Abstract
Context
A single-nucleotide polymorphism (SNP) in the human glucocorticoid receptor (hGR) N363S (rs6195) has been the focus of several clinical studies, and some epidemiological data link this SNP to increased glucocorticoid sensitivity, coronary artery disease, and increased body mass index. However, molecular studies in vitro using reporter gene expression systems have failed, for the most part, to define a link between this polymorphism and altered glucocorticoid receptor function.
Objective
The objective of this study was to address the biological relevancy of N363S SNP in GR function by establishing stable U-2 OS (human osteosarcoma) cell lines expressing wild-type hGR or N363S and examining these receptors under a variety of conditions that probe for GR activity including human gene microarray analysis.
Design
Functional assays with reporter gene systems, Western blotting, and human microarray analysis were used to evaluate the activity of wild-type and N363S GR in both transiently and stably expressing cells. In addition, quantitative RT-PCR was used to confirm the microarray analysis.
Results
Functional assays with reporter gene systems and homologous down-regulation revealed only minor differences between the wild-type hGR and N363S receptors in both transiently and stably expressing cell lines. However, examination of the two receptors by human gene microarray analysis revealed a unique gene expression profile for N363S.
Conclusions
These studies demonstrate that the N363S SNP regulates a novel set of genes with several of the regulated genes supporting a potential role for this GR polymorphism in human diseases.
GLUCOCORTICOID RECEPTORS (GR) ARE expressed in almost all cells and are necessary for life. GR signaling at the physiological level is regulated predominantly by the hypothalamic-pituitary-adrenal (HPA) axis where endocrine, neural, and cytokine signaling converge in the periventricular nucleus of the hypothalamus and regulate the synthesis and release of CRH. CRH stimulates the release of ACTH from the anterior pituitary, which in turn induces synthesis and secretion of cortisol by the adrenal cortex. Although most cortisol is bound to corticosteroid-binding globulin in the blood, approximately 10% remains free and is the biologically active form of the hormone (1). Upon exposure to stress, cortisol levels are increased and affect almost all physiological systems including carbohydrate, lipid, and protein metabolism as well as the cardiovascular system, the immune system, behavior, bone density, and cell growth in addition to the regulation of the HPA axis itself (2). Cortisol also has been found to be elevated in obese individuals, and dysregulation of the HPA axis was found to be more pronounced in individuals with central obesity (3). In addition, sensitivity to glucocorticoids, as measured clinically by dexamethasone suppression tests, varies greatly among individuals (4).
Both natural and synthetic glucocorticoids exert their physiological and pharmacological effects through binding to the intracellular GR, which upon activation by glucocorticoids, activate or repress the transcription of target genes. Several polymorphisms in the GR gene (NR3C1) have been reported in the normal population, and these genetic variations may influence an individual’s response to glucocorticoids. One such single-nucleotide polymorphism (SNP) at amino acid 363, which changes the codon from asparagine (N) to serine (S), was initially identified in a study of Dutch kindred who presented with hypercortisolism and half the number of GR per cell (5). In another Dutch study, a group of 216 elderly individuals were examined for the N363S polymorphism, and 13 heterozygotes (6% of the group) were identified (6). Interestingly, these carriers exhibited an increased sensitivity to exogenously administered glucocorticoids as well as an increased insulin response and increased body mass index (BMI) (6). In addition, a study using the Trier Social Stress Test, which assesses cortisol and ACTH responses to psychosocial stress, showed that N363S carriers had significantly increased salivary cortisol responses to stress (7). In two studies of Australians (all Caucasian of British descent), Lin et al. (8, 9) confirmed this association of increased BMI with the N363S polymorphism. In a separate study by Lin et al. (10) on subjects of Anglo-Celtic descent with coronary artery disease (CAD), they reported that the frequency of the S363 allele was 0.04 in a healthy normal-weight control group but rose to 0.15 in patients with CAD. This association rose even higher in patients with unstable angina (0.45), suggesting a role for the N363S polymorphism in the underlying cause of CAD (10). In a severely obese Italian population, N363S was associated with increased BMI, resting energy expenditure, and food intake (11). A study by Roussel et al. (12) on French subjects with type 2 diabetes mellitus also reported an increase in BMI in subjects carrying the N363S polymorphism. Furthermore, Dobson et al. (13) also showed an association with increased waist-to-hip ratio in male N363S carriers but no associations with BMI, blood pressure, or serum cholesterol levels. However, other reports by Halsall et al. (14), Echwald et al. (15), Rosmond (16), and Buemann et al. (17) have shown no association between N363S and increased BMI. Interestingly, the N363S variant did not occur in Japanese subjects (18) and was of extremely low prevalence in a study performed on South Asians living in the United Kingdom (19).
Biochemical and molecular biological studies examining the ability of N363S to bind ligand or mediate transcriptional activation of transfected glucocorticoid-responsive promoters have been largely negative with no differences observed between the variant and the wild-type receptor (5, 6, 20, 21), although recently, Russcher et al. (22) found a significant but small (8%) increase in transactivation of a GRELuc reporter gene by the N363S GR. Additionally, Russcher et al. (22) showed that there was no difference between wild-type and the variant in dexamethasone-mediated repression of the NF-κB p65 subunit. Thus, the molecular basis for the altered phenotype seen in some patients carrying the N363S SNP remains undefined.
In this study, we employed functional assays to analyze potential differences between wild-type human GR (hGR) and the N363S variant on gene activation and gene repression in both transiently transfected and stable cell lines expressing either wild-type or N363S GR. Under all conditions evaluated, only minimal differences were observed in the ability of N363S and wild-type hGR to regulate transiently transfected reporter genes. In contrast, examination of this polymorphism by microarray analysis showed, for the first time, that there are significant differences between wild-type hGR and the N363S SNP in their ability to regulate gene expression selectively. Several of these genes may define the link between the N363S SNP and human disease.
Materials and Methods
Cell culture and transfection
COS-1 cells (African Green Monkey kidney cells) were cultured in DMEM-H (GIBCO, Grand Island, NY) supplemented with a mixture of 10% FCS:CS (fetal calf serum:calf serum), 2 mm glutamine, 100 IU penicillin, and 100 mg/ml streptomycin. U-2 OS (human osteosarcoma) cells were maintained in DMEM/F-12 supplemented with 10% FCS:CS, 2 mm glutamine, and penicillin/streptomycin, and selected clones were maintained in the same media with the addition of 200 μg/ml Geneticin and 200 μg/ml hygromycin. All cells were maintained in a humidified, 5% CO2 atmosphere.
U-2 OS cell lines stably expressing wild-type hGR and N363S
To establish the U-OFF parental cell line, the well characterized BD Clontech regulatory plasmid pTet-Off was transfected into U-2 OS cells (human osteosarcoma). These cells were chosen because they do not have detectable levels of GR (23). Tet-Off regulates genes responsive to tetracycline, so to generate the tetracycline-responsive plasmid, pTRE2hGRα, MluI and EcoRV ends were generated onto the coding region of hGRα using PCR amplification of the pCMVhGRα plasmid. The pTRE2hyg vector was digested with MluI and EcoRV, and the two DNAs were ligated (23). Site-directed mutagenesis was then performed to make pTRE2N363S. The wild-type pTRE2hGRα and the pTRE2N363S plasmids were individually transfected into the U-OFF cells, and selection was initiated by supplementing the U-OFF growth media with 500 μg/ml hygromycin. After 4 wk, several colonies, selected for expression of wild-type hGR or N363S, were transferred to 12-well plates and maintained in growth media containing 200 μg/ml hygromycin. The receptor levels for each of these cell lines were then compared using Western blot analyses with the well-characterized anti-GR antibody no. 57 (24). One cell line expressing wild-type hGR and one cell line expressing N363S GR at comparable levels were chosen for functional assays, microarray analysis, and initial quantitative RT-PCR studies. In addition, two other cell lines expressing wild-type hGR and two other cell lines expressing N363S GR were analyzed using quantitative RT-PCR. All cell lines were maintained as described above.
Transient transfections and luciferase assays
Cells were plated in six-well plates at approximately 80% confluency 1 d before transfection. Transient transfections were carried out with FuGene 6 reagent (Roche Diagnostics Corp., Indianapolis, IN). After 18-24 h, the transfection media were removed, replaced with fresh medium supplemented with FCS:CS stripped of endogenous glucocorticoids, and treated with dexamethasone or vehicle (H2O) at the concentrations described. Twenty-four hours after treatment, cells were harvested in 400 μl Reporter Gene Assay Lysis buffer (Roche) per well. Cellular debris was pelleted at maximal speed in a refrigerated micro-centrifuge, and the supernatant was assayed for total protein content by Bradford assay (Bio-Rad Laboratories, Hercules, CA). The luciferase activity was measured by using the 96-well plate format with an MLX automated microtiter plate luminometer from Dynex (Thermo Lab-systems, Helsinki, Finland) and corrected to the amount of total protein.
Western blotting
Cell extracts prepared for luciferase assays were denatured in a sodium dodecyl sulfate buffer and then separated on precast 8% Trisglycine gels (Invitrogen, San Diego, CA) and transferred to nitrocellulose membrane (0.2 μm). The membranes were blocked in Tris-buffered saline/0.5% Tween (TBS-T) containing 10% nonfat milk for a 1 h at room temperature, washed in TBS-T, then incubated with anti-GR antibody no. 57 (1:1000) and anti-β-actin (Chemicon, Temecula, CA) (1:10,000) for 1 h at room temperature (24). After additional washing in TBS-T, the blots were incubated with peroxidase-conjugated antirabbit and anti-mouse secondary antibodies (1:20,000) for 1 h at RT. Bands were visualized using ECL reagents (Amersham, Piscataway, NJ). To quantitate the amount of receptor in each band, the wild-type hGR and N363S GR signals were quantitated densitometrically using NIH Image analysis software and normalized to the β-actin signal for each band.
Microarray analysis
Microarray analysis was carried out using Agilent human 1Av2 arrays (Agilent Technologies, Palo Alto, CA). U-2 OS cells stably expressing either wild-type hGR or N363S were cultured for 24 h in media containing charcoal-stripped serum and subsequently treated for 6 h with 10 nm dexamethasone or vehicle (H2O), and total RNA was isolated using the QIAGEN RNeasy midi kit (QIAGEN, Valencia, CA). RNA samples were then amplified using the Agilent low-RNA-input fluorescent linear amplification kit protocol. Total RNA (0.5 μg) was then labeled with Cy3 or Cy5 following the manufacturer’s protocol. Two-color comparison was then carried out using 750 ng each of the Cy3- and Cy5-labeled cRNA, which were mixed and fragmented using the Agilent in situ hybridization kit protocol. Hybridizations were performed for 16 h in a rotating hybridization oven using the Agilent 60-mer oligo microarray processing protocol. After this same protocol, slides were washed and then scanned with an Agilent scanner. Data were obtained using the Agilent Feature Extraction software (version 7.1), using defaults for all parameters. Each RNA sample was analyzed in duplicate (for a total of eight chips per analysis), and two experimental samples of RNA, isolated at different times, were analyzed separately. The gene lists from each analysis were then combined and further analyzed for genes common between the two individual microarray analyses. The microarray data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) (25) and is accessible through GEO accession no. GSE5796.
Real-time PCR analysis
Three stably expressing cell lines each of wild-type hGR and N363S were cultured for 24 h in charcoal-stripped serum medium and then treated with 10 nm dexamethasone or vehicle (H2O) for 6 h, and total RNA was isolated using the QIAGEN RNeasy mini kit. Real-time PCR was performed using the 7900HT sequence detection system predesigned primer/probe sets available from Applied Biosystems (Foster City, CA) and following the manufacturer’s instructions. The signal obtained from each gene primer/probe set was normalized to that of the unregulated housekeeping gene cyclophilin B primer/probe set (also available from Applied Biosystems). Each primer/probe set was analyzed with at least three different sets of RNA.
Transient transfections and quantitative RT-PCR analysis
U-2 OS cells were plated in 100-mm dishes at approximately 80% confluency 1 d before transfection. Cells were transfected with TransIt-LT1 reagent (Mirus, Madison, WI) as described by the manufacturer using 25 μl TransIt-LT1 and 2.5 μg DNA per dish. After 18-24 h, the transfection medium was removed, replaced with charcoal-stripped serum medium, and further incubated for 24 h at 37 C. The cells were then treated with 10 nm dexamethasone or vehicle (H2O) for 6 h. Total RNA was isolated, and quantitative RT-PCR analysis was performed as described.
Results
Analysis of the N363S hGR polymorphism
To analyze the function of N363S compared with wild-type hGR, we took a two-prong experimental approach. First, to recapitulate and confirm previous studies in several laboratories (5, 6, 20-22), we transiently transfected COS-1 cells with either hGR or N363S. Second, we generated stable human cell lines expressing either wild-type hGR or the N363S polymorphism by stably transfecting the receptors into U-OFF cells (U-2 OS cells, human osteosarcoma), which do not have detectable levels of GR (23). After culturing the transfected cells for up to 4 wk in selective media containing 500 μg/ml hygromycin, several colonies of cells expressing either hGR or N363S were transferred to 12-well plates and maintained in 200 μg/ml hygromycin. These stable cell lines were sequenced to confirm that the cells were expressing either hGR or N363S, and one wild-type (hGR-B5) and one N363S (N363S-A2) cell line expressing comparable GR protein levels as assessed by Western blot were chosen for further evaluation.
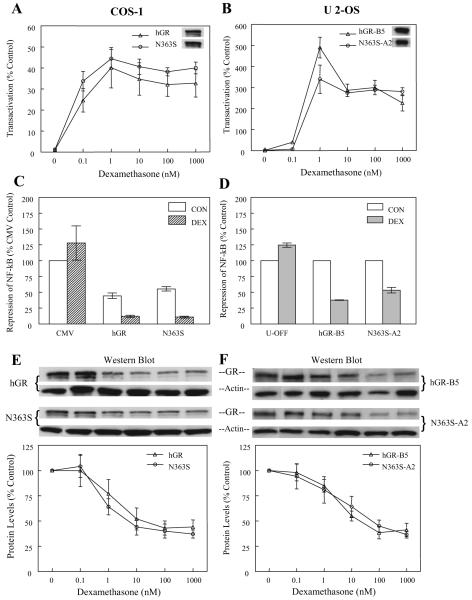
Both the transiently transfected COS-1 cells and the stably expressing U-2 OS cell lines were then assayed for their ability to activate gene expression. The cell lines were transfected with the glucocorticoid-responsive promoter GRE2-TATA-Luc and treated with increasing concentrations of dexamethasone (0-1000 nm) for 24 h. Figure 1, A and B, shows that both hGR and N363S activated this transient reporter gene to similar levels compared with the 0 nm dexamethasone (control) for each type of receptor in both the transiently expressing COS-1 cells and the stably expressing U-2 OS, although the activity levels of the transiently expressed GRE2-TATA-Luc were 10-fold higher in the U-2 OS stable cell lines (Fig. 1B). The U-2 OS cell lines also showed maximal activity at 1 nm dexamethasone. This appears to be cell type specific or could reflect down-regulation of the GR protein at the higher concentrations. Western blots of the hGR and N363S protein extracts probed with the anti-GR antibody (no. 57) (24) are shown in the insets and demonstrate that the expression levels of wild-type hGR and the N363S polymorphism were comparable, suggesting that both transiently and stably expressed wild-type and N363S GR function similarly.
Fig. 1.
Functional analysis of N363S. A, COS-1 cells were transfected with either wild-type hGR or N363S and the glucocorticoid-responsive promoter GRE2-TATA-Luc. Eighteen hours after transfection, the cells were treated with concentrations of dexamethasone ranging from 0-1000 nm for 24 h. Data are presented as percent activity over the 0 nm dexamethasone (vehicle) control (n = 3). The inset shows a representative Western blot of wild-type hGR and the variant probed with anti-GR antibody (no. 57). B, U-2 OS stable cell lines hGR-B5 and N363S-A2 were transfected with the glucocorticoid-responsive promoter GRE2-TATA-Luc and treated with dexamethasone as for COS-1 (n = 3). C, COS-1 cells were transfected with the NF-κB-responsive promoter, 3XMHCLuc, the NF-κB subunit p65, and wild-type hGR, N363S, or empty vector (CMV). Eighteen hours after transfection, the cells were treated with 100 nm dexamethasone or vehicle for 24 h. Data are presented as 100% of the vehicle-treated control (p65 with the empty CMV vector) (n = 3). D, Repression of NF-κB was assayed in the stable cell lines by transfecting the parental cell line U-OFF or hGR-B5 or N363S-A2 with the NF-κB-responsive promoter 3XMHCLuc and the NF-κB subunit p65 and then treated as for COS-1. Here the hormone-mediated repression of p65 is compared with the vehicle controls set at 100% (n = 3). E, Down-regulation of the GR protein was assessed by transfecting COS-1 cells with either wild-type hGR or N363S. After treatment with increasing concentrations of dexamethasone ranging from 0-1000 nm for 24 h, proteins were isolated, and protein levels were assessed by Western blotting with anti-GR antibody (no. 57) and anti-β-actin. The Western blot pictured is one blot representative of three. To generate the graph, protein levels were normalized to actin and the 0 nm dexamethasone (vehicle) controls set at 100% (n = 3). F, Down-regulation of the hGR-B5 and N363S-A2 stably expressed proteins was assayed by Western blot after treatment of the cells with increasing concentrations of dexamethasone (0-1000 nm) for 24 h. Protein extracts were prepared, and Western blotting and data analysis were performed as for COS-1 (n = 3).
The ability of the GR to repress the action of the proin-flammatory cytokine, signal transduction mediator nuclear factor-κB (NF-κB) is well known (26, 27). To determine whether the N363S polymorphism would have the same repressive effect as wild-type hGR in this model system, COS-1 cells were transfected with the NF-κB-responsive promoter 3XMHCLuc, the NF-κB subunit p65, and hGR, N363S, or empty vector (CMV) (Fig. 1C). The U-2 OS stable cell lines, including the parental cell line U-OFF, were transfected with 3XMHCLuc and the NF-κB subunit, p65 (Fig. 1D). For the COS-1 transiently transfected cells, cells transfected with 3XMHCLuc, empty vector (CMV) and the NF-κB subunit p65 and treated with vehicle served as the control. The U-2 OS GR stable cell line control is represented by the U-OFF cells transfected with 3XMHCLuc and p65 and treated with 100 nm dexamethasone. Figure 1C demonstrates that there is no significant difference in the repressive effect of hGR or N363S on p65 NF-κB when this evaluation is made in cells harboring transiently expressed GR. In addition, neither the transiently expressing COS-1 cells (Fig. 1C) nor the stably expressing U-2 OS cells (Fig. 1D) show a significant difference in dexamethasone-mediated transrepression between hGR and N363S.
Clinical studies of patients carrying the N363S SNP revealed that carriers have a higher sensitivity to glucocorticoids (6). Thus, it is possible that this increased sensitivity to glucocorticoids may result in a greater level of N363S GR homologous down-regulation. To determine whether there was any difference in glucocorticoid-mediated down-regulation of the wild-type or N363S expressed proteins, both hGR and N363S transiently expressing COS-1 cells and stably expressing U-2 OS cells were treated with increasing amounts of dexamethasone (0-1000 nm) for 24 h. Figure 1, E and F, shows a representative Western blot of hGR and N363S proteins that have been probed with the anti-GR antibody and anti-β-actin as well as a graphic representation of the data from at least three experiments with the protein levels normalized to β-actin. The 0-nm dexamethasone treatments served as the controls set at 100%. The data were generated by densitometrically quantitating the GR signal for wild-type and N363S GR and using the NIH Image software program for analysis of this signal. Together, these studies demonstrate that there is no difference in homologous down-regulation of the wild-type hGR or N363S when this physiological process is examined in either transiently or stably expressing cells.
Gene regulation analysis of U-2 OS cells stably expressing hGR and N363S
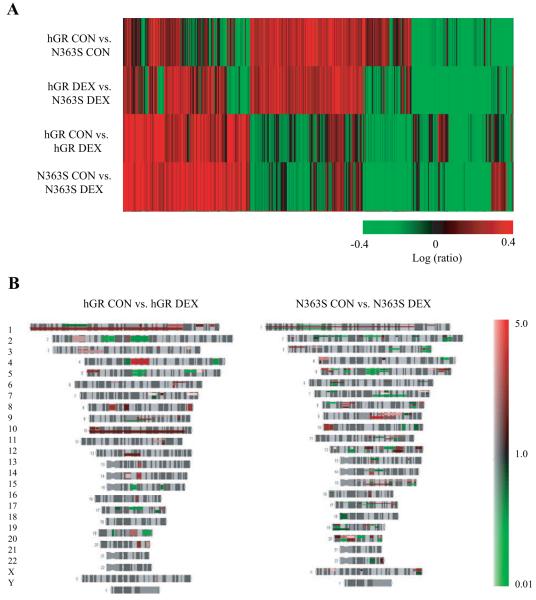
Because assays using synthetic reporter genes comprised of consensus DNA elements revealed no differences in the ability of hGR and N363S to transactivate or transrepress expression of well characterized synthetic genes, nor did they reveal an altered sensitivity to glucocorticoids, we used whole-genome microarray analysis to directly compare genes induced or repressed between hGR and N363S both in the presence and absence of dexamethasone. The U-2 OS stably expressing cell lines, hGR-B5 and N363S-A2, were cultured for 24 h in charcoal-stripped serum media and then treated with 10 nm dexamethasone or vehicle for 6 h, and total RNA was isolated and subjected to microarray analysis. Each experimental hybridization consisted of four comparison groups and was run in duplicate: hGR CON (vehicle-treated control) vs. N363S CON, hGR DEX (treated with 10 nm dexamethasone) vs. N363S DEX, hGR CON vs. hGR DEX, and N363S CON vs. N363S DEX. The gene lists generated from two distinct, independent experiments (biological replicates) were analyzed by selecting genes that were differentially regulated at P < 0.001 and common between two of two biological replicates. These common gene lists are represented in Figs. 2 and 3. Figure 2A shows the cluster analysis of these common genes. Those genes shown in green are significantly repressed and those in red are significantly induced. The mean range represented is 2.5-fold induced to 2.5-fold repressed. This analysis revealed that when N363S is compared with wild-type (hGR CON vs. N363S CON and hGR DEX vs. N363S DEX), we see a striking change in the pattern of gene regulation reflecting that expression of the N363S polymorphism is regulating a novel set of genes even in the absence of added glucocorticoid. Furthermore, treatment with dexamethasone affects this gene regulation and reveals additional differences between genes regulated by wild-type hGR- and N363S-expressing cells in the presence of dexamethasone (hGR CON vs. hGR DEX and N363S CON vs. N363S DEX). Additional analysis of these data using human chromosome mapping and comparing genes affected by the glucocorticoid-treated wild-type-expressing cells (hGR CON vs. hGR DEX) and the treated N363S-expressing cells (N363S CON vs. DEX) shows the physical position of the genes with known loci (Fig. 2B). This figure illustrates that there are differences in the genes affected by the expression of these receptors across the entire human genome. The structure of each chromosome is depicted in gray, up-regulated genes are red, and down-regulated genes are green. The color bar on the right shows the expression level of these genes ranging from 5.0 (highly up-regulated) to 0.1 (highly down-regulated). Differences in the genes affected by these two different GR-expressing cell lines are apparent on every chromosome.
Fig. 2.
Microarray analysis of N363S. hGR-B5 and N363S-A2 cells were treated with 10 nm dexamethasone or vehicle (control) for 6 h. Total RNA was isolated and submitted for microarray analysis. Each experimental hybridization consisted of four comparison groups: hGR CON (vehicle control) vs. N363S CON; hGR DEX (treated with 10 nm dexamethasone) vs. N363S DEX; hGR CON vs. hGR DEX; and N363S CON vs. N363SDEX. The data generated were analyzed by selecting genes that were differentially regulated at P < 0.001 and common between two independent experiments. A, Cluster analysis of these common genes. Those in green are down-regulated, and those in red are up-regulated. B, Chromosome mapping.
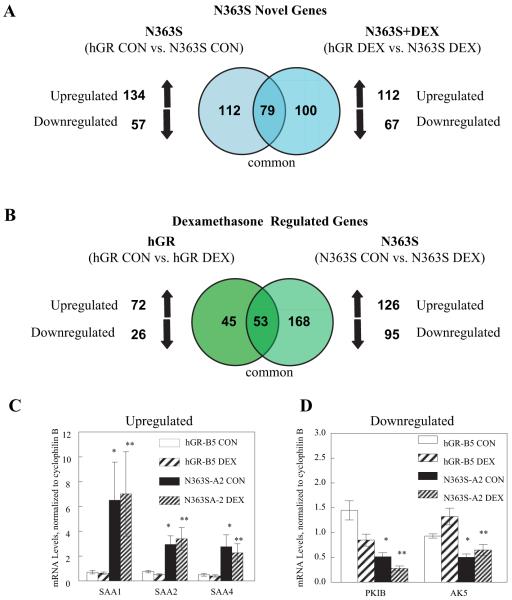
Fig. 3.
Venn diagram analysis and quantitative RT-PCR confirmation of N363S-regulated genes. A, Common gene lists generated from the microarray analysis were compared using Venn diagrams. Genes regulated by N363S over wild-type hGR (hGR CON vs. N363S CON) (left-hand circle) are compared with those treated with 10 nm dexamethasone (hGR DEX vs. N363S DEX) (right-hand circle). The overlapping circle represents genes that are common to N363S and N363S and dexamethasone. B, This Venn diagram compares the effects of dexamethasone on genes regulated by the expression of wild-type hGR vs. N363S. The left-hand circle shows wild-type hGR-expressing cells treated with 10 nm dexamethasone compared with the vehicle-treated (control) cells (hGR CON vs. hGR DEX). The right-hand circle displays genes regulated by N363S-expressing cells treated with 10 nm dexamethasone compared with vehicle-treated (N363S CON vs. N363S DEX). The overlapping circle represents genes common to both the wild-type and N363S-treated cells. C and D, Total RNA was isolated from either wild-type hGR-B5 or N363S-A2 stably expressing cell lines treated with 10 nm dexamethasone or vehicle for 6 h. Quantitative RT-PCR was performed using the 7900HT sequence detection system predesigned primer/probe sets available from Applied Bio-systems. Each primer/probe set was analyzed in duplicate or triplicate and with three different sets of RNAs. mRNA levels were normalized to the endogenous gene cyclophilin B and averaged with sem. Independent t tests were performed comparing N363S-A2 CON to hGR-B5 CON (*) and N363S-A2 DEX to hGR-B5 DEX (**). C, Verification of N363S highly up-regulated genes, which included the SAA gene family members SAA1 (*, P < 0.09; **, P < 0.09), SAA2 (*, P < 0.009; **, P < 0.009), and SAA4 (*, P < 0.03; **, P < 0.03). D, Verification of two of the N363S down-regulated genes including PKIB (*, P < 0.001; **, P < 0.001) and AK5 (*, P < 0.001; **, P < 0.01).
This diversity in the ability of the N363S to selectively signal is further illustrated in the Venn diagrams. Figure 3A reveals that expression of N363S (left-hand circle, hGR CON vs. N363S CON) significantly regulated a total of 191 novel genes compared with wild-type hGR (the comparison control). Of these genes, 134 were up-regulated and 57 were down-regulated. When treated with 10 nm dexamethasone and compared with wild-type hGR treated with dexamethasone (right-hand circle, hGR DEX vs. N363S DEX), 179 genes were detected with 112 up-regulated and 67 down-regulated. Of these, 179 genes, 100 were novel to the expression of N363S in the presence of dexamethasone. The other 79 genes, common between the overlapping circles, represent those genes that were regulated by N363S but were not further regulated by treatment with dexamethasone. Figure 3B displays a Venn diagram comparing the effects of dexamethasone on genes regulated by the expression of wild-type hGR vs. N363S. On the left, wild-type hGR-expressing cells regulated 98 genes when treated with 10 nm dexamethasone (hGR CON vs. hGR DEX). Of these 98 genes, 72 genes were up-regulated (73%) and 26 genes were down-regulated (27%). On the right, we see that N363S-expressing cells treated with dexamethasone (N363S CON vs. N363S DEX) regulated a total of 221 genes with 126 genes up-regulated (57%) and 95 down-regulated (43%). Fifty-three genes were commonly regulated by dexamethasone in both the hGR-expressing cells and the N363S-expressing cells. Additionally, it is interesting to note that DEX treatment of N363S-expressing cells caused down-regulation of more genes than wild-type hGR (43 vs. 27%), whereas the majority of genes in the dexamethasone-treated wild-type-expressing cells were up-regulated (73 vs. 57%). Thus, our microarray analysis revealed 1) that N363S could regulate a novel set of genes when compared with hGR, 2) that an additional 100 novel genes were regulated in the N363S cells after treatment with dexamethasone, and 3) that dexamethasone treatment commonly regulated a number of genes in both hGR- and N363S-expressing cells. All of the genes regulated by both the wild-type hGR and N363S polymorphism have been deposited in NCBI’s Gene Expression Omnibus (GEO, accession no. GSE5796).
We next verified several genes from the microarray data that were highly up-regulated or highly down-regulated in the N363S-expressing cells compared with wild-type hGR-expressing cells and confirmed these genes using quantitative RT-PCR analysis (Fig. 3, C and D). Interestingly, the serum amyloid A (SAA) family of proteins, which are increased in plasma concentration during acute inflammatory reactions and involved in cholesterol metabolism (28) and includes SAA1, SAA2, and SAA4, were among some of the genes highly up-regulated by the N363S polymorphism but were unaffected by dexamethasone. This result was confirmed by quantitative RT-PCR. Figure 3C shows that the mRNA levels of SAA1, SAA2, and SAA4 were significantly up-regulated by expression of N363S both in the absence and presence of hormone (N363S-A2 CON and N363S-A2 DEX), but neither wild-type hGR nor N363S was further regulated by the addition of dexamethasone. One of the genes most highly down-regulated by N363S was protein kinase inhibitor B (PKIB), which is a member of the human cAMP-dependent protein kinase inhibitor gene family. Another highly down-regulated gene was adenylate kinase 5 (AK5), which plays an important role in the synthesis of adenine nucleotides that are required for cellular metabolism. This down-regulation of gene expression was confirmed by RT-PCR. Figure 3D demonstrates that both PKIB and AK5 were significantly down-regulated by N363S both in the absence and presence of hormone (N363S-A2 CON and N363S-A2 DEX) and, furthermore, that dexamethasone down-regulated PKIB in both wild-type and N363S-expressing cells. However, AK5 was not significantly regulated by DEX in either wild-type or N363S-expressing cells.
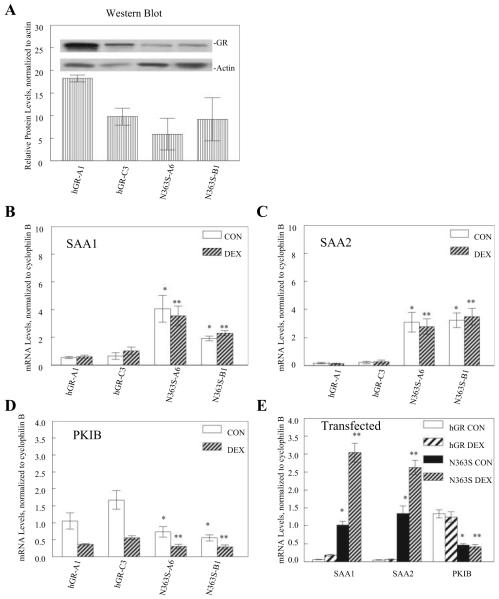
To further determine whether the N363S effect on gene regulation was specific to the expression of N363S, two other cell lines of either wild-type hGR (hGR-A1 and hGR-C3) or N363S (N363S-A6 and N363S-B1) were assayed for their affects on SAA1, SAA2, and PKIB using quantitative RT-PCR. We first analyzed the receptor protein expression levels of each cell line by Western blot. Figure 4A graphically depicts the relative protein levels of each GR-expressing cell line when normalized to actin (Fig. 4A, inset). Total RNA was then prepared from each cell line treated with 10 nm dexamethasone or vehicle for 6 h, and gene regulation was assayed using quantitative RT-PCR. Figure 4, B-D, shows the effect of N363S expression on the regulation of SAA1 (Fig. 4B), SAA2 (Fig. 4C), and PKIB (Fig. 4D). Despite the differences in expression level of each of the cell lines, SAA1 and SAA2 were significantly up-regulated and PKIB was significantly down-regulated by the expression of N363S (N363S-A6 and N363S-B1 compared with hGR-A1 and hGR-C3). This consistent pattern of gene regulation occurred despite the fact that these different cell lines contain diverse levels of hGR (Fig. 4A).
Fig. 4.
Quantitative RT-PCR analysis of other U-2 OS cell lines stably or transiently expressing wild-type hGR or N363S. A, GR expression levels in the stable cell lines hGR-A1, hGR-C3, N363S-A6, and N363S-B1 were assessed by Western blotting protein extracts with anti-GR antibody (no. 57) and anti-β-actin and is representative of two (inset). The protein levels were quantitated using the NIH Image program, and GR protein levels were normalized to those of β-actin. The graph depicts the relative protein levels of each cell line (n = 2). B-D, Total RNA was isolated from cell lines treated with 10 nm dexamethasone (DEX) or vehicle (CON), and quantitative RT-PCR was performed as described. mRNA levels were normalized to the endogenous gene cyclophilin B and averaged with sem. Independent t tests were performed comparing N363S-A6 CON and N363S-B1 CON to hGR-A1 CON and hGR-C3 CON (*) and N363S-A6 DEX and N363S-B1 DEX to hGR-A1 DEX and hGR-C3 DEX (**). B and C, Verification of up-regulation of SAA1 (*, P < 0.001; **, P < 0.005) (B) and SAA2 (*, P < 0.005; **, P < 0.001) (C); D, down-regulation of PKIB (*, P < 0.01; **, P < 0.01 compared with hGR-C3) by N363S stably expressing cell lines. E, U-2 OS cells were transiently transfected with either wild-type hGR or N363S and treated with 10 nm dexamethasone or vehicle for 6 h. Total RNA was isolated, and quantitative RT-PCR and analyses were performed as described above: SAA1 (*, P < 0.0001; **, P < 0.0001), SAA2 (*, P < 0.0001; **, P < 0.0001), and PKIB (*, P < 0.001; **, P < 0.005).
Finally, we wished to determine whether genes identified by the microarray and confirmed in all of our stable cell lines could be studied in the context of a heterologous system using transfected receptors. Thus, we transiently transfected U-2 OS cells with either wild-type hGR or N363S, treated the cells with 10 nm dexamethasone or vehicle for 6 h, and isolated total RNA. The samples were then assayed for gene regulation using quantitative RT-PCR. Figure 4E clearly shows that even the transient expression of N363S can affect gene regulation of SAA1, SAA2, and PKIB. Interestingly, in the transiently transfected U-2 OS cells, there was a further up-regulation of SAA1 and SAA2 in the presence of dexamethasone that was not seen in the stably expressing cell lines. In addition, dexamethasone had no further effect on the down-regulation of PKIB in either the wild-type or N363S transiently expressing cells compared with those stably expressing GR. This difference in dexamethasone-mediated regulation in the stable vs. transiently transfected cells is most likely due to differences in the high expression level of the GR that is well known to occur during transient transfection.
These data confirm that the expression of N363S GR regulates a novel set of genes in comparison with wild-type hGR. Thus, when one uses the correct endogenous genes selectively regulated by the N363S polymorphism, one can recapitulate data from the stable cell lines in a transient system. This discovery will allow us to elucidate the molecular basis for this polymorphism’s altered gene expression.
Discussion
In this study, we have analyzed the differences between wild-type hGR and the hGR SNP N363S that has been associated with higher glucocorticoid sensitivity, increased insulin response to dexamethasone, CAD, and obesity, specifically, with an increase in BMI (6, 8, 10-12). Under our experimental conditions, the data presented here revealed no major differences between wild-type hGR and the N363S polymorphism when transactivation and transrepression were assayed in cells transiently transfected with synthetic reporter genes comprised of consensus hormone or transcription factor elements (Figs. 1, A and C). These results support most of the previous reports (5, 6, 20, 21), although recently Russcher et al. (22) demonstrated that N363S had a small but significant increase in transactivating capacity on a GRE-LUC reporter. In addition to the transactivation and transrepression assays, we analyzed each GR for its ability to undergo hormone-mediated down-regulation in a dose-dependent manner, and again, we found no difference between the N363S polymorphism and wild-type hGR (Fig. 1E). Because transient transfection assays can be variable from one experiment to the next, they may not reveal subtle differences when comparing the expression and function of two different proteins when one uses synthetic reporter elements. Therefore, we created stable cell lines overexpressing either hGR or N363S to eliminate some of this potential variability.
Lu and Cidlowski (23) in our laboratory analyzed the differences between the wild-type human GR and its isoforms using stable cell lines expressing either wild-type hGR or each isoform. Using this same expression system, we produced stable cell lines expressing either wild-type hGR or the N363S SNP. Even though creation of the stable cell lines gave us cells expressing equal amounts of hGR and N363S proteins (Fig. 1B), thus reducing some of the variability associated with transient transfection assays, the functional assays with the stable cell lines revealed little or no difference between wild-type hGR and N363S (Fig. 1, B, D, and F). These data also argue against the 363 asparagine to serine change becoming an additional site for phosphorylation of the GR protein because our laboratory has previously shown that the greatest differences in transactivation of the mGR phosphorylation site mutants was seen using the simple GRE2-TATA promoter (29). In addition, the amino acid sequences surrounding the serine site do not correspond to the published consensus sequence (30).
Therefore, we used gene array analysis to determine whether there was differential regulation between the two cell lines in the absence and presence of dexamethasone. Although we cannot rule out that residual glucocorticoids were still present in the cells at the time of our analysis, our microarray results show, for the first time, that the N363S polymorphism can regulate a novel set of genes in comparison to wild-type hGR both in the absence and presence of dexamethasone (Figs. 2 and 3). These data were confirmed using several populations of cells expressing different levels of receptor (Fig. 4). Together these data suggest that the N363S phenotype may be the result of differential effects on gene regulation. Interestingly, the effect of the polymorphism, in the absence of dexamethasone, could be recapitulated in a transient transfection system which will now allow us to dissect the molecular basis for altered gene expression.
One interesting family of genes involved in the response to external stimulus (which includes immune response) is the SAAs. These acute-phase proteins, precursors of amyloid A (AA), which is involved in the pathogenesis of AA amyloidosis, are produced primarily in the liver and increase many-fold during inflammation. During the acute inflammatory response, the circulating levels of SAA rise dramatically within 24 h of an inflammatory stimulus (28). SAA along with C-reactive protein (CRP) are the major acute-phase protein responders and are regulated by cytokines such as IL-1, IL-6, and TNF, although how they are regulated is not precisely known (28). Because SAA increases where tissue damage has occurred with subsequent inflammation, patients who have suffered myocardial infarctions also have elevated SAA levels (28). This has led to SAA being investigated as a prognostic indicator, along with CRP, for acute myocardial infarction (31). Interestingly, the SAAs were among the genes most highly up-regulated in the N363S-expressing cells. However, it remains unclear whether the increased SAA levels in the U-2 OS cell lines expressing the N363S SNP can be associated with the increase in CAD in carriers of the SNP as reported by Lin et al. (10), although clearly, further study is now warranted.
Although CRP and IL-6 have been previously associated with parameters of obesity, only recently has SAA also been investigated in relation to body composition (32). In a large study in both men and women in the area of Augsburg, Germany, both total adiposity (fat mass, BMI) and abdominal adiposity (waist circumference and waist-to-hip ratio) were measured, and these measurements correlated with markers of systemic inflammation CRP, SAA, fibrinogen, and IL-6. Their study discovered that adiposity was strongly associated with these markers of systemic inflammation in both men and women and that this correlation, especially for CRP, was even stronger in women. Other studies have not only associated SAA and CRP with obesity but also found that weight loss significantly decreased the levels of SAA and CRP (33-35). Of these studies, Poitou et al. (34) found that SAA1, SAA2, and SAA4 transcripts were overexpressed in the sc white adipose tissue of obese women compared with the lean controls. In addition, Gomez-Ambrosi et al. (35) found high levels of SAA mRNA transcripts in the omental adipose tissue of morbidly obese patients undergoing gastric bypass surgery. In all three of these studies, weight loss whether by diet (33, 34) or gastric bypass (35) was associated with a significant decrease in SAA. Finally, O’Brien and Chait (36) suggest that SAA and CRP may be chronically elevated in individuals with conditions such as atherosclerosis, insulin resistance, diabetes, and obesity leading to an increased risk of cardiovascular disease. Again, although these findings are tantalizing, we cannot yet draw a direct correlation between the obesity phenotype observed in N363S carriers with an increase in SAA levels observed in our stably expressing U-2 OS cells.
There were many other genes that were differentially regulated by the N363S SNP compared with wild-type GR, with the majority of the genes involved in biological processes such as cell growth/maintenance and nucleic acid metabolism. However, the most notable aspect of our study is that the N363S SNP does regulate a novel set of genes in comparison with wild-type hGR. Additional studies will be needed to correlate specific genes with the N363S phenotype and provide a molecular understanding of the ability of the polymorphism to selectively regulate these genes.
Acknowledgments
We thank Jennifer Collins, Danica Ducharme, and Sherri Grissom of the National Institute of Environmental Health Sciences microarray facility for their generation and analysis of the microarray data.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
Abbreviations
- BMI
Body mass index
- CAD
coronary artery disease
- CRP
C-reactive protein
- FCS:CS
fetal calf serum:calf serum
- GR
glucocorticoid receptor
- hGR
human GR
- HPA
hypothalamic-pituitary-adrenal
- NF-κB
nuclear factor-κB
- SAA
serum amyloid A
- SNP
singlenucleotide polymorphism.
Footnotes
Disclosure Statement: C.M.J. and J.A.C. have nothing to disclose.
References
- 1.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 2.DeRijk RH, Schaaf M, de Kloet ER. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81:103–122. doi: 10.1016/s0960-0760(02)00062-6. [DOI] [PubMed] [Google Scholar]
- 3.Rosmond R, Dallman MF, Bjorntorp P. Stress-related cortisol secretion in men: relationships with abdominal obesity and endocrine, metabolic, and hemodynamic abnormalities. J Clin Endocrinol Metab. 1998;83:1853–1859. doi: 10.1210/jcem.83.6.4843. [DOI] [PubMed] [Google Scholar]
- 4.Huizenga NA, Koper JW, de Lange P, Pols HA, Stolk RP, Grobbee DE, de Jong FH, Lamberts SW. Interperson variability but intraperson stability of baseline plasma cortisol concentrations, and its relation to feedback sensitivity of the hypothalamo-pituitary-adrenal axis to a low dose of dexamethasone in elderly individuals. J Clin Endocrinol Metab. 1998;83:47–54. doi: 10.1210/jcem.83.1.4498. [DOI] [PubMed] [Google Scholar]
- 5.Karl M, Lamberts SW, Detera-Wadleigh SD, Encio IJ, Stratakis CA, Hurley DM, Accili D, Chrousos GP. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J Clin Endocrinol Metab. 1993;76:683–689. doi: 10.1210/jcem.76.3.8445027. [DOI] [PubMed] [Google Scholar]
- 6.Huizenga NA, Koper JW, De Lange P, Pols HA, Stolk RP, Burger H, Grobbee DE, Brinkmann AO, De Jong FH, Lamberts SW. A polymorphism in the glucocorticoid receptor gene may be associated with and increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab. 1998;83:144–151. doi: 10.1210/jcem.83.1.4490. [DOI] [PubMed] [Google Scholar]
- 7.Wust S, Van Rossum EF, Federenko IS, Koper JW, Kumsta R, Hellhammer DH. Common polymorphisms in the glucocorticoid receptor gene are associated with adrenocortical responses to psychosocial stress. J Clin Endocrinol Metab. 2004;89:565–573. doi: 10.1210/jc.2003-031148. [DOI] [PubMed] [Google Scholar]
- 8.Lin RC, Wang WY, Morris BJ. High penetrance, overweight, and glucocorticoid receptor variant: case-control study. BMJ. 1999;319:1337–1338. doi: 10.1136/bmj.319.7221.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin RC, Wang XL, Dalziel B, Caterson ID, Morris BJ. Association of obesity, but not diabetes or hypertension, with glucocorticoid receptor N363S variant. Obes Res. 2003;11:802–808. doi: 10.1038/oby.2003.111. [DOI] [PubMed] [Google Scholar]
- 10.Lin RC, Wang XL, Morris BJ. Association of coronary artery disease with glucocorticoid receptor N363S variant. Hypertension. 2003;41:404–407. doi: 10.1161/01.HYP.0000055342.40301.DC. [DOI] [PubMed] [Google Scholar]
- 11.Di Blasio AM, van Rossum EF, Maestrini S, Berselli ME, Tagliaferri M, Podesta F, Koper JW, Liuzzi A, Lamberts SW. The relation between two polymorphisms in the glucocorticoid receptor gene and body mass index, blood pressure and cholesterol in obese patients. Clin Endocrinol (Oxf) 2003;59:68–74. doi: 10.1046/j.1365-2265.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 12.Roussel R, Reis AF, Dubois-Laforgue D, Bellanne-Chantelot C, Timsit J, Velho G. The N363S polymorphism in the glucocorticoid receptor gene is associated with overweight in subjects with type 2 diabetes mellitus. Clin Endocrinol (Oxf) 2003;59:237–241. doi: 10.1046/j.1365-2265.2003.01831.x. [DOI] [PubMed] [Google Scholar]
- 13.Dobson MG, Redfern CP, Unwin N, Weaver JU. The N363S polymorphism of the glucocorticoid receptor: potential contribution to central obesity in men and lack of association with other risk factors for coronary heart disease and diabetes mellitus. J Clin Endocrinol Metab. 2001;86:2270–2274. doi: 10.1210/jcem.86.5.7465. [DOI] [PubMed] [Google Scholar]
- 14.Halsall DJ, Luan J, Hales N, Wareham NJ, O’Rahilly S. Glucocorticoid receptor variant and body mass index. Rapid Response to BMJ. 2000;7221:1337–1338. [Google Scholar]
- 15.Echwald SM, Sorensen TI, Andersen T, Pedersen O. The Asn363Ser variant of the glucocorticoid receptor gene is not associated with obesity or weight gain in Danish men. Int J Obes Relat Metab Disord. 2001;25:1563–1565. doi: 10.1038/sj.ijo.0801744. [DOI] [PubMed] [Google Scholar]
- 16.Rosmond R. Association studies of genetic polymorphisms in central obesity: a critical review. Int J Obes Relat Metab Disord. 2003;27:1141–1151. doi: 10.1038/sj.ijo.0802397. [DOI] [PubMed] [Google Scholar]
- 17.Buemann B, Black E, Holst C, Toubro S, Echwald S, Pedersen O, Astrup A, Sorensen T. The N363S polymorphism of the glucocorticoid receptor and metabolic syndrome factors in men. Obes Res. 2005;13:862–867. doi: 10.1038/oby.2005.99. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda Y, Suehiro T, Tsuzura S, Shiinoki T, Kaneda T, Kumon Y, Hashimoto K. A polymorphism in the promoter region of the glucocorticoid receptor gene is associated with its transcriptional activity. Endocr J. 2001;48:723–726. doi: 10.1507/endocrj.48.723. [DOI] [PubMed] [Google Scholar]
- 19.Syed AA, Irving JA, Redfern CP, Hall AG, Unwin NC, White M, Bhopal RS, Alberti KG, Weaver JU. Low prevalence of the N363S polymorphism of the glucocorticoid receptor in South Asians living in the United Kingdom. J Clin Endocrinol Metab. 2004;89:232–235. doi: 10.1210/jc.2003-030995. [DOI] [PubMed] [Google Scholar]
- 20.de Lange P, Koper JW, Huizenga NA, Brinkmann AO, de Jong FH, Karl M, Chrousos GP, Lamberts SW. Differential hormone-dependent transcriptional activation and -repression by naturally occurring human glucocorticoid receptor variants. Mol Endocrinol. 1997;11:1156–1164. doi: 10.1210/mend.11.8.9949. [DOI] [PubMed] [Google Scholar]
- 21.Gaitan D, DeBold CR, Turney MK, Zhou P, Orth DN, Kovacs WJ. Glucocorticoid receptor structure and function in an adrenocorticotropin-secreting small cell lung cancer. Mol Endocrinol. 1995;9:1193–1201. doi: 10.1210/mend.9.9.7491111. [DOI] [PubMed] [Google Scholar]
- 22.Russcher H, Smit P, van den Akker EL, van Rossum EF, Brinkmann AO, de Jong FH, Lamberts SW, Koper JW. Two polymorphisms in the glucocorticoid receptor gene directly affect glucocorticoid-regulated gene expression. J Clin Endocrinol Metab. 2005;90:5804–5810. doi: 10.1210/jc.2005-0646. [DOI] [PubMed] [Google Scholar]
- 23.Lu NZ, Cidlowski JA. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol Cell. 2005;18:331–342. doi: 10.1016/j.molcel.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Cidlowski JA, Bellingham DL, Powell-Oliver FE, Lubahn DB, Sar M. Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Mol Endocrinol. 1990;4:1427–1437. doi: 10.1210/mend-4-10-1427. [DOI] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol Cell Biol. 1995;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKay LI, Cidlowski JA. Cross-talk between nuclear factor-κB and the steroid hormone receptors: mechanisms of mutual antagonism. Mol Endocrinol. 1998;12:45–56. doi: 10.1210/mend.12.1.0044. [DOI] [PubMed] [Google Scholar]
- 28.Kisilevsky R, Tam SP. Acute phase serum amyloid A, cholesterol metabolism, and cardiovascular disease. Pediatr Pathol Mol Med. 2002;21:291–305. doi: 10.1080/02770930290056523. [DOI] [PubMed] [Google Scholar]
- 29.Webster JC, Jewell CM, Bodwell JE, Munck A, Sar M, Cidlowski JA. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J Biol Chem. 1997;272:9287–9293. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]
- 30.Ismaili N, Garabedian MJ. Modulation of glucocorticoid receptor function via phosphorylation. Ann NY Acad Sci. 2004;1024:86–101. doi: 10.1196/annals.1321.007. [DOI] [PubMed] [Google Scholar]
- 31.Katayama T, Nakashima H, Takagi C, Honda Y, Suzuki S, Iwasaki Y, Yano K. Prognostic value of serum amyloid A protein in patients with acute myocardial infarction. Circ J. 2005;69:1186–1191. doi: 10.1253/circj.69.1186. [DOI] [PubMed] [Google Scholar]
- 32.Thorand B, Baumert J, Doring A, Herder C, Kolb H, Rathmann W, Giani G, Koenig W. Sex differences in the relation of body composition to markers of inflammation. Atherosclerosis. 2006;184:216–224. doi: 10.1016/j.atherosclerosis.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien KD, Brehm BJ, Seeley RJ, Bean J, Wener MH, Daniels S, D’Alessio DA. Diet-induced weight loss is associated with decreases in plasma serum amyloid a and C-reactive protein independent of dietary macronutrient composition in obese subjects. J Clin Endocrinol Metab. 2005;90:2244–2249. doi: 10.1210/jc.2004-1011. [DOI] [PubMed] [Google Scholar]
- 34.Poitou C, Viguerie N, Cancello R, De Matteis R, Cinti S, Stich V, Coussieu C, Gauthier E, Courtine M, Zucker JD, Barsh GS, Saris W, Bruneval P, Basdevant A, Langin D, Clement K. Serum amyloid A: production by human white adipocyte and regulation by obesity and nutrition. Diabetologia. 2005;48:519–528. doi: 10.1007/s00125-004-1654-6. [DOI] [PubMed] [Google Scholar]
- 35.Gomez-Ambrosi J, Salvador J, Rotellar F, Silva C, Catalan V, Rodriguez A, Jesus Gil M, Fruhbeck G. Increased serum amyloid A concentrations in morbid obesity decrease after gastric bypass. Obes Surg. 2006;16:262–269. doi: 10.1381/096089206776116525. [DOI] [PubMed] [Google Scholar]
- 36.O’Brien KD, Chait A. Serum amyloid A: the “other” inflammatory protein. Curr Atheroscler Rep. 2006;8:62–68. doi: 10.1007/s11883-006-0066-0. [DOI] [PubMed] [Google Scholar]