Abstract
Arf GAP2 is one of four Arf GAPs that function in the Golgi apparatus. We characterized the kinetics of Arf GAP2 and its regulation. Purified Arf GAP2 had little activity compared to purified Arf GAP1. Of the potential regulators we examined, coatomer had the greatest effect, stimulating activity one to two orders of magnitude. The effect was biphasic, with half-maximal activation observed at 50 nM coatomer and activation peaking at ≈150 nM coatomer. Activation by coatomer was greater for Arf GAP2 than has been reported for Arf GAP1. The effects of phosphoinositides and changes in vesicle curvature on GAP activity were small compared to coatomer; however, both increased coatomer-dependent activity. Peptides from p24 cargo proteins increased Arf GAP2 activity by an additional 2- to 4-fold. The effect of cargo peptide was dependent on coatomer. Overexpressing the cargo protein p25 decreased cellular Arf1•GTP levels. The differential sensitivity of Arf GAP1 and Arf GAP2 to coatomer could coordinate their activities. Based on the common regulatory features of Arf GAP1 and 2, we propose a mechanism for cargo selection in which GTP hydrolysis triggered by cargo binding to the coat protein is coupled to coat polymerization.
Keywords: ADP-ribosylation factor, GTPase-activating protein, COPI, Golgi, p24 Cargo protein
1. Introduction
The Golgi apparatus is central to post translational processing of a diverse group of transmembrane and secreted proteins [1–3]. The function of the Golgi depends on membrane traffic to selectively transport materials between the Golgi apparatus and target organelles, including the retrograde transport from the Golgi apparatus to the endoplasmic reticulum. The machinery that mediates membrane traffic from the Golgi has been identified and includes vesicle coat proteins, phosphoinositides, Arf family GTP-binding proteins and the guanine nucleotide exchange factors and GTPase-activating proteins (GAPs) that regulate Arf proteins [2,4–8].
ADP-ribosylation factor 1 (Arf1) is a critical regulator of the Golgi apparatus and membrane traffic from the Golgi [9,10]. Arf1 is a myristoylated GTP-binding protein. When bound to GTP, Arf1 tightly associates with membranes. In the prevailing models of membrane traffic, vesicle coat proteins bind to membranes and drive the formation of transport intermediates [2,4,11,12]. Coat proteins dissociate from the intermediates, which enables them to interact with a target membrane. The cycle of coat protein binding and dissociation is linked to the cycle of Arf1 binding to and hydrolyzing GTP. Arf1•GTP, which binds to membranes and to coat proteins simultaneously, recruits coat proteins to membranes. Hydrolysis of GTP yields Arf1•GDP, which does not tightly associate with membranes and does not bind coat proteins. Coat proteins consequently dissociate from the membranes. Arf1, which does not have detectable intrinsic GTPase activity, depends on GTPase-activating proteins (GAPs) for the hydrolysis of GTP [8,13–15].
Arf GAPs induce the hydrolysis of GTP bound to Arf [8,10,14,15]. The reaction occurs on a membrane surface to which Arf1•GTP is tightly bound. Humans have 31 genes that encode proteins with Arf GAP domains [10,14,16]. Four Arf GAPs associate with the Golgi apparatus: Arf GAP1, Arf GAP2, Arf GAP3 and ARAP1. Three of these, Arf GAP1, Arf GAP2 and Arf GAP3, function with the coat protein coatomer [17–22]. Arf GAP1 differs from Arf GAP2 and Arf GAP3 in two ways. First, Arf GAP1 contains Arf GAP lipid packing sensor (ALPS) motifs, which sense the curvature of the membrane [23,24]. Second, Arf GAP2 and Arf GAP3 bind coatomer more tightly than does Arf GAP1 [18].
The regulation of Arf GAP1 has been extensively studied [21,25–27]. Two mechanisms that ensure GTP on Arf is hydrolyzed by Arf GAP1 after the polymerization of vesicle coat and formation of a transport intermediate have been proposed. In one mechanism, Arf GAP1 activity is dependent on the vesicle coat protein coatomer and is inhibited by cargo. When cargo is present, GTP hydrolysis is slowed allowing formation of a transport intermediate [17,28]. In another proposed mechanism, Arf GAP1 senses defects in membrane surfaces [23,24,29]. ALPS motifs are amphipathic helices that insert into the hydrophobic center of a lipid bilayer. The hydrophobic center becomes available when the packing density of the lipid head groups is decreased, which occurs when the curvature increases. In the model, Arf GAP1 is recruited to membranes when the curvature increases consequent to forming a transport intermediate. Recruitment to the surface containing the substrate Arf1•GTP is the primary means of regulating the GAP. This proposed regulatory mechanism does not link cargo sorting to GAP activity. Phosphatidylinositol 4-phosphate (PI4P) is a lipid important to regulation of the Golgi [3,5,30]. Although some Arf GAPs, such as ASAP1 and ARAP1, are regulated by other phosphoinositides, Arf GAP1 has not been found to be affected by this class of lipid [8,31].
Arf GAP2 has recently been reported to have coatomer dependent GAP activity [32,33]. Here, we examined the effects of cargo, PI4P and curvature on coatomer-dependent Arf GAP2 activity. As previously described, GAP activity of Arf GAP2 was highly dependent on coatomer [32,33], which increased activity by over 300-fold. The activation was biphasic as described for Arf GAP1 and predicted based on coatomer functioning as an allosteric modifier that can also sequester the substrate Arf1•GTP [22]. The magnitude of activation was greater than for Arf GAP1 and was increased by both PI4P and increasing vesicle curvature. Different than reported for Arf GAP1, cargo also stimulated activity. The effect of cargo was dependent on coatomer. We conclude that cargo modulates coatomer-dependent Arf GAP activity and we discuss a model for the formation of transport intermediates enriched in cargo.
2 Materials and Methods
2.1 Plasmids
[1–521] Arf GAP2 His6 was generated by PCR cloning of ArfGAP2 from Ultimate™ ORF Clone IOH23017 (Invitrogen) into NdeI and BamHIsite of pET21b. Plasmids for cargo protein pCB6-myc p23, pCB6-myc p24, pCB6-myc p25 are generous gifts from Dr. Jean Gruenberg.
2.2 Protein purification
Human myrArf1 protein was expressed in and purified from bacteria as described [34]. His10 [325–724]ASAP1 and [1–415]Arf GAP1-His was expressed and purified asdescribed [22]. Coatomer was purified from rat liver according to [35]. [1–521] Arf GAP2 His6 was expressed in E. coli and purified from inclusion bodies. One liter of the bacterial pellet was suspended in 50 ml of 20 mM TrisHCl pH 8.0 with one protease inhibitor tablet (Complete™, RocheApplied Science), and then lysed using a French press operated at 12,000 psi. The lysate was spun at 10,000 × g for 10 min at 4° C. The pellet containing inclusion bodies was washed twice with 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM DTT. The inclusion bodies were collected by centrifugation at 10,000 × g for 10 min. The protein was solubilized in 24 ml of 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole and 6 M guanidine-HCl at room temperature. After centrifuging the lysate at 10,000 × g for 10 min at 4° C, the supernatant was applied to a 1ml His column (GE Healthcare), eluted using an imidazole gradient from 20 to 600 mM in 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM DTT and 6 M guanidine-HCl. The fractions containing Arf GAP2 protein were dialyzed against 20 mM Tris-HCl, pH 8.0, 500 mM NaCl and 2 mM DTT at room temperature for 3 hrs. The precipitated protein was removed by centrifugation at 100,000 × g at 4° C for 15 min. The supernatant was applied to a 1 ml mono Q column (GE Healthcare) and proteins were eluted with a NaCl gradient from 0–1000 mM in 25 mM Tris-HCl, pH 8.0 and 2 mM DTT.
2.3 Large Unilamellar Vesicles (LUVs)
Lipids were purchased from Avanti Polar Lipids. LUVs for most experiments were formed by serial extrusion through filters with pore sizes of 1.0, 0.4, 0.1, 0.05, and 0.03 μm as described [22,34,36] and consisted of 40% phosphatidylcholine (PC), 25% phosphatidylethanolamine(PE), 15% phosphatidylserine (PS), 7.5% phosphatidylinositol(PI),10% cholesterol with 2.5% phosphatidylinositol 4-phosphate (PI4P) or 2.5% phosphatidylinositol 4,5-bisphosphate (PIP2). In some experiments, 10% PI was used without PI4P or PIP2.
2.4 GAP assays and kinetic analyses
To determine the C50 (the concentration of [1–521] Arf GAP2 His6 necessary for 50% of the maximal observed hydrolysis of GTP bound to myrArf1 in a fixed time), Arf GAP was titrated into a reaction mixture containing a total phospholipid concentration of 500 μM. For most experiments, 0.6 μM Arf1•[α-32P]GTP were used as the substrate and the reaction was stopped after 3 min. Coatomer titrations and cargo peptide titrations were done with a fixed concentration of Arf GAP, [α32P]GTP•Arf1 as a substrate and a fixed time point assay. For time courses, the amount Arf GAP2 and other additions are indicated in the figure legends. For experiments using [α32P]GTP, the extent of hydrolysis was analyzed as described [13,37,38]. Saturation kinetics were performed as described following the reaction by changes in fluorescence or by using [α32P]GTP•Arf1 as a substrate [22,34,37,39]. In the assay using fluorescence, corrections were made for a time dependent change in fluorescence of Arf GAP2 and Arf GAP2 with coatomer.
2.5 Determination of Arf1•GTP levels in vivo
Cos7 cells were cultured in DMEM with 10% FBS and were transfected with plasmids directing expression of Arf1-HA and myc tagged p25 as indicated. 24 hours after transfection, cellular levels of Arf1•GTP were determined as described [40]. Briefly, purified recombinant GST (30 μg) or GST-GGA3 (30 μg) were immobilized on glutathione-Sepharose 4B beads for 1 hour at room temperature. After 3 washes with ice-cold PBS, the immobilized GST fusion proteins were incubated overnight at 4°C with equal amounts of lysates from Cos7 cells. The beads were collected by centrifugation and washed 3 times with ice-cold PBS. Proteins were eluted from the beads by boiling in SDS-PAGE sample buffer and detected by standard immunoblotting using rabbit anti-HA (Covance, PRB-101P) and rabbit anti-Myc (Covance PRB-150P) antisera. Signals from the immunoblots were quantified by densitometric scanning and the signals normalized to the Arf1 signal from a fixed quantity of the cell lysates.
2.6 Protein binding to LUVs
A coatomer enriched soluble fraction from rat liver was prepared. Frozen rat liver was lysed in 25 mM Tris-HCl, pH 8.0, 500 mM KCl, 250 mM sucrose, 2 mM EGTA, 1mM DTT with protease inhibitors using a Polytron™ homogenizer for 2 X 30 seconds. The lysate was clarified by centrifugation at 15,000 × g at 4° C for 30 min. The supernatant was collected and clarified by centrifugation at 100, 000 × g at 4° C for 1 hr. The supernatant from the 100,000 × g centrifugation (30 mg/ml protein) was fractionated on a Superdex 200 column equilibrated and run in 20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM DTT. Fractions containing coatomer were pooled and used for the experiments. Sucrose loaded LUVs were prepared as described in [36]. The LUVs (total lipid concentration was 1 mM) were incubated with 26 μg protein of the coatomer enriched soluble fraction from rat liver and either no Arf or 0.5 μM myrArf1•GTPγS in a total volume of 100 μl. The samples were incubated for 5 min at 30°C and then the vesicles were separated from the bulk solution by centrifugation at 100,000 × g for 15 min at 4°C. β-COP associated with the vesicles was determined by immunoblotting. Purified Arf GAP2, at a concentration of 1 μM, was incubated with the LUVs under the same conditions. Protein associated with the LUVs was fractionated by SDS-PAGE and visualized with Coomassie Blue dye. Arf association with the vesicles was determined both by immunoblotting and by staining with Coomassie Blue dye.
2.7 Peptides
2.7.1 Synthesis of palmitic acid-WRMRHLKSAAEAKKLV and WRMRHLKSFFEAKKLV
N-α-9-fluorenylmethyloxycarbonyl (Fmoc) protected amino acids and preloaded Fmoc-Valine Wang resin were purchased from Nobabiochem (San Diego CA). Chemicals including palmitic acid and solvents were obtained from Sigma-Aldrich (St. Louis, MO) and American bioanalytical (Natick, MA). Peptide syntheses were carried out by solid-phase synthesis with an Applied Biosystem 431A peptide synthesizer. Peptides were assembled on Wang resin using Fmoc synthetic strategy. Coupling was accomplished using the HBTU-HOBt method. Coupling of palmitic acid at the N-terminus was performed with diisopropylcarbodiimide, N-hydroxysuccinimide, and N-methylmorphorine. Cleavage of the peptide from the resin was achieved with trifluoroacetic acid/water/ethanthiol/triisopropylsilane (94/2.5/2.5/1, v/v%) for 3 hours at room temperature. After removing the resin by filtration, the filtrate was concentrated by nitrogen gas flashing, and crude peptides were precipitated by diethylether. Crude peptides were purified by using preparative RP-HPLC. The homogeneities and structures of synthetic peptides were verified by analytical RP-HPLC and MALDI-TOF MS.
2.7.2 Other peptides
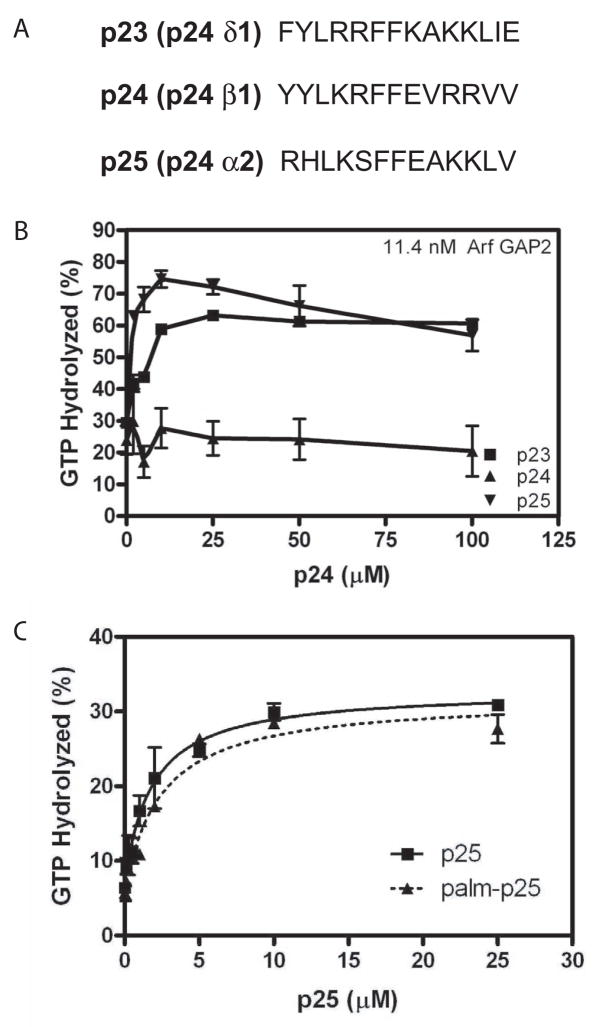
Sequences of the peptides from p23, p24 and p25 cytoplasmic tails are shown in Figure 6A. They were synthesized by AnaSpec, Inc (San Jose, CA).
Figure 6. Effect of peptides from p24 cargo proteins on Arf GAP2 activity. A. Sequence of peptides used. B. Peptide titration.
Peptides fromp23, p24 and p25 were titrated into a reaction containing LUVs containing PI4P and extruded through 0.03 μm pores, 0.6 μM [α32P]GTP•myrArf1, 11.4 nM [1–521] Arf GAP2 His6 and 124 nM coatomer. Reactions were terminated after 3 min at 30 °C. C. Effect of palmitoylated peptide. P25 peptide with palmitic acid- WRM or WRM modifying the N-terminus was titrated into a GAP reaction as described in B. The data were fit to a one site binding model with an offset.
3 Results
3.1 Purified Arf GAP2 is less active than purified Arf GAP1
We started our analysis by comparing enzymatic activity of Arf GAP1 and Arf GAP2. All of our assays used large unilamellar vesicles (LUVs) as a reaction surface. LUVs were extruded through either 1.0 μm or 0.03 μm pores, resulting in different size distributions [41]. Arf GAP2 and Arf GAP1 were titrated into reactions containing myrArf1 to determine the maximum hydrolysis of GTP bound to myrArf1 and the amount of Arf GAP to achieve 50% of the maximum observed hydrolysis (referred to as the C50) in a fixed period of time. Arf GAP2 had little activity compared to purified Arf GAP1 (Figure 1A and Table 1). Arf GAP1 was greater than 100-fold more active than Arf GAP2 (Figure 1A)1. The time dependence for GTP-hydrolysis catalyzed by 3.4 μM Arf GAP2 was determined. The data were fit to a first order rate equation. The rate constant was three-fold greater on the smaller vesicles than on the larger vesicles (Figure 1B) but was less than 1/100th the kcat that has been determined for Arf GAP1 [22]. The effect of vesicle size on activity was not the result of differences in recruitment to vesicles; we did not detect an effect of vesicle size on either Arf GAP2 or Arf1•GTP binding to vesicles (not shown).
Figure 1. Arf GAP2 activity.
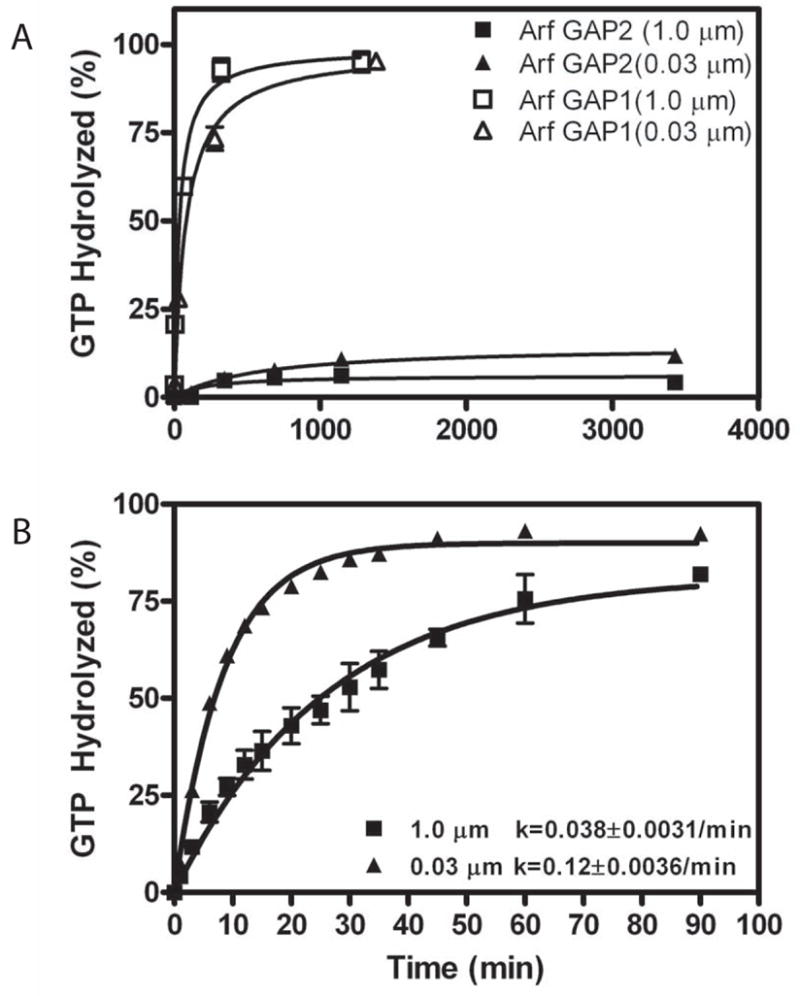
GAP activity using myrArf1•GTP as a substrate was determined in the presence of LUVs formed by extrusion through membranes with pores of the indicated diameters. Total phospholipid concentration was 500 μM. A. Comparison of recombinant purified Arf GAP1 and Arf GAP2. [1–521] Arf GAP2 His6 and [1–415]Arf GAP1-His6 were titrated into a reaction containing [α32P]GTP•myrArf1. The fraction of GTP converted to GDP in 3 min was determined. B. Time dependence of GTP hydrolysis. 3.4 μM of [1–521] Arf GAP2-His6 was incubated with 0.6 μM myrArf1•GTP for the indicated times and extent of GTP hydrolysis was determined as described in “Materials and Methods.”
Table 1.
Maximum GTP hydrolysis and C50 for Arf GAP1 and Arf GAP2
| Maximum GTP Hydrolysis (%) and C50 (nM) | ||||||
|---|---|---|---|---|---|---|
| Vesicles | Lipid content | 0% PIPx | 2.5% PIP2 | 2.5% PI4P | ||
| Extrusion pore | 1.0 μm | 1.0 μm | 0.03 μm | 1.0 μm | 0.03 μm | |
| ArfGAP1 | Max Hydrolysis (%) C50 (nM) |
ND | 98.9±3.0 31.5±2.2 |
98.4±2.3 35.2±14.8 |
ND | ND |
| ArfGAP2 | Max Hydrolysis (%) C50 (nM) |
11 ± 1.6 >500 |
25 ± 2.0*** >500 |
37 ± 5.2***,+ >500 |
6.2 ± 1.2 >500 |
15 ± 1+++ >500 |
| ArfGAP2+coatomer | Max Hydrolysis (%) C50 (nM) |
22 ± 2.0 15 ± 8.6 |
45 ± 2.4^^^ 26 ± 8.4 |
81 ± 3.3^^^,+++ 19 ± 4.8 |
55 ± 3.9^^^ 5.7 ± 1.8@@ |
86 ± 3.2^^^,+++ 7.4 ± 1.2@ |
| ArfGAP2+coatomer+p25 | Max Hydrolysis (%) C50 (nM) |
ND | ND | ND | 47 ± 2.5 1.9 ± 0.7%%% |
84 ± 1.6 0.78 ± 0.2%%% |
The indicated Arf GAPs were titrated into a reaction mixture containing 0.6 μM [α32P]GTP•Arf1 with LUVs containing 10% PI (column labeled 0% PIPx), 7.5% PI and 2.5% PIP2 (column labeled 2.5% PIP2) or 7.5% PI and 2.5% PI4P (column labeled PI4P) and extruded through membranes with pores 0.03 μm or 1.0 μm in diameter, and, as indicated, 124 nM coatomer and 25 μM p25 peptide. The top entry is the maximum percentage of GTP, bound to Arf1, hydrolyzed in a 3 min reaction at 30 °C. The bottom entry is the amount of the indicated Arf GAP needed to achieve 50% of the maximum observed hydrolysis. Arf GAP2 was titrated to a concentration of 3.4 μM in those experiments in which a clear maximum was not achieved. The means ± SD from three experiments are presented. ND, not done. Three ANOVA analysis with Bonferroni post tests were performed. In the first, Arf GAP1 and Arf GAP2 using PIP2 containing vesicles of two different sizes were compared.
indicates less than Arf GAP1 under the same condition, p<0.001. A second ANOVA followed by a Bonferroni post test compared maximum hydrolysis observed with Arf GAP2 and Arf GAP2+ coatomer.
indicates greater than Arf GAP2 with no coatomer, p<0.001
greater than hydrolysis with 1.0 μm, p<0.05
greater than hydrolysis with 1.0 μm vesicle, p<0.001. A third ANOVA analysis compared the C50 values for Arf GAP2 + coatomer with Arf GAP2 + coatomer + p25.
indicates less than C50 with vesicles containing PIP2, p< 0.05;
indicates less than with vesicles containing PIP2, p<0.01;
indicates less than Arf GAP2 + coatomer, p<0.001.
3.2 GAP activity of Arf GAP2 is dependent on coatomer
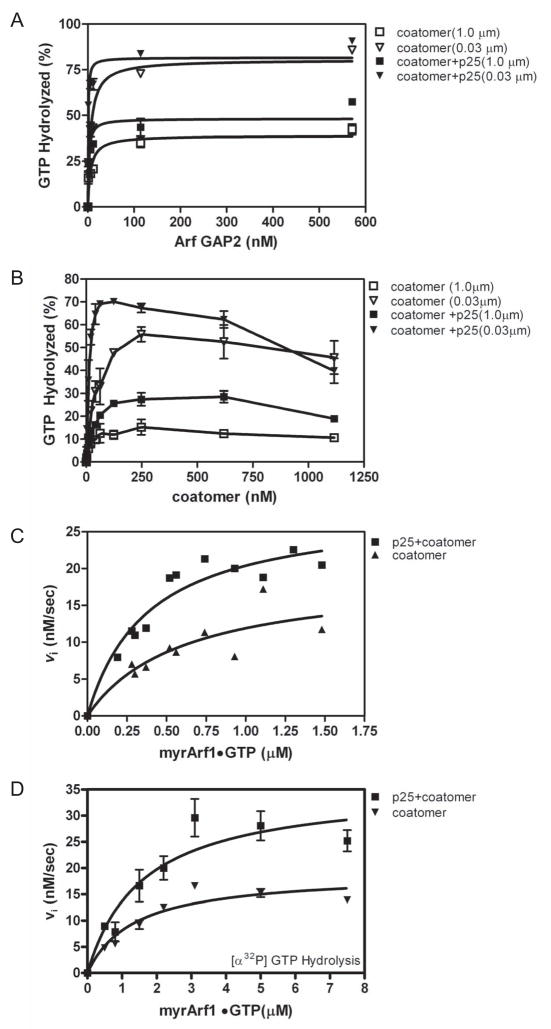
Purified Arf GAP2 might have low activity because activity is dependent on a regulatory interaction. Arf GAPs have been found to be regulated by coat proteins and phospholipids. We examined coatomer, a positive regulator of Arf GAP1 [17,22] that also binds to and activates Arf GAP2 [18,32,33], and phosphoinositides as regulators of Arf GAP2 (Figure 2 and Table 1). Coatomer significantly stimulated activity, reducing the C50 and increasing the extent of GTP hydrolysis (Figure 2A and Table 1). Phosphatidylinositol 4-phosphate (PI4P), which is found in the transGolgi, had no effect by itself (Figure 2A and Table 1). Phosphatidylinositol 4,5-bisphosphate (PIP2) affected the extent of hydrolysis but not the C50 (Table 1); however, PIP2 is not abundant in the Golgi and may not be relevant to Golgi function [5,30].
Figure 2. Effect of vesicle size, coatomer, PI4P and PIP2 on GAP-induced hydrolysis of GTP bound to Arf1.
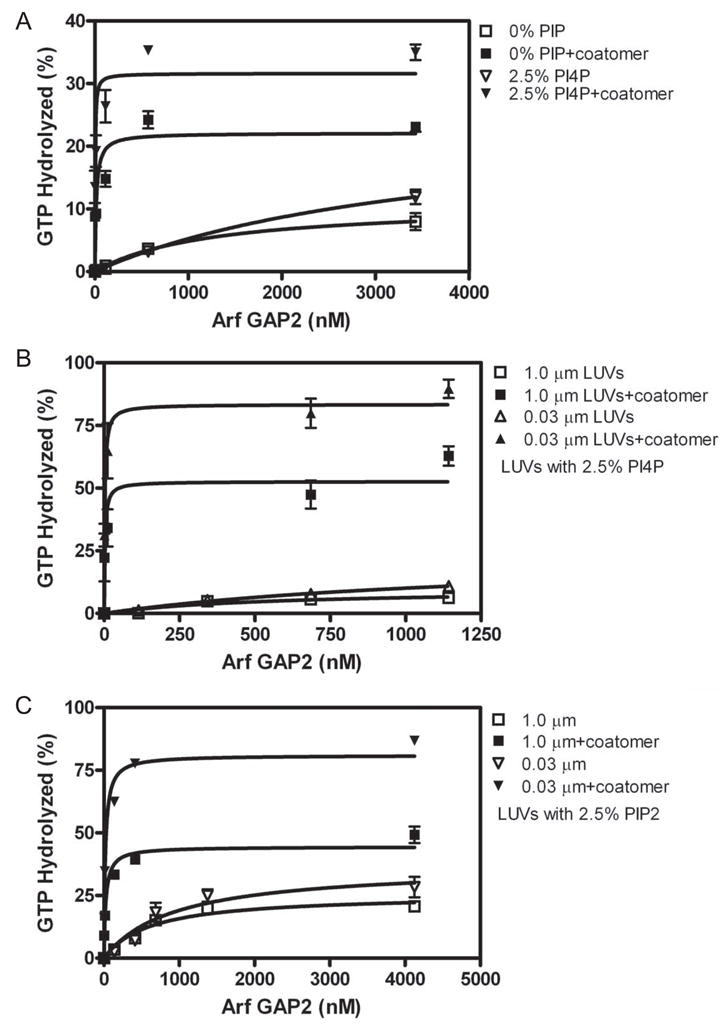
[1–521]Arf GAP2-His6 was titrated into a reaction with LUVs containing either 10% PI, 7.5% PI and 2.5% PI4P (indicated +PI4P) or 7.5% PI and 2.5% PIP2 (indicated +PIP2). A. Interaction of coatomer and PI4P. Reaction contained LUVs extruded through 1.0 μm pores with PI4P where indicated and 124 nM coatomer where indicated. B. Effect of vesicle size on PI4P- and coatomer-dependent GAP activity. GAP activity was determined in reaction mixtures containing 124 nM coatomer and LUVs containing PI4P and extruded through membranes with pores of the indicated diameters. C. Effect of vesicle size on PIP2- and coatomer-dependent GAP activity. GAP activity was determined in reaction mixtures containing 124 nM coatomer and LUVs containing PIP2 and extruded through membranes with pores of the indicated diameters.
The possibility of synergy between coatomer, phosphoinositides and curvature was examined (Figure 2B and C and Table 1). Phosphoinositides had small effects on coatomer-dependent activity, increasing the extent of GTP hydrolysis by 2- to 4-fold. PI4P also reduced the C50 by 2-fold when coatomer was present. Although the magnitude of the effects was not sufficient to conclude that the phosphoinositides have a regulatory role, we included PI4P in subsequent assays to study the GAP reaction under optimal conditions. Similarly, curvature had a small effect on coatomer dependent activity. The extent of GTP-hydrolysis was 2-fold greater with LUVs extruded through 0.03 μm pores than with LUVs extruded through 1 μm pores.
The dependence of Arf GAP2 enzymatic activity on coatomer concentration was determined. Consistent with the predictions from a model in which coatomer is an allosteric modifier of the GAP activity and, at higher concentrations, can sequester the substrate Arf1•GTP [22], the effect of coatomer was biphasic (Figure 3). The half maximal stimulatory effect was observed with ≈ 50 nM coatomer. Activity peaked at 100 – 200 nM coatomer and diminished at coatomer concentrations of greater than 500 nM. Maximum hydrolysis was greater and the second inhibitory phase was smaller in the presence of the smaller LUVs than with the large LUVs.
Figure 3. Coatomer dependence of Arf GAP2 activity.
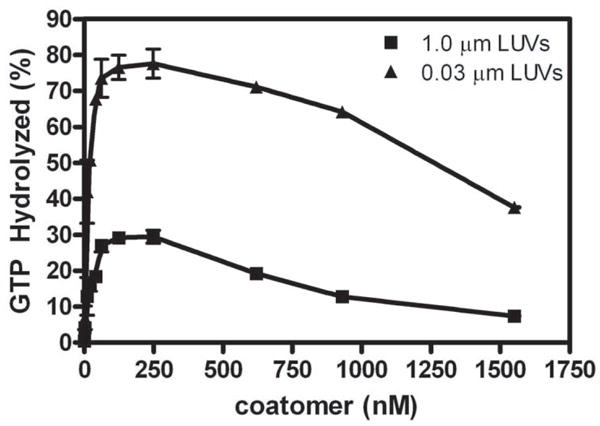
Coatomer was titrated into reactions containing LUVs extruded through 0.03 and 1 μm pore filters, 11.4nM of [1–521] Arf GAP2 His6, and 0.6 μM myrArf1•[α32P]GTP. The reaction was followed by the conversion of [α32P]GTP to [α32P]GDP as described in “Materials and Methods.”
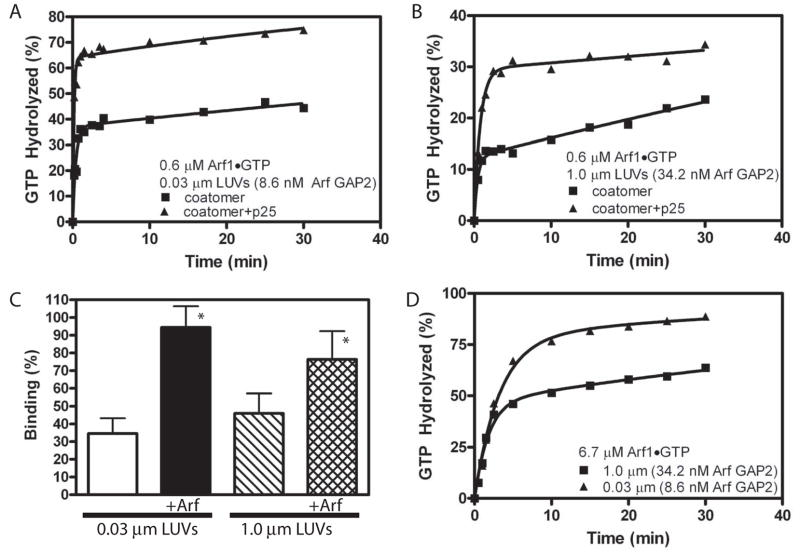
Using optimal coatomer concentrations and lipid conditions, the kinetics of the Arf GAP2/coatomer induced GTP hydrolysis were determined. First, the time dependence of hydrolysis of GTP bound to Arf1 was examined. LUVs of two sizes were used (Figure 4A and B, Table 2. Note, the figure also includes data examining the effect of the cargo p25, which is discussed in section 3.3). The rate of hydrolysis was greater with smaller vesicles. The data were fit to a second order rate equation. The slow and fast rates were similar with both vesicle sizes; however, a greater fraction of the GTP was hydrolyzed with the faster rate with the smaller vesicles. The faster rate was 20- to 60-fold greater than the rate observed in the absence of coatomer.
Figure 4. Analysis of the time dependence of coatomer-dependent Arf GAP2 activity A. Time dependence of GTP hydrolysis and effect of p25 cargo peptide with small LUVs.
The reaction mixture contained LUVs, with PI4P and extruded through membranes with 0.03 μm diameter pores, 0.6 μM Arf1•[α32P]GTP, 8.6 nM [1–521] Arf GAP2 His6, 62 nM coatomer and, where indicated, 25 μM p25. B. Time dependence of GTP hydrolysis and effect of p25 cargo peptide with large LUVs. The reactions contained LUVs with PI4P and extruded through membranes with 1.0 μm pores, 0.6 μM Arf1•[α32P]GTP, 34.2 nM [1–521] Arf GAP2 His6, 62 nM coatomer and, where indicated, 25 μM p25. C. Effect of Arf1•GTP and curvature on coatomer binding to LUVs. Sucrose filled LUVs extruded through either 0.03 or 1 μm pore membranes were incubated with a gel filtered soluble fraction from rat liver containing coatomer but excluding Arf and either no Arf1•GTP or 0.5 μM Arf1•GTPγS for 5 min at 30°C. The LUVs were separated from bulk solution by centrifugation and coatomer associated with the LUVs measured by immunoblotting. The data are the means and standard deviations from 7 determinations. *, different than binding to 0.03 μm LUVs, p<0.05. D. Time dependence and effect of p25 cargo peptide with small LUVs and 6.7 μM Arf1•GTP. Experiment similar to that described in A except Arf1•GTP concentration was in 100 fold excess of coatomer.
Table 2.
Effect of coatomer and cargo on kinetics of GTP hydrolysis
| Size of LUVs | 0.03 μm | 1.0 μm | ||
|---|---|---|---|---|
| constant | Coatomer | coatomer+p25 | coatomer | coatomer+p25 |
| A1 (%) | 38.1±2.0 | 60.1±1.4*** | 11.9±0.4+++ | 24.8±1.1***,+++ |
| A2 (%) | 58.8±2.0 | 35.0±1.4 | 83.1±0.35 | 70.24±1.1 |
| k1 (min−1) | 2.3±0.39 | 6.7±0.87*** | 2.2±0.26 | 1.7±0.29 |
| k2 (min−1) | 0.012±0.0026 | 0.027±0.005*** | 0.005±0.0005 | 0.004±0.0012 |
Progress curves from Figure 4 were fit to the 2nd order rate equation % GTPHydrolyzed = A1 · (1 − e −k1t) + A2 · (1− e−k2t). Reactions contained 0.6 μM [α32P]GTP•Arf1 as a substrate, LUVs containing PI4P and extruded through 0.03 μm or 1.0 μm pores, 124 nM coatomer and 25 μM p25 peptide as indicated. Values are the means ± SD from three experiments. Three ANOVA analyses (one for each of A1, k1 and k2) followed by Bonferroni post tests were performed.
indicates greater than coatomer without p25, p<0.001.
indicates less than 0.03 μm vesicles, p<0.001.
The second order rate observed in the time dependence might be related to the effect of vesicle size. For instance, coatomer recruitment and productive interaction with Arf GAP2 might depend on vesicle size. In this case, the heterogeneities in the vesicles [41] could result in multiple rates. To test this possibility, coatomer association with vesicles of two different sizes was measured. We found that coatomer binding to vesicles was dependent on Arf1•GTP, but we did not detect a significant effect of vesicle size on coatomer binding (Figure 4C). Given the heterogeneity of the sizes within a preparation of vesicles [41], we cannot completely exclude differential binding based on size.
The two phases comprising the second order rate could also result from Arf1•GTP being sequestered by coatomer or by being recruited to vesicles that did not contain Arf GAP2 with coatomer. If sequestered by coatomer, slow release would account for the slow phase in the progress curve. To test for sequestration, the time dependence of GTP hydrolysis was examined at a concentration of Arf1•GTP that was much greater than the concentration of coatomer (Figure 4D). If Arf1•GTP sequestration by coatomer were the reason for the slower rate, increasing Arf concentration to significantly exceed the coatomer and Arf GAP2 concentrations should increase the fraction of GTP bound to Arf that was hydrolyzed with the faster rate, which is what we found2. Together with the biphasic coatomer dependence, this result supports the idea that some Arf is sequestered by coatomer. Also consistent with sequestration by coatomer was the first order rate of hydrolysis observed in the absence of coatomer (see Figure 1B). However, there were still two clearly distinguished phases at high Arf concentrations, especially with larger LUVs, which favors the idea that coatomer, Arf GAP2 and Arf1•GTP may be differentially recruited to vesicles based on size. Although we were not able to conclusively address the molecular basis for the effect of vesicle size, the results indicate that optimal activity is observed with the smaller vesicles, which we have used to further characterize the enzyme.
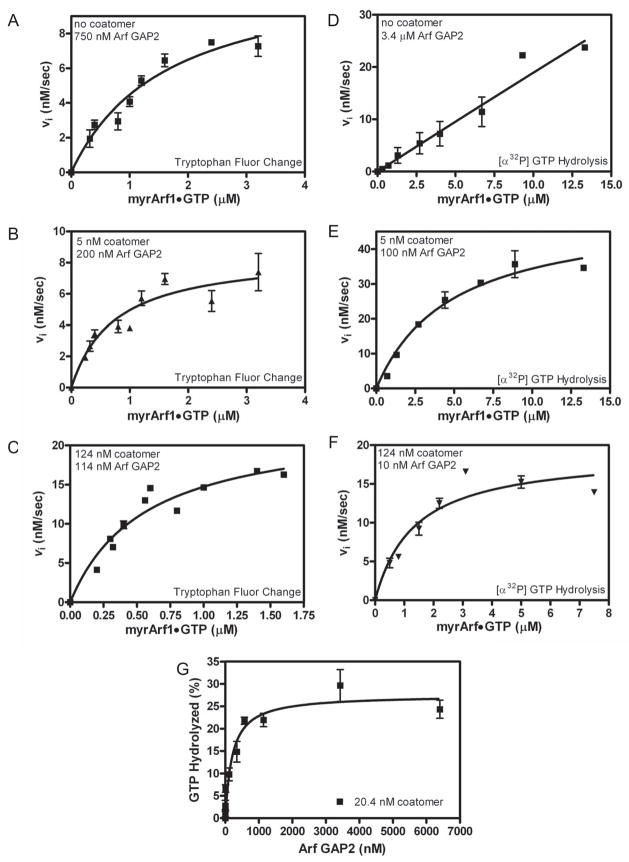
We carried out saturation kinetics under optimal reaction conditions, ie. LUVs containing PI4P and extruded through 0.03 μm pores (Figure 5 and Table 3). We used two different assays for measuring GAP activity, which gave qualitatively similar results although there were quantitative differences. In one assay, which is frequently used in the literature (eg. in ref [22–24,32,33]), the conversion of Arf1•GTP to Arf1•GDP was followed by tryptophan fluorescence. For this assay, changes in fluorescence on mixing Arf GAP2 with coatomer independent of Arf1 were observed. At high concentrations of 2 In panels A and B, 100% hydrolysis is 0.6 μM Arf1•GTP, which is less than the Km for the reaction (see Table 3). In panel D, 100% is 6.7 μM Arf1•GTP, which is greater than the Km. Because the enzyme is saturated, the fractional rates are slower in panel D than in panels A and B; however, the absolute rate (ie. μM Arf1•GTP converted/sec) is faster at the higher Arf concentration. Arf GAP2, its fluorescence changed on the time scale of the experiments. We corrected for these changes and were able to follow a change in fluorescence dependent on Arf1. From those plots, initial rates were determined, which were plotted against Arf concentration (Figure 5A–C and Table 3). The data were fit to the Michaelis-Menten and Hill equations (apparent cooperativity results from coatomer binding both the enzyme and the substrate, as described in [22]). The Hill coefficient was not significantly greater than 1; therefore, we report the analysis using the Michaelis-Menten equation. We found that coatomer decreased the Km and increased the kcat (the kcat calculated from the Vmax is presented in Table 3) with an increase in enzymatic power of approximately 30-fold. We also followed the reaction using [α32P]GTP•Arf1 as a substrate (Figure 5D-F and Table 3). This method directly follows the hydrolysis of GTP. In the absence of coatomer, we titrated up to 13 μM Arf1•GTP without achieving saturation. The reaction saturated in the presence of coatomer. The Km was smaller and the kcat greater at higher coatomer concentrations. We could not estimate the kcat in the absence of coatomer because we could not reach a saturating concentration of Arf1•GTP; however, by comparing the amount of Arf GAP2 required to achieve a rate at a given Arf1•GTP concentration, we were able to estimate the increase in enzymatic power due to coatomer to be greater than 300-fold. As a second means of determining if coatomer affected the Km, Arf GAP2 was titrated into a reaction containing a fixed amount of coatomer and limiting Arf1•GTP (equations 1–3 in appendix) (Figure 5G). In this case, if the effect of coatomer were only on the kcat, ArfGAP2 that was not in complex with coatomer would bind to and sequester the substrate. Consequently, the titration would be biphasic with an initial increase in GAP activity until all the coatomer was in complex with Arf GAP2. At higher concentrations, activity would decrease. If there were an effect on Km, the dependence on enzyme concentration would be hyperbolic. These predictions are independent of substrate binding by coatomer. We found that the dependence on Arf GAP2 was hyperbolic. Taken together, our results indicate coatomer decreases the Km and increases the kcat for Arf GAP2 catalyzed hydrolysis of GTP bound to Arf1.
Figure 5. Effect of coatomer on kinetics of Arf GAP2-catalyzed GTP hydrolysis. Panels A – C. Saturation kinetics performed with fluorescent based assay.
The conversion of Arf1•GTP to Arf•GDP was followed using tryptophan fluorescence in a reaction containing LUVs with PI4P and extruded through 0.03 μm pores and either (A) 750 nM [1–521] Arf GAP2 His6, (B) 5 nM coatomer with 200 nM [1–521] Arf GAP2 His6 or (C) 124 nM coatomer with 114 nM [1–521] Arf GAP2 His6. Initial rates were estimated, and the plot of initial rate versus myrArf1•GTP was fit to the Michaelis-Menten equation to estimate the Km and Vmax. The Km and the kcat, calculated from the Vmax, are presented in Table 3. Panels D – F. Saturation kinetics performed following [α32P]GTP hydrolysis. [α32P]GTP•Arf1 was titrated into reactions containing LUVs with PI4P and extruded through 0.03 μm pores and either (D) 3.4 μM [1–521] Arf GAP2 His6, (E) 5 nM coatomer with 100 nM [1–521] Arf GAP2 His6 or (F) 124 nM coatomer with 10 nM [1–521] Arf GAP2 His6 as indicated. Initial rates were estimated, and the plot of initial rate versus myrArf1•GTP was fit to the Michaelis-Menten equation to estimate the Km and Vmax. The Km and kcat, calculated from the Vmax, are presented in Table 3. Panel G. Arf GAP2 concentration dependence in the presence of limiting coatomer. [1–521] Arf GAP2 His6 was titrated into a reaction containing LUVs containing PI4P and extruded through 0.03 μm pores, 0.6 μM of [α32P]GTP•myrArf1 and 20 nM coatomer. Reactions were stopped after 3 min at 30°C. Three experiments are summarized.
Table 3.
Effect of coatomer on kinetic parameters of Arf GAP2
| A. GTP hydrolysis followed with tryptophan fluorescence | ||||
|---|---|---|---|---|
| 0 coatomer | 5 nM coatomer | 124 nM coatomer | 124 nM coatomer + p25 peptide | |
| Km (μM) | 1.7 ± 0.4 (4) | 0.74 ± 0.22 (3) ** | 0.6 ± 0.2 (6)*** | 0.38 ± 0.1 (3) *** |
| kcat (sec−1) | 0.014 ± 0.001 (4) | 0.043 ± 0.005 (3) | 0.19 ± 0.03 (6)*** | 0.36 ± 0.03 (3)***,+++ |
| B. GTP hydrolysis followed using radioisotopic tracer | ||||
| 0 coatomer | 5 nM coatomer | 124 nM coatomer | 124 nM coatomer + p25 peptide | |
| Km (μM) | >13# (3) | 4.8 ± 0.97 (4) | 1.3 ± 0.39 (3) *** | 1.6 ± 0.24 (3) *** |
| kcat (sec−1) | NC (3) | 0.50 ± 0.043 (4) | 1.9 ± 0.18 (3) *** | 3.5 ± 0.4 (3) ***, +++ |
Initial rate as a function of Arf1•GTP concentration was fit to the Michaelis-Menten equation to estimate the Km and Vmax. The kcat was calculated from the Vmax. The reactions contained the indicated concentration of coatomer and, where indicated, 25 μM p25 peptide. A. GAP assayed by changes in fluorescence. Arf1•GTP to Arf1•GDP conversion was followed using tryptophan fluorescence. The parameters are the means and SD from fitting data presented in Figures 5 and 8. The number of experiments is indicated in parentheses. ** indicates different than 0 coatomer, p<0.01; *** indicates different than 0 coatomer, p< 0.001; +++, indicates different than 124 nM coatomer alone, p<0.001. B. GAP reaction followed by conversion of [α32P]GTP to [α32P]GDP. Data from three experiments presented in Figures 5 and 8 were fit to the Michaelis-Menten equation. The parameter estimates are presented as means ± SD for the number of experiments indicated in parentheses. #, did not saturate with reaction velocity linearly related to Arf1•GTP concentration in the range of 0.5 to 13 μM. NC, Not calculated because did not achieve saturation. ***, different than 5 nM coatomer, p<0.001, +++, indicates different than 124 nM coatomer alone, p<0.001.
3.3 Peptides from p24 cargo proteins increase coatomer-dependent activity of Arf GAP2
Peptides derived from p24 cargo proteins have been reported to inhibit the activity of Arf GAP1. We examined cargo peptides (Figure 6A) derived from three p24 family proteins: p23 (also called p24δ1), p24 (also called p24β1) and p25 (also called p24α2). p24 has been the focus of previous studies on inhibition of Arf GAPs [28,42]. It has the lowest affinity for coatomer among the three [43]. The peptide from p24, at concentrations greater than 100 to 200 μM, has previously been reported to inhibit Arf GAP1 [22,28,42,43]. The peptide from p24 did not affect Arf GAP2. In contrast, peptides from p23 and p25 stimulated GAP activity with half maximal effects at 0.6 ± 0.3 μM (mean ± range for duplicate experiments) p23 peptide and 0.5 ± 0.2 μM p25 peptide (Figure 6B). The effect of p25 decreased at concentrations greater than 25 to 50 μM. In cells, the short cytoplasmic tails of the p25 proteins bind to coatomer at the membrane surface. We also examined the effect of a p25 peptide with a three amino acid extension on the N-terminus to which a palmitate was covalently linked. The palmitoylated peptide associates with the lipid bilayer. The peptide with the three amino acid extension stimulated activity with the half maximal effect observed at 1.8 ± 0.5 μM peptide. Like the free peptide, the palmitoylated peptide stimulated activity (Figure 6C) with the half maximal effect observed at 2.4 ± 0.6 μM peptide.
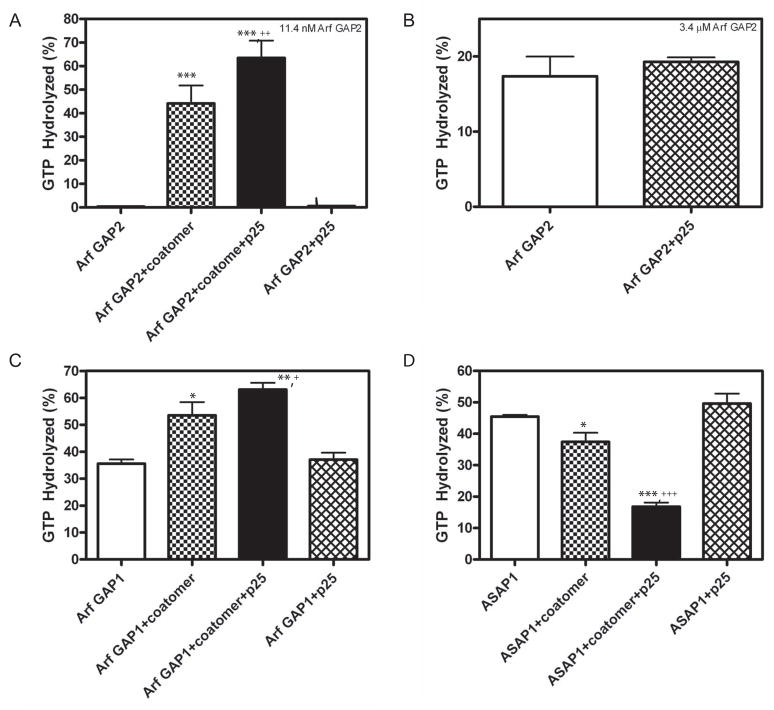
Stimulation of GAP activity by p25 depended on the presence of coatomer. The effect of p25 on 11.4 nM Arf GAP2 with and without 124 nM coatomer was determined (Figure 7A). p25 increased the activity in the samples containing both Arf GAP2 and coatomer. No activity was detected in the absence of coatomer. We also examined the effect of p25 on 3.4 μM Arf GAP2 (Figure 7B). At the higher concentration of Arf GAP2, activity was detected in the absence of coatomer. p25 had no effect. The enzymatic activity of Arf GAP1, which also binds to coatomer, was also stimulated by p25 when coatomer was present (Figure 7C). In contrast, the activity of ASAP1, which does not bind coatomer, was inhibited by coatomer, which was likely due to sequestration of Arf1•GTP. The coatomer-dependent inhibition was increased by p25. p25 had no effect in the absence of coatomer (Figure 7D).
Figure 7. Effect of cargo peptide on Arf GAP depends on coatomer.
GTP hydrolysis in a 3 min reaction at 30°C was determined. Reaction mixtures contained LUVs with PI4P and extruded through 0.03 μm pores, 0.6 μM [α32P]GTP•Arf1 and, where indicated, 124 nM coatomer and 25 μM p25 peptide. The type and concentration of Arf GAPs used are: A. 11 nM [1–521] Arf GAP2 His6; B. 3.4 μM of [1–521] Arf GAP2 His6; C. 28 nM [1–415]Arf GAP1 His6; and D. 1 nM of His10[325–724]ASAP1. The data from each panel were analyzed by one way ANOVA followed by a Newman-Kuels post test. *, different than GAP alone, p<0.05; ***, different than GAP alone, p<0.001; +, different than GAP+coatomer, p<0.05; ++, different than GAP+coatomer, p<0.01; +++, different than GAP+coatomer, p<0.001.
We further analyzed the effect of p25 on the kinetics of Arf GAP2 enzymatic activity to confirm that it stimulated activity. Arf GAP2 was titrated into a reaction containing a fixed concentration of coatomer and p25. p25 decreased the C50 by approximately 10-fold (Table 1 and Figure 8A). We also titrated coatomer into reactions containing Arf GAP2 and p25 (Figure 8B). Two sizes of LUVs were used. p25 increased the extent of hydrolysis. With the larger LUVs, peptide increased activity at every coatomer concentration we examined. With the smaller LUVs, the greatest effect of the peptide was observed when the concentration of coatomer was less than 250 nM. p25 peptide shifted the peak activity from 250 nM to 100 nM coatomer.
Figure 8. Effects of p25 peptide on the kinetics of coatomer/Arf GAP2 catalyzed GTP hydrolysis. A. Titration of Arf GAP2 into reaction with fixed concentration of p25 peptide and coatomer.
[1–521] Arf GAP2 His6 was titrated into a reaction mixture with LUVs containing PI4P and extruded through membranes with either 0.03 or 1.0 μm pores, 0.6 μM of [α32P]GTP•myrArf1, 0.124 μM coatomer and 25 μM p25 peptide at 30° C. Reactions were terminated after 3 min. B. Effect of p25 on coatomer dependence of Arf GAP2 activity. Coatomer was titrated into a reaction mixture with LUVs containing PI4P and extruded through either 1.0 μm or 0.03 μm pores, 0.6 μM of [α32P]GTP•myrArf1, 11.4 nM [1–521] Arf GAP2 His6 and 25 μM p25 peptide at 30° C. Reactions were terminated after 3 min. C. Effect of p25 on Arf1•GTP dependence of Arf GAP2 activity determined using fluorescent based assay. The conversion of Arf1•GTP to Arf•GDP was followed using tryptophan fluorescence in a reaction mixture with 114 nM[1–521] Arf GAP2 His6, LUVs containing PI4P and extruded through 0.03 μm pores, 124 nM coatomer and, where indicated, 25 μM p25 peptide. D. Effect of p25 on Arf1•GTP dependence of Arf GAP2 activity determined following [α32P]GTP hydrolysis. The conversion of [α32P]GTP bound to Arf1 to [α32P]GDP was followed. The reaction mixture contained the indicated concentration of Arf1•GTP, 10 nM [1–521] Arf GAP2 His6, LUVs containing PI4P and extruded through 0.03 μm pores, 124 nM coatomer and, where indicated, 25 μM p25 peptide.
The effect of p25 on the time dependence of Arf GAP2/coatomer-dependent GTP hydrolysis was also consistent with a stimulatory effect of the peptide. p25 accelerated hydrolysis. As previously described, the progress curve was fit to a second order rate equation. The fraction of GTP hydrolyzed at the faster rate was increased and the magnitude of the rate constant was increased when using smaller vesicles (Figure 4 and Table 2).
We also determined the effect of cargo peptide on kinetic parameters. In these experiments, myrArf1•GTP was titrated into a reaction containing 25 μM peptide, 124 nM coatomer and LUVs extruded through 0.03 μm pores. Consistent with the effect on the rate of GTP hydrolysis in progress curves, analysis of saturation kinetics both using the fluorescent based assay and by following hydrolysis of [α32P]GTP revealed that p25 increased the kcat of the reaction (Table 3 and Figure 8C and D).
The result that cargo accelerated GAP activity was unexpected. To test whether cargo had the same effect in vivo, we expressed myc-p25 in cells and measured Arf1•GTP levels. If cargo accelerated GAP activity, relative Arf1•GTP levels should be less than in control cells. As predicted, we found that Arf1•GTP levels were lower in the cells expressing myc-p25 than in the controls (Figure 9).
Figure 9. Effect of overexpressing p25 on cellular Arf1•GTP levels.
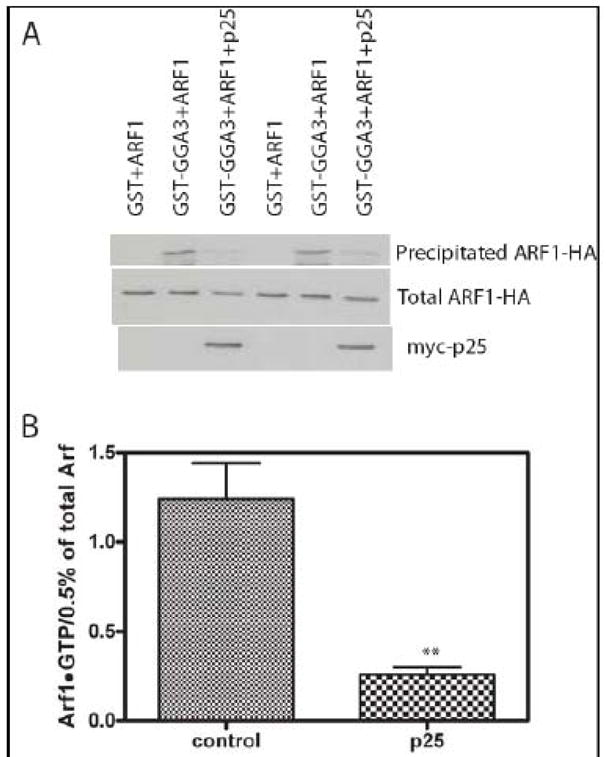
Cos7 cells were transfected with plasmids directing the expression of Arf1-HA and myc-p25 as indicated. 24 hours after transfection cells were lysed and cellular levels of Arf1•GTP were determined by selective precipitation of Arf1•GTP with GST-GGA as described in “Materials and Methods.” The primary data from one experiment is shown in panel A and a summary of three experiments in which signal was quantified as described in “Materials and Methods” is shown in panel B. **, different than control, p<0.01 by Student t-test.
4 Discussion
We examined the effects of known regulators of Arf GAPs on the activity of Arf GAP2, whose kinetics had not been characterized previously. In common with Arf GAP1, Arf GAP2 was regulated by coatomer. However, Arf GAP2 was more sensitive to coatomer than Arf GAP1 and, different than previous reports describing Arf GAP1, peptides from cargo proteins accelerated GAP activity dependent on the presence of coatomer. Based on these properties of Arf GAP2, we propose a model for membrane traffic that accounts for a number of observations in the literature that have not been accommodated by prevailing paradigms.
We were able to detect increases in Arf GAP2 activity due to coatomer of 30- fold when examined by changes in Arfs tryptophan fluorescence and more than 300-fold when GAP activity was followed by hydrolysis of radioactive GTP. The effect of coatomer was consistent with its function as an allosteric modifier that can also bind to the substrate. Coatomer dependence, as previously reported for Arf GAP1 [22], was biphasic, with an initial stimulatory phase, consequent to coatomer binding the Arf GAP, followed by a phase of diminishing activation, which is thought to be a consequence of coatomer binding and sequestering the substrate Arf1•GTP. Arf GAP2 has higher affinity for coatomer, which could explain a stimulatory phase of greater amplitude. More coatomer – Arf GAP complex could be formed at concentrations of coatomer that did not significantly sequester Arf1•GTP in the in vitro assay. The difference in sensitivity to coatomer may be involved in differential regulation of these two Arf GAPs.
Peptides derived from p24 cargo proteins were found to accelerate catalytic activity of Arf GAP2. Relatively high concentrations of free peptides were used for most experiments. However, the half maximal effects were observed with less than 1 μM peptide. A palmitoylated p25 peptide, which partitions to the membrane mimicking the cytoplasmic tail extending from the membrane for the full length protein, also stimulated activity. The effect of cargo was dependent on the presence of coatomer and shifted the coatomer dependence to the left. Expressing full length cargo protein reduced Arf1•GTP levels in cells. Taken together, these results support the idea that cargo is a positive regulator of Arf GAP2 activity. Based on these results, we hypothesize that cargo affects GAP activity by changing the conformation of coatomer to a form that more efficiently binds to and/or activates Arf GAP2. Consistent with this idea, cargo has been previously reported to affect the conformation of coatomer [44,45] and of clathrin adaptor protein-1 [46]. Cargo peptide also stimulated Arf GAP1 when coatomer was present leading us to postulate that the effect of cargo we have described for Arf GAP2 and coatomer may extend to other Arf GAP/coat pairs.
Cargo did not stimulate the activity of ASAP1, an Arf GAP that does not bind coatomer; instead it increased the inhibition of ASAP1 GAP activity by coatomer. Inhibition is likely the result of Arf1•GTP sequestration by coatomer as has been observed with GGA [47]. Cargo may have increased the affinity of Arf1•GTP for coatomer, similar to the cargo dependent change in affinity of Arf1•GTP for clathrin adaptor protein-1 [46]. Cargo may not inhibit Arf GAP2 activity because it has a different effect on Arf GAP-coatomer-Arf1 complex. For instance, coatomer may form part of the substrate binding pocket with Arf GAP, similar to a model previously proposed [17], with cargo acting as an allosteric modifier increasing the activity of the coatomer/Arf GAP complex. The effect of p25 to increase the affinity of coatomer for Arf1•GTP could account for the diminishing activity that was observed in titrations when coatomer concentration was greater than Arf GAP2 concentration; coatomer could then bind Arf1•GTP independently of Arf GAP2. The second phase in the coatomer dependence curve was not as steep in the absence of p25.
Although the saturation kinetic data were qualitatively consistent, we found quantitative differences between kinetics assayed using fluorescence to follow changes in Arf1•GTP levels and kinetics in which [α32P]GTP•Arf1 was used as a substrate. The fluorescent assay is complicated by a number of factors. The tryptophan fluorescence of all the proteins in the assay could change due to conformational changes or microaggregation. Because a high mass of coatomer is present in the reactions, the fluorescent signal from coatomer is significant compared to the signal from Arf1. Although we could partly correct for these changes, we do not know if there were changes in the GAP or coatomer fluorescence that were dependent on Arf1. In addition, protein association and dissociation from vesicles can cause changes in light scattering, which can interfere with fluorescent signals. Blunting of the fluorescent signal from Arf1 by these factors could account for difference in parameter estimates from those determined by the assay following hydrolysis of radioisotopically labeled GTP. Nevertheless, the data using either assay supported the conclusion that coatomer decreased the Km and that coatomer and p25 increased the kcat of the reaction and the sum of experiments in the paper are consistent with coatomer and p25 increasing enzymatic activity.
Some cargo peptides, when present at high concentrations, were found to inhibit Arf GAP activity. The effect is nonspecific [22], seen with Arf GAP1 and ASAP1 and was independent of coat protein [22,42]. One possible mechanism for the inhibition is that cargo peptide, at high concentrations, can bind to and sequester Arf1•GTP [48]. Another possibility is that the peptide at high concentrations is a chaotrope, nonspecifically disrupting protein structure.
Arf GAP2 does not have an ALPS domain; therefore, curvature sensitivity was not anticipated. In a recent paper, Arf GAP2 was reported to be stimulated by coatomer but was not affected by curvature [33]. The differences could be attributable to the assays used. Wieland and colleagues used only an assay based on tryptophan fluorescence, which has some confounding factors as described above, whereas we primarily used an assay using radioactive GTP. Even if we can explain the difference in results from the two groups, the effects we observed were relatively small, which raises a concern of physiological relevance of this particular in vitro result. We have speculated about mechanisms that can explain the result and that may be related to possible cellular function. First, the GAP may more efficiently penetrate a highly curved bilayer, and be more active in the hydrophobic environment. This explanation is consistent with our observation of an effect of vesicle size on Arf GAP2 activity when no coatomer was present. Second, Arf1•GTP may be more available or exposed on a curved surface than on a flat surface, also consistent with the effect on Arf GAP2 with no coatomer present. Third, there may be differential recruitment, with Arf GAP2 and/or coatomer preferentially recruited to smaller vesicles. Fourth, surface curvature may influence the conformation of coatomer. Because coatomer binds directly to both Arf GAP2 and Arf1•GTP, the change in conformation may affect one or both proteins. On going studies are focused on first determining whether the small curvature dependence is physiologically relevant and second discerning among the possible explanations for the curvature dependence.
Based on our results and those in the literature in which GTP hydrolysis has been found to be required for generation of cargo laden transport vesicles [20,42,49], we propose a model for the function of Arf GAPs in cargo sorting and generation of transport vesicles [50]. Two current models for the regulation of Arf GAPs in membrane traffic do not offer complete explanations for reported observations related to these processes. In one model, cargo inhibits Arf GAP activity, slowing hydrolysis of GTP on Arf1•GTP, which maintains coat protein on the membrane, allowing it to polymerize. Although this model accounts for cargo sorting, it cannot explain the control of GAP activity within the transport vesicle in which the inhibitory cargo is concentrated nor does it explain the effect of cargo to accelerate Arf GAP1 hydrolytic activity in vivo (this paper and [51]). In a second model, Arf GAP activity is regulated by the recruitment of Arf GAP to the curved membrane surface induced by formation of the transport vesicle [23,24]. This model also fails to explain the requirement for GTP hydrolysis for sorting and, except by extensions of the model, does not account for sorting. We have previously described a model, consistent with the results reported here [8,50], that can explain the requirement of GTP hydrolysis for sorting. In the model, Arf1 in membrane traffic has a function analogous to EfTu function in one model for proofreading in protein synthesis. EfTu has been proposed to hold a tRNA in the acyl site for a finite period of time. If the tRNA matches the codon, a conformational change in the ribosome leads to hydrolysis of GTP on EfTu with dissociation of EfTu and elongation of the polypeptide chain [52]. We propose that in membrane traffic, Arf1•GTP holds the coatomer/GAP complex on the membrane for a finite period of time during which cargo can be recognized. If the appropriate cargo is present, a conformational change in the coatomer/GAP complex leads to GTP hydrolysis on Arf and simultaneous polymerization of coatomer into COPI. This mechanism would provide a proofreading function, contributing to concentration of cargo into transport intermediates, and is consistent with reports in the literature of vesicle formation [19,20] and cargo concentration [19,20,42,53–55] requiring GTP hydrolysis. It is also consistent with reports that cargo accelerates AP-1 polymerization and Arf GAP1-mediated GTP hydrolysis at the trans Golgi [51] and is required for the formation of COPI retrograde transport vesicles [56]. This model may apply to other Arf GAP families such as the AGAPs which bind to clathrin adaptor proteins [57–59].
5 Conclusions
We examined the kinetics and regulation of Arf GAP2. We consider two findings particularly relevant for understanding the role of Arf GTP-binding proteins and Arf GAPs in membrane traffic. First, Arf GAP activity was highly dependent on the vesicle coat protein coatomer. Second, peptides from proteins that are transported by coatomer (cargo) stimulate coatomer-dependent GAP activity. Based on these results together with previous work, we propose that cargo binding to coatomer triggers a conformational change in both Arf GAP and coatomer, leading to increased rates of GTP hydrolysis and coat polymerization. This model departs from the current paradigms for the role of Arf in membrane traffic.
Acknowledgments
We thank Dr. Jean Gruenberg for expression vectors for p24 family members and Dr. Jonathan Goldberg for insightful discussion. This work was supported by the Intramural Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services
Abbreviations
- ALPS
Arf GAP lipid packing sensor
- Arf
ADP-ribosylation factor
- GAP
GTPase-activating protein
- PI4P
phosphatidylinositol 4-phosphate
- PIP2
phosphatidylinositol 4,5-bisphosphate
6 Appendix
Reference [22] presents the derivation of equations assuming rapid equilibrium and assuming that Arf GAP concentration are much less than the substrate myrArf1•GTP and less than Km. Here we derive an equation describing the dependence on Arf GAP concentration assuming the substrate and coatomer are limiting.
Abbreviations:
E=ArfGAP2
S=myrArf1•GTP
C=coatomer
Kd=affinity constant for coatomer-Arf GAP2 complex
Km1= Michaelis constant of Arf1•GTP for uncomplexed ArfGAP2
Km2= Michaelis constant of Arf1•GTP for Arf GAP2 in complex with coatomer
kcat1= catalytic constant for Arf GAP2 that is not bound to coatomer
kcat2= catalytic constant for Arf GAP2 that is in complex with coatomer
We assume S<C<E. Then
Consider two extremes. If kcat1 ≪kcat2, then the equation becomes , which describes a biphasic dependence on E.
If Km1≫Km2, then the equation reduces to the hyperbola:
Footnotes
The estimate of 100 fold more activity was based on the amount of enzyme required to achieve a fixed amount of hydrolysis. The C50 for Arf GAP1 was 31 and 35 nM for the two vesicle sizes. Therefore between 15 and 25 nM enzyme was required to achieve 25% – 40% hydrolysis, the extent of hydrolysis observed with the highest concentration of Arf GAP2 used in the reaction, 3.4 μM. 3.4μM is more than 100 fold greater than 25 nM.
In panels A and B, 100% hydrolysis is 0.6 μM Arf1•GTP, which is less than the Km for the reaction (see Table 3). In panel D, 100% is 6.7 μM Arf1•GTP, which is greater than the Km. Because the enzyme is saturated, the fractional rates are slower in panel D than in panels A and B; however, the absolute rate (ie.μM Arf1•GTP converted/sec) is faster at the higher Arf concentration.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donaldson JG, Lippincott-Schwartz J. Cell. 2000;101:693–696. doi: 10.1016/s0092-8674(00)80881-8. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Glick BS. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 3.De Matteis MA, Luini A. Nature Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino JS, Lippincott-Schwartz J. Nature Rev Mol Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- 5.D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 6.Jackson CL, Casanova JE. Trends in Cell Biology. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 7.Casanova JE. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 8.Nie ZZ, Randazzo PA. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- 9.Souza-Schorey C, Chavrier P. Nature Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 10.Gillingham AK, Munro S. Ann Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 11.Rothman JE. Nature Med. 2002;8:1059–1062. doi: 10.1038/nm770. [DOI] [PubMed] [Google Scholar]
- 12.Nie ZZ, Hirsch DS, Randazzo PA. Curr Opin Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 13.Randazzo PA, Kahn RA. J Biol Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- 14.Inoue H, Randazzo PA. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 15.Randazzo PA, Inoue H, Bharti S. Biol Cell. 2007;99:583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- 16.Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg J. Cell. 1999;96:893–902. doi: 10.1016/s0092-8674(00)80598-x. [DOI] [PubMed] [Google Scholar]
- 18.Frigerio G, Grimsey N, Dale M, Majoul I, Duden R. Traffic. 2007;8:1644–1655. doi: 10.1111/j.1600-0854.2007.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, Yang JS, Hong WJ, Premont RT, Hsu VW. J Cell Biol. 2005;168:281–290. doi: 10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang JS, Lee SY, Gao MG, Bourgoin S, Randazzo PA, Premont RT, Hsu VW. J Cell Biol. 2002;159:69–78. doi: 10.1083/jcb.200206015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber I, Cukierman E, Rotman M, Aoe T, Hsu VW, Cassel D. J Biol Chem. 1998;273:24786–24791. doi: 10.1074/jbc.273.38.24786. [DOI] [PubMed] [Google Scholar]
- 22.Luo RB, Randazzo PA. J Biol Chem. 2008;283:21965–21977. doi: 10.1074/jbc.M802268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigay J, Gounon P, Robineau S, Antonny B. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 25.Cassel D. In: Arf Family GTPases. Kahn R, editor. Kluwer Academic Publishers; Boston: 2003. pp. 137–158. [Google Scholar]
- 26.Cukierman E, Huber I, Rotman M, Cassel D. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 27.Aoe T, Cukierman E, Lee A, Cassel D, Peters PJ, Hsu VW. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg J. Cell. 2000;100:671–679. doi: 10.1016/s0092-8674(00)80703-5. [DOI] [PubMed] [Google Scholar]
- 29.Antonny B, Huber I, Paris S, Chabre M, Cassel D. J Biol Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- 30.De Matteis MA, Di Campli A, Godi A. Biochim Biophys ACTA. 2005;1744:396–405. doi: 10.1016/j.bbamcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Randazzo PA, Hirsch DS. Cell Signal. 2004;16:401–413. doi: 10.1016/j.cellsig.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Kliouchnikov L, Bigay J, Mesmin B, Parnis A, Rawet M, Goldfeder N, Antonny B, Cassel D. Mol Biol Cell. 2008:E08–E10. doi: 10.1091/mbc.E08-10-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weimer C, Beck R, Eckert P, Reckmann I, Moelleken J, Brugger B, Wieland F. J Cell Biol. 11182008;183:725–735. doi: 10.1083/jcb.200806140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo R, Ahvazi B, Amariei D, Shroder D, Burrola B, Losert W, Randazzo PA. Biochem J. 2007;402:439–447. doi: 10.1042/BJ20061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavel J, Harter C, Weiland FT. Proc Nat’l Acad Sci USA. 1998;95:2140–2145. doi: 10.1073/pnas.95.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie Z, Hirsch DS, Luo R, Jian X, Stauffer S, Cremesti A, Andrade J, Lebowitz J, Marino M, Ahvazi B, Hinshaw JE, Randazzo PA. Curr Biol. 2006;16:130–139. doi: 10.1016/j.cub.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 37.Che MM, Nie ZZ, Randazzo PA. Method Enzymol. 2005;404:147–163. doi: 10.1016/S0076-6879(05)04015-2. [DOI] [PubMed] [Google Scholar]
- 38.Randazzo PA, Miura K, Jackson TR. Method Enzymol. 2001;329:343–354. doi: 10.1016/s0076-6879(01)29096-x. [DOI] [PubMed] [Google Scholar]
- 39.Luo RB, Jacques K, Ahvazi B, Stauffer S, Premont RT, Randazzo PA. Curr Biol. 2005;15:2164–2169. doi: 10.1016/j.cub.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 40.Yoon HY, Bonifacino JS, Randazzo PA. Methods Enzymol. 2005;404:316–332. doi: 10.1016/S0076-6879(05)04028-0. [DOI] [PubMed] [Google Scholar]
- 41.Kunding AH, Mortensen MW, Christensen SM, Stamou D. Biophys J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanoix J, Ouwendijk J, Stark A, Szafer S, Cassel D, Dejgaard K, Weiss M, Nilsson T. J Cell Biol. 2001;155:1199–1212. doi: 10.1083/jcb.200108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bethune J, Kol M, Hoffmann J, Reckmann I, Brugger B, Wieland F. Mol Cell Biol. 2006;26:8011–8021. doi: 10.1128/MCB.01055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langer JD, Roth CM, Bethune J, Stoops EH, Brugger B, Herten DP, Wieland FT. Traffic. 2008;9:597–607. doi: 10.1111/j.1600-0854.2007.00697.x. [DOI] [PubMed] [Google Scholar]
- 45.Langer JD, Stoops EH, Bethune J, Wieland FT. FEBS Lett. 2007;581:2083–2088. doi: 10.1016/j.febslet.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 46.Lee I, Doray B, Govero J, Kornfeld S. J Cell Biol. 2008;180:467–472. doi: 10.1083/jcb.200709037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacques KM, Nie ZZ, Stauffer S, Hirsch DS, Chen LX, Stanley KT, Randazzo PA. J Biol Chem. 2002;277:47235–47241. doi: 10.1074/jbc.M208875200. [DOI] [PubMed] [Google Scholar]
- 48.Gommel DU, Memon AR, Heiss A, Lottspeich F, Pfannstiel J, Lechner J, Reinhard C, Helms JB, Nickel W, Wieland FT. EMBO J. 2001;20:6751–6760. doi: 10.1093/emboj/20.23.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, Nilsson T. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirsch DS, Stanley KT, Chen LX, Jacques KM, Puertollano R, Randazzo PA. Traffic. 2003;4:26–35. doi: 10.1034/j.1600-0854.2003.40105.x. [DOI] [PubMed] [Google Scholar]
- 51.Meyer DM, Crottet P, Maco B, Degtyar E, Cassel D, Spiess M. Mol Biol Cell. 2005;16:4745–4754. doi: 10.1091/mbc.E05-06-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramakrishnan V. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 53.Malsam J, Gommel D, Wieland FT, Nickel W. FEBS Lett. 1999;462:267–272. doi: 10.1016/s0014-5793(99)01543-4. [DOI] [PubMed] [Google Scholar]
- 54.Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, Wieland FT. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- 55.Weiss M, Nilsson T. Traffic. 2003;4:65–73. doi: 10.1034/j.1600-0854.2003.40202.x. [DOI] [PubMed] [Google Scholar]
- 56.Aguilera-Romero A, Kaminska J, Spang A, Riezman H, Muniz M. J Cell Biol. 2008;180:713–720. doi: 10.1083/jcb.200710025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nie ZZ, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, Randazzo PA. J Biol Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 58.Nie ZZ, Fei J, Premont RT, Randazzo PA. J Cell Sci. 2005;118:3555–3566. doi: 10.1242/jcs.02486. [DOI] [PubMed] [Google Scholar]
- 59.Nie Z, Boehm M, Boja ES, Vass WC, Bonifacino JS, Fales HM, Randazzo PA. Dev Cell. 2003;5:513–521. doi: 10.1016/s1534-5807(03)00234-x. [DOI] [PubMed] [Google Scholar]