Abstract
Here we demonstrate a reagentless, electrochemical platform for the specific detection of proteins that bind to single- or double-stranded DNA. The sensor is composed of a double- or single-stranded, redox-tagged DNA probe which is covalently attached to an interrogating electrode. Upon protein binding the current arising from the redox tag is suppressed, indicating the presence of the target. Using this approach we have fabricated sensors against the double-stranded DNA binding proteins TATA-box binding protein and M.HhaI methyltransferase, and against the single-strand binding proteins Escherichia coli SSBP and replication protein A. All four targets are detected at nanomolar concentrations, in minutes, and in a convenient, general, readily reusable, electrochemical format. The approach is specific; we observed no significant cross-reactivity between the sensors. Likewise the approach is selective; it supports, for example, the detection of single strand binding protein directly in crude nuclear extracts. The generality of our approach (including its ability to detect both double- and single-strand binding proteins) and a strong, non-monotonic dependence of signal gain on probe density support a collisional signaling mechanism in which binding alters the collision efficiency, and thus electron transfer efficiency, of the attached redox tag. Given the ubiquity with which protein binding will alter the collisional dynamics of an oligonucleotide, we believe this approach may prove of general utility in the detection of DNA and RNA binding proteins.
The reagentless, electrochemical E-DNA (electrochemical DNA) [reviewed in Ricci and Plaxco, 2008]1 and E-AB (electrochemical, aptamer-based) [reviewed in Xiao et al., 2008]2 sensing platforms are a promising approach for the detection of a wide range of molecular analytes.3–5 Composed of an electrode-bound, redox-modified probe oligonucleotide, E-DNA and E-AB sensors require only that target binding alters the rates with which the probe-attached redox-tag collides with, and thus transfers electrons to, the interrogating electrode.6,7 Because all of the sensing components in the E-DNA/E-AB platform are tightly linked to the electrode, these sensors are reagentless and readily reusable.8 Likewise, because their signaling is linked to a binding-specific change in the properties of the probe DNA, and not simply to adsorption to the sensor surface, E-DNA and E-AB sensors have proven remarkably robust against the non-specific adsorption of contaminants and perform well even when challenged in complex sample matrixes such as undiluted blood serum, soil, crude cellular extracts, and foodstuffs.9–13
Recent studies suggest that E-DNA and E-AB signaling requires only that a target binds to an oligonucleotide probe and, in doing so, changes the efficiency with which the attached redox tag strikes the electrode.6,7 This, in turn, accounts for the approach’s generalizability. For example, E-DNA architectures have been described to date in which hybridization with a target oligonucleotide leads to changes in the flexibility of DNA probes adopting stem-loop, single-stranded, double-stranded, or pseudoknot configurations.7,14–16 Likewise, more than a half-dozen E-AB sensors have been reported to date that employ DNA or RNA aptamers re-engineered to undergo folding or strand displacement, and concomitantly changes in flexibility, upon target binding.9,13,17,18 Those latter, aptamer-based sensors have enabled the detection of proteins,9,17,19,20 small molecules,10,13,18 and inorganic ions11,21 via this simple, general mechanism.
Another broad and important class of analytes that can be detected via their interactions with DNA are naturally occurring DNA binding proteins. Such proteins are abundant and essential in cells, interacting with DNA to organize its packing, regulate transcription, and perform replication and repair. A clinically important subset of these naturally occurring proteins are the transcription factors, which have proven diagnostic of developmental status and cancer.22,23 Thus motivated, Barton and co-workers have recently described an E-DNA-like platform for the detection of a protein that binds to a specific, double-stranded, redox-tagged DNA sequence, the TATA-box binding protein, by employing a double-stranded DNA probe containing the sequence naturally targeted by the protein.24 Here we expand on this theme by converting a number of naturally occurring, protein-targeting DNA sequences into a similar class of E-DNA-like sensors for the detection of specific DNA binding proteins.
MATERIALS AND METHODS
Probe DNA Sequences
We have employed the following probe DNA sequences:
TATA probe: 5′-HS-(CH2)6-CGGGCTATAT*(MB)AAGGGGCGTTTTCTTATATAG-3′
M.HhaI probe: 5′-HS-(CH2)6- AAGACGAGCGCATGTT*(MB)-TATGCGCTC-3′
Poly-T20 probe: 5′-HS-(CH2)6- T20-(CH2)7-NH-MB-3′
Poly-T40 probe: 5′-HS-(CH2)6- T40-(CH2)7-NH-MB-3′
Poly-T70 probe: 5′-HS-(CH2)6- T70-(CH2)7-NH-MB-3′
where -(CH2)7-NH-MB-3′ represents a methylene blue (MB) added to the terminal phosphate via a C-7 amino linker and T*(MB) represents a thymine nucleotide modified by the addition of MB to a 6-carbon, amino-terminated linker attached at the 5 position of the nucleobase. All probes were synthesized, labeled, and purified by BioSearch, Tech (Novato, CA) and used as received. The methylene blue was conjugated to either the 3′ end of the probe or the internal linker-modified thymine via succinimide ester coupling. The modified oligonucleotides were purified via C18 Reverse-Phase HPLC and PAGE and confirmed by mass spectrometry. Reagent grade chemicals, including 6-mercapto-1-hexanol (C6-OH), guanidine hydrochloride, sulfuric acid, potassium phosphate monobasic, dibasic, and sodium chloride (all from Sigma-Aldrich, St. Louis, MO), were used without further purification.
Sensor Fabrication
The sensors were fabricated as previously described,25 which we reiterate in brief here. E-DNA sensors were fabricated on rod gold disk electrodes (2.0 mm diameter, BAS, West Lafayette, IN). And while we note that these electrodes are not readily amenable to mass production, E-DNA fabrication is also supported on screen printed gold electrodes [ref 26 and Ricci et al., 2008, manuscript in preparation]. The disk electrodes were prepared by polishing with diamond and alumina (BAS), followed by sonication in water, and electrochemical cleaning (a series of oxidation and reduction cycles in 0.5 M H2SO4, 0.01 M KCl/0.1MH2SO4, and 0.05 M H2SO4). Effective electrode areas were determined from the charge associated with the gold oxide reduction peak obtained after the cleaning process.
The relevant probe DNA was immobilized onto these freshly cleaned electrodes by incubating for 1 h in a solution of 1 mM TCEP (Tris(2-carboxyethyl) phosphine hydrochloride) in 100 mM NaCl/10 mM potassium phosphate pH 7 buffer containing the appropriate concentrations of probe DNA. Different probe densities were obtained by controlling the concentration of probe DNA employed during the fabrication process ranging from 0.01 µM to 1 µM. Optimal response for TBP and M.HhaI is obtained by using a probe DNA concentration of 0.05 µM which leads to probe density of about 2 × 1011 molecules/cm2. For the detection of single-strand binding proteins a probe DNA concentration of 0.5 µM was used obtaining a probe density of about 5 × 1012 molecule/cm2. Following probe immobilization the electrode surface was rinsed with distilled, di-ionized water passivated with 1 mM 6-mercaptohexanol in 1 M NaCl/10 mM potassium phosphate buffer, pH 7, for 2 h and followed by further rinsing with deionized water. Sensors were stored in dark in buffer, under air, conditions that allows multi-month storage stability.27
Probe density (i.e., the number of electroactive probe DNA molecules per unit area of the electrode surface) was determined using a previously established relationship with ACV peak current (for details, see refs 6 and 7).
Electrochemical Measurements
All experiments were performed at room temperature using a CHI 730C Electrochemical Workstation (CH Instruments, Austin, TX). Square Wave Voltammetry (SWV) were recorded at 60 Hz frequency, 50 mV amplitude, and with an increment potential of 1 mV over a potential range of −0.1 to −0.45V in a standard cell with a platinum counter electrode and Ag/AgCl (3 M NaCl) reference electrode. The measurements of TBP, RPA, and SSBP were carried out in 0.1 M NaCl/10 mM potassium phosphate buffer/10 mM MgCl2, pH 7.55. M.HhaI, in contrast, was measured in a solution of 0.1 M Tris buffer/10 mM EDTA, 0.1 mM S-adenosylhomocysteine, pH 7. Of note, the S-adenosylhomocysteine was included because this cofactor (or S-adenosylmethionine) is required to form a M.HhaI-DNA complex. Consistent with this, we do not observe any detectable M.HhaI binding in the absence of this cofactor (data not shown). After the addition of the target protein the sensors were equilibrated for 5 min (for M.HhaI, RPA, and SSBP) or 10 min (TBP). In the case of RPA detection in Raji extract, the sensor based on a 70-base probe was first equilibrated in pure buffer solution and sequential injections of Raji extract were made to obtain the desired concentration. After each injection 5 min were allowed to record the signal. To recover the initial signal a high concentration (10 µM) of a competitor sequence (polyT-70) was injected in the solution. All reported values represent the mean and standard deviations of measurements conducted using three or more independently fabricated electrodes.
Protein Expression and Purification
TBP and M.HhaI were obtained by expression of recombinant, his-tagged proteins in Escherichia coli, as described previously.28,29 Briefly, T7 promoter-driven constructs were cloned into BL-21 derivative E. coli cells and induced with isopropyl β-D-1-thiogalactopyranoside. Following this the cells were lysed via French press and the recombinant protein purified via nickel affinity (Ni-NTA) and cation exchange (BioRex) chromatography. Protein activity was evaluated by electrophoretic mobility shift assay with cognate DNA targets and concentrations determined via Bradford assay. The RPA used in this study was a kind gift of Prof. Marc Wold (Department of Biochemistry Carver College of Medicine, University of Iowa) and was purified as described previously.30 The Raji nuclear extracts were obtained from Millipore (Bedford, MA). The SSBP employed here was purchased from Epicenter Biotechnologies (Madison, WI, USA) and used without further purification.
RESULTS
We have previously argued that E-DNA and E-AB signal generation likely requires only that target binding changes the efficiency with which the probe’s redox tag strikes the electrode. 6,7,31 This, in turn, suggests that the approach would also support the detection of DNA binding events that lead to the formation of bulky and/or rigid complexes. To test this suggestion we have fabricated and characterized E-DNA-like sensors for the detection of two double-strand binding proteins, the eukaryotic TATA-box Binding Protein (TBP, a core component of the eukaryotic transcriptional machinery) and the prokaryotic M.HhaI methyltransferase (M.HhaI, involved in the restriction-modification system of bacteria), and two single-strand binding proteins involved in the replication machinery, the prokaryotic Single-Strand Binding Protein (SSBP) and the eukaryotic Replication Protein A (RPA).
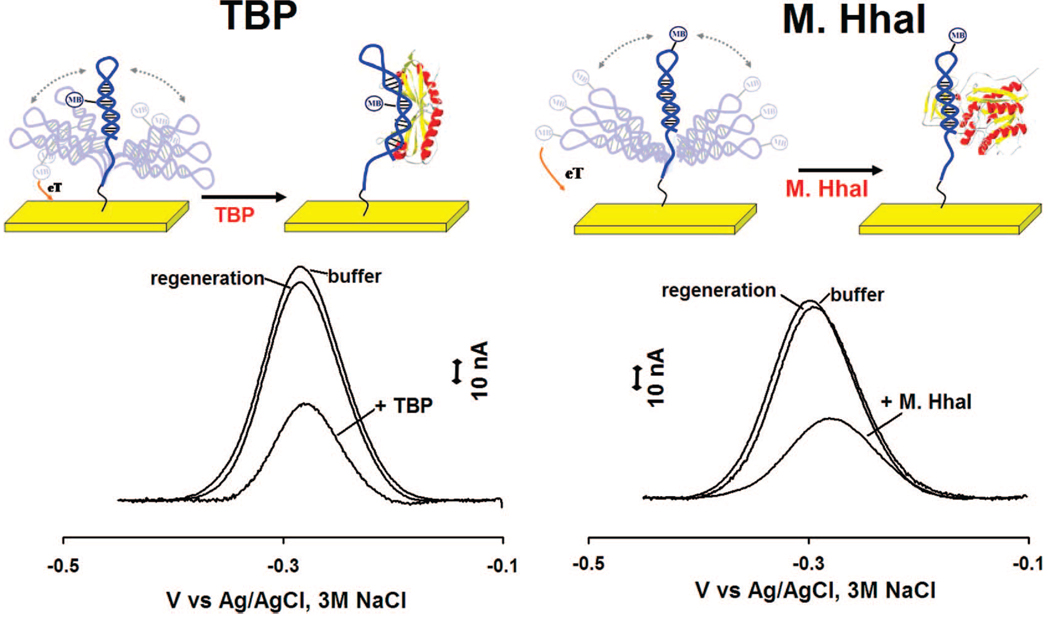
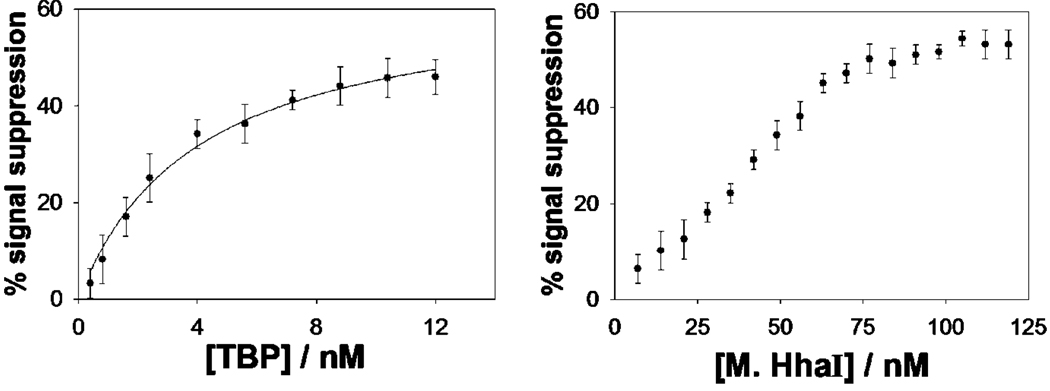
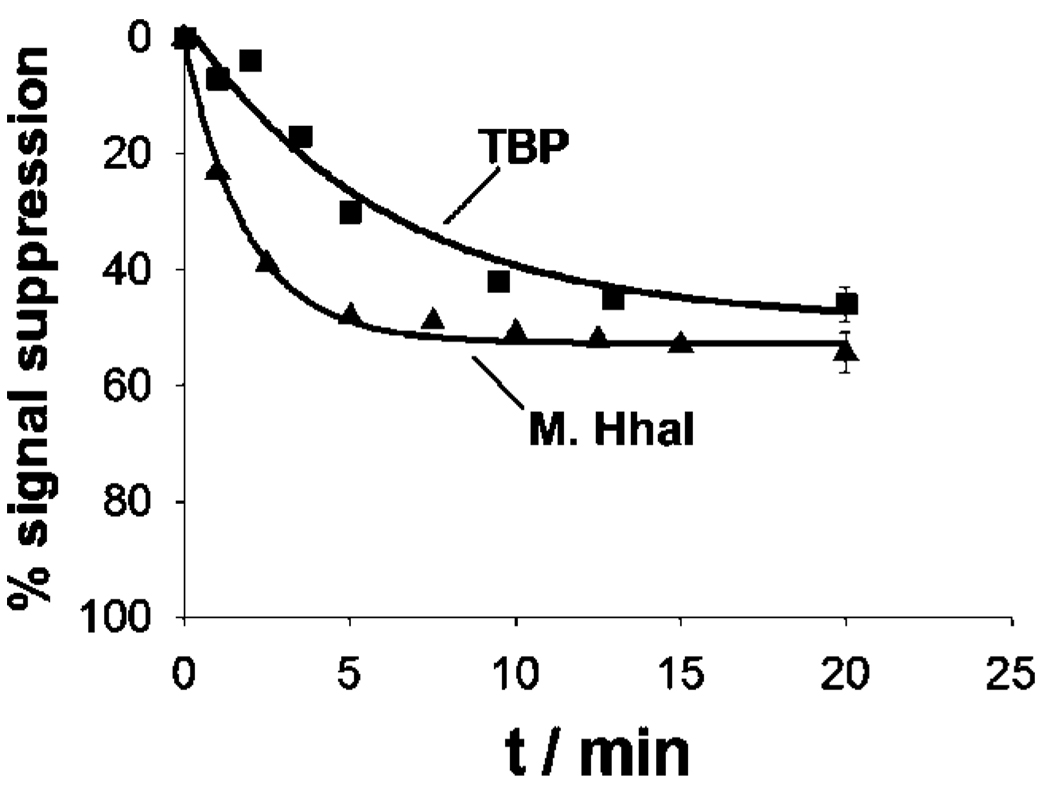
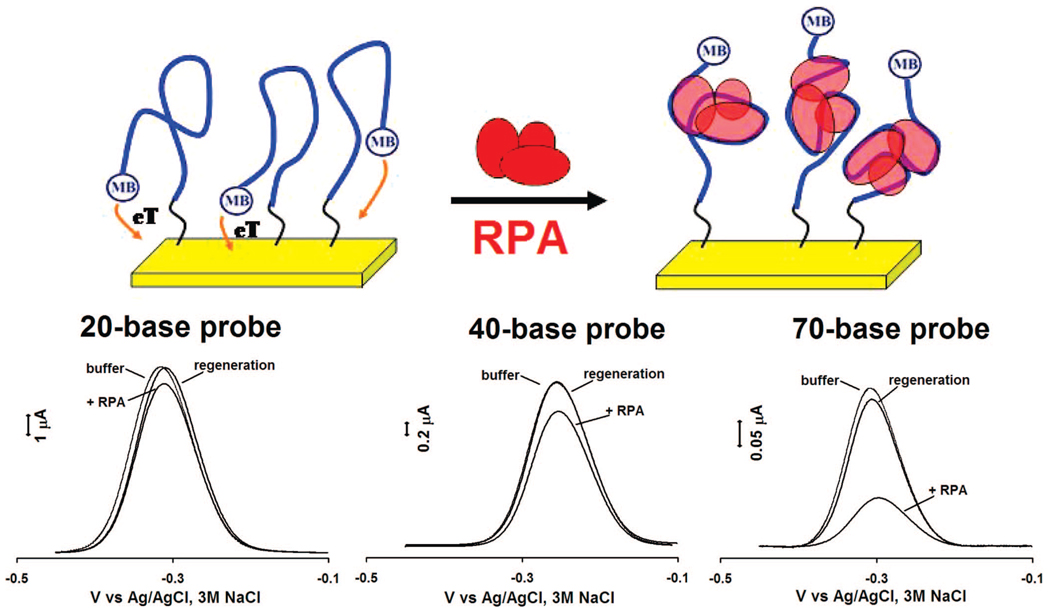
We have fabricated sensors against the double-strand binding proteins using short, stem-loop probe DNAs in which the relevant recognition sequences are contained within the double-stranded stem. These probes were modified with a 3′ thiol group, supporting strong chemisorption to an interrogating electrode, and a methylene blue redox tag pendant on a thymine base along the double-stranded stem (for TBP, Figure 1, left) or within the single-stranded loop (for M.HhaI, Figure 1, right). In the absence of target both probes produce a large faradaic peak at the potential expected for the methylene blue redox tag (Figure 1, bottom). In the presence of saturating TBP and M.HhaI these currents are reduced by 45 and 55%, respectively. Upon titration with its target the dose-response curve of the TBP sensor is hyperbolic (Figure 2, left), as expected for single-site, saturable binding, and exhibits a dissociation constant of 4.0 ± 0.6 nM, which is comparable to previously reported values.32 The dose-response curve of the M.HhaI sensor exhibits a bilinear shape (Figure 2, right) presumably arising because of the extremely low (pM) dissociation constant of this protein.33 The two sensors support the ready detection of their target proteins at concentrations as low as 2 and 25 nM for TBP and M.HhaI, respectively (Figure 2). The response times of both sensors are likewise rapid, with the TBP and M.HhaI sensors exhibiting equilibration time constants of 5.9 ± 0.6 and 1.9 ± 0.2 min, respectively (Figure 3). Finally, because all of the sensing components are strongly adsorbed to the electrode surface, this sensing architecture is readily regenerable; a short wash (30 s) in 8 M guanidine chloride is sufficient to regenerate 98% of the original signaling current of both sensors (Figure 1, bottom), allowing multiple cycles of detection and regeneration (data not shown).
Figure 1.
Schematics of E-DNA-like sensors for the detection of DNA binding proteins. (Top) The sensor is composed of a DNA hairpin covalently attached to a classic gold rod electrode using thiol-gold self-assembled monolayer chemistry and containing an internal methylene blue redox tag. (Bottom) In the absence of target relatively efficient collision between the label and the electrode produces a large faradic current. Upon target binding this faradic current is significantly reduced, presumably because the bulky structure of the protein reduces the collision rate. Because all of the sensing components are strongly chemisorbed to the interrogating electrode, the sensor is readily regenerated via a 30 s wash in 8 M guanidine chloride.
Figure 2.
Both the TBP (left) and M.HhaI (right) sensors detect their target proteins at low nanomolar concentrations. The TBP dose–response curve is hyperbolic with a dissociation constant comparable to previous literature values.30 The dose–response curve of the M.HhaI sensor, in contrast, exhibits a bilinear shape, presumably because of the low (pM) dissociation constant of this protein.31 The data points and error bars represent the average and standard deviations of measurements conducted using three independently fabricated electrodes.
Figure 3.
Sensor equilibration is rapid. Shown, in this example, are the response curves obtained with TBP and M.HhaI at 10 and 80 nM respectively.
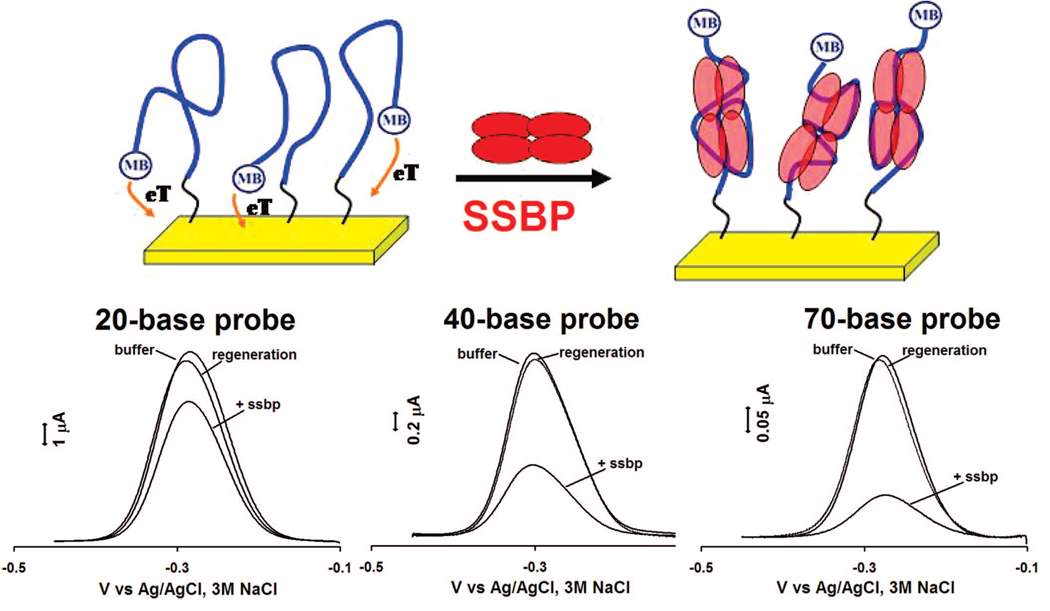
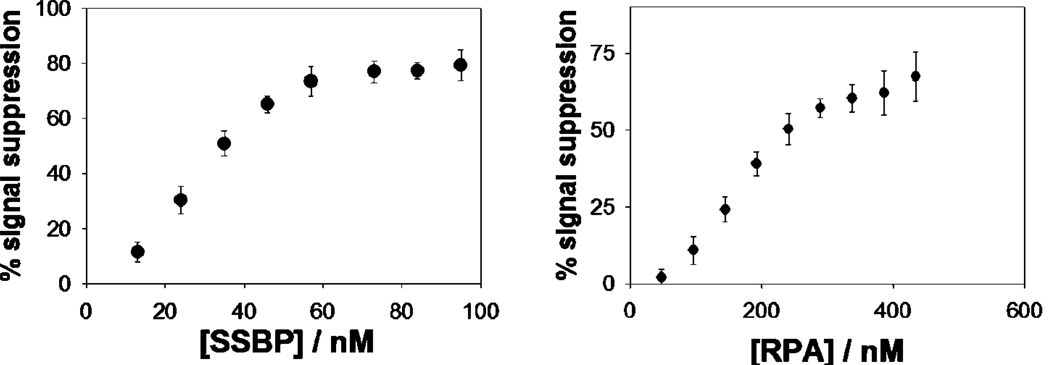
Single-stranded DNA probes also support this analytical approach, enabling the sensitive, convenient detection of proteins that bind to such targets. Using single-stranded, poly-thymine probes (neither of our target proteins exhibits any significant sequence specificity), we have fabricated sensors for the detection of the single-strand binding proteins SSBP (Figure 4, 6) and RPA (Figure 5, 6). Of note, because the single-stranded DNA is thought to wrap entirely around SSBP, the gain of these sensors is dependent on the length of the single-stranded probe employed: when targeting this protein the observed signal suppression (at saturating target concentration) increases from 20 to 70% as the probe is lengthened from 20 to 70 bases (Figure 4, bottom). The same change in probe length is associated with a 5% to 70% increase in gain in the presence of RPA (Figure 5, bottom), an effect that could arise when multiple RPAs bind to the longer probe.34
Figure 4.
Sensing mechanism also holds for single-stranded probes, thus enabling the detection of E. coli single-strand binding protein (SSBP). (Top) The Faradaic current arising from such a single-stranded probe is significantly reduced upon binding with this target. (Bottom) Of note, because the single-stranded probe wraps around the protein target, different length probes (20, 40, and 70 bases) give rise to different responses upon protein binding. Shown are responses to 80 nM of SSBP.
Figure 6.
Both E. coli single-strand binding protein (SSBP, left) and eukaryotic replication protein A (RPA, right) are readily detected at nanomolar concentrations using the high-gain, 70-base probe DNA. The data points and error bars represent the average and standard deviation of measurements taken with three independent electrodes.
Figure 5.
E-DNA platform for the detection of single strand binding protein is general: the measurement of human Replication Protein A (RPA) using a linear probe DNA as recognition element leads to results similar of those observed with E. coli SSBP and shares a similar dependence on probe length. Shown are responses to 300 nM of RPA.
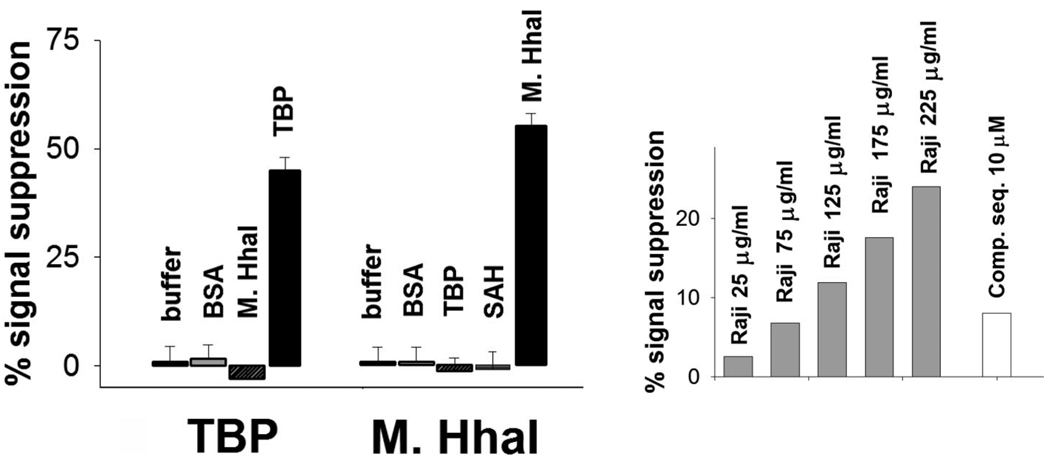
All three sensors are as specific as the DNA probes from which they are fabricated. For example, we do not detect any significant cross-reactivity between the two sensors directed against double-strand-binding proteins (Figure 7, left). (Neither of the two single-strand binding proteins investigated here exhibits any significant sequence specificity34,35 and thus, while the signal gain produced by the two proteins is not identical, the ability of our sensor to discriminate between these two targets is limited.) Likewise, because their signaling is linked to a binding-specific change in the probe DNA (and not simply to adsorption of target to the sensor surface), our sensors are effective in rejecting false positives arising because of the non-specific adsorption of interferents and can be employed directly in complex samples. For example, our single-strand sensor supports the specific detection of exogenous levels of RPA directly in crude Raji cell nuclear extracts (Figure 7, right).
Figure 7.
DNA binding protein sensors are as specific as the DNA probes from which they are fabricated. (Left) For example, no significant cross-reactivity is observed between the two sensors directed against double-strand binding proteins in the presence of 1.5 µM BSA and saturating concentrations of the non-targeted double-strand binding protein (80 nM M.HhaI or 10 nM TBP). (Right) Likewise our sensors are also effective in rejecting false positives arising because of interferents and perform well when challenged with realistically complex sample matrixes. For example, we can detect exogenous level of RPA in crude Raji nuclear extracts and partially restore initial signal by adding high concentration of a competitor sequence (polyT-70).
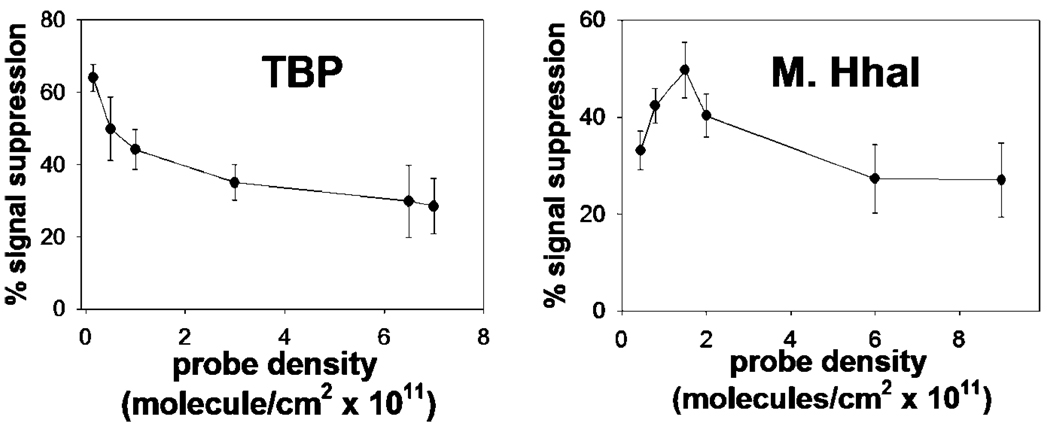
Previous studies of E-DNA and E-AB sensors indicate that their signaling is sensitive to the packing density of the probe DNAs on the electrode surface.6,7 Thus, in an effort to optimize the gain of our sensors, we have studied the effect of probe density (number of probe strands per unit area) on the signaling of our TBP and M.HhaI sensors. We find that the performance of both sensors is sensitive to this parameter. Enhanced signal suppression of TBP sensor is seen, for example, at the lowest measurable packing densities we can achieve (4.5 × 1010 molecules/cm2, Figure 8, left). The M.HhaI sensor, in contrast, achieves optimal performance at intermediate probe densities (2 × 1011 molecules/ cm2, Figure 8, right).
Figure 8.
Consistent with the collisional mechanism that we have proposed, the performance of the M.HhaI and TBP sensors are sensitive to probe packing density on sensor surface. For example, while maximum signal suppression is obtained for the TBP sensor (left) at very low probe densities, the optimal density for the M.HhaI sensor (right) is achieved at intermediate densities.
DISCUSSION
Here we have demonstrated a reagentless, electrochemical method for the detection of specific DNA binding proteins. Our approach is rapid (minutes), convenient, and quantitative. It is also selective enough to deploy directly in realistically complex sample matrixes, such as crude nuclear extracts. Finally, our approach appears to be quite general; we have achieved a three-successes-out-of-three-attempts success rate in fabricating sensors targeting both sequence-specific, double-strand biding proteins and non-specific, single-strand binding proteins.
As noted above, Barton and co-workers have previously described a similar sensor for the detection of TBP and attributed its signaling to binding-induced changes in electron transfer through the probe DNA.24 It appears unlikely, however, that this mechanism is the dominant contributor to signaling in our sensors. First, Barton has argued that through-DNA electron transfer requires close electronic coupling of the redox tag and the base pair stack,36 which is unlikely to occur in our system because its redox tag is attached via a simple alkane chain to the backbone of the probe DNA. Second, Barton has argued that enhanced through-DNA electron transfer occurs only for double-stranded DNA,37,38 and thus our single-stranded probes are unlikely to support a through-DNA electron transfer mechanism and to show a signal change upon target binding. Finally, while steric blocking of the target at higher probe densities could lead to poorer affinity, it appears unlikely that a through-DNA electron transfer mechanism would produce the complex relationships between gain and probe density (including optimal gain at intermediate densities) we have observed (Figure 8). Instead we believe that signaling in our sensors is dominated by binding-induced changes in the efficiency with which the redox tag collides with, and thus transfers electrons to, the interrogating electrode. Single-stranded probes, for example, readily support this signaling mechanism.7,39 Likewise, the collisional mechanism readily accounts for a strong relationship between signal gain and probe-density: whereas high probe densities may reduce the collision efficiency of the unbound state (reducing the signal change observed upon binding), low probe densities enhance collisions from both the bound and unbound states.6,7 Optimal gain represents a compromise between these two factors, leading to either monotonically improving signaling with decreasing probe density (as seen for TBP, Figure 8, left) or optimal gain at intermediate probe densities (as seen for M.HhaI, Figure 8, right) depending on the sizes and geometries of the probe and the probe-target complex.
The potential applications of a convenient method for monitoring protein-DNA interactions are widespread. For example, many essential cellular processes rely on sequence- and protein-specific protein-DNA interactions, including the control of transcription and, consequently, the control of growth, development, and many environmental responses.22 Probes for the detection of specific protein-DNA interactions thus provide sensitive assays for many clinically relevant cellular processes, such as the expression of transcription factors in response to a disease state or in the presence of a drug.23 Specific examples include transcription factors that are up-regulated in clinically relevant cell states and can serve as markers for these state, such as for the identification of stem cells via the detection of Oct-440 or the up-regulation of the Stat proteins in many cancers.41 Current methods for the detection of such factors, however, such as chromatin immunoprecipitation, are complex, multistep processes often requiring extensive sample processing.42 As such these traditional approaches contrast sharply with the reagentless, electrochemical method documented here which, because of its convenience, generality, and sensitivity, may prove a valuable approach to the detection of specific oligonucleotide binding proteins.
ACKNOWLEDGMENT
The authors wish to thank Prof. Marc Wold of the Department of Biochemistry Carver College of Medicine, University of Iowa for helpful suggestions about RPA detection. This work was supported by the Institute for Collaborative Biotechnologies through Grant No. R01EB002046-07 (KWP) and DAAD19-03-D-0004, U.S. Army Research Office (NOR).
References
- 1.Ricci F, Plaxco KW. Microchim. Acta. 2008;163:149–155. [Google Scholar]
- 2.Xiao Y, Plaxco KW. In: Functional nucleic acids for sensing and other analytical applications. Lu Y, Li Y, editors. Norwell, MA: Kluwer/Springer; 2008. in press. [Google Scholar]
- 3.Palecek E. Trends Biotechnol. 2004;22:55–58. doi: 10.1016/j.tibtech.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Thorp HH. Trends Biotechnol. 2003;21:522–524. doi: 10.1016/j.tibtech.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Eisenstein M. Nat. Methods. 2006;3:244. doi: 10.1038/nmeth0406-244b. [DOI] [PubMed] [Google Scholar]
- 6.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci F, Lai RY, Plaxco KW. Chem. Commun. 2007;36:3768–3770. doi: 10.1039/b708882e. [DOI] [PubMed] [Google Scholar]
- 8.Lubin AA, Lai RY, Heeger AJ, Plaxco KW. Anal. Chem. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew. Chem., Int. Ed. 2005;44:2–5. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 10.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Rowe AA, Plaxco KW. J. Am. Chem. Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 12.Lubin AA, Fan C, Schafer M, Clelland CT, Bancroft C, Heeger AJ, Plaxco KW. For. Sci. Commun. 2008;10(1):1. [Google Scholar]
- 13.Zuo X, Song S, Zhang J, Pan D, Wang L, Fan C. J. Am. Chem. Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 14.Fan C, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Lubin AA, Baker BR, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16677–16680. doi: 10.1073/pnas.0607693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, Qu X, Plaxco KW, Heeger AJ. J. Am. Chem. Soc. 2007;129:11896–11897. doi: 10.1021/ja074218y. [DOI] [PubMed] [Google Scholar]
- 17.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J. Am. Chem. Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 18.Ferapontova EE, Olsen EM, Gothelf KV. J. Am. Chem. Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 19.Acero Sanchez JL, Baldrich E, Radi A, Dondapati S, Sanchez PL, Katakis I, O’Sullivan CK. Electroanalysis. 2006;18:1957–1962. [Google Scholar]
- 20.Lai RY, Plaxco KW, Heeger A. J. Anal. Chem. 2007;79:229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 21.Radi AE, Acero Sanchez JL, Baldrich E, O’Sullivan CK. J. Am. Chem. Soc. 2006;128:117–124. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 22.Kadonaga JT. Cell. 2004;116(2):247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 23.Bulyk ML. Curr. Opin. Biotech. 2006;17(4):422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorodetsky AA, Ebrahim A, Barton JK. J. Am. Chem. Soc. 2008;130:2924–2925. doi: 10.1021/ja7106756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Lai RY, Plaxco KW. Nat. Prot. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins DM, Chami B, Kreuzer M, Presting G, Alvarez AM, Liaw BY. Anal. Chem. 2006;78(7):2314–2318. doi: 10.1021/ac051619s. [DOI] [PubMed] [Google Scholar]
- 27.Phares N, White RJ, Plaxco KW. Anal. Chem. 2008 doi: 10.1021/ac8021983. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart JJ, Stargell LA. J. Biol. Chem. 2001;276:30078–30084. doi: 10.1074/jbc.M105276200. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H, Shatz W, Purdy MM, Fera N, Dahlquist FW, Reich NO. Biochemistry. 2007;46(24):7261–7268. doi: 10.1021/bi602662e. [DOI] [PubMed] [Google Scholar]
- 30.Henricksen LA, Umbricht CB, Wold MS. J. Biol. Chem. 1994;269(15):11121–11132. [PubMed] [Google Scholar]
- 31.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Howard GM, Weil PA, Beechem JM. Biochemistry. 1995;34:8005–8017. doi: 10.1021/bi00025a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindstrom WM, Flynn J, Reich NO. J. Biol. Chem. 2000;275:4912–4919. doi: 10.1074/jbc.275.7.4912. [DOI] [PubMed] [Google Scholar]
- 34.Kim C, Paulus BF, Wold MS. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 35.Roy R, Kozlov A, Lohman TM, Ha T. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorodetsky AA, Green O, Yavin E, Barton JK. Bioconjugate Chem. 2007;18:1434–1441. doi: 10.1021/bc0700483. [DOI] [PubMed] [Google Scholar]
- 37.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Nucleic Acids Res. 1999;27:4830–4837. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley S, Holmlin RE, Stemp EDA, Barton JK. J. Am. Chem. Soc. 1997;119:9861–9870. [Google Scholar]
- 39.Lubin AA, Vander Stoep Hunt B, Plaxco KW. Anal. Chem. 2008 in press. [Google Scholar]
- 40.Pesce M, Scholer HR. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 41.Bowman T, Garcia R, Turkson J, Jove R. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 42.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]