Abstract
Post-transcriptional regulation is a key feature controlling gene expression in the protozoan parasite Leishmania. The nine-nucleotide paraflagellar rod regulatory element (PRE) in the 3′UTR of L. mexicana PFR2 is both necessary and sufficient for the observed ten-fold higher level of PFR2 mRNA in promastigotes compared to amastigotes. It is also found in the 3′UTRs of all known PFR genes. A search of the L. major Friedlin genomic database revealed several genes that share this cis element including a homolog of a heterotrimeric kinesin II subunit, and a gene that shares identity to a homolog of a Plasmodium antigen. In this study, we show that genes that harbor the PRE display promastigote-enriched transcript accumulation ranging from 4 – 15 fold. Northern analysis on episomal block substitution constructs revealed that the regulatory element is necessary for the proper steady-state accumulation of mRNA in L. mexicana paraflagellar rod gene 4 (PFR4). Also we show that the PRE plays a major role in the proper steady-state mRNA accumulation of PFR1, but may not account for the full regulatory mechanism acting on this mRNA. Our evidence suggests that the PRE coordinately regulates the mRNA abundance of not only the PFR family of genes, but in a larger group of genes that have unrelated functions. Although the PRE alone can regulate some mRNAs, it may also act in concert with additional elements to control other RNA transcripts.
Keywords: Leishmania, protozoan parasite, post-transcriptional gene regulation, mRNA stability, regulatory element, 3′ UTR, developmental gene regulation
Introduction
Protozoan parasites of the genus Leishmania cause widespread diseases whose human manifestations range from self-healing cutaneous lesions to fatal visceral infections [1–4]. The Phlebotomine sandfly transmits the parasite to human and other mammalian hosts, and the parasites adapt to their environments by cycling between insect-stage flagellated promastigotes and mammalian-stage spherical amastigotes [2, 5, 6]. This dramatic morphological change is accompanied by changes in the parasite’s biochemistry and it is likely orchestrated by gene regulatory events [6]. Studying how the parasite accomplishes and controls these changes may lead to the elucidation of novel therapeutic targets [7, 8].
Leishmania and other trypanosomatid protozoans do not share the extensively-studied gene expression pathways of bacteria or eukaryotes such as yeast and mammals which regulate primarily at the level of transcription [9]. Although transcription of T. brucei PARP and VSG gene expression can be controlled by specialized RNA polymerase I promoters [10–13], canonical RNA polymerase II promoters are not known to be intimately associated with protein coding genes in Trypanosomatids. RNA polymerase II promoters may be present in strand-switch regions of the Leishmania chromosomes where they transcribe the genome into long polycistronic pre-mRNA [14, 15]. Mature mRNAs are chimeric molecules formed by processing polycistronic pre-mRNAs via the trans-splicing of a mini-exon leader sequence to the 5′ end of open reading frames (ORFs) in a process that is coupled with polyadenylation of the adjacent upstream ORF [16, 17]. The parasites are reported to produce pre-mRNAs at a uniform rate across the genome, however these polycistronic units often contain mRNAs whose steady-state levels are significantly different and differentially accumulate in specific life cycle stages. This has led to a paradigm in which post-transcriptional mechanisms for regulation of gene expression predominate [12, 18], including mRNA processing [19–22], translational efficiency [22–29] and mRNA stability. Genes whose mRNAs are controlled at the level of mRNA stability include T. brucei procyclin [13, 30, 31] and hexose transporter [32]; T. cruzi amastin [33], mucin [24], and FL169 surface antigen [22]; and Leishmania A2 [34], A600 [35, 36], HSP83 [25, 29, 37, 38], GP63/major surface protease [39–44], amastin [26, 45], CBP proteases [19], glucose transporters [46], paraflagellar rod genes [47, 48], and S-phase enriched genes [49].
Several studies have begun to illuminate post-transcriptional regulatory mechanisms in Trypanosomatids. Researchers have identified nucleotide sequences that contribute to the regulation of mRNA within the 5′ untranslated region (UTR) [49, 50], the coding sequence [22], intercistronic region [19–22], but most commonly in the 3′ UTR [22, 24, 26, 30, 32–35, 42, 47, 51, 52]. These regulatory sequences influence mRNA maturation, translational efficiency, and frequently mRNA abundance. In T. cruzi, AU-rich repeat regions that are structurally and functionally related to mammalian adensoine- and uridine-rich elements (AREs) [53, 54] control the differentially regulated expression of the mucin gene family [24].
In Leishmania, mRNA regulatory sequences have been reported that are greater than 100 nt in length [19, 42, 45] and as short as 8 – 9 nucleotides. Examples of short regulatory sequences include the putative cycling sequence octamer that directs the accumulation of S-phase enriched mRNAs [49] and a short element that signals for decay of mRNAs which harbor it in amastigotes termed the paraflagellar rod regulatory element (PRE).
Previously, we reported that expression of the L. mexicana paraflagellar rod 2 (PFR2) genes is regulated post-transcriptionally by modulation of mRNA decay rates by a negative regulatory mechanism [47]. PFR2 encodes a structural component of the paraflagellar rod (PFR), a cytoskeletal structure essential for motility in promastigotes [55, 56]. The steady-state level of PFR2 mRNA is 10-fold greater in flagellated promastigotes than in amastigotes [57]. This differential accumulation of PFR2 mRNA depends on the PRE. This novel AU-rich RNA element, is a short sequence ( AUGUAnAGU) contained within the 3′ UTR of the PFR2 mRNA that accelerates its decay in amastigotes. Mutation of the PRE results in an increase in amastigote mRNA half-life and steady-state abundance to levels that coincide with PFR2 mRNA levels in promastigotes [47]. Insertion of the PRE into an unregulated transcript that harbors the PFR2 ORF flanked by a 3′UTR consisting solely of vector-derived sequences confers regulation on the chimeric transcript by decreasing mRNA levels in amastigotes. Thus, the PRE appears to be both necessary and sufficient for post-transcriptional regulation of PFR2 mRNA accumulation.
The PRE is present in the 3′ UTR of all known L. mexicana PFR gene family members, including all three copies of PFR2, the two sequenced copies of PFR1, PFR4, and in other genes that have a putative flagellar function [47]. The recent completion of sequencing of the L. major genome [58] has made possible a genome-wide search for the PRE in L. major. This, in addition to several reports of Leishmania transcript profiling [59–66], has allowed candidate genes that display PRE-dependent regulation to be identified.
In this report, we show that L. mexicana mRNAs that harbor the PRE display promastigote-enriched transcript accumulation, including genes not related to the PFR. We provide evidence that the PRE is necessary for proper mRNA regulation of PFR4 and plays a major role in the proper steady-state mRNA accumulation of PFR1. This evidence suggests that the PRE coordinately regulates mRNA abundance not only in the PFR family, but in a larger group of genes with diverse or unknown function.
Materials and Methods
Parasite strains and culture
Promastigotes of Leishmania mexicana (WHO strain MNYC/BZ/62/m379) were cultured in M199 medium containing 5% (v/v) fetal bovine serum, and 5% (v/v) bovine embryonic fluid at 26°C as described previously [67]. All amastigotes of L. mexicana used in this study were from axenic cultures and were obtained by shifting the incubation conditions of promastigotes from 26°C to 33°C and pH 5.5 in a modified UM 54 medium as described previously [48]. Promastigote and amastigote cultures were maintained by serial dilutions of 1:100 and 1:25, respectively, every three to four days. Logarithmic phase cultures (5 × 106 to 9 × 106 cells/ml) were used in all the experiments described. RNA isolated from axenic amastigote cultures was harvested at least five days after initial differentiation. The wild-type strain used is this work is referred to as line LM16. The Δpfr1 line used in this work is the L. mexicana PFR1 knockout line, 167.2, that has been described previously [68]. The Δpfr2 line used in this work is the L. mexicana PFR2 knockout line, 13.2, that has been described previously [57].
Gene names, accession numbers and genome analysis
NCBI accession numbers and GeneDB systematic names (www.genedb.org) for genes used or discussed in this work are as follows: LmexPFR1, AY198411; LmexPFR2, U45884; LmexPFR4, AY198410; LmexKLP, AY496942; Lmex3040, DQ768431; LmexAnt, DQ526428; Hypothetical protein-encoding gene, LmjF15.0020. The last gene copy in the PFR2 array, PFR2C, described in previous articles [47, 48, 57], has been renamed to PFR2-3 following standardized guidelines [69]. Likewise, genes PFR1C and PFR1D have been renamed to PFR1-3 and PFR1-4, respectively.
To search the complete Leishmania genome for candidate genes that harbor the PRE, we analyzed the Leishmania major Friedlin database (http://www.genedb.org/genedb/leish/) which contained the most complete genome annotation of any Leishmania species [58]. We used the ARTEMIS tool to identify matches to the sequence ATGTAnAGT for each of the 36 chromosomes. Genes were classified into biological or molecular functions according to the annotation in the database (see supplemental information). The ARTEMIS tool at the L. infantum database (http://www.genedb.org/genedb/linfantum/) and the L. braziliensis database (http://www.genedb.org/genedb/lbraziliensis/) was used to locate the PRE in the genomic sequences of the orthologs of the L. mexicana genes used in this study. Gene prefixes signify species: Lmex, L. mexicana; LmjF, L. major; LinJ, L. infantum; and LbrM, L. braziliensis.
PCR
To amplify DNA fragments for cloning, L. mexicana promastigote genomic DNA was used as a template for PCR using Taq polymerase (Promega). Briefly, 50 μl reactions contained 10 U Taq enzyme, 1X reaction buffer (10mM Tris-HCl, pH 9.0 at 25°C; 50mM KCl; 0.1% Triton® X-100), 0.4 mM of each dNTP, 2 mM MgCl2, approximately 200 ng DNA template, 200 nM gene-specific reverse oligonucleotide primer, and 200 nM gene-specific forward oligonucleotide primer. A three stage thermal profile was used to amplify products: 1) Denaturation (94°C for 3 min), 2) PCR amplification (20 – 30 cycles of: Denaturation, 94°C for 20 s; Annealing, 55°C for 40 s; Extension, 72°C for 1 min/kb), 3) Final extension (72°C for 7 min).
To obtain L. mexicana mRNA sequence, L. mexicana promastigote total RNA was used as a template for reverse transcriptase-PCR using SuperScript™ One-Step RT-PCR with Platinum® Taq (Invitrogen). Briefly, 50 μl reactions contained 1 μl RT/Taq Mix, 25 μl 2X Reaction Mix (0.4 mM of each dNTP, 2.4 mM MgSO), approximately 200 ng of total promastigote RNA template, 200 nM poly d(T) reverse oligonucleotide primer, and 200 nM gene-specific forward oligonucleotide primer. A three stage thermal profile was used to make cDNA and to amplify products: 1) cDNA synthesis and pre-denaturation (45–55°C for 15–30 min, 94°C for 2 min), 2) PCR amplification (40 cycles of: Denaturation, 94°C for 15 s; Annealing, 55°C for 30 s; Extension, 72°C for 1 min/kb), 3) Final extension (72°C for 7 min).
PCR products were resolved on a 1% agarose gel, purified using the QIAquick Gel Extraction Kit (Qiagen), ligated into the pGEM-T Easy vector system (Promega) for sequencing or into the indicated cloning vector. Plasmids were sequenced at the Purdue University Genomics Core Facility or Iowa State University DNA Facility.
Plasmid construction
All transfected plasmids were constructed in the pX63PAC backbone, a Leishmania expression vector which confers resistance to puromycin [67]. Construct pKM1, a derivative of the p7NEO targeting plasmid described previously [57], was made by removing the 5.5 kb insert, consisting of the neomycin phosphotransferase (NEO) coding sequence (identical to NCBI Accession No. AJ627603, region: 2662 to 3465) flanked by PFR2 sequence, by digesting with SmaI and XhoI. The fragment was treated with the Klenow fragment (New England Biolabs, NEB) to create blunt ends and ligated into Klenow-treated, BamHI-linearized pX63PAC using T4 DNA ligase (NEB).
Plasmids pXPFR4-WT, pXPFR4-Sub, pXPFR1-WT, pXPFR1-Sub, and pXPFR1-Δ3′ were created using unique PCR cloning strategies. In the schematic diagrams in this report, PCR primers used to construct the inserts are indicated with a symbol (lowercase letter, arrow). The oligonucleotide sequences of the primers are shown in Table 1. Nucleotides that are identical to Leishmania genomic DNA are capitalized, and restriction endonuclease recognition sites used for cloning are underlined. The pXPFR1-Δ3′ construct is not a total deletion of the PFR1 3′UTR. The first 646 nt, up to the endogenous NcoI site remain intact, however the PRE sequence is not present. Details of the cloning strategies used to create the plasmids are provided in Supplemental Information A.
Table 1.
Oligonucleotide primer sequences
| Primer symbol | Sequencea |
|---|---|
| a | 5′-ccgcccaagcttCCAACGGCGGCTCTGGCTCTT-3′ |
| b | 5′-gccggaattctcgagcgCACAGCTCCAGTCAGGGTTTTG-3′ |
| c | 5′-gtgtcagtacgtattgcggaaGGCCGAGAAGGACGACGAG-3′ |
| d | 5′-ACCTTTTTTGCTCTCTCGtgcggccgcACATTTTTTGTTTTTTGT-3′ |
| e | 5′-cccaagcttgggACCGTGCTTTAGTCAGGGA-3′ |
| f | 5′-ggaattccatatgCGTGGCAGGGGAGGAGAGATGA-3′ |
| g | 5′-AAGATGTGCGTGTGGCGCGCCATACCA-3′ |
| h | 5′-gctctagagcATGGCTGCGATGGGGAAGTTGG-3′ |
| i | 5′-CGGGATCCCGACCGTGCTTTAGTCAGGGA-3′ |
| j | 5′-CGGGATCCCGACCCACCTCTCCAGCACCAA-3′ |
| k | 5′-tccccgcggggaAGATGTGCGTGTTGCGCGCCATA-3′ |
| l | 5′-AGAGAAAAGAAAGAGAGACCgcggccgcaCGTTCCATTTGCCCGCTGTG-3′ |
| m | 5′-CACAGCGGGCAAATGGAACGtgcggccgcGGTCTCTCTTTCTTTTCTCT-3′ |
Sequence identical to Leishmania genomic DNA are in uppercase. Restriction endonuclease recognition sites used for cloning are underlined.
Transfection of parasites
Methods for electroporation of plasmid DNA into Leishmania and selection of transfectants were described previously [67]. Briefly, 10 μg of circular DNA was transfected into parasites and they were allowed to recover in M199 growth media overnight. Cultures were spread on 1% noble agarose plates. Colonies were selected approximately 1 week after plating. Puromycin was the selective drug for all experiments described in this work and was maintained at a concentration of 10 μM in liquid culture and 20 μM on selective plates. Leishmania lines generated from three to five independent transfectants were assayed for regulation; figures show representative experiments. An exception to this is the experimentation on Δpfr1 parasites transfected with pXPFR1-Sub, in which 12 independent transfectants were selected for experimentation. The replacement of endogenous PFR2 coding sequences with NEO by homologous recombination was described previously [57].
RNA hybridization
Total Leishmania RNA was isolated by using the RNeasy Mini Kit (Qiagen). 2 – 5 μg of RNA was fractionated by electrophoresis on 1.2% (w/v) SeaKem LE Agarose (Cambrex) gels containing formaldehyde as described previously [17]. The RNA was transferred to Hybond-N nylon membranes (Amersham) in 20X SSPE (0.2 M Sodium phosphate pH 7.4, 0.2 M EDTA, 3 M NaCl). Prehybridization and hybridization were carried out at 65°C in a hybridization oven using a buffer containing 50% formamide, 5X Denhart’s solution (0.1% Ficol, 0.1% polyvinylpyrrolidone and 0.1% bovine serum albumin), 5X SSPE, 0.1% SDS, 0.2 mg/ml denatured salmon sperm DNA and 0.3 mg/ml yeast RNA (Ambion). Radio-labeled probes used to hybridize to blots were generated by in vitro transcription of linerized plasmids harboring appropriate L. mexicana gene coding sequences with bacteriophage T3 or T7 RNA polymerase as described previously [47], or by random primer labeling of dsDNA. Unincorporated nucleotides were removed by passing labeled products through a NucAway Spin column (Ambion). mRNA levels were determined with a Molecular Dynamics Typhoon phosphorimager and were normalized to ribosomal RNA by reprobing membranes with a radiolabeled ribosomal small subunit RNA probe prepared by in vitro transcription of EcoRI-linearized pLmrRNA which contains the L. mexicana small rRNA subunit DNA sequences whose construction was described previously [47]. Methylene blue staining was completed by incubating blots at room temperature in 5% acetic acid for 15 min, then incubating in methylene blue buffer (0.04% methylene blue, 0.5 M sodium acetate) for 10 min, and rinsing with water.
Results
Identification of PRE-harboring mRNAs
An initial interspecies analysis of sequences of the PFR family of genes showed that the PRE consensus sequence ( ATGTAnAGT) is conserved among L. mexicana and L. major orthologs [47]. We reasoned that we could identify PRE-harboring L. mexicana mRNAs by identifying instances of the PRE within or adjacent to predicted L. major open reading frames. We identified 343 occurrences of the element in the entire genome. Because consensus sequences for neither a 5′ spliced leader addition signal nor a 3′ polyadenylation signal have been defined [70], analysis of sequences in the genome database cannot discern if any of the instances of the PRE are located within the 5′UTR or 3′UTR, likely locations for a functional regulatory element. We systematically categorized each instance of the PRE in relation to their orientation and proximity to the nearest predicted ORF. 127 (37%) of the instances of the PRE are on the minus strand relative to the nearest ORF (Fig. 1; additional data available in the supplemental information). It is unlikely that these genomic occurrences of the PRE sequence correspond to mRNAs because ORFs are transcribed as polycistronic precursors arranged in the same relative orientation and these PREs are on the minus strand. Similarly, 28 instances (8%) where the PRE sequence was found on the plus strand were likely in intergenic regions (not mRNA) because they were either greater than 500 bp upstream of the start codon or greater than 2 kb downstream of the stop codon. There are instances where the PRE was located within 500 bp upstream of the start codon (predicted 5′UTR, n = 6, 2%) or in the coding sequence of the gene (n = 43, 13%). However, 139 of the 343 occurrences (40%) are on the coding strand within 2 kb downstream of the ORF, which makes it possible that these PREs are present in the 3′UTR of the corresponding mature transcript as the average size of the region between coding sequences is approximately 2 kb [58]. More than half of these PREs, 78 (56% of 139), are within 500 bp downstream of the stop codon of the adjacent ORF, increasing the probability that these PREs are present in the 3′UTR. The annotated function of the 139 PRE-harboring genes do not fall into a single functional category, but include annotated biological or molecular functions such as calcium/calmodulin binding, cellular metabolism, flagellum organization and biogenesis, intracellular transport, and amino acid phosphorylation/dephosphorylation (www.genedb.org).
Figure 1. Location of the PRE in the L. major genome.
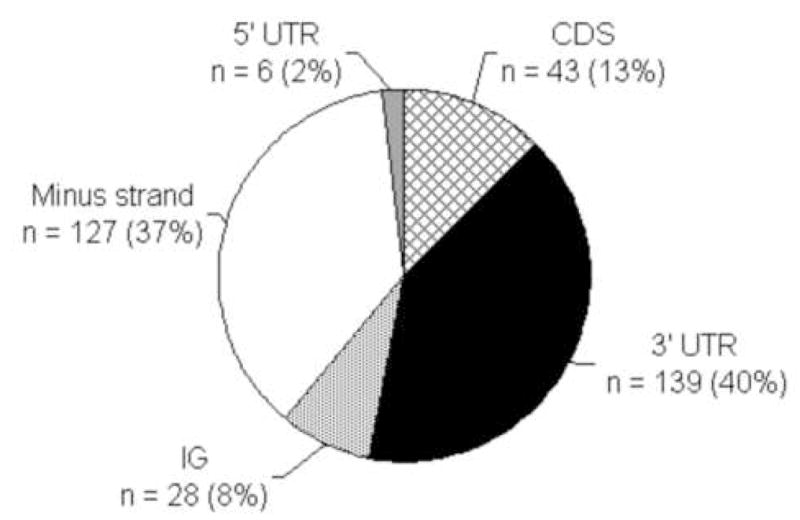
The pie charts shows the results of a search for the base sequence (ATGTAnAGT) in the L. major Friedlin genomic database using the ARTEMIS tool. Each of the 343 instances were systematically categorized by their position and orientation relative to the nearest predicted ORF in the genome. Categories: Minus strand, the PRE was in the opposite orientation relative to the nearest predicted ORF (white), all other categories indicate that the PRE was found in the same orientation as the nearest predicted ORF; 5′UTR, PRE is located within 0.5 kb of the ORF (gray); CDS, the PRE is found within the predicted coding sequences (gridded); 3′UTR, the PRE is found within 2 kb downstream of the nearest stop codon (black); IG, the element lies in the predicted intergenic regions, more than 0.5 kb 5′ and 2 kb 3′ of the nearest predicted ORF (gray dotted).
We chose several candidate genes for further study including LmexKLP, which encodes a homolog to the motor subunit of a kinesin-like protein; LmexAnt, a gene that encodes a product of unknown function but shares identity to a Plasmodium falciparum putative liver stage antigen; and Lmex3040, another gene that encodes a protein of unknown function. Lmex3040 was initially brought to our attention in the results of an interspecies microarray experiment in which its transcript profile fell only slightly outside of the strict criteria defining a promastigote-enriched gene (www.purdue.edu/biochem/holzer, [65]). Sequencing RT-PCR products using an oligo d(T) reverse primer and a gene-specific forward primer, showed that the L. mexicana ortholog of each mRNA harbors the PRE (Table 2). Two genes in which the L. mexicana 3′UTRs had already been sequenced, paraflagellar rod gene 1 (LmexPFR1), a gene that encodes a structural component of the PFR [68, 71]; and paraflagellar rod gene 4 (LmexPFR4), a gene that has ~40% identity at the amino acid level to PFR4 in T. cruzi (S.M. Moore, J.H. LeBowitz, unpublished data; [72]) also harbor the PRE in each respective 3′ UTR. As noted previously, casual observation of the location of each PRE across different L. mexicana genes suggests that its position in the 3′UTR in relation to the ORF is not conserved and ranges from 117 to 1354 bp downstream of the stop codon of the ORF.
Table 2.
Location of the PRE in the 3′ UTR for regulated L. mexicana transcripts studied in this report and their orthologs in L. major, L. infantum, and L. braziliensis.
| Gene name or Systematic IDa | Genbank accession | mRNA length (nt) | ORF (bp) | 3′ UTR (nt) | Position of PRE in3 ′ region (nt) | DNA sequence of PRE region | |
|---|---|---|---|---|---|---|---|
| PFR1 | LmexPFR1-3b | AY198411 | 3700c | 1788 | 1700c | 1354 | ATGTAtAGTg |
| LmjF29.1770 | -- | -- | 1788 | -- | 1359 | ATGTAtAGTg | |
| LmjF29.1760 | -- | -- | 1788 | -- | 1360 | ATGTAtAGTg | |
| LinJ29.2040 | -- | -- | 1788 | -- | 1369 | ATGTAtAGTg | |
| LbrM29.1800 | -- | -- | 1788 | -- | 1315 | ATGTAtAGTg | |
| LmexPFR1-4b | AY198411 | 3700c | 1788 | 1700c | 778 | ATGTAcAGTg | |
| LmjF29.1750 | -- | -- | 1788 | -- | 324 | ATGTAcAGTa | |
| LinJ29.2030 | -- | -- | 1788 | -- | 320 | ATGTAcAGTa | |
| “ | “ | 751 | ATGTAcAGTa | ||||
| LbrM29.1790 | -- | -- | 1032 | -- | No match | -- | |
| PFR4 | LmexPFR4b | AY198410 | 2378 | 1746 | 518 | 117 | ATGTAaAGTa |
| LmjF05.0040 | -- | -- | 1746 | -- | 141 | ATGTAaAGTa | |
| LinJ05.0040 | -- | -- | 1746 | -- | 138 | ATGTAaAGTa | |
| LbrM05.0050 | -- | -- | 1746 | -- | 134 | ATGTAaAGTa | |
| KLP | LmexKLP | AY496942 | 3681 | 2190 | 1391 | 518 | ATGTAcAGTc |
| LmjF13.0130 | -- | -- | 2190 | -- | 519 | ATGTAcAGTc | |
| LinJ13.0130 | -- | -- | 2181 | -- | 527 | ATGTAcAGTc | |
| LbrM13.0120 | -- | -- | 1971 | -- | 482 | ATGTAcAGTc | |
| Ant | LmexAnt | DQ526428 | 3800b | -- | 1034 | 926 | ATGTAaAGTa |
| LmjF36.5340 | -- | -- | 2610 | -- | 910 | ATGTAgAGTa | |
| LinJ36.0030 | -- | -- | 2592 | -- | 910 | ATGTAaAGTa | |
| LbrM35.5050 | -- | -- | 2559 | -- | 907 | ATGTAaAGTa | |
| 3040 | Lmex3040 | DQ768431 | 2200c | -- | 200c | 170 | ATGTAcAGTt |
| LmjF34.3040 | -- | -- | 1794 | -- | 170 | ATGTAcAGTt | |
| LinJ34.2650 | -- | -- | 1800 | -- | 170 | ATGTAcAGTt |
Systematic IDs on www.genedb.org
Modified from reference [47]
Estimated by comparisons to molecular weight standards on a northern blot.
The PRE is also present in gene orthologs across L. mexicana, L. major, L. infantum and L. braziliensis (Table 2). The position of the element in the 3′UTRs varies by no more than 54 bp for orthologs in these four species, except in the case of PFR1-4. The L. mexicana version of this gene harbors the PRE at position 778 bp in the 3′ UTR, however the L. major version of this gene (LmjF29.1750) harbors the element at position 324 bp. Further, the L. infantum ortholog of PFR1-4 (LinJ29.2030) contains two copies of the PRE at positions 320 and 751 bp in its predicted 3′ UTR, while the L. braziliensis version (LbrM29.1790) contains no matches at all.
A complementary sequence to each mRNA described above was used to probe a northern blot of total L. mexicana RNA from both promastigotes and axenic amastigotes (Fig. 2). A band of the predicted size of the mRNA was detected in each blot, and each displayed promastigote-enriched transcript accumulation. The ratios of the particular mRNA in promastigotes to amastigotes (P:A ratios) were calculated after normalization to rRNA and range from 15, in the case of LmexPFR1 (lanes 1, 2); to 4, in the case of Lmex3040 (lanes 5, 6). In order to show that our selection and blotting techniques were not biased toward promastigote transcript enrichment, a hypothetical protein-encoding gene from the database (LmjF15.0020) that neither possesses the PRE in L. major nor L. mexicana was tested and displayed no differential regulation (lanes 11, 12).
Figure 2. Genes selected by the presence of the PRE display promastigote-enriched mRNA accumulation.
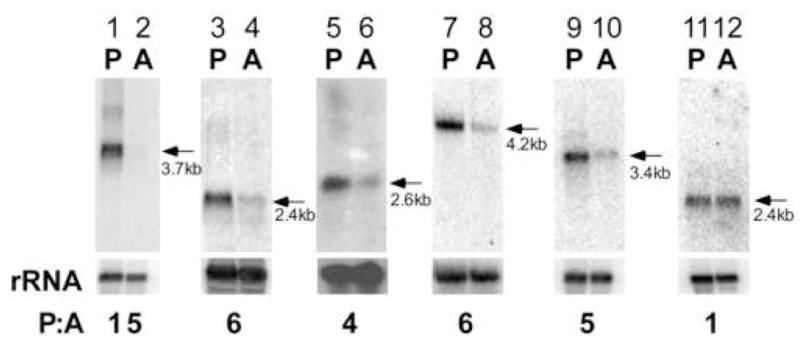
Northern analysis of L. mexicana total RNA from promastigotes, P, and amastigotes, A, probed with a labeled RNA complementary to coding sequences (lanes 1,2, PFR1; lanes 3, 4, PFR4; lanes 5,6, Lmex3040; lanes 7,8 LmexKLP; lanes 9,10, LmexANT; lanes 11, 12, hypothetical open reading frame gene, no PRE). The small ribosomal subunit RNA probed with its labeled complement is shown below each blot for lanes 1–4, 7–12. rRNA is shown in a methylene blue staining for lanes 5,6. P:A is the ratio of mRNA signal in promastigotes to that in amastigotes as after normalization with rRNA.
The PRE is necessary for the promastigote-enriched mRNA accumulation of LmexPFR4
The PRE sequence was delineated using episomally expressed PFR2 3′UTR deletion and block substitution constructs in a L. mexicana strain lacking endogenous PFR2 [47, 57]. However, parasite knockout strains are available for only one other gene discussed in this report, PFR1 [68]. To study the regulation of other mRNAs, chimeric genes were constructed that contained the neomycin phosphotransferase (NEO) ORF under the control of 3′ flanking sequences from the corresponding Leishmania genes. To test whether chimeric constructs were able to recapitulate promastigote-enriched transcript accumulation, we used a Leishmania strain in which the NEO gene was integrated into the PFR2 locus by homologous recombination. This resulted in a single NEO ORF flanked by PFR2 UTR sequence in the PFR2 locus [57]. Probing a northern blot of promastigote and amastigote RNA isolated from this heterozygous line with the complement to the NEO ORF, we showed that the integrated chimeric gene accumulates transcripts approximately five-fold higher in the promastigote stage (Fig. 3A; lanes 3, 4). When the same NEO ORF flanked by PFR2 UTR sequences was transcribed episomally in a wild-type cell, an equivalent ratio was calculated (pKM1; lanes 5, 6). Based on the predicted 2.4 kb size of transcript produced from both the endogenous locus and the episomal construct, it appears that endogenous trans-splicing and polyadenylation site were maintained and used for proper processing of both integrated and episomal NEO chimeric transcript [48]. The episomal chimeric NEO transcripts were more abundant than the corresponding NEO transcripts produced from the PFR2 genomic locus. This is most likely because the chromosomal NEO gene is present in a single copy, whereas the episomal copy number is higher. A similar observation has been reported in Leishmania previously [34]. These results show that sequences present in the open reading frame of PFR2 are not required for differential transcript accumulation — consistent with the PRE being wholly responsible for the observed transcript regulation [47], and that chimeric constructs can be used to study differential transcript accumulation regulated by 3′UTR elements.
Figure 3. Chimeric episomal constructs reveal that the PRE in the 3′UTR of LmexPFR4 is required for the promastigote-enriched transcript accumulation.
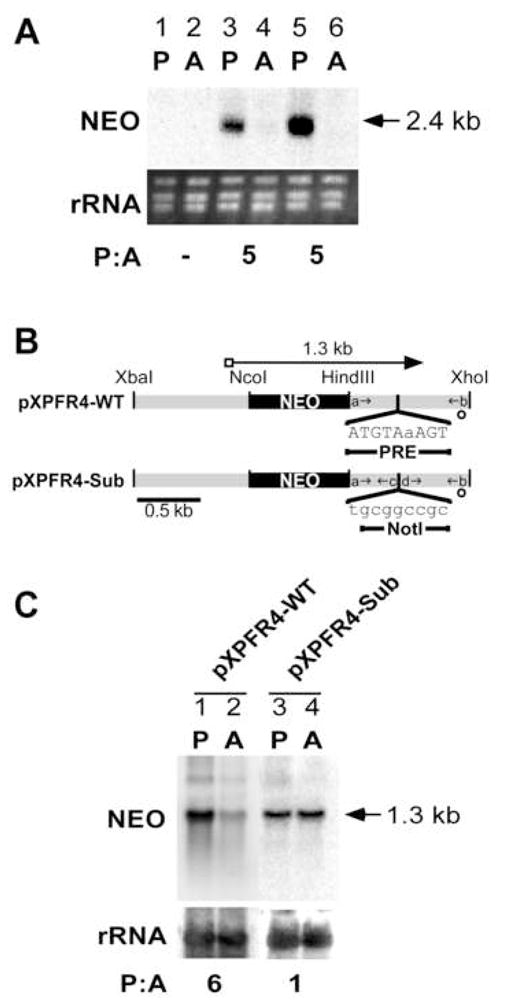
A) Episomal expression of a PFR2 chimeric gene recapitulates chromosomal expression. Northern analysis of total RNA probed with a labeled RNA that is complementary to the NEO coding sequence is shown for (lanes 1,2, wild-type parasites, no transfection; lanes 3, 4, Δpfr2 parasites in which one PFR2 allele has been replaced by NEO yet flanked by PFR2 UTR sequence; lanes 5, 6, episomal expression of a NEO/PFR2 UTR chimeric transcript in wild-type parasites, pKM1. rRNA bands are visualized by ethidium bromide staining. B) Schematic of two DNA constructs used in stable transfection experiments representing a chimeric construct that harbors the PRE (pXPFR4-WT) and one with the PRE sequences mutated (pXPFR4-Sub). Gray bars represent the intergenic flanking regions of the L. mexicana PFR4 gene, and black bars represent the NEO coding region. Lowercase letters and small arrows represent the oligonucleotide primers used to create the plasmid constructs (Table 1). Long arrows at the top of the genes represent the position and length of mRNA synthesized from respective constructs. A rectangular box at the 5′ end of each transcript denotes the mini-exon. Open circles represent the predicted AG dinucleotide splice acceptor site downstream of the indicated coding sequence. Restriction endonuclease recognition sites used in the plasmid construction are indicated. The sequence and position of the PRE and a NotI site substitution are shown under each 3′UTR. C) Northern analysis of L. mexicana total RNA containing the indicated plasmids (lanes 1,2, pXPFR4-WT; lanes 3,4, pXPFR4-Sub) probed with a labeled RNA complementary to the NEO coding sequence. rRNA bands are visualized by methylene blue-stained blots. All P:A values were determined as described in Fig. 2.
A chimeric episomal construct was used to study LmexPFR4, a PRE-harboring gene. Two DNA constructs were made, one that contains the wild-type 3′ UTR sequence and another with a block substitution of the PRE sequence (pXPFR4-WT and pXPFR4-Sub, repectively; Fig. 3B). When pXPFR4-WT was transfected into wild-type parasites, the episomal construct produced a regulated 1.3 kb chimeric NEO transcript with a P:A ratio of 6 (Fig. 3C; lanes 1, 2), thus recapitulating the observed regulation of the endogenous transcript (Fig. 2). However, when the PRE region was substituted (pXPFR4-Sub), the transcripts accumulated to the same level in both life cycle stages and exhibit a P:A ratio of 1.0 (Fig. 3C; lanes 3, 4).
Substitution of the PRE in the 3′UTR of LmexPFR1 disrupts stage-specific transcript regulation
The results of the molecular analysis on LmexPFR4 show that the presence of the PRE is necessary to direct the stage-specific expression of mRNA from a gene that harbors it. However, the results from substitution of the PRE in the 3′ UTR of PFR1 are more complex. To test the influence of the PRE on the regulation of PFR1, three DNA constructs containing the PFR1 ORF and selected portions of the PFR1 flanking intergenic sequences were created. Two of these plasmids were identical except for the presence or the substitution of the PRE (pXPFR1-WT and pXPFR1-Sub, respectively; Fig. 3A). When pXPFR1-WT was transfected into parasites lacking endogenous PFR1 (Δpfr1), the episomal construct produced appropriately regulated PFR1 transcripts. Northern blots of total RNA isolated from these parasites probed with a complement to the PFR1 coding sequence showed a transcript P:A ratio of approximately 15 (Fig. 4B, lanes 1–4). Plasmid pXPFR1-Δ3′ includes approximately 1 kb of genomic sequence upstream of the coding region, the complete ORF, and contains the first 646 bp of sequence immediately downstream. This plasmid retains the same 5′UTR and upstream transcript processing sites, however it lacks all sequences downstream of position 646 in the 3′UTR including the PRE. After transfection of pXPFR1-Δ3′ into pfr1 parasites, the episomal construct produced PFR1 transcripts that accumulated to similar levels in both stages with a P:A ratio of 1 (Fig. 4B, lanes 5–8). However, the episomal construct pXPFR1-Sub produced moderately regulated PFR1 transcripts (Fig. 4B, lanes 9–12). This unexpected result led us to examine the P:A ratios from 12 independent pXPFR1-Sub transfectants from which we calculated an average P:A ratio of approximately 6 (Fig. 4C). Although it is clear that the PRE regulates the relative abundance of the PFR1 mRNA, the results suggest that it is not the only determinant of stability.
Figure 4. Substitution of the PRE in the 3′UTR of PFR1 disrupts stage-specific transcript regulation.

A) Schematic of the three DNA constructs used in stable transfection experiments representing a construct containing wild-type sequence (pXPFR1-WT), a deletion construct that contains 1 kb of intergenic sequence upstream and 646 bp of sequence downstream of the PFR1 ORF and does not contain the PRE sequence (pXPFR1-Δ3′), and a substitution construct that contains NotI restriction site in place of the PRE (pXPFR1-Sub). Symbols are as described in the legend to Fig. 3. B) Northern analysis of total RNA from a L. mexicana PFR1 knockout strain (Δpfr1) containing the indicated plasmids (lanes 1–4, pXPFR1-WT; lanes 5–8, pXPFR1-Δ3′; lanes 9–12, pXPFR1-Sub) probed with an RNA complementary to the PFR1 coding sequence. The same membrane probed with small ribosomal subunit RNA is shown below each PFR1 blot. C) Graphical representation of normalized P:A ratios showing averages from at least three independent transfectants each. Error bars show the standard deviation. Relative P:A for pXPFR1-WT, pXPFR1-Δ3′, and pXPFR1-Sub are 16.7±1.7, 1.4±0.7, and 7.6±1.8, respectively.
Discussion
PRE Family: Coordinate regulation of a family of promastigote-enriched genes by a common regulatory element
The PRE signals for amastigote-specific decay when present in the 3′UTR of Leishmania mRNAs. It was originally studied in the paraflagellar rod 2 gene, is both necessary and sufficient for the accelerated decay of PFR2 in amastigotes, and is present in the 3′ UTRs of all PFR genes known to date. In this report, we demonstrate by mutational analysis that the PRE is also necessary for proper regulation of PFR4 and PFR1, thus showing that this element is befitting of its given name, the paraflagellar rod regulatory element.
To date, nine L. mexicana genes have been shown to contain a copy of the PRE in the 3′UTR, and in each case the corresponding mRNA displays promastigote-enriched accumulation. However, not every promastigote-enriched mRNA has a copy of the PRE. For example, the promastigote-enriched mRNA LmGT2, a member of the glucose transporter family of genes [46]; and a promastigote-enriched calmodulin mRNA [65] do not contain the PRE sequence (data not shown). L. mexicana is the only species in which the PRE has been shown to be functional, however for each PRE-harboring gene studied in L. mexicana, the L.major, L. infantum, and L. braziliensis gene orthologs also contain the PRE in approximately the same position in the respective predicted 3′ UTRs (Table 2).
Based on the data accumulated, we propose that: 1. the PRE acts as a signal for the accelerated decay in the amastigote stage for mRNAs in which it is present; 2. the PRE can be used to identify promastigote-enriched mRNAs; and 3. the PRE regulatory pathway is conserved across all Leishmania species.
Other studies have identified RNA regulatory regions in Leishmania. For example, a 202 nt region containing conserved or nearly conserved 7 and 8 nt repeats within the L. chagasi MSPL 3′UTR is responsible for the suppression of MSPL mRNA in logarithmic promastigotes, however the role of these repeated sequences are not known [42]. The 120 nt InS element, located downstream of the polyadenylation site, is both necessary and sufficient for developmental regulation of certain metacyclic-enriched CBP transcripts in L. mexicana [19]. Also, flanking regions of several hundred nucleotides of L. amazonensis HSP83 transcripts and of L. donovani A2 genes play a role in the increase of the stability of corresponding mRNAs during incubation in low pH conditions and/or elevated temperature [34, 37].
Studies on the PRE contrast with these previous reports in two ways: the RNA sequence that is a short nine nucleotide element that is located in the 3′ UTR, and unlike others it is involved in promastigote-enriched and amastigote-depleted mRNA regulation. In addition, like the ~450 nt 3′UTR element originally identified in the large Leishmania amastin gene family [45], the PRE is found in a diverse group of genes that is conserved across species.
It is tempting to speculate that all genes that harbor the PRE have a role in flagellar structure or biology, however two genes that contain the PRE lack significant homology to PFR genes and lack experimental characterization of their gene products. These genes both harbor the PRE in their 3′ UTR and produce promastigote-enriched mRNAs. One of these genes, LmexKLP, is homologous to the motor subunit of a heterotrimeric kinesin II which functions in anterograde intraflagellar transport (IFT) in Chlamydomonas [73]. If the protein encoded by the L. mexicana homolog of this gene plays a role in IFT, it stands to reason that it should be regulated similarly to other flagellar genes, specifically in a PRE-dependent manner. The PRE-harboring LmexANT shares partial identity to a Plasmodium liver-stage antigen, but lacks any other substantial homology to known genes in the available databases. It is possible that these stage-specific genes encode proteins that have a function in the flagellum like the PFR genes, however the PRE may influence genes with more diverse range of functions. The PRE consists of eight conserved nucleotides within the region of 3′ UTR mutated in each analysis (ATGTAnAGT). We note, however, that the element has not yet been mapped to the nucleotide level. It may be possible to refine the sequence in order to determine if any of the nucleotides are dispensable for a functional element, and allow more candidate PRE-regulated genes to be identified.
The PRE may act in concert with other RNA elements that signal for decay
The results of the PRE deletion and substitution studies in PFR1 deviated from the results obtained with PFR2 and PFR4. While a deletion of all PFR1 sequences downstream of nucleotide position 646 in the 3′ UTR including the PRE completely inhibits differential transcript accumulation, the block substitution of the PRE shows an intermediate result — differential accumulation is partial between wild type and the entire 3′ UTR deletion. Plasmid copy numbers are unlikely to account for these results for two reasons. First, plasmid pXPFR1-Δ3′ and the block substitution construct using PFR4 (pXPFR4-Sub) show a 1:1 promastigote to amastigote ratio of the respective mRNA produced from the plasmids in each case. Second, the expression constructs used to test PFR2 levels were made from the same pX63 parent vectors, and the observed developmental regulation is attributed wholly to the alteration of mRNA half-life [47]. Unlike the complete abatement of regulation observed in PRE block substitutions, the results of the PFR1 experiment suggest that another RNA regulatory element may be involved.
A complex post-transcriptional gene regulation system has been reported for a family of Leishmania genes with a conserved ~450 nt element in the 3′ UTR region that stimulates translation initiation [23, 45]. In approximately 20% of these mRNAs, another conserved region of ~100 nt is found downstream of the first element and functions in an additive manner to stimulate mRNA translation. Further, amastigote-enriched mRNA accumulation of genes in this family may be triggered by a distinct and yet undefined element located elsewhere in the 3′ UTR [26]. It is possible that a variant or closely related version of the PRE may function synergistically with the complete PRE, similar to the AREs that are found in multiple copies on the same mRNA. In the orthologs of PFR1-4 across four species of Leishmania, the presence and position of the PRE was not as well conserved as the other PRE-harboring genes (Table 2). The PRE sequence is found at position 778 bp in L. mexicana. However, a 7 out of 8 nucleotide match for the PRE sequence is found 424 nt upstream in a similar position as the PRE in other species. Partial activity of this PRE related sequence could account for the incomplete effect of the PFR1 block substitution construct.
Identification of RNA regulatory elements and assigning function to genes
Inspired by the identification of a Crithidia fasciculata RNA element that functions in cell cycle-dependent post-transcriptional control, Zick et al identified several novel S-phase expressed genes in L. major by searching the genome ORFs that contain a similar element in close proximity and in the same orientation [49]. We followed a similar strategy to identify members of the family of genes displaying PRE-dependent regulation. Because of the relatively low complexity of the PRE, many of the 343 occurrences are expected by chance and may not function in mRNA stability [47]. Some PRE sequences are located on the minus strand and others are likely in intergenic regions. Nevertheless, 40% of PRE hits in the L. major genome are within 2 kb of a downstream ORF and therefore possibly in 3′UTRs, yet only 9% of the corresponding L. mexicana genes met at least one criteria for a promastigote-enriched mRNA in a previous microarray study [65]. It is possible that many of these PREs lie outside the 3′ UTR, but others may be located in mRNAs yet fail to regulate mRNA stability because their effect is masked by other sequence elements.
Lmex3040 was originally identified as a differentially regulated transcript in a microarray analysis. The PRE was observed in the L. major genome and confirmed in the L. mexicana 3040 3′ UTR (Table 1), hence we identified a putative PRE gene family member through a combination of genomic and bioinformatic tools. These two technologies together not only provide insights about known regulatory elements, but also may hasten the discovery of novel elements. It may be possible to identify additional regulatory elements in the 3′ UTRs using a bioinformatic search of regulated mRNAs, and then test these using traditional or emerging biochemical experimental techniques [74].
Conclusion
We have identified a family of genes that harbor the PRE instability sequence and have shown that it is functional in several contexts, suggesting coordinate expression of genes by the presence of this element. The identification of the PRE in the 3′ UTRs of Leishmania genes may be a rapid way of identifying developmentally regulated transcripts which can hasten the discovery of additional novel therapeutic targets [7].
Supplementary Material
Acknowledgments
The authors thank Zane Bergman and Jane Kinney (Department of Biochemistry, Purdue University) for their contributions to this work, and the Forney and Broyles lab for their helpful input. This work was supported by National Institute of Health grant A147909. J.H.L. was a Burroughs Welcome Fund new investigator in Molecular Parasitology. This is journal paper 2007-18165 from Purdue Agriculture Research Programs.
List of Abbreviations
- PFR
paraflagellar rod
- PRE
paraflagellar rod regulatory element
- ORF
open reading frame
- UTR
untranslated region
- NEO
neomycin phosphotransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chang KP, Bray RS. Leishmaniasis. New York: Elsevier; 1985. [Google Scholar]
- 2.Herwaldt BL. Leishmaniasis. Lancet. 1999;354:1191–9. doi: 10.1016/S0140-6736(98)10178-2. [DOI] [PubMed] [Google Scholar]
- 3.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–77. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 4.Roberts LS, Janovy J, Schmidt GD. Gerald D. Schmidt & Larry S. Roberts’ foundations of parasitology. 5. Dubuque, IA: Wm. C. Brown; 1996. [Google Scholar]
- 5.Bogitsh BJ, Cheng TC. Human parasitology. Philadelphia: Saunders College Pub; 1990. [Google Scholar]
- 6.Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Annu Rev Microbiol. 1994;48:449–70. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 7.Almeida R, Norrish A, Levick M, Vetrie D, Freeman T, Vilo J, Ivens A, Lange U, Stober C, McCann S, Blackwell JM. From genomes to vaccines: Leishmania as a model. Philos Trans R Soc Lond B Biol Sci. 2002;357:5–11. doi: 10.1098/rstb.2001.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stober CB, Lange UG, Roberts MT, Gilmartin B, Francis R, Almeida R, Peacock CS, McCann S, Blackwell JM. From genome to vaccines for leishmaniasis: screening 100 novel vaccine candidates against murine Leishmania major infection. Vaccine. 2006;24:2602–16. doi: 10.1016/j.vaccine.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Stiles JK, Hicock PI, Shah PH, Meade JC. Genomic organization, transcription, splicing and gene regulation in Leishmania. Ann Trop Med Parasitol. 1999;93:781–807. doi: 10.1080/00034989957781. [DOI] [PubMed] [Google Scholar]
- 10.Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–40. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Palenchar JB, Bellofatto V. Gene transcription in trypanosomes. Mol Biochem Parasitol. 2006;146:135–41. doi: 10.1016/j.molbiopara.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Clayton CE. Life without transcriptional control? From fly to man and back again. Embo J. 2002;21:1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotz HR, Biebinger S, Flaspohler J, Clayton C. PARP gene expression: control at many levels. Mol Biochem Parasitol. 1998;91:131–43. doi: 10.1016/s0166-6851(97)00196-5. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–17. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–9. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 16.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–35. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 18.Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–40. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks DR, Denise H, Westrop GD, Coombs GH, Mottram JC. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J Biol Chem. 2001;276:47061–9. doi: 10.1074/jbc.M108498200. [DOI] [PubMed] [Google Scholar]
- 20.Curotto de Lafaille MA, Laban A, Wirth DF. Gene expression in Leishmania: analysis of essential 5′ DNA sequences. Proc Natl Acad Sci U S A. 1992;89:2703–7. doi: 10.1073/pnas.89.7.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluck C, Salomone JY, Kurath U, Roditi I. Cycloheximide-mediated accumulation of transcripts from a procyclin expression site depends on the intergenic region. Mol Biochem Parasitol. 2003;127:93–7. doi: 10.1016/s0166-6851(02)00310-9. [DOI] [PubMed] [Google Scholar]
- 22.Weston D, La Flamme AC, Van Voorhis WC. Expression of Trypanosoma cruzi surface antigen FL-160 is controlled by elements in the 3′ untranslated, the 3′ intergenic, and the coding regions. Mol Biochem Parasitol. 1999;102:53–66. doi: 10.1016/s0166-6851(99)00079-1. [DOI] [PubMed] [Google Scholar]
- 23.Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J Biol Chem. 2002;277:19511–20. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- 24.Di Noia JM, D’Orso I, Sanchez DO, Frasch AC. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J Biol Chem. 2000;275:10218–27. doi: 10.1074/jbc.275.14.10218. [DOI] [PubMed] [Google Scholar]
- 25.Larreta R, Soto M, Quijada L, Folgueira C, Abanades DR, Alonso C, Requena JM. The expression of HSP83 genes in Leishmania infantum is affected by temperature and by stage-differentiation and is regulated at the levels of mRNA stability and translation. BMC Mol Biol. 2004;5:3. doi: 10.1186/1471-2199-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNicoll F, Muller M, Cloutier S, Boilard N, Rochette A, Dube M, Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J Biol Chem. 2005;280:35238–46. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- 27.Nozaki T, Cross GA. Effects of 3′ untranslated and intergenic regions on gene expression in Trypanosoma cruzi. Mol Biochem Parasitol. 1995;75:55–67. doi: 10.1016/0166-6851(95)02512-x. [DOI] [PubMed] [Google Scholar]
- 28.Zeiner GM, Sturm NR, Campbell DA. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J Biol Chem. 2003;278:38269–75. doi: 10.1074/jbc.M304295200. [DOI] [PubMed] [Google Scholar]
- 29.Zilka A, Garlapati S, Dahan E, Yaolsky V, Shapira M. Developmental regulation of heat shock protein 83 in Leishmania. 3′ processing and mRNA stability control transcript abundance, and translation id directed by a determinant in the 3′-untranslated region. J Biol Chem. 2001;276:47922–9. doi: 10.1074/jbc.M108271200. [DOI] [PubMed] [Google Scholar]
- 30.Furger A, Schurch N, Kurath U, Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol. 1997;17:4372–80. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hehl A, Vassella E, Braun R, Roditi I. A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1994;91:370–4. doi: 10.1073/pnas.91.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotz HR, Lorenz P, Fischer R, Krieger S, Clayton C. Role of 3′-untranslated regions in the regulation of hexose transporter mRNAs in Trypanosoma brucei. Mol Biochem Parasitol. 1995;75:1–14. doi: 10.1016/0166-6851(95)02503-0. [DOI] [PubMed] [Google Scholar]
- 33.Coughlin BC, Teixeira SM, Kirchhoff LV, Donelson JE. Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3′-untranslated region position-dependent cis-element and an untranslated region-binding protein. J Biol Chem. 2000;275:12051–60. doi: 10.1074/jbc.275.16.12051. [DOI] [PubMed] [Google Scholar]
- 34.Charest H, Zhang WW, Matlashewski G. The developmental expression of Leishmania donovani A2 amastigote- specific genes is post-transcriptionally mediated and involves elements located in the 3′-untranslated region. J Biol Chem. 1996;271:17081–90. doi: 10.1074/jbc.271.29.17081. [DOI] [PubMed] [Google Scholar]
- 35.Murray A, Fu C, Habibi G, McMaster WR. Regions in the 3′ untranslated region confer stage-specific expression to the Leishmania mexicana a600-4 gene. Mol Biochem Parasitol. 2007;153:125–32. doi: 10.1016/j.molbiopara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Bellatin JA, Murray AS, Zhao M, McMaster WR. Leishmania mexicana: identification of genes that are preferentially expressed in amastigotes. Exp Parasitol. 2002;100:44–53. doi: 10.1006/expr.2001.4677. [DOI] [PubMed] [Google Scholar]
- 37.Aly R, Argaman M, Halman S, Shapira M. A regulatory role for the 5′ and 3′ untranslated regions in differential expression of hsp83 in Leishmania. Nucleic Acids Res. 1994;22:2922–9. doi: 10.1093/nar/22.15.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argaman M, Aly R, Shapira M. Expression of heat shock protein 83 in Leishmania is regulated post-transcriptionally. Mol Biochem Parasitol. 1994;64:95–110. doi: 10.1016/0166-6851(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 39.Kelly BL, Nelson TN, McMaster WR. Stage-specific expression in Leishmania conferred by 3′ untranslated regions of L. major leishmanolysin genes (GP63) Mol Biochem Parasitol. 2001;116:101–4. doi: 10.1016/s0166-6851(01)00307-3. [DOI] [PubMed] [Google Scholar]
- 40.Myung KS, Beetham JK, Wilson ME, Donelson JE. Comparison of the post-transcriptional regulation of the mRNAs for the surface proteins PSA (GP46) and MSP (GP63) of Leishmania chagasi. J Biol Chem. 2002;20:20. doi: 10.1074/jbc.M200174200. [DOI] [PubMed] [Google Scholar]
- 41.Brittingham A, Miller MA, Donelson JE, Wilson ME. Regulation of GP63 mRNA stability in promastigotes of virulent and attenuated Leishmania chagasi. Mol Biochem Parasitol. 2001;112:51–9. doi: 10.1016/s0166-6851(00)00346-7. [DOI] [PubMed] [Google Scholar]
- 42.Purdy JE, Donelson JE, Wilson ME. Regulation of genes encoding the major surface protease of Leishmania chagasi via mRNA stability. Mol Biochem Parasitol. 2005;142:88–97. doi: 10.1016/j.molbiopara.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Ramamoorthy R, Donelson JE, Wilson ME. 5′ sequences essential for trans-splicing of msp (gp63) RNAs in Leishmania chagasi. Mol Biochem Parasitol. 1996;77:65–76. doi: 10.1016/0166-6851(96)02581-9. [DOI] [PubMed] [Google Scholar]
- 44.Wilson ME, Paetz KE, Ramamoorthy R, Donelson JE. The effect of ongoing protein synthesis on the steady state levels of Gp63 RNAs in Leishmania chagasi. J Biol Chem. 1993;268:15731–6. [PubMed] [Google Scholar]
- 45.Rochette A, McNicoll F, Girard J, Breton M, Leblanc E, Bergeron MG, Papadopoulou B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol Biochem Parasitol. 2005;140:205–20. doi: 10.1016/j.molbiopara.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Burchmore RJ, Landfear SM. Differential regulation of multiple glucose transporter genes in Leishmania mexicana. J Biol Chem. 1998;273:29118–26. doi: 10.1074/jbc.273.44.29118. [DOI] [PubMed] [Google Scholar]
- 47.Mishra KK, Holzer TR, Moore LL, LeBowitz JH. A negative regulatory element controls mRNA abundance of the Leishmania mexicana Paraflagellar rod gene PFR2. Eukaryot Cell. 2003;2:1009–17. doi: 10.1128/EC.2.5.1009-1017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore LL, Santrich C, LeBowitz JH. Stage-specific expression of the Leishmania mexicana paraflagellar rod protein PFR-2. Mol Biochem Parasitol. 1996;80:125–35. doi: 10.1016/0166-6851(96)02688-6. [DOI] [PubMed] [Google Scholar]
- 49.Zick A, Onn I, Bezalel R, Margalit H, Shlomai J. Assigning functions to genes: identification of S-phase expressed genes in Leishmania major based on post-transcriptional control elements. Nucleic Acids Res. 2005;33:4235–42. doi: 10.1093/nar/gki742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahmood R, Hines JC, Ray DS. Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata. Mol Cell Biol. 1999;19:6174–82. doi: 10.1128/mcb.19.9.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Orso I, Frasch AC. Functionally different AU- and G-rich cis-elements confer developmentally regulated mRNA stability in Trypanosoma cruzi by interaction with specific RNA-binding proteins. J Biol Chem. 2001;276:15783–93. doi: 10.1074/jbc.M010959200. [DOI] [PubMed] [Google Scholar]
- 52.Hotz HR, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–26. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 55.Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 56.Vickerman K. The mechanism of cyclical development in trypanosomes of the Trypanosoma brucei sub-group: an hypothesis based on ultrastructural observations. Trans R Soc Trop Med Hyg. 1962;56:487–95. doi: 10.1016/0035-9203(62)90072-x. [DOI] [PubMed] [Google Scholar]
- 57.Santrich C, Moore L, Sherwin T, Bastin P, Brokaw C, Gull K, LeBowitz JH. A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol Biochem Parasitol. 1997;90:95–109. doi: 10.1016/s0166-6851(97)00149-7. [DOI] [PubMed] [Google Scholar]
- 58.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O’Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–42. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akopyants NS, Matlib RS, Bukanova EN, Smeds MR, Brownstein BH, Stormo GD, Beverley SM. Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol Biochem Parasitol. 2004;136:71–86. doi: 10.1016/j.molbiopara.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Cohen-Freue G, Holzer TR, Forney JD, McMaster WR. Global gene expression in Leishmania. Int J Parasitol. 2007;37:1077–86. doi: 10.1016/j.ijpara.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 62.Almeida R, Gilmartin BJ, McCann SH, Norrish A, Ivens AC, Lawson D, Levick MP, Smith DF, Dyall SD, Vetrie D, Freeman TC, Coulson RM, Sampaio I, Schneider H, Blackwell JM. Expression profiling of the Leishmania life cycle: cDNA arrays identify developmentally regulated genes present but not annotated in the genome. Mol Biochem Parasitol. 2004;136:87–100. doi: 10.1016/j.molbiopara.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 63.Duncan R. DNA microarray analysis of protozoan parasite gene expression: outcomes correlate with mechanisms of regulation. Trends Parasitol. 2004;20:211–5. doi: 10.1016/j.pt.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Duncan RC, Salotra P, Goyal N, Akopyants NS, Beverley SM, Nakhasi HL. The application of gene expression microarray technology to kinetoplastid research. Curr Mol Med. 2004;4:611–21. doi: 10.2174/1566524043360221. [DOI] [PubMed] [Google Scholar]
- 65.Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Saxena A, Worthey EA, Yan S, Leland A, Stuart KD, Myler PJ. Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol Biochem Parasitol. 2003;129:103–14. doi: 10.1016/s0166-6851(03)00100-2. [DOI] [PubMed] [Google Scholar]
- 67.LeBowitz JH. Transfection experiments with Leishmania. Methods Cell Biol. 1994;45:65–78. doi: 10.1016/s0091-679x(08)61846-4. [DOI] [PubMed] [Google Scholar]
- 68.Maga JA, Sherwin T, Francis S, Gull K, LeBowitz JH. Genetic dissection of the Leishmania paraflagellar rod, a unique flagellar cytoskeleton structure. J Cell Sci. 1999;112:2753–63. doi: 10.1242/jcs.112.16.2753. [DOI] [PubMed] [Google Scholar]
- 69.Clayton C, Adams M, Almeida R, Baltz T, Barrett M, Bastien P, Belli S, Beverley S, Biteau N, Blackwell J, Blaineau C, Boshart M, Bringaud F, Cross G, Cruz A, Degrave W, Donelson J, El-Sayed N, Fu G, Ersfeld K, Gibson W, Gull K, Ivens A, Kelly J, Vanhamme L, et al. Genetic nomenclature for Trypanosoma and Leishmania. Mol Biochem Parasitol. 1998;97:221–4. doi: 10.1016/s0166-6851(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 70.Liang XH, Haritan A, Uliel S, Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot Cell. 2003;2:830–40. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maga JA, LeBowitz JH. Unravelling the kinetoplastid paraflagellar rod. Trends Cell Biol. 1999;9:409–13. doi: 10.1016/s0962-8924(99)01635-9. [DOI] [PubMed] [Google Scholar]
- 72.Fouts DL, Stryker GA, Gorski KS, Miller MJ, Nguyen TV, Wrightsman RA, Manning JE. Evidence for four distinct major protein components in the paraflagellar rod of Trypanosoma cruzi. J Biol Chem. 1998;273:21846–55. doi: 10.1074/jbc.273.34.21846. [DOI] [PubMed] [Google Scholar]
- 73.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webb H, Burns R, Kimblin N, Ellis L, Carrington M. A novel strategy to identify the location of necessary and sufficient cis-acting regulatory mRNA elements in trypanosomes. Rna. 2005;11:1108–16. doi: 10.1261/rna.2510505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


