type 4 (PAD-4) is a frequent, specific target in rheumatoid arthritis (RA). A, 35S-methionine–labeled PAD-4 generated by in vitro transcription and translation (IVTT) was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and visualized by fluorography. B, For each immunoprecipitation, 1 μl of PAD-4 generated by IVTT was mixed with 1 μl of patient serum. Results obtained using sera from 17 patients in the RA pilot group (lanes 1–17) are shown. C, Immunoprecipitation with PAD-4 generated by IVTT was performed as described above, using sera from 16 normal controls (lanes 3–18). The RA sera used in lanes 1 and 2 in B were included as reference sera in the immunoprecipitations shown in lanes 1 and 2 of C.
To define possible associations between RA phenotypes, patient genotype, and anti–PAD-4 antibodies, we screened a second, larger patient population from a prospective, observational cohort study of RA (the ESCAPE RA cohort) (Table 1), for which extensive clinical and serologic data are being gathered, and DNA was available. Anti–PAD-4 autoantibodies were found in a similar proportion of these patients (46 [36%] of 129). Using a semiquantitative scale (0−3+) based on densitometry of scanned immunoprecipitation autoradiograms, 83 of 129 sera were found to be negative for anti–PAD-4, while 29 (63%) of 46 antibody-positive sera fell into the highest group (3+). Of 64 randomly selected RA patients from the ESCAPE RA cohort, only 1 serum sample (1.6%) immunoprecipitated PAD-2, which shares 50% homology with PAD-4 (data not shown). The single anti–PAD-2-positive serum sample did not have antibodies against PAD-4. Anti–PAD-4 autoantibodies detected by immunoprecipitation were therefore highly specific for RA.
Recognition of PAD-4 by anti–PAD-4 autoantibodies requires the N-terminal polymorphic domain
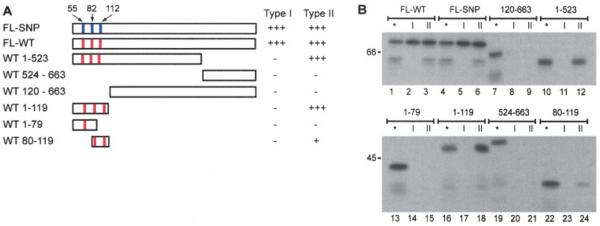
In initial experiments, 2 distinct patterns of antibody recognition of PAD-4 could be distinguished, based on ability to immunoprecipitate full-length PAD-4 and a 58/59-kd truncated product. This short doublet is produced by ribosomal slippage and premature translational termination toward the C-terminus along a stretch of 9 adenine nucleotide residues in the PAD-4 transcript, which encodes 3 lysine residues (K520, 521, and 522) (data not shown). While all antibody-positive sera recognized full-length PAD-4, only a subset of these (5 [31%] of 16 in the pilot study, and 25 [54%] of 46 in the ESCAPE RA cohort) also precipitated the 58/59-kd doublet (Figure 1B). We have designated sera that recognize exclusively full-length PAD-4 as type I sera, and those that recognize both the full-length and truncated forms as type II sera (Figure 2B).
Figure 2.
Both subtypes of PAD-4 autoantibodies require the N-terminal domain encompassing the polymorphic residues for recognition. A, Schematic representation of the PAD-4 constructs used for the immunoprecipitation studies shown in B. Red vertical lines denote S55, A82, and A112 in wild-type (WT) PAD-4. Blue vertical lines denote G55, V82, and G112 in polymorphic PAD-4. The ability of type I and type II PAD-4 antibodies to immunoprecipitate the different constructs is summarized on the right. B, Equally labeled amounts (assessed by densitometry) of different 35S-methionine–labeled PAD-4 products were immunoprecipitated with representative type I or type II PAD-4 autoantibody–positive RA sera. All constructs in the lower panel were fusion proteins containing glutathione S-transferase (GST). None of the sera recognized GST alone (results not shown). For each of the different IVTT constructs, a gel sample consisting of one-fifth of the volume used for immunoprecipitation (lanes labeled with an asterisk) was electrophoresed adjacent to the paired immunoprecipitations. Type I sera recognized only full-length (FL) PAD-4 constructs, and removal of either the N- or C-terminal domains prevented recognition. Type II sera immunoprecipitated all constructs containing the 119 N-terminal amino acids of PAD-4. SNP = single-nucleotide polymorphism (see Figure 1 for other definitions).
To define the epitopes recognized by anti–PAD-4 autoantibodies, we generated constructs encoding polymorphic PAD-4 and PAD-4 truncations, and used these for immunoprecipitation studies with type II and type I sera from the ESCAPE RA cohort (Figure 2). We initially generated a construct (PAD-4 120–663) that did not include the 119 N-terminal amino acids, which represent the first Ig-like domain of PAD-4 (29). Of note, the deleted region also contains the 3 SNPs (padi4_89, padi4_90, and padi4_92) encoding amino acid substitutions (S55G, A82V, and A112G) which segregate the major nonsusceptibility wild-type (WT) and susceptibility haplotypes of PAD-4 (16). Both type I and type II sera failed to immunoprecipitate PAD-4 120–663, demonstrating that the first 119 amino acids are required for recognition by both groups of antibodies (Figures 2A and B, lanes 7–9).
We next investigated whether sera recognized the N-terminal 119–amino acid domain (PAD-4 1–119) independently. All type II sera immunoprecipitated this construct strongly, demonstrating that this domain is necessary and sufficient for recognition by type II sera. In contrast, type I sera failed to recognize the 119 N-terminal amino acids alone (Figures 2A and B, lanes 16–18). Twenty type II serum samples were used to further analyze the epitope recognized by type II sera. These experiments used 2 shorter N-terminal constructs, containing either the first 79 amino acids (PAD-4 1–79) or the 40 amino acids from methionine-80 through alanine-119 (PAD-4 80–119). No sera immunoprecipitated PAD-4 1–79 (Figure 2B, lanes 13–15). In contrast, 13 (65%) of 20 samples immunoprecipitated the PAD-4 80–119 construct (albeit significantly less strongly than PAD-4 1–119), which includes amino acids 82 and 112, which are polymorphic in the susceptible form of PAD-4 (Figure 2B, lanes 22–24).
Since type I (but not type II) antibodies failed to recognize the 58/59-kd form of PAD-4 and PAD-4 1–523, which lacks the 141 C-terminal amino acids (Figure 2B, lanes 10–12), we addressed whether type I sera could recognize the C-terminal region of PAD-4 (amino acids 524–663) when expressed alone. Type I sera failed to immunoprecipitate PAD-4 524–663, a construct containing the 141 C-terminal amino acids of PAD-4 fused to GST (Figure 2B, lanes 19–21). Taken together, these data demonstrate that recognition of PAD-4 by autoantibodies requires the 119 N-terminal amino acids of PAD-4, either exclusively (type II) or together with additional areas of PAD-4 at the C-terminus (type I). Type I autoantibodies therefore require contributions from both N- and C-terminal domains of PAD-4. This is notable because when crystallized, PAD-4 exists as a dimer with head-to-tail contact between the N-terminus of one molecule and the C-terminus of the second (29).
To define whether polymorphisms in the PADI4 gene influenced recognition by anti-PAD-4 autoanti-bodies, we immunoprecipitated equal amounts of IVTT WT or polymorphic PAD-4 with 46 anti–PAD-4– positive sera. No differences in recognition of WT and polymorphic PAD-4 were observed (Figure 2B, lanes 1–6). We also established a competitive immunoprecipitation assay for type II sera, using mixtures of full-length PAD-4 (WT or polymorphic) or PAD-4 1–119 (WT or polymorphic). All constructs behaved equivalently in these assays, demonstrating that autoantibodies do not distinguish the various polymorphic forms of PAD-4 (results not shown).
Anti–PAD-4 antibodies are associated with the PADI4 susceptibility haplotype.
Since the epitopes recognized by both type I and type II antibodies require the region containing the SNPs of PADI4 that have previously been associated with RA (16,18,19), we investigated whether the susceptibility haplotype was associated with anti–PAD-4 antibodies. We genotyped the ESCAPE RA patients for the 3 SNPs that encode nonsynonymous changes within the N-terminus of PAD-4: rs11203366 (padi4_89), rs11203367 (padi4_90), and rs874881 (padi4_92). To avoid confounding effects resulting from population stratification, we included only the white subjects (n = 111) in these analyses. Of the 4 haplotypes observed, 2 were most frequent (Table 2), consistent with previous studies (16,18). Haplotype 1 is the haplotype defined as “nonsusceptible” and haplo-type 2 as “susceptible” by Suzuki et al (16). The presence of anti–PAD-4 antibodies was associated with PADI4 haplotype 2 (OR 2.59 [95% CI 1.02–6.08], P = 0.04) (Table 2). Interestingly, when diplotypes were examined, patients who were heterozygous for the presence of haplotypes 1 and 2 had an increased likelihood of having anti–PAD-4 antibodies compared with patients who were homozygous for haplotype 1 (OR 4.02 [95% CI 1.43–11.3], P = 0.009) (Table 3). In contrast, the OR for the detection of anti–PAD-4 antibodies was not significantly increased among patients who were homozygous for haplotype 2.
Table 2.
Association of anti–PAD-4 autoantibodies with the susceptibility haplotype of PADI4 in the 111 white patients in the ESCAPE RA cohort*
| SNP/amino acid substitution |
Odds ratio |
||||||
|---|---|---|---|---|---|---|---|
| Haplotype | No. of alleles |
Allele frequency |
padi4 89/ S55G |
padi4 90/ A82V |
padi4 92/ A112G |
Anti–PAD-4 antibodies |
Anti-CCP antibodies |
| 1 | 121 | 0.55 | A | C | C | 1.13 | 1.10 |
| 2 | 95 | 0.43 | G | T | G | 2.59† | 1.46 |
| 3 | 5 | 0.023 | A | C | G | – | – |
| 4 | 1 | 0.0045 | G | C | C | – | – |
Patients were genotyped for the single-nucleotide polymorphisms (SNPs) that encode amino acid changes within the N-terminus of PADI4: padi4_89 (rs11203366), padi4_90 (rs11203367), and padi4_92 (rs874881). The respective amino acid changes encoded by these SNPs are shown with the change in the direction from haplotype 1 (the nonsusceptibility haplotype) to haplotype 2 (the susceptibility haplotype). Odds ratios for the presence of anti–PAD-4 or anti–cyclic citrullinated peptide (anti-CCP) antibodies among patients with either PADI4 haplotype 1 or PADI4 haplotype 2, compared with the rest of the population, were calculated. See Table 1 for other definitions.
P = 0.04.
Table 3.
Association of anti–PAD-4 antibodies with the heterozygous diplotype in the 111 white patients in the ESCAPE RA cohort*
| Diplotype | No. of patients | OR (95% CI) |
|---|---|---|
| 1 and 1 | 33 | 1 |
| 1 and 2 | 53 | 4.02 (1.43–11.3)† |
| 2 and 2 | 19 | 2.63 (0.726–9.49) |
Patients were grouped according to the presence of 1 or 2 alleles of either haplotype 1 (the nonsusceptibility haplotype) or haplotype 2 (the susceptibility haplotype). Odds ratios (ORs) and 95% confidence intervals (95% CIs) for the presence of anti–PAD-4 autoantibodies, compared with patients who were homozygous for haplotype 1, were calculated. A similar analysis evaluating associations between diplotypes and anti–cyclic citrullinated peptide antibodies did not yield statistically significant results. See Table 1 for other definitions.
P = 0.009.
Anti–PAD-4 antibody levels are independently associated with radiographic severity in RA
To define whether anti–PAD-4 autoantibodies were associated with disease severity, we examined the relationship between the strongest anti–PAD-4 autoantibody responses and joint damage, measured radiographically by modified Sharp score (25), in the 129 patients in the ESCAPE RA study. We compared anti–PAD-4– negative patients (n = 83) with those with anti–PAD-4 autoantibodies scored as 3+ in the immunoprecipitation assay (n = 26). Mean unadjusted Sharp scores were 57 (95% CI 43.6–70.9) in the anti–PAD-4–negative group, compared with 132 (95% CI 90.6–173.7) in the group with high anti–PAD-4 scores. These differences were statistically significant (P < 0.001).
Since there are many potential confounding variables that might influence total Sharp score, a multivariate analysis was performed, adjusting for age, disease duration, swollen joint count, current use of biologic and nonbiologic disease-modifying antirheumatic drugs (DMARDs), rheumatoid factor (RF) seropositivity, and shared epitope status. After these adjustments, the difference in the mean Sharp score remained significant (mean Sharp score 64 versus 113 in anti–PAD-4 antibody–negative versus–positive groups; P = 0.001) (Table 4), demonstrating that strong anti–PAD-4 autoantibody responses are independently associated with severe RA.
Table 4.
Association of anti–PAD-4 antibodies with higher mean Sharp/van der Heijde scores in patients with RA*
| Unadjusted Sharp/van der Heijde score, mean (95% CI) |
Adjusted Sharp/van der Heijde score, mean (95% CI) |
|
|---|---|---|
| PAD-4–negative patients (n = 83) | 57 (44–71) | 64 (51–77) |
| PAD-4–positive patients (n = 26)† | 132 (91–174) | 113 (89–138) |
| P | <0.001 | 0.001 |
Multivariable linear regression was used to model the association of PAD-4 autoantibody level with radiographic damage (total Sharp/van der Heijde score) in subjects enrolled in the ESCAPE RA cohort study, with covariate adjustment for confounding demographic and RA disease and treatment characteristics (age, disease duration, swollen joint count, current use of biologic and nonbiologic disease-modifying antirheumatic drugs, rheumatoid factor seropositivity, and shared epitope status). 95% CI = 95% confidence interval (see Table 1 for other definitions).
Patients with an anti–PAD-4 antibody score of 3+ (on a scale of 0–3+), determined by densitometry of scanned immunoprecipitation autoradiograms.
Anti–PAD-4 autoantibodies mark a distinct subset of anti-CCP–positive patients
Since PAD-4 can generate the antigens recognized by anti-CCP antibodies, we examined the relationship of these 2 antibody systems. The frequency of anti-CCP antibodies in the ESCAPE RA cohort was consistent with that observed in previous studies (8,30), and was almost double that of anti–PAD-4 antibodies. In addition, the presence of anti–PAD-4 antibodies was strongly associated with anti-CCP positivity (OR 6.17 [95% CI 1.88–16.6], P = 0.0005), such that approximately half of anti-CCP– positive patients were also anti–PAD-4–positive. However, in contrast to the association seen between anti–PAD-4 antibodies and the PADI4 susceptibility haplotype, we did not observe an association between the presence of anti-CCP antibodies and PADI4 variants (Table 2). This result is consistent with the findings of other studies, in which associations between the PADI4 susceptibility SNPs and anti-CCP antibodies were not detected (20,23,31-33).
Anti-CCP antibodies have, however, been found to be strongly associated with class II major histocompatibility complex shared epitope alleles (18,34,35), a finding that also held true in the ESCAPE RA cohort. The OR for the association of anti-CCP with the presence of ≥1 shared epitope allele was 5.06 (95% CI 2.05–11.8) (P = 0.0002). In contrast, anti–PAD-4 did not appear to be associated with the shared epitope, revealing a second distinction between anti-CCP and anti–PAD-4. Thus, univariate analysis showed only a borderline association of anti–PAD-4 antibodies with the presence of ≥1 shared epitope allele (OR 2.44 [95% CI 0.961–5.73], P = 0.052). Further analysis by multiple logistic regression that included anti-CCP along with the presence of any shared epitope allele, and with anti–PAD-4 as the dependent variable, showed that anti–PAD-4 antibodies were not independently associated with the shared epitope. Thus, the shared epitope effect on anti–PAD-4 status appears to be mediated through anti-CCP.
Taken together, the data confirm that anti-CCP antibodies are found in the majority of patients with RA, and are strongly associated with the presence of the shared epitope, but not with the PADI4 susceptibility allele. In contrast, anti–PAD-4 antibodies identify a subgroup of anti-CCP–positive patients that is enriched for the PADI4 susceptibility haplotype and more severe disease. Although PAD-4 can autocitrullinate, extensive experiments failed to demonstrate that such citrullination was required for recognition by PAD-4 autoantibodies (results not shown), indicating that recognition of PAD-4 by autoantibodies is not simply due to recognition by a subgroup of anti-CCP antibodies.
DISCUSSION
These findings show that anti–PAD-4 antibodies are novel, highly specific (99%), and frequent (36–42%) seromarkers in RA. Although they occur less frequently than anti-CCP antibodies, anti–PAD-4 antibodies appear to be more specific for RA than anti-CCP antibodies; the latter were found in up to 10% of the rheumatic disease controls used in our study (data not shown) and in previous reports (36-40). Interestingly, the presence or absence of anti–PAD-4 antibodies divides anti-CCP– positive patients into 2 relatively equal groups, with the anti-CCP, anti–PAD-4-double positive group having higher disease severity and enrichment of the disease-susceptible PADI4 genotype.
Although others have used ELISAs to detect anti–PAD-4 and anti–PAD-2 antibodies (41,42), these assays had high background, and the antibodies detected were not specific for RA; they were found in patients with other rheumatic diseases, as well as in normal controls. In contrast, we found immunoprecipitation of IVTT PAD-4 to be highly specific for RA. The reasons for these assay discrepancies are unclear, but numerous examples exist where screening ELISAs for clinically relevant antibodies have substantial false-positive rates, which disappear with more specific testing (e.g., by Western blotting to detect Lyme disease and human immunodeficiency virus antibodies [43,44]). In preliminary studies, we have established a screening capture ELISA for anti–PAD-4 antibodies, which is 97% sensitive and has a false-positive rate of 12–15% in RA (data not shown). When used in conjunction with the immunoprecipitation assay, such a screening test may be useful to identify patients with anti–PAD-4 antibodies.
Identification of the subgroup of RA patients with anti–PAD-4 antibodies is potentially important, because these autoantibodies appear to be useful markers of RA disease severity. Patients with the strongest anti-PAD-4 antibody responses had higher mean modified Sharp scores, a relationship that was preserved after adjusting for age, disease duration, swollen joint count, current use of biologic and nonbiologic DMARDs, RF seropositivity, and shared epitope status.
The mechanisms underlying the relationship of anti–PAD-4 with more severe joint damage remain unknown at this time. The association may be causal (i.e., the anti–PAD-4 immune response may participate directly in joint damage) or anti–PAD-4 may be a downstream product of more aggressive joint inflammation. Future studies defining the temporal relationship of antibodies and joint damage will clarify the use of such antibodies as potential markers of disease activity and predictors of joint damage, and will provide additional insights into mechanism. Additionally, defining the effects of anti–PAD-4 antibodies on PAD-4 function and specificity, and potential proinflammatory properties of PAD-4 immune complexes will further clarify the role of antibodies in pathogenesis. Regardless of their exact role, anti–PAD-4 antibodies identify a group of patients with higher disease severity, in which early and aggressive intervention may be particularly important.
Polymorphisms in PADI4 have recently been demonstrated to be associated with RA in several different populations, including Japanese, Korean, and US cohorts (16,18-20). Several studies have failed to confirm this association in various European cohorts (21-23). When we addressed whether the PAD-4 genotype was associated with anti–PAD-4 antibodies in the white patients in this study (n = 111), we found that the previously defined “susceptible” haplotype was strikingly associated with anti–PAD-4 antibodies (OR 2.59 [95% CI 1.02–6.08], P = 0.04). This was particularly evident upon analysis of diplotypes, in which heterozygotes had the highest likelihood of having anti–PAD-4 antibodies (OR 4.02 [95% CI 1.43–11.3], P = 0.009). Of note, the relatively low number of patients homozygous for haplotype 2 (n = 19) may have provided insufficient power to detect an increased likelihood of developing an anti–PAD-4 immune response among these homozygous patients (Table 3). Nevertheless, the diplotype results are striking, and suggest that haplotype 2 may have a dominant effect on the production of anti–PAD-4 antibodies. The observation that PAD-4 exists as a dimer is highly relevant in this regard. It raises the possibility that the PAD-4 heterodimer may have unique structure and/or function not present in the homodimers (e.g., different stability, antigen processing, or potential susceptibility to intradimer modification such as citrullination), which makes it more likely to become an autoantigen. Defining these mechanisms will require using the appropriate dimeric forms of PAD-4.
There was no significant association between anti-CCP antibodies and the PADI4 susceptibility haplotype (Table 2). Although Suzuki et al (16) detected an association of homozygosity for the PADI4 susceptibility haplotype with antibodies against citrullinated filaggrin, other groups have failed to find associations between the susceptibility SNPs and elevated levels of synovial intracellular citrullinated proteins (32) or anti-CCP antibodies (20,23,31-33). The lack of association of anti-CCP antibodies with the PADI4 susceptibility haplotype is particularly interesting, in light of the strong association between anti–CCP antibodies and anti–PAD-4 antibodies (OR 6.17 [95% CI 1.88–16.6], P = 0.0005). These findings highlight that anti–PAD-4 antibodies mark a subgroup of anti–CCP-positive patients enriched for the PADI4 susceptibility haplotype.
It is noteworthy that anti-CCP antibodies frequently precede the development of clinical disease in RA, and that other autoantibodies occur during the propagation phase when disease becomes symptomatic (11). In a preliminary analysis, we have demonstrated that anti–PAD-4 antibodies occur less frequently in early disease (12%) compared with established disease (data not shown). We therefore hypothesize that anti-CCP antibodies precede the anti–PAD-4 immune response, and that the latter (autoantibodies or potentially T cells) may participate in amplification pathways in RA and contribute to joint erosion.
The fact that the epitope recognized by the majority of anti–PAD-4 antibody–positive individuals is located within the 119 N-terminal amino acids encompassing the polymorphisms associated with disease susceptibility suggests that the 2 properties (polymorphism and immune response) are mechanistically related. Interestingly, autoantibodies do not appear to discriminate between the WT and polymorphic versions of PAD-4, suggesting that polymorphisms may rather play a role at the level of antigen processing and T cell responses.
Previous studies have demonstrated that minor differences in autoantigen structure may have profound effects on antigen processing and T cell recognition. For example, an amino acid change occurring at a site critically required in early proteolytic processing can result in generation of a completely different set of epitopes from that antigen (45). Similarly, a minor posttranslational modification (isoaspartyl formation) can initiate an immune response in which responding T cells are specific for the modified antigen, whereas antibodies do not distinguish between the modified and the unaltered forms of the antigen (46). We propose that during conversion from the CCP-positive, asymptomatic phase of RA to the amplifying propagation phase of the disease, novel processing of the polymorphic form of PAD-4 allows generation of unique epitopes not previously tolerized, making initiation of an anti–PAD-4 immune response more likely.
The novel findings presented in this report demonstrate that autoimmunity in RA is directed against a polymorphic molecule genetically associated with the disease. The epitopes targeted include the polymorphic region itself, and autoantibodies to this molecule are associated with the susceptibility polymorphisms. Lastly, PAD4 autoantibodies are associated with increased disease severity. The data suggest that the generation of an anti–PAD-4 immune response may mediate some of the genetic effect of the PADI4 susceptibility allele in RA.
ACKNOWLEDGMENTS
We thank Michelle Jones and Marilyn Towns for providing technical and data management support, Brian Iglehart for performing the DNA extractions, and Dr. John Hall for generous assistance with preparing the figures.
Supported by the Maryland Chapter of the Arthritis Foundation, the Within Our Reach program of the American College of Rheumatology Research and Education Foundation, and the Donald B. and Dorothy L. Stabler Foundation. Dr. Lam is recipient of an Arthritis Foundation Senior Fellow award. Drs. Gao and Barnes' work was supported by NIH grant P30-AR-053503; Dr. Barnes' work also was supported in part by the Mary Beryl Patch Turnbull Scholar Program. Dr. Casciola-Rosen's work was supported by NIH grant R01-AR-44684. Dr. Bathon's work was supported by NIH grant AR-050026. Dr. Rosen's work was supported by NIH grant DE-12354; he is also a Cosner Scholar in Translational Research.
Footnotes
Dr. Scott owns stock in Pfizer. Dr. Bathon has received consulting fees, speaking fees, and/or honoraria from Abbott, Amgen, Centocor, and Novartis (less than $10,000 each); she has received research support from Amgen, Biogen Idec, and Bristol-Myers Squibb.
REFERENCES
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–61. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Edwards JC, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350:2572–81. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 3.Genovese MC, Becker JC, Schiff M, Luggen M, Sherrer Y, Kremer J, et al. Abatacept for rheumatoid arthritis refractory to tumor necrosis factor α inhibition. N Engl J Med. 2005;353:1114–23. doi: 10.1056/NEJMoa050524. [DOI] [PubMed] [Google Scholar]
- 4.Zendman AJ, van Venrooij WJ, Pruijn GJ. Use and significance of anti-CCP autoantibodies in rheumatoid arthritis. Rheumatology (Oxford) 2006;45:20–5. doi: 10.1093/rheumatology/kei111. [DOI] [PubMed] [Google Scholar]
- 5.Vossenaar ER, Despres N, Lapointe E, van der Heijden A, Lora M, Senshu T, et al. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res Ther. 2004;6:R142–50. doi: 10.1186/ar1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon M, Girbal E, Sebbag M, Gomes-Daudrix V, Vincent C, Salama G, et al. The cytokeratin filament-aggregating protein filaggrin is the target of the so-called “antikeratin antibodies,” autoantibodies specific for rheumatoid arthritis. J Clin Invest. 1993;92:1387–93. doi: 10.1172/JCI116713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, et al. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the α- and β-chains of fibrin. J Immunol. 2001;166:4177–84. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 8.Nijenhuis S, Zendman AJ, Vossenaar ER, Pruijn GJ, vanVenrooij WJ. Autoantibodies to citrullinated proteins in rheumatoid arthritis: clinical performance and biochemical aspects of an RA-specific marker. Clin Chim Acta. 2004;350:17–34. doi: 10.1016/j.cccn.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Van Boekel MA, Vossenaar ER, van den Hoogen FH, van Venrooij WJ. Autoantibody systems in rheumatoid arthritis: specificity, sensitivity and diagnostic value. Arthritis Res. 2002;4:87–93. doi: 10.1186/ar395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Helm-van Mil AH, Verpoort KN, Breedveld FC, Toes RE, Huizinga TW. Antibodies to citrullinated proteins and differences in clinical progression of rheumatoid arthritis. Arthritis Res Ther. 2005;7:R949–58. doi: 10.1186/ar1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielen MM, van Schaardenburg D, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MH, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004;50:380–6. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen KT, Wiik A, Pedersen M, Hedegaard CJ, Vestergaard BF, Gislefoss R, et al. Cytokines, autoantibodies, and viral antibodies in premorbid and postdiagnostic sera from patients with rheumatoid arthritis—case-control study nested in a cohort of Norwegian blood donors. Ann Rheum Dis. 2007 doi: 10.1136/ard.2007.073825. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Majka DS, Deane KD, Parrish LA, Lazar AA, Baron AE, Walker CW, et al. The duration of pre-clinical rheumatoid arthritis-related autoantibody positivity increases in subjects with older age at time of disease diagnosis. Ann Rheum Dis. 2007 doi: 10.1136/ard.2007.076679. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–18. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 15.Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 2005;44:40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 17.Ikari K, Kuwahara M, Nakamura T, Momohara S, Hara M, Yamanaka H, et al. Association between PADI4 and rheumatoid arthritis: a replication study. Arthritis Rheum. 2005;52:3054–7. doi: 10.1002/art.21309. [DOI] [PubMed] [Google Scholar]
- 18.Kang CP, Lee HS, Ju H, Cho H, Kang C, Bae SC. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum. 2006;54:90–6. doi: 10.1002/art.21536. [DOI] [PubMed] [Google Scholar]
- 19.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–60. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppe B, Haupl T, Gruber R, Kiesewetter H, Burmester GR, Salama A, et al. Detailed analysis of the variability of peptidylarginine deiminase type 4 in German patients with rheumatoid arthritis: a case-control study. Arthritis Res Ther. 2006;8:R34. doi: 10.1186/ar1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez A, Valdivia A, Pascual-Salcedo D, Lamas JR, Fernandez-Arquero M, Balsa A, et al. PADI4 polymorphisms are not associated with rheumatoid arthritis in the Spanish population. Rheumatology (Oxford) 2005;44:1263–6. doi: 10.1093/rheumatology/kei008. [DOI] [PubMed] [Google Scholar]
- 22.Barton A, Bowes J, Eyre S, Spreckley K, Hinks A, John S, et al. A functional haplotype of the PADI4 gene associated with rheumatoid arthritis in a Japanese population is not associated in a United Kingdom population. Arthritis Rheum. 2004;50:1117–21. doi: 10.1002/art.20169. [DOI] [PubMed] [Google Scholar]
- 23.Harney SM, Meisel C, Sims AM, Woon PY, Wordsworth BP, Brown MA. Genetic and genomic studies of PADI4 in rheumatoid arthritis. Rheumatology (Oxford) 2005;44:869–72. doi: 10.1093/rheumatology/keh614. [DOI] [PubMed] [Google Scholar]
- 24.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 25.Van der Heijde DM. How to read radiographs according to the Sharp/van der Heijde method. J Rheumatol. 1999;26:743–5. corrected and republished in J Rheumatol 2000;27:261–3. [PubMed] [Google Scholar]
- 26.Goldbach-Mansky R, Lee J, McCoy A, Hoxworth J, Yarboro C, Smolen JS, et al. Rheumatoid arthritis associated autoantibodies in patients with synovitis of recent onset. Arthritis Res. 2000;2:236–43. doi: 10.1186/ar93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casciola-Rosen LA, Pluta AF, Plotz PH, Cox AE, Morris S, Wigley FM, et al. The DNA mismatch repair enzyme PMS1 is a myositis-specific autoantigen. Arthritis Rheum. 2001;44:389–96. doi: 10.1002/1529-0131(200102)44:2<389::AID-ANR58>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca2+-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–83. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 30.Schellekens GA, Visser H, de Jong BA, van den Hoogen FH, Hazes JM, Breedveld FC, et al. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum. 2000;43:155–63. doi: 10.1002/1529-0131(200001)43:1<155::AID-ANR20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Barton A, Bowes J, Eyre S, Symmons D, Worthington J, Silman A. Investigation of polymorphisms in the PADI4 gene in determining severity of inflammatory polyarthritis. Ann Rheum Dis. 2005;64:1311–5. doi: 10.1136/ard.2004.034173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantaert T, Coucke P, De Rycke L, Veys EM, De Keyser F, Baeten D. Functional haplotypes of PADI4: relevance for rheumatoid arthritis specific synovial intracellular citrullinated proteins and anticitrullinated protein antibodies. Ann Rheum Dis. 2005;64:1316–20. doi: 10.1136/ard.2004.033548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki M, Miyagi J, Kuribayashi M, Negishi E, Ueno K, Moriya H. Evaluation of allele frequencies in the PADI4 gene and anti-cyclic citrullinated peptide antibodies of patients with rheumatoid arthritis in a Japanese population. Ann Rheum Dis. 2006;65:1399–400. doi: 10.1136/ard.2006.052431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huizinga TW, Amos CI, van der Helm-van Mil AH, Chen W, van Gaalen FA, Jawaheer D, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA–DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–8. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 35.Van Gaalen FA, van Aken J, Huizinga TW, Schreuder GM, Breedveld FC, Zanelli E, et al. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 2004;50:2113–21. doi: 10.1002/art.20316. [DOI] [PubMed] [Google Scholar]
- 36.Sene D, Ghillani-Dalbin P, Limal N, Thibault V, van Boekel T, Piette JC, et al. Anti-cyclic citrullinated peptide antibodies in hepatitis C virus associated rheumatological manifestations and Sjögren's syndrome. Ann Rheum Dis. 2006;65:394–7. doi: 10.1136/ard.2005.038042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vander Cruyssen B, Hoffman IE, Zmierczak H, Van den Berghe M, Kruithof E, De Rycke L, et al. Anti-citrullinated peptide antibodies may occur in patients with psoriatic arthritis. Ann Rheum Dis. 2005;64:1145–9. doi: 10.1136/ard.2004.032177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamali S, Polat NG, Kasapoglu E, Gul A, Ocal L, Aral O, et al. Anti-CCP and antikeratin antibodies in rheumatoid arthritis, primary Sjögren's syndrome, and Wegener's granulomatosis. Clin Rheumatol. 2005;24:673–6. doi: 10.1007/s10067-005-1104-y. [DOI] [PubMed] [Google Scholar]
- 39.Caspi D, Anouk M, Golan I, Paran D, Kaufman I, Wigler I, et al. Synovial fluid levels of anti–cyclic citrullinated peptide antibodies and IgA rheumatoid factor in rheumatoid arthritis, psoriatic arthritis, and osteoarthritis. Arthritis Rheum. 2006;55:53–6. doi: 10.1002/art.21691. [DOI] [PubMed] [Google Scholar]
- 40.Riedemann JP, Munoz S, Kavanaugh A. The use of second generation anti-CCP antibody (anti-CCP2) testing in rheumatoid arthritis—a systematic review. Clin Exp Rheumatol. 2005;23(5 Suppl 39):S69–76. [PubMed] [Google Scholar]
- 41.Nissinen R, Paimela L, Julkunen H, Tienari PJ, Leirisalo-Repo M, Palosuo T, et al. Peptidylarginine deiminase, the arginine to citrulline converting enzyme, is frequently recognized by sera of patients with rheumatoid arthritis, systemic lupus erythematosus and primary Sjögren syndrome. Scand J Rheumatol. 2003;32:337–42. doi: 10.1080/03009740410004990. [DOI] [PubMed] [Google Scholar]
- 42.Takizawa Y, Sawada T, Suzuki A, Yamada R, Inoue T, Yamamoto K. Peptidylarginine deiminase 4 (PADI4) identified as a conformation-dependent autoantigen in rheumatoid arthritis. Scand J Rheumatol. 2005;34:212–5. doi: 10.1080/03009740510026346-1. [DOI] [PubMed] [Google Scholar]
- 43.Dressler F, Whalen JA, Reinhardt BN, Steere AC. Western blotting in the serodiagnosis of Lyme disease. J Infect Dis. 1993;167:392–400. doi: 10.1093/infdis/167.2.392. [DOI] [PubMed] [Google Scholar]
- 44.Mylonakis E, Paliou M, Lally M, Flanigan TP, Rich JD. Laboratory testing for infection with the human immunodeficiency virus: established and novel approaches. Am J Med. 2000;109:568–76. doi: 10.1016/s0002-9343(00)00583-0. [DOI] [PubMed] [Google Scholar]
- 45.Antoniou AN, Blackwood SL, Mazzeo D, Watts C. Control of antigen presentation by a single protease cleavage site. Immunity. 2000;12:391–8. doi: 10.1016/s1074-7613(00)80191-0. [DOI] [PubMed] [Google Scholar]
- 46.Mamula MJ, Gee RJ, Elliott JI, Sette A, Southwood S, Jones PJ, et al. Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. J Biol Chem. 1999;274:22321–7. doi: 10.1074/jbc.274.32.22321. [DOI] [PubMed] [Google Scholar]