Abstract
The DNA-binding inorganic compound cisplatin is one of the most successful anticancer drugs. The detailed mechanism by which cells recognize and process of cisplatin-DNA damage is of great interest. Although the family of proteins that bind cisplatin 1,2- and 1,3-intrastrand cross-links has been identified, much less is known about cellular protein interactions with cisplatin interstrand cross-links (ICLs). In order to address this question, a photoreactive analogue of cisplatin, PtBP6, was used to construct a DNA duplex containing a site-specific platinum ICL. This DNA probe was characterized and used in photo-cross-linking experiments to separate and identify nuclear proteins that bind to the ICL by peptide mass fingerprint analysis. Several such proteins were discovered, including PARP-1, hMutSβ, DNA ligase III, XRCC1, and PNK. The photo-cross-linking approach was independently validated by an electrophoretic mobility shift assay demonstrating hMutSβ binding to a cisplatin ICL. Proteins that recognize the platinum ICL were also identified in cisplatin resistant cells, cells halted at various phases of the cell cycle, and in different carcinoma cells. Nuclear proteins that bind to the platinum ICL differ from those binding to intrastrand cross-links, indicating different mechanisms for disruption of cellular functions.
cis-Diamminedichloroplatinum(II) (cisplatin) is widely used in cancer chemotherapy, a key feature of its mechanism being the binding to nuclear DNA and subsequent blockage of transcription while eluding repair (1, 2). A better understanding of the details of these processes has the potential to advance development of better platinum-based anticancer drug strategies. An important step in the process is to identify cellular factors that recognize and facilitate the processing of Pt-DNA adducts (3).
Cisplatin binding to DNA begins with platination of the N(7) atoms of purine residues, mainly guanine but also adenine, under physiological conditions. The major products (approximately 90%) are 1,2- and 1,3-intrastrand cross-links involving these nucleosides, with a minor product being interstrand cross-links (ICLs) (1). The frequency of ICLs is noticeably higher (approximately 30%) in supercoiled plasmid DNA than in linear DNA (4). A transcription mapping study revealed that, in linear DNA, cis-platin preferentially forms an ICL between the N-7 atoms of two guanine residues at 5′-GC/5′-GC sites, whereas DNA ICLs of the anticancer inactive molecule trans-diamminedichloroplatinum(II) (trans-DDP) are preferentially formed between guanine and its cytosine complement (5, 6).
Although intrastrand cross-links comprise the major cisplatin-DNA adducts responsible for its biological activity, the ICL has long been a topic of discussion (7). Cisplatin ICLs strongly inhibit DNA transcription elongation by RNA polymerases (8). Moreover, cellular resistance to cisplatin has been associated with increased gene-specific repair DNA efficiency of ICLs (9), and such repair may be one reason for the less prominent role of ICLs in evoking cytotoxicity and anticancer activity.
Structural studies of cisplatin ICLs were first performed by using chemical probes of DNA conformation, gel electrophoresis mobility shift assays, and molecular modeling (10, 11). The general conclusion was that the cisplatin ICL induces a 45° bending and a 79° unwinding of the duplex, distortions larger than those caused by intrastrand cross-links. NMR solution structures of cisplatin ICLs solved for double-stranded DNA decamers of different sequences (12, 13) revealed the deoxyguanosine-bridged cis-diammineplatinum(II) unit to lie in the minor groove and the double helix to be locally reversed, forming a left-handed, Z-DNA structure. The cross-linked guanine residues were no longer paired to the complementary cytosines. The crystal structure of a cisplatin ICL has also been solved (14). Two cytosine residues in the central 5′-GC/5′-GC sequence are extruded from the double-helix, and one participates in a cross-strand base-base stacking interaction.
Nuclear proteins that interact with platinum-DNA ICLs are poorly understood. Intrastrand 1,2-d(GpG) and 1,3-d(GpNpG) DNA adducts are removed by the excision repair pathway, and the high mobility group box 1 (HMGB1) protein can bind to the former cross-link and shield it from this process (15, 16). Electrophoretic mobility shift assays (EMSAs) also reveal very weak recognition of DNA containing trans-DDP ICLs by HMGB1 (17). When 110-bp DNA probes containing five equally spaced ICLs of cisplatin or trans-DDP were examined, only the former was recognized by HMGB1, with an affinity similar to that of the 1,2-d(GpG) intrastrand cross-link (17). Replication protein A (RPA), a protein involved in excision repair, and E. coli MutS, a protein involved in mismatch repair (MMR), display lower affinity for cisplatin ICLs compared to non-platinated duplexes (18-20). Ku70 and Ku86 also bind with lower affinity to cisplatin DNA ICLs compared to intrastrand cross-links. As a consequence, the activity of Ku-dependent DNA-PKcs decreases with ICL DNA probes (21).
To identify nuclear proteins that recognize cisplatin DNA damage, we previously employed a cis-platin analog containing a photoreactive benzophenone moiety, PtBP6 (Figure 1a) (22-24). Short oligonucleotide duplexes containing a PtBP6 1,2-d(GpG) or 1,3-d(GpTpG) intrastrand cross-link were used to capture proteins in cancer cell nuclear extract that interact with the DNA probe (22, 24). One of the advantages of this photo-cross-linking system is that irradiation at 365 nm does not labilize Pt-guanine bonds, as previously reported for an earlier construct (25). With the use of a biotinylated form of this intrastrand cross-linked DNA, proteins in the nuclear extract that bind to the probe have sufficient proximity to the benzophenone moiety to form covalent bonds upon UV irradiation. The resulting covalent protein-DNA adducts are then separated on streptavidin-coated magnetic beads, resolved by gel electrophoresis, and the products identified by peptide mass fingerprinting analysis. By this method, the family of nuclear proteins have been identified that interact with the 1,2-d(GpG) and 1,3-d(GpTpG) intrastrand DNA cross-links, including HMGB1, HMGB2, and poly(ADP-ribose) polymerase 1 (PARP-1) (22, 24).
Figure 1.
(a) The structure of the cisplatin analogue, PtBP6, used in photo-cross-linking experiments. (b) The sequence of a 25-bp duplex containing PtBP6 ICL and a biotin moiety.
In the present study, we have prepared and characterized a 25-bp DNA probe containing a PtBP6 ICL (Figure 1b) and used it in a photo-cross-linking study to identify the nuclear proteins that recognize this adduct. The information provides the first comprehensive analysis of this family of proteins. The potential implications of our findings for the mechanism of cisplatin ICL recognition and processing in cancer cells are discussed.
EXPERIMENTAL PROCEDURES
Material and Methods
All chemicals and solvents were purchased from commercial sources. Enzymes were obtained from New England Biolabs (Ipswich, MA). UV-vis spectra were recorded on a HP 8453 spectrometer. Platinum analysis was performed by flameless atomic absorption spectrometry on a Perkin-Elmer AAnalyst 300 system. Matrix-assisted laser desorption/ionization (MALDI) measurements were conducted on a Bruker Omniflex mass spectrometer at the MIT Department of Chemistry Instrumentation Facility. Analytical and preparative HPLC was performed on an Agilent 1200 HPLC system. SequaGel concentrate used in the preparation of the denaturing polyacrylamide gels was purchased from National Diagnostics. DNA concentrations were obtained with the assistance of an oligonucleotide calculator at http://www.basic.northwestern.edu/biotools/oligocalc.html. Polyacrylamide gel electrophoresis (PAGE) was performed on either a Life Technologies S2 sequencing gel electrophoresis apparatus or a Protean II xi cell from Biorad. Electrophoresis gels with radioactive samples were dried and documented on a Storm 840 Phosphorimager system from Amersham or exposed wet to a Kodak Biomax MS film. Radioactive samples were quantitated on a Beckman LS 6500 scintillation counter. A2780 and A2780/CP70 cells were obtained from by Dr. Thomas Hamilton (Fox Chase Cancer Center, Jenleintown, PA). hMutSβ protein was kindly provided by Dr. Paul Modrich (Duke University, Durham, NC).
Synthesis of 25-mer 5′-CCTCTCCTCTCCTGCTCTTCTCTCC-3′ (ts) and 5′-GGAGAGAAGAGCAGGAGAGGAGAGG-3′ (bs)
These oligonucleotides were synthesized by using standard phosphoramidite methods on an Applied Biosystems 392 DNA/RNA synthesizer on a 1.0 μmol scale. Phosphoramidites were obtained from Glen Research. After automated synthesis, deprotection was performed in ammonium hydroxide at 55 °C for 16 h, according to the manufacturer’s directions. Single-stranded oligodeoxyribonucleotides were purified by 12% denaturing PAGE (7.5 M urea, 29:1 acrylamide: bisacrylamide, 90 mM Tris-borate, 2.0 mM EDTA, pH 8.3) gels run for 4 h at 300 V. Oligodeoxyribonucleotide bands were visualized under UV light, cut from the gel, and soaked at 37 °C overnight in buffer containing 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 1 mM EDTA. The eluent was cleared by filtration (0.2 μm syringe filter, Pall Corp.) and ethanol precipitated. The purity of material was checked by ion-exchange HPLC and the amount of oligodeoxyribonucleotide quantitated spectrophotometrically.
Platination of ts with PtBP6
cis-[Pt(NH3)BP6ClI] was synthesized as reported (22). To a solution of cis-[Pt(NH3)BP6ClI] (8.73mM in DMF) was added 0.9 equiv of AgNO3 in H2O. The resulting mixture was vortexed in dark for 4 h and centrifuged. The ts (0.25 mM) was allowed to react with activated PtBP6 in a buffer of 10 mM NaClO4, pH 5.6. The reaction mixture was incubated at 37 °C for 10 min. The reaction was stopped by adding NaCl to 0.4 M. The PtBP6-modified ts was purified by reverse phase HPLC (Agilent ZORBAX SC300-C18, linear gradient, 9% acetonitrile in 0.1M NaCl to 12% acetonitrile in 0.1M NaCl over 17 min). After purification, the platinated DNA solution was lyophilized and dialyzed against 0.1 M NaCl. The products were characterized by UV-vis spectroscopy, atomic absorption spectroscopy, and mass spectrometry.
PtBP6-modified single-stranded oligodeoxyribonucleotides (50 pmol) were radiolabeled with [γ-32P]-ATP (Dupont/NEN) in a solution (30 μL) containing 70 mM Tris-HCl, pH 7.6, 10 mM MgCl2, and 10 units of T4 polynucleotide kinase (PNK) at 37 °C. Another 10 units of T4 PNK were added to the solution after 45 min. After a total of 90 min, the reaction solutions were passed through spin dialysis columns (G-25 Sephadex Quickspin column, Roche). The PtBP6-modified ts was further characterized by nuclease S1 digestion assay and by Maxam-Gilbert A/G digestion (Figure S1, S2) (26).
Synthesis of 25-bp DNA Probe Containing a Site-Specific PtBP6 ICL
The top strand containing the monofunctional adduct (5 μM) was annealed with 1 equiv of the complementary strand (bs) in 0.45 M NaCl, 2 mM MgCl2, and 20 mM Tris-HCl, pH 7.4, at room temperature for 2 h and then at 4 °C for 2.5 h. The duplex solution was dialyzed against 0.1 M NaClO4 at 4 °C for 6 h and incubated in the dark at 37 °C for 64 h to form interstrand cross-links. The resulting product was purified by ion-exchange HPLC (Dionex DNAPac PA-100, 9 × 250 mm, 10 mM NaOH pH 12, linear gradient if 0.7 - 0.85 M NaCl over 20 min). Using this denaturing gradient, non-interstrand cross-linked strands were eluted first, whereas the interstrand cross-linked products were eluted later in a single peak as a high-molecular-mass species. This single peak was collected, lyophilized, dialyzed against 0.1 M NaClO4 at 4 °C for 6 h, and stored at -20 °C. The product was radiolabeled with [γ-32P]-ATP as described above. Alternatively, the ts containing the monofunctional adduct was annealed with [γ-32P]-ATP labeled complementary bs and the interstrand cross-linked material was synthesized as described above. The duplexes containing the interstrand cross-links were separated on 12% polyacrylamide/7.5 M urea denaturing gels. Bands corresponding to interstrand cross-linked duplexes were visualized under UV light, cut from the gel, and soaked at 37 °C overnight in a buffer containing 10mM Tris-HCl, pH 7.5, 50 mM NaCl, and 1 mM EDTA. The interstrand cross-linked products were ethanol precipitated, resuspended in 0.1 M NaClO4, and stored in -20 °C. For NaCN reversal experiments, DNA samples containing the ICL were treated with 0.2 M NaCN pH 11-12 and were incubated at 50 °C overnight.
Preparation of HeLa Nuclear Extract
HeLa nuclear extract was prepared as described (22).
Analytical Scale Photo-Cross-Linking Reactions with HeLa Nuclear Extracts
The radiolabeled double-stranded DNA probe (1 pmol) was incubated with a HeLa nuclear extract (2 mg/mL) in a solution (total volume 25 μL) containing 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 50 mM KCl, 1 mM EDTA, 0.05% Nonidet-P40, 0.2 μg/mL BSA on ice for 30 min to allow protein-DNA binding. After incubation, the reaction mixtures were irradiated with UV light (λmax = 365 nm) for 2 h on ice using a UV Stratalinker (Stratagene). Following irradiation, the reaction mixture was combined with 20 μL of gel-loading solution containing 125 mM Tris-HCl, pH 6.8, 40% glycerol, 4% SDS, and 10% β-mercaptoethanol. The DNA was denatured by heating at 90 °C for 5 min followed by rapid cooling on ice and then resolved on a 10% SDS-PAGE gel. The gel was dried and analyzed on a Bio-Rad GS-525 Molecular Imager using the Multi-Analyst software package (Bio-Rad).
Preparative-Scale Photo-Cross-Linking
A 100 pmol quantity of unradiolabeled double-stranded ICL DNA probe as well as a negative control, unradiolabeled double-stranded DNA lacking the platinum cross-link, were each incubated with 2 mg of HeLa nuclear extract in a solution (total volume 500 μL) containing 2 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 10 mM KCl, 0.2 mM EDTA, 0.01% NP-40, 0.04 μg/mL BSA on ice for 30 min to allow protein-DNA binding. The reaction mixtures were irradiated for 2 h on ice. A 5 mg portion of streptavidin-coated magnetic beads (Dynabeads, Invitrogen) was then added to each reaction mixture, and the solutions were incubated at 25 °C for 15 min. The supernatant was then removed from the beads by using a magnet. The beads were washed 3 times with a buffer of 5 mM Tris-HCl, 0.5 mM EDTA, 1M NaCl, 0.05% Tween-20, pH 7.5, each time removing the supernatant with the magnet. After the final wash, the beads were combined with 40 μL of 2X SDS-PAGE loading buffer (100 mM Tris-HCl, 20 mM DTT, 2% SDS, 30% glycerol, 0.1% bromophenol blue, pH 7.9) and heated to 90 °C for 5 min to dissociate the biotinylated DNA from the magnetic beads. After heating, the supernatant was quickly removed with the help of a magnet. The supernatant from a preparative-scale photo-cross-linking experiment, along with reaction mixtures from an analytical-scale photo-cross-linking were resolved by a 10% SDS-PAGE with a 4% stacking gel. After electrophoresis, the lanes containing the preparative-scale photo-cross-linked products were separated from the rest of the gel. These lanes were stained with Coomassie Blue R-250 for 12 min and destained twice in 40% methanol, 10% glacial acetic acid, and 50% deionized H2O for 30 min. The result from the radioactive analytical-scale photo-cross-linking experiment was used as a guide to excise bands from the gel loaded with preparative-scale photo-cross-linked material. A protein marker (BenchMark™ Pre-Stained Protein Ladder, Invitrogen) was included on both analytical and preparative gels to help locate the band more precisely. Bands were excised from lanes containing DNA with an ICL as well as the negative controls. The excised bands were then analyzed by trypsin digest-coupled LC-MS/MS. The proteins were identified by SEQUEST protein analysis software.
HeLa Cell Synchronization and Fluorescence-Activated Cell Sorting (FACS) Analysis
HeLa cells were maintained as exponentially growing cultures in a DMEM medium with 10% FBS, 1% penicillin, and 1% glutamine. Cells were synchronized to different phases before obtaining nuclear extracts. To obtain cells in S phase, HeLa cells were treated with 2 mM thymidine for 19 h, released in fresh medium for 12 h, treated with 2 mM thymidine for 12 h and then released in fresh medium for 4 h. To obtain cells in G2/M phase, HeLa cells were treated with 2 mM thymidine for 12 h, released in fresh medium for 4 h, and then treated with 100 ng/mL nocodazol for 16 h. To obtain G1/S phased cells, the cells were treated with 2 mM thymidine for 17 h, released in DMEM for 8 h, and treated with 2 mM thymidine for 17 h. After synchronization, cells were collected, then washed twice with PBS. A total of 5×106 cells were examined by FACS analysis to confirm the synchronization, and the rest were used to obtain nuclear extracts. For FACS analysis, cells were fixed with 70% ethanol at 4 °C overnight, then washed with PBS, and subsequently stained with 20 ng/mL propidium iodide in PBS containing 0.1% Triton X-100, 0.2 mg/mL Ribonuclease A (Sigma) at 37 °C for 15 min. The cells were then analyzed with a BD FACScan™ Flow Cytometer (Figure S4).
Electrophoretic Mobility Shift Assays
Binding assays were performed according to a protocol reported before (27) with modification. A 5 fmol quantity of radiolabeled 25-bp DNA strand containing cisplatin ICL (5′-CCTCTCCTCTCCTG*CTCTTCTCTCC/5′-GGAGAGAAGAG*CAGGAGAGGAGAGG, where the asterisks denote the platinated bases) was synthesized following a reported protocol (28) and incubated in buffer (25 mM HEPES-KOH, pH 7.8, 5 mM MgCl2, 80 mM KCl, 1mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1 mg/mL bovine serum albumin) with carrier/competitor poly(dGdC) DNA at a 10:1 ratio with platinated DNA on a per nucleotide basis and the indicated concentrations of hMutSβ at room temperature for 10 min. A negative control of a 25-bp DNA strand without ICL (sequence the same as for the ICL above) and a positive control of a 25-bp DNA containing a 5′-CA insert (5′-CCTCTCCTCTCCTGCACTCTTCTCTCC/5′-GGAGAGAAGAGCAGGAGAGGAGAGG), were also included. Reaction products were separated by electrophoresis on 5% polyacrylamide gels with a 37.5:1 ratio of acrylamide:bisacrylamide at 4 °C in TBE buffer (89 mM Tris borate, 2 mM EDTA). The gels were dried for autoradiography.
RESULTS
Design of DNA Sequences
The choice of the 25-bp oligonucleotide duplex containing a site-specific PtBP6 ICL (Fig. 1b) was based on the sequences of our related probes containing a single 1,2-d(GpG) (22) or 1,3-d(GpTpG) intrastrand cross-link (24), the difference being that only a single guanosine was contained in the top strand. In this manner we were able to obtain the best comparison between proteins identified here and those for the related 1,2-d(GpG) and 1,3-d(GpTpG) PtBP6-modified DNA probes.
Synthesis and Characterization of the 25-bp PtBP6 ICL Probe
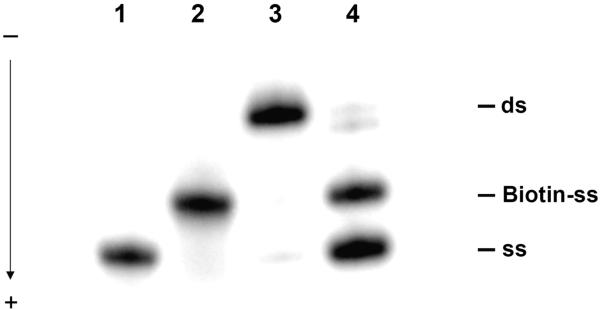
The overall strategy for the synthesis of a PtBP6 ICL probe was first to obtain a monofunctional [Pt(BP6)(NH3)(DNA)Cl] product and then to cross-link it to the annealed complementary strand. [Pt(BP6)(NH3)ClI] was used to platinate the 25mer top strand DNA fragment. The product, in which the iodide ligand was replaced by N7 of the guanine base, was purified by reverse phase HPLC and fully characterized by UV-vis spectroscopy, atomic absorption spectroscopy, MALDI-MS/MS, a nuclease S1 digestion assay, and a Maxam-Gilbert A/G digestion by standard methods (Tables S1 and S2; Figures S1 and S2) (25, 29). Two orientational isomers (30), were readily separated under the current HPLC conditions with Pt/DNA ratios of 1.02 ± 0.12 and 1.01 ± 0.01, respectively. The monofunctional adducts were annealed with the complementary strand containing a biotin moiety at the 3′ end. ICLs were formed in NaClO4 buffer and purified by ion-exchange HPLC. From the two isomers of monofunctional product, two ICL products, ICL-1 and ICL-2, were obtained. The yields from the monofunctional products were 8.9% and 15.8%, respectively. An analytical denaturing PAGE gel revealing the starting materials and ICL-2 is shown in Figure 2. Removal of the cross-link by treatment with NaCN confirmed the presence of the platinum ICL. Since ICL-2 formed in better yield and had higher photo-cross-linking efficiency, as discovered in subsequent assays, we used it as the DNA probe for identifying proteins in nuclear extracts. The orientation of the photoreactive group relative to DNA in each isomer was not determined.
Figure 2.
Autoradiogram of the PtBP6 ICL DNA probe. The oligonucleotides were 5′-end labeled and revealed on a 12% denaturing polyacrylamide gel. 1, top strand; 2, bottom strand with 3′-biotin moiety; 3, PtBP6 ICL with both top and bottom strand labeled; 4, PtBP6 ICL after NaCN treatment.
Analytical Scale Photo-Cross-Linking of the DNA ICL with HeLa Nuclear Extracts
The 25-bp ICL DNA probe was radiolabeled and incubated with HeLa nuclear extracts under UV irradiation to form protein-DNA cross-links. The relative amounts of DNA and nuclear extracts were optimized. A 10% SDS-PAGE gel readily showed cross-linked protein bands of high molecular weight (Figure 3, Lane 3). A radiolabeled duplex lacking the ICL used as a negative control exhibited no cross-linking activity with HeLa nuclear extracts (Figure 3, Lane 1). The ICL DNA probe was also irradiated in the absence of a nuclear extract and showed no self-cross-linking activity (Figure 3, Lane 2). We also carried out the following control studies. The monofunctional PtBP6-modified ts and the PtBP6 ICL probes were incubated in HeLa nuclear cell extracts without irradiation (Figure S3). SDS-PAGE analysis revealed the absence of DNA-Pt-protein cross-links, even though the guanine base was modified by PtBP6 (Figure S3, Lanes 1 and 2). In order to rule out the possibility that the DNA probe might be unstable under the UV irradiation condition (λ 365 nm), or that the ICL might either dissociate or rearrange into an intrastrand cross-link, the ICL DNA probe was UV-irradiated without nuclear extracts in the protein binding buffer for 0, 1, 2 h. A denaturing gel confirmed that the ICL probe migrates identically upon UV irradiation for different time periods (Figure 4), confirming its stability under the photo-cross-linking conditions.
Figure 3.
Photo-cross-linking result for ICL DNA and HeLa nuclear extracts. Lane 1, control probe; Lane 2, PtBP6 ICL probe without HeLa nuclear extract; Lane 3, PtBP6 ICL probe with HeLa nuclear extract
Figure 4.

Stability of the interstrand cross-linked probe under UV irradiation. The probe was revealed on a 12% denaturing polyacrylamide gel.
Preparative Scale Photo-Cross-Linking of DNA ICLs in HeLa Nuclear Extracts
To identify proteins cross-linked by the ICL DNA probe, preparative scale photo-cross-linking was performed. The DNA-to-platinum ratio remained the same as in the analytical scale experiment. Proteins that bind to the biotin-conjugated DNA probe were pulled down by streptavidin-coated magnetic beads and washed stringently with high salt buffer to remove any non-covalently bound material. A 25-bp DNA duplex with no ICL was used as a negative control. The protein-DNA complexes were separated by SDS-PAGE. An analytical scale experiment was simultaneously performed and the protein-DNA complex loaded onto the same gel. Protein-DNA bands from preparative scale experiments were excised using results from the analytical scale experiment as a guide. Proteins in the various bands (Figure 5) were then identified by peptide mass fingerprinting analysis as described below.
Figure 5.

Preparative-scale photo-cross-linking of the ICL in a HeLa nuclear extract. Bands were excised for peptide mass fingerprinting analysis.
Protein Identification
Bands from the photo-cross-linking experiment as well as a negative control were analyzed by trypsin-digestion coupled LC-MS/MS (Table 1) (31). The “probability” column in the table is a number assigned to each protein based on the likelihood that the identification is an artifact due to background. Any protein with a probability higher than 1 × 10-4 was excluded from the results. Proteins that appeared in both a photo-cross-linking and a negative control experiment were considered to be background or non-covalently bound and also excluded from the results. The “unique peptides” column reports the number of different unique peptides that match a known protein sequence from a data bank. The listed values come from two repeats of preparative scale photo-cross-linking experiments. The proteins listed either appeared in both experiments or had a low probability number. The results indicate that several categories of proteins have an affinity for a cisplatin ICL. Included are the DNA-dependent protein kinase (DNA-PK) complex DNA-PKcs, Ku86 and Ku70, which is involved in the non-homologous end joining (NHEJ) DNA repair pathway; PARP-1, DNA ligase III, X-ray repair cross complementing 1 (XRCC1), and PNK, which are involved in the single-strand-break repair (SSBR) pathway; mismatch repair proteins hMSH2 and hMSH3; HMG-domain proteins HMGB1, HMGB2, and HMGB3; and other proteins such as nuclear mitotic apparatus protein 1 (NUMA1) and xeroderma pigmentosum group E binding factor (XPE-BF).
Table 1.
Proteins identified by photo-cross-linking of a 25-bp DNA duplex containing a PtBP6 ICL adduct
| Band[a] | Protein[b] | Probability[c] | Unique peptides[d] | Mw [Da][e] |
|---|---|---|---|---|
| 1 | DNA-PKcs | 1.11 × 10-6 | 4 | 468786.9 |
| NUMA1 | 2.09 × 10-6 | 2 | 238712.5 | |
| 2 | PARP-1 | 7.31 × 10-11 | 20 | 113012.4 |
| hMSH2 | 6.72 × 10-9 | 4 | 104676.8 | |
| DNA Ligase III | 3.35 × 10-7 | 7 | 112834.9 | |
| hMSH3 | 6.15 × 10-7 | 10 | 127376.1 | |
| 3 | hMSH2 | 3.40 × 10-10 | 7 | 104676.8 |
| PARP-1 | 6.58 × 10-10 | 18 | 113012.4 | |
| Ku86 | 3.72 × 10-9 | 4 | 82652.4 | |
| hMSH3 | 3.99 × 10-7 | 13 | 127376.1 | |
| DNA Ligase III | 7.69 × 10-7 | 6 | 112834.9 | |
| Ku70 | 1.78 × 10-6 | 3 | 69799.2 | |
| XPE-BF | 3.35 × 10-6 | 3 | 126887.4 | |
| XRCC1 | 2.79 × 10-5 | 1 | 70970.6 | |
| 4 | Ku86 | 1.42 × 10-10 | 8 | 82652.4 |
| PARP-1 | 1.10 × 10-8 | 2 | 113012.4 | |
| DNA Ligase III | 1.23 × 10-6 | 6 | 112834.9 | |
| Ku70 | 2.71 × 10-6 | 3 | 69799.2 | |
| XRCC1 | 2.84 × 10-5 | 1 | 70970.6 | |
| hMSH2 | 1.44 × 10-4 | 1 | 104676.8 | |
| hMSH3 | 1.80 × 10-4 | 1 | 127376.1 | |
| 5 | Ku70 | 7.88 × 10-11 | 11 | 69799.2 |
| PARP-1 | 1.17 × 10-8 | 4 | 113012.4 | |
| XRCC1 | 4.52 × 10-5 | 1 | 70970.6 | |
| hMSH2 | 4.59 × 10-5 | 2 | 104676.8 | |
| PNK | 1.54 × 10-5 | 2 | 57040.6 | |
| 6 | Ku86 | 4.84 × 10-4 | 1 | 82652.4 |
| 7 | HMGB1 | 1.24 × 10-5 | 2 | 24878.2 |
| HMGB2 | 5.31 × 10-4 | 1 | 24018.7 | |
| HMGB3 | 7.71 × 10-8 | 1 | 14597.2 | |
| 8 | Ku70 | 1.28 × 10-6 | 1 | 69799.2 |
The mass fingerprint analysis was carried out on eight gel slices as indicated in Figure 5.
Protein identified as described in the Experimental Procedures.
Probability as described in the text.
Unique Peptides as described in the text.
Masses listed are those of the full length proteins.
hMutSβ EMSA
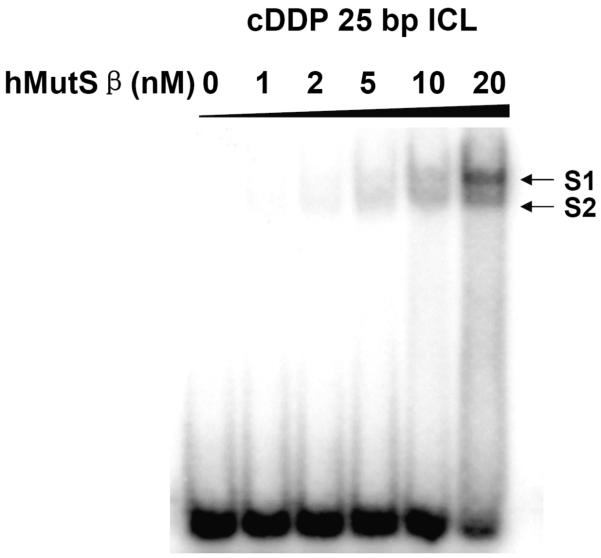
We validated these photo-cross-linking results by demonstrating that a protein identified in the assay has a measurable affinity for a DNA duplex containing a cisplatin ICL. Since hMSH2 and hMSH3 form a heterodimer of hMutSβ protein (32), we obtained a purified sample of the protein and performed an EMSA to determine whether it recognizes the cisplatin ICL. A 25-bp duplex oligonucleotide containing a site-specific cisplatin ICL was synthesized as reported previously (28) and characterized by denaturing PAGE (Figure S5). The cisplatin ICL probe had the same sequence and platination site as the PtBP6 ICL probe used in the photo-cross-linking study (Figure 1b). EMSAs were performed with different concentrations of protein in the presence of radiolabeled duplex oligonucleotide as well as a competitor to reduce non-specific binding. As shown in Figure 6, hMutSβ results in the formation of two specific bands (S1 and S2). These results indicate that hMutSβ recognizes and bind a cisplatin ICL, a finding not previously reported to the best of our knowledge. This discovery thus confirms the photoaffinity labeling strategy for identifying nuclear proteins that recognize a cisplatin ICL. The introduction of benzophenone moiety apparently does not significantly affect the binding of nuclear proteins to and ICL containing this cisplatin analogue.
Figure 6.
EMSA analysis of the binding of hMutSβ to a 25-bp DNA duplex containing a cisplatin ICL. The radiolabeled 25-bp cisplatin ICL DNA (0.25 nM) was incubated with 0, 1, 2, 5, 10, or 20 nM hMutSβ.
Photo-Cross-Linking with Different Cancer Cell Nuclear Extracts
Nuclear extracts from different mammalian carcinoma cells HeLa (cervical), BxPC3 (pancreatic), NTera 2 (testicular), and U2OS (bone), as well as cell-free extracts of CHO (Chinese hamster ovary) cells were used in analytical scale photo-cross-linking studies with the PtBP6 ICL probe (Figure 7). The same amount of nuclear extract and DNA were used in each case. Different human cell nuclear extracts showed similar proteins bound to the ICL, although the quantities differed. For example, there is more PARP1 bound to the ICL in extracts from HeLa compared to the other cells. More XRCC1/Ku70 is bound to the ICL in HeLa and NTera, but not in BxPC3 and U2OS cells. The binding proteins in hamster cells are quite different from those in the human cell lines. Taken together, these results demonstrate that the PtBP6 probe is a useful tool for distinguishing the binding of nuclear proteins to the ICL in cells of various origin.
Figure 7.
Photo-cross-linking of the ICL probe with nuclear extracts from different cells. In each case, 1 pmol of DNA probe was incubated with 20 μg of nuclear extract for photo-cross-linking analysis, and the same amount of the mixture was loaded onto the gel in each lane.
Photo-Cross-Linking with Cisplatin Resistant Cells
We purified nuclear extracts from a cisplatin resistant cell line A2780/CP70, as well as the parental line A2780, to investigate whether different nuclear proteins would bind to the PtBP6 ICL. Identical amounts of DNA and nuclear extracts were incubated to form DNA-Pt-protein cross-links (Figure 8). The cross-linking efficiency was essentially the same in nuclear extracts from A2780/CP70 and A2780 cells, the percentages of cross-linked DNA being 9.3% and 8.8%, respectively. The results revealed the same protein binding to the PtBP6 ICL, although the quantities differed. More PARP-1 protein bound to platinum ICL in A2780/CP70 cells than in A2780 cells. In the case of HMGB1/2/3, when the total amount of bound proteins was normalized, 9.1% of the bound proteins were HMGB1/2/3 in the A2780/CP70 cells, but only 4.5% were HMGB1/2/3 in the A2780 cells. The PtBP6 ICL is therefore valuable for determining different levels of nuclear protein binding to an ICL in cisplatin resistant vs. sensitive cells.
Figrue 8.

Photo-cross-linking of the ICL probe with nuclear extracts from cisplatin resistant (A2780/CP70) and sensitive (A2780) cells. In both cases, 1 pmol of DNA probe was incubated with 20 μg of nuclear extract for photo-cross-linking study, and the same amount of the mixture was loaded onto the gel in each lane.
Photo-Cross-Linking with Nuclear Extracts from Different Phases of the Cell Cycle
In order to evaluate recognition of cisplatin ICLs by nuclear proteins at different phases of the cell cycle, HeLa nuclear extracts from synchronized cells were purified. The cells were synchronized at G1/S, S, or G2/M by a thymidine or thymidine+nocodazole block and the DNA contents were characterized by FACS analysis (Figure S4). Nuclear extracts from unsynchronized cells (U) and synchronized cells at different phases were used in the photo-cross-linking study with the duplex oligonucleotide containing a PtBP6 ICL (Figure 9). In each case, the same amounts of nuclear extract and ICL DNA probe were incubated, and the same amount of this mixture was loaded into each lane of gel in Figure 9. The bound proteins are the same in the nuclear extracts from unsynchronized cells and synchronized cells, but again the levels differ. For example, more PARP-1 and HMGB1/2/3 proteins bind to the PtBP6 ICL at G2/M than in other phases or with unsynchronized cells. The results illustrate the ability of the PtBP6 ICL probe to determine the recognition of the platinum DNA damage at different stages of the cell cycle.
Figure 9.
Photo-cross-linking study of the ICL probe with nuclear extracts from different phase of the cell cycle. The ICL probe was photo-cross-linked with HeLa cell nuclear extracts from unsynchronized (U), G1/S phase (G1/S), S phase (S) and G2/M phase (G2/M) cells. In each case, 1 pmol of DNA probe was incubated with 20 μg of nuclear extract for photo-cross-linking study, and the same amount of the mixture was loaded onto the gel in each lane.
DISCUSSION
The identification of nuclear proteins that recognize a cisplatin ICL in previous work has mainly involved experiments comparing its binding affinity for a specific protein with that of a related probe carrying an intrastrand cross-link (18, 19, 21). The present investigation offers a more systematic approach for identifying ICL recognition proteins. We prepared and characterized a DNA probe containing a site-specific PtBP6 ICL for this purpose. Upon near-UV irradiation of nuclear extracts containing the probe, the benzophenone moiety forms covalent bonds to proteins that bind in the vicinity of the platinum cross-link. The resulting covalent DNA-platinum-protein ternary complexes were then separated and subsequently analyzed by peptide mass fingerprinting.
Identification of Proteins that Bind to ICLs
The preparative scale photo-cross-linking study identified a variety of bands in the gel that were excised and subjected to trypsin digestion-coupled mass spectrometric analysis. As anticipated, and for the reasons previously discussed (24), many proteins appeared in more than one region of the gel. During photo-cross-linking analysis, despite precautions, protein degradation during the experimental procedures is inevitable. Fragments from proteins with higher molecular weight appear as contaminants in the lower molecular weight region, and there is some carry-over during the mass spectrometric analysis, since the method is extremely sensitive.
hMutSβ Recognizes the Cisplatin ICL
DNA mismatch repair is a major pathway for correcting mistakes in replication (32). In the first step of DNA mismatch recognition, two heterodimers containing hMSH2 play very important roles. These are hMustSα, a heterodimer of hMSH2 and hMSH6, and hMustSβ, a heterodimer of hMSH2 and hMSH3. Previous work revealed that E. coli MutS has a lower affinity for a cisplatin ICL than it has for intrastrand cross-links (18). The human hMSH2 binds specifically to DNA globally modified by cisplatin (33), and hMutSα has an affinity for cisplatin 1,2-intrastrand d(GpG) but not 1,3-intrastrand d(GpTpG) cross-links (34, 35). hMutSβ binds to psoralen ICLs, and MMR is involved in repair of psoralen ICLs in an error-free manner (27, 36). Here we identified hMutSβ recognition of the cisplatin ICL by photo-cross-linking, and the result was independently confirmed by EMSA using a cisplatin probe. The results indicate that mismatch repair is involved in repairing the cisplatin ICL.
PARP-1 and XRCC1 Bind to the ICL-Damaged DNA
PARP-1 functions as a molecular sensor of DNA single-strand breaks. PARP-1 consumes NAD+ as substrate to covalently ribosylate itself or other acceptor proteins (37). The roles of PARP-1 in DNA repair include alteration of chromatin architecture and gene expression; induction of cell death by promoting nuclear translocation of apoptosis-inducing factor (AIF); and recruitment of SSBR and base-excision repair (BER) factors such as XRCC1 and DNA ligase III (38). PARP-1 inhibitors potentiate cisplatin activity in both cell-based assays and mouse breast cancer xenograft models (39, 40). The activity of PARP-1 bound to DNA containing cisplatin intrastrand cross-links results in its dissociation as well as that of other DNA repair proteins (41). The present results implicate a role for PARP-1 in mediating repair of ICL platinum-DNA damage.
XRCC1 acts as a coordinator of SSBR and BER. Genetic polymorphisms of XRCC1 are associated with decreased overall survival in non-small cell lung cancer (NSCLC) patients treated with platinum drugs (42). XRCC1 transcript abundance levels correlate with cisplatin resistance in NSCLC cell lines (43). Therefore, XRCC1 may also play a role in repair of platinum-DNA damage.
Previous work revealed that PARP-1 binds to cisplatin intrastrand cross-links (22, 24), but there is no prior evidence that XRCC1 recognizes platinum ICL adducts or any other kind of ICL DNA damage. Here we find that PARP-1, together with its partners XRCC1 and DNA ligase III, bind to a DNA duplex containing a platinum ICL. This result implies that SSBR proteins may be important for platinum ICL repair.
HMG-Domain Proteins Bind to the ICL
HMGB1 is an abundant non-histone nuclear protein that regulates chromatin structure and DNA metabolism. It binds to distorted DNA structures such as four-way junctions and damaged DNA (44). Convincing evidence has established that HMGB1 recognizes platinum 1,2-d(GpG) and 1,2-d(ApG) intrastrand cross-links rather than the 1,3-d(GpTpG) cross-link (16). HMGB1 also recognizes psoralen and cisplatin ICLs (17, 45). Our data reveal that HMGB1/2/3 proteins bind to a site-specific PtBP6 ICL, but the strength of the bands on the gel is significantly weaker than for the 1,2-intrastrand cross-link, similar to that of the 1,3-d(GpTpG) adduct (cf. Fig. 9 and Fig. 4 of ref. (24)). This result is consistent with prior data that HMGB1 recognizes mainly cisplatin intrastrand 1,2-cross-links, where it shields the damage from nucleotide excision repair (15).
Nuclear Proteins that Bind to ICLs are Identical in Different Mammalian Cells
We investigated nuclear extracts from HeLa cells as the major source for identifying proteins that binds to the cisplatin ICL. We then could compare the results of analytical photo-cross-linking studies for nuclear extracts from different sources. The results indicate a very similar pattern of binding proteins in the various mammalian nuclear extracts. Since these cells have different sensitivities to cisplatin (24), binding of damage-response proteins, involved in the first step for processing the adduct, may not be the major determinant of the differential cytotoxicity.
The PtBP6 ICL DNA Probe is a Useful Tool to Study the Cellular Response to Platinum Damage
Although a deficiency in MMR seems to confer cisplatin cellular resistance (46), there is little evidence to relate HMGB1/PARP-1 to cisplatin resistance. With our PtBP6 ICL DNA probe, we could differentiate the binding levels of these proteins in cisplatin sensitive and resistant cells. In particular, greater amounts of PARP-1 and HMGB1 are cross-linked to the ICL probe in the cisplatin resistant than in the sensitive cells. It thus appears that PARP-1 and HMGB1 may play a role in the cellular resistance to cisplatin ICL damage.
In some cell lines, cisplatin induces arrest of the cell cycle at G2, like many other chemotherapeutic agents, and it has been reported to cause an S-phase block in human ovarian carcinoma and hepatoma cell lines (47, 48). Cisplatin activity also differs with cell cycle perturbations in resistant cells (49). Little is known about the recognition of cisplatin DNA cross-links at different stage of cell cycle. By utilizing nuclear extracts from synchronized cells, we were able to apply the PtBP6 ICL probe to compare protein recognition of ICLs at different phases of the cell cycle. We find similar nuclear protein binding patterns to the ICL probe at the different G1/S, S, and G2/M phases, implying that the recognition of cisplatin ICL damage is consistent throughout the cell cycle, at least for HeLa. The level of binding proteins varies, however. For example, the amount of HMGB1/2/3 and PARP-1 photo-cross-linked to the ICL is greatest at G2/M. Either the binding properties of these proteins change throughout the cell cycle, or the levels of expression are higher at G2/M. In either case, the PtBP6 ICL probe has identified key proteins to be evaluated in future studies.
Comparison of Proteins Bound to Cisplatin Intrastrand Cross-links versus ICLs; Mechanistic Implications
In Figure 10 we compare the photo-cross-linking results for Pt-BP6 probes containing a 1,2-d(GpG) cross-link, 1,3-d(GpTpG) cross-link, or an ICL. The identified proteins fall into the following categories: excision repair proteins, include RPA1; MMR proteins, including hMSH3 and hMSH2; SSBR proteins, including PARP1/DNA Ligase III, XRCC1 and PNK; NHEJ proteins, including DNA-PKcs, Ku80/86 and Ku70; HMG-domain proteins, including HMGB1/2/3 and upstream binding factor 1 (UBF1). From the results, we can clearly distinguish different platinum-DNA damage recognition patterns (Fig. 10C). Moreover, the photo-cross-linking strategy has allowed several ICL-binding proteins to be identified for the first time, namely, hMSH2, hMSH3, PARP-1/DNA ligase III, XRCC1, PNK, NUMA1 and XPE-BF. Several proteins that bind to the ICL do not recognize intrastrand cross-links, including hMSH3, XRCC1, and PNK. More importantly, the protein binding pattern to 1,2-d(GpG), 1,3-d(GpTpG) and ICL platinum damage is significantly different. In the case of 1,2-d(GpG) recognition, the most abundant proteins are in the HMG-domain class, followed by SSBR proteins. In the case of 1,3-d(GpG) recognition, excision repair proteins, SSBR proteins, NHEJ proteins, and HMG-domain proteins share the identical abundance. However, in the case of ICL recognition, the most abundant proteins are in the MMR family, followed by NHEJ proteins and SSBR proteins. Considering that the DNA probes used for the photo-cross-link experiments share similar sequences and the experimental condition are identical, the results imply that the recognition and processing of cisplatin DNA damage is distinctive for different platinum-DNA adducts. The results also indicate different repair mechanisms for cisplatin intrastrand cross-links than for ICLs. The present research opens avenues for future detailed study of different nuclear protein binding to cisplatin ICLs and suggests that the differential recognition and subsequent processing of the various Pt-DNA adducts by these proteins dictates their relative contributions to the anticancer activity.
Figure 10.
Comparison of proteins that bind to different cisplatin DNA adducts. (a) assignments of different bands by molecular weight from peptide mass fingerprint; (b) quantification of different proteins when the total amount of bound proteins being normalized to 100% in each case. Note the quantification results for 1,2-d(GpG) and 1,3-d(GpTpG) probes are from gel images published elsewhere (ref. (24)). (c) Pie charts showing the binding of different categories of protein to cisplatin 1,2-d(GpG), 1,3-d(GpTpG) and ICL DNA cross-linked PtBP6 probes.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grant CA034992 from the National Cancer Institute. We thank Dr. Evan R. Guggenheim for many helpful suggestions, Rick Schiavoni and Dr. Ioannis Papayannopoulos from the MIT Biopolymers and Proteomics laboratories for their assistance with trypsin digestion-coupled mass spectrometry experiments, and Drs. Ravi Iyer and Paul Modrich for providing us with a sample of hMutSβ.
ABBREVIATIONS
- AIF
apoptosis-inducing factor
- BER
base-excision repair
- DNA-PK
DNA-dependent protein kinase
- EMSA
electrophoretic mobility shift assay
- FACS
fluorescence-activated cell sorting
- HMGB1
high mobility group protein B1
- ICL
interstrand cross-link
- MALDI
matrix-assisted laser desorption/ionization
- MMR
mismatch repair
- NHEJ
non-homologous end joining
- NSCLC
non-small cell lung cancer
- NUMA1
nuclear mitotic apparatus protein 1
- PAGE
polyacrylamide gel electrophoresis
- PARP-1
poly(ADP-ribose) polymerase 1
- PNK
polynucleotide kinase
- RPA
replication protein A
- SSBR
single-strand-break repair
- trans-DDP
trans-diammine-dichloroplatinum(II)
- XPE-BF
xeroderma pigmentosum group E binding factor
- XRCC1
X-ray repair cross complimenting 1
- UBF1
upstream binding factor 1
REFERENCES
- 1.Jamieson ER, Lippard SJ. Structure, Recognition, and Processing of Cisplatin-DNA Adducts. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 2.Jung Y, Lippard SJ. Direct Cellular Responses to Platinum-Induced DNA Damage. Chem. Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 4.Vrána O, Boudny V, Brabec V. Superhelical torsion controls DNA interstrand cross-linking by antitumor cis-diamminedichloroplatinum(II) Nucleic Acids Res. 1996;24:3918–3925. doi: 10.1093/nar/24.20.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabec V, Leng M. DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5345–5349. doi: 10.1073/pnas.90.11.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemaire MA, Schwartz A, Rahmouni AR, Leng M. Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum (II) and DNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1982–1985. doi: 10.1073/pnas.88.5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts JJ, Friedlos F. Quantitative estimation of cisplatin-induced DNA interstrand cross-links and their repair in mammalian cells: relationship to toxicity. Pharmacol. Ther. 1987;34:215–246. doi: 10.1016/0163-7258(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 8.Corda Y, Job C, Anin MF, Leng M, Job D. Spectrum of DNA-platinum adduct recognition by prokaryotic and eukaryotic DNA-dependent RNA polymerases. Biochemistry. 1993;32:8582–8588. doi: 10.1021/bi00084a027. [DOI] [PubMed] [Google Scholar]
- 9.Zhen W, Link CJ, Jr., O’Connor PM, Reed E, Parker R, Howell SB, Bohr VA. Increased gene-specific repair of cisplatin interstrand cross-links in cisplatin-resistant human ovarian cancer cell lines. Mol. Cell Biol. 1992;12:3689–3698. doi: 10.1128/mcb.12.9.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sip M, Schwartz A, Vovelle F, Ptak M, Leng M. Distortions induced in DNA by cis-platinum interstrand adducts. Biochemistry. 1992;31:2508–2513. doi: 10.1021/bi00124a010. [DOI] [PubMed] [Google Scholar]
- 11.Malinge JM, Perez C, Leng M. Base sequence-independent distorsions induced by interstrand cross-links in cis-diamminedichloroplatinum (II)-modified DNA. Nucleic Acids Res. 1994;22:3834–3839. doi: 10.1093/nar/22.19.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842–1845. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- 13.Paquet F, Perez C, Leng M, Lancelot G, Malinge JM. NMR solution structure of a DNA decamer containing an interstrand cross-link of the antitumor drug cis-diamminedichloroplatinum (II) J. Biomol. Struct. Dyn. 1996;14:67–77. doi: 10.1080/07391102.1996.10508930. [DOI] [PubMed] [Google Scholar]
- 14.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 A resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pil PM, Lippard SJ. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 17.Kašpárková J, Brabec V. Recognition of DNA interstrand cross-links of cis-diamminedichloroplatinum(II) and its trans isomer by DNA-binding proteins. Biochemistry. 1995;34:12379–12387. doi: 10.1021/bi00038a035. [DOI] [PubMed] [Google Scholar]
- 18.Fourrier L, Brooks P, Malinge JM. Binding discrimination of MutS to a set of lesions and compound lesions (base damage and mismatch) reveals its potential role as a cis-platin-damaged DNA sensing protein. J. Biol. Chem. 2003;278:21267–21275. doi: 10.1074/jbc.M301390200. [DOI] [PubMed] [Google Scholar]
- 19.Patrick SM, Turchi JJ. Replication protein A (RPA) binding to duplex cisplatin-damaged DNA is mediated through the generation of single-stranded DNA. J. Biol. Chem. 1999;274:14972–14978. doi: 10.1074/jbc.274.21.14972. [DOI] [PubMed] [Google Scholar]
- 20.Patrick SM, Tillison K, Horn JM. Recognition of cisplatin-DNA interstrand cross-links by replication protein A. Biochemistry. 2008;47:10188–10196. doi: 10.1021/bi800460d. [DOI] [PubMed] [Google Scholar]
- 21.Turchi JJ, Patrick SM, Henkels KM. Mechanism of DNA-dependent protein kinase inhibition by cis-diamminedichloroplatinum(II)-damaged DNA. Biochemistry. 1997;36:7586–7593. doi: 10.1021/bi963124q. [DOI] [PubMed] [Google Scholar]
- 22.Zhang CX, Chang PV, Lippard SJ. Identification of nuclear proteins that interact with platinum-modified DNA by photoaffinity labeling. J. Am. Chem. Soc. 2004;126:6536–6537. doi: 10.1021/ja049533o. [DOI] [PubMed] [Google Scholar]
- 23.Sadler PJ. Protein recognition of platinated DNA. ChemBioChem. 2009;10:73–74. doi: 10.1002/cbic.200800733. [DOI] [PubMed] [Google Scholar]
- 24.Guggenheim ER, Xu D, Zhang CX, Chang PV, Lippard SJ. Photoaffinity isolation and identification of proteins in cancer cell extracts that bind to platinum-modified DNA. ChemBioChem. 2009;10:141–157. doi: 10.1002/cbic.200800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kane SA, Lippard SJ. Photoreactivity of platinum(II) in cisplatin-modified DNA affords specific cross-links to HMG domain proteins. Biochemistry. 1996;35:2180–2188. doi: 10.1021/bi952240a. [DOI] [PubMed] [Google Scholar]
- 26.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutS β is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofr C, Brabec V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J. Biol. Chem. 2001;276:9655–9661. doi: 10.1074/jbc.M010205200. [DOI] [PubMed] [Google Scholar]
- 29.Mikata Y, He Q, Lippard SJ. Laser-induced photo-cross-linking of cisplatin-modified DNA to HMG-domain proteins. Biochemistry. 2001;40:7533–7541. doi: 10.1021/bi010318j. [DOI] [PubMed] [Google Scholar]
- 30.Hartwig JF, Lippard SJ. DNA binding properties of cis-[Pt(NH3)(C6H11NH2)Cl2], a metabolite of an orally active platinum anticancer drug. J. Am. Chem. Soc. 1992;114:5646–5654. [Google Scholar]
- 31.MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 32.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 33.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem. Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 34.Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, Modrich P. Human MutS α recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 39.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, Klein-Szanto A, Dionne C, Ruggeri B. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol. Cancer. Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 40.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, McGonigal TP, Meulbroek JA, Olson AM, Palma JP, Rodriguez LE, Shi Y, Stavropoulos JA, Tsurutani AC, Zhu GD, Rosenberg SH, Giranda VL, Frost DJ. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 41.Guggenheim ER, Ondrus AE, Movassaghi M, Lippard SJ. Poly(ADP-ribose) polymerase-1 activity facilitates the dissociation of nuclear proteins from platinum-modified DNA. Bioorg. Med. Chem. 2008;16:10121–10128. doi: 10.1016/j.bmc.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J. Clin. Oncol. 2004;22:2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 43.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol. Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 45.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 46.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 47.Qin LF, Ng IO. Induction of apoptosis by cisplatin and its effect on cell cycle-related proteins and cell cycle changes in hepatoma cells. Cancer Lett. 2002;175:27–38. doi: 10.1016/s0304-3835(01)00720-0. [DOI] [PubMed] [Google Scholar]
- 48.Chu G. Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA repair. J. Biol. Chem. 1994;269:787–790. [PubMed] [Google Scholar]
- 49.Wilkins DE, Ng CE, Raaphorst GP. Cell cycle perturbations in cisplatin-sensitive and resistant human ovarian carcinoma cells following treatment with cisplatin and low dose rate irradiation. Cancer Chemother. Pharmacol. 1997;40:159–166. doi: 10.1007/s002800050641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.