Abstract
Vasa is a DEAD-box RNA helicase that functions in translational regulation of specific mRNAs. In many animals it is essential for germ line development and may have a more general stem cell role. Here we identify vasa in two sea urchin species and analyze the regulation of its expression. We find that vasa protein accumulates in only a subset of cells containing vasa mRNA. In contrast to vasa mRNA, which is present uniformly throughout all cells of the early embryo, vasa protein accumulates selectively in the 16 cell stage micromeres, and then is restricted to the small micromeres through gastrulation to larval development. Manipulating early embryonic fate specification by blastomere separations, exposure to lithium, and dominant-negative cadherin each suggest that, although vasa-protein accumulation in the small micromeres is fixed, accumulation in other cells of the embryo is inducible. Indeed, we find that embryos in which micromeres are removed, respond by significant up-regulation of vasa protein translation, followed by spatial restriction of the protein late in gastrulation. Overall, these results support the contention that sea urchins do not have obligate primordial germ cells determined in early development, that vasa may function in an early stem cell population of the embryo, and that vasa-expression in this embryo is restricted early by translational regulation to the small micromere lineage.
Keywords: Vasa, sea urchin, small micromeres
Introduction
Vasa is an ATP-dependent DEAD box helicase, similar to the eukaryotic initiation factor 4A (eIF4A). Vasa unwinds double-stranded RNA, though in vivo, DEAD-box proteins appear to unwind only local RNA-RNA interactions of a few base pairs or to dissociate proteins from the RNA, which in turn allows other interactions to occur (Linder and Lasko, 2006). The exact mechanism of function is not known although the recent crystal structure of Drosophila vasa suggests that the winding activity is actually the result of contortion of the dsRNA by bending (Sengoku et al., 2006). Extensive work in Drosophila indicates that vasa acts as a translational regulator of two mRNAs that are localized in the oocyte: gurken (grk), which directs both anterior/posterior and dorsal/ventral polarity, and oskar (osk), which directs germ plasm assembly at the posterior of the oocyte (Mahowald, 2001; Riechmann and Ephrussi, 2001). Both grk and osk mRNAs are translationally repressed by the well-conserved RNA-binding protein bruno until they reach their destination in the oocyte resulting in localized protein accumulation (Castagnetti et al., 2000; Kim-Ha et al., 1995; Webster et al., 1997; Yan and Macdonald, 2004). Bruno binds the BRE (bruno response element) in the 3′UTR of the osk message and recruits the protein Cup which then binds the cap-binding protein eIF4E (eukaryotic initiation factor 4E; Nakamura et al., 2004). With Cup bound to eIF4E, the translational machinery cannot be recruited and translation is blocked. Vasa appears to function in this process by relieving the bruno/cup repression of gurken and oskar mRNA (Johnstone and Lasko, 2004). Mutations in the helicase domain of vasa cause a lack of gurken and oskar translation, and result in germ line defects and female sterility in Drosophila (Lasko and Ashburner, 1988; Styhler et al., 1998). Vasa mutations in mice result in male sterility, largely due to defective primordial germ cell proliferation and differentiation (Tanaka et al., 2000). Although the regulation of expression and the molecular function of vasa is not understood in most animals, given the wide conservation of vasa, bruno, and the translational machinery, it is likely that vasa’s role as a translational activator is conserved.
In most of the deuterostomes studied, including fish, frogs, and Ascidians, vasa mRNA is localized to discreet regions of oocytes and/or early embryos. For example, in the ascidian Ciona intestinalis, for which vasa is a stringent marker of the germ line, vasa mRNA is present in eggs and by the 2nd cleavage division it is concentrated at the posterior region of the embryo (Fujimura and Takamura, 2000). Vasa mRNA further concentrates to the posterior region of the B3 blastomeres at the 4-cell stage, and is subsequently inherited by the posterior-most blastomeres, B4.1, at the 8-cell stage, B5.2 at the 16-cell stage, B6.3 at the 64-cell stage, and B7.6 at the early gastrula stage. In Xenopus, the vasa-like genes DEADsouth (RNA) and Xvlg1 (protein) are localized to the granules of the germ plasm at the vegetal pole of oocytes, are segregated to the four vegetal cells during the initial cell divisions and accumulate specifically in primordial germ cells (PGCs) of the tadpole (Komiya et al., 1994; MacArthur et al., 2000). Zebrafish vasa RNA is also localized to germ plasm granules, but these granules are associated with the cortex of the animal pole of late-stage oocytes. During the first two cleavage divisions in zebrafish embryos, vasa RNA granules are concentrated to the distal parts of the cleavage furrows, resulting in four vasa RNA containing aggregates. These aggregates are asymmetrically segregated during every cell division such that only four PGCs are present in the late zebrafish blastula. Shortly before gastrulation, these four cells begin to divide symmetrically and start to migrate to the future site of the gonads (Knaut et al., 2002; Yoon et al., 1997). In each case, vasa serves as a specific marker for the birth, development, and migration of primordial germ cells.
Mice and sea urchins, however, have no detectable localized germ line determinants in oocytes or early embryos, either functionally (e.g. Ransick et al., 1996; Tam and Zhou, 1996) or by molecular markers (e.g. Hayashi et al., 2007; Juliano et al., 2006). In mice, removal of the proximal epiblast region, the source of precursors for the primordial germ cells, results in a re-specification of the germ cells and normally gravid adults (Hayashi et al., 2007; Tam and Zhou, 1996). The mRNA of the mouse vasa homolog is present in oocytes and early embryos, but is then not detectable until the primordial germ cells begin to migrate into the genital ridge at 10.5 days post-coitum (d.p.c.) (Toyooka et al., 2000). Thereafter it remains specific for the primordial germ cells in this animal. Similarly, removal of the cells thought to contribute to the germ line in sea urchins did not prevent fertility of the resulting animals (Ransick et al., 1996). The small micromeres were hypothesized to contribute to the primordial germ cells in sea urchins because they had characteristically enlarged nuclei, slow cell divisions (Pehrson and Cohen, 1986; Tanaka and Dan, 1990), and specifically accumulated mRNA for nanos, vasa and piwi (Fujii et al., 2006; Juliano et al., 2006). That embryos lacking small micromeres still developed germ cells argues that the small micromeres could not be the obligatory primordial germ cell line in this animal, and/or that the primordial germ cells are not actually determined until later in development (Ransick et al., 1996).
Vasa may also function in a broader context of stem cells in development. For example, in polychaetes (annelid worms), vasa and its close family member PL10, are each present in a large population of mesodermal cells, only a fraction of which become primordial germ cells (Rebscher et al., 2007). The multipotent i-cells of hydrozoans and the neoblasts of flatworms similarly express germ plasm components along with stem cell markers (Mochizuki et al., 2001; Mochizuki et al., 2000; Shibata et al., 1999). Setting aside a population of undifferentiated multipotent cells, from which the PGCs are segregated later, may constitute an ancestral mechanism that has been lost in species exhibiting premature specification of the adult body plan by localized maternal determinants (Johnson et al., 2003). Further, Hayashi et al., (2007) have speculated a stem cell model for germ cell determination in mice in which only a few of the precursor cells actually become primordial germ cells, through progressive restriction, whereas other progeny diversify into somatic stem cell fates.
Here we examine the expression and regulation of vasa protein in the sea urchin, a basal deuterostome. We find that although all cells of the embryo contain vasa mRNA, protein expression is under specific cellular control, and that its translation is inducible throughout the embryo. The cells that normally accumulate vasa are the small micromeres, and results herein suggest that these cells may fit the two step (Rebscher et al., 2007), or progressive restriction mechanism (Hayashi et al., 2007) of stem cells that contribute to several adult cell types, including the germ line.
Materials and Methods
Reagents were purchased from Sigma (St. Louis, MO), unless otherwise noted.
Animals
Adult husbandry was managed as described (Voronina et al., 2003). Treatment of embryos with LiCl (30 mM) was performed as described previously (Logan et al., 1999), as was rearing of advanced larvae and juveniles (George et al., 2004). Microinjections of in-vitro transcribed mRNA were performed as described (Oliveri et al., 2003) and were done three times, in different batches of eggs. Micromere removal experiments were performed three times as described (Ransick et al., 1996), with different batches of embryos. Controls included untreated embryos, and embryos treated with dissociation medium but not surgically altered.
cDNA cloning
A partial S. purpuratus vasa sequence was found in the genome (http://sugp.caltech.edu/). Gene-specific primers (5′-GGTCGAGACAGGCCCAAAAATATAC – 3′and 5′-CACAGGTCGTATGGTGGTGC – 3′) were used to screen an S. purpuratus ovary cDNA library (described in Wessel et al., 1998) by PCR and the amplification product was cloned into pGEMT-Easy (Promega, Madison, WI). DNA sequencing was performed by the macromolecular sequencing facility at Brown University, using ABI 377 prism automated sequencers (Perkin-Elmer, Foster City, CA). This sequence was identified as vasa based on homology of the encoded protein to vasa in the region conserved specifically in vasa, as opposed to other DEAD-box helicases. Initial isolated fragment was used to design gene-specific primers for 5′ and 3′ RACE procedures. The consensus sequence of the resulting cDNAs is entered in the GenBank (accession numbers to be obtained) and the actual S. purpuratus vasa gene (http://annotation.hgsc.bcm.tmc.edu/Urchin/) is a modified version of SPU_08908. To isolate vasa-like sequences from L. variegates, we utilized degenerate PCR with primers specific for DEAD-box motifs using a gastrula cDNA library. The screen resulted in a single sequence containing vasa-specific DEAD box motifs, which was then extended to full open reading frame by 5′ and 3′ RACE protocol, from both gastrula and ovary cDNA.
Cell and Embryo Labeling
We designed and obtained several vasa-specific antibodies. To design peptide-specific vasa antigens, we utilized the S. purpuratus vasa sequence information. The vasa-specific motif within the DEAD-box region, KPTPVQKYGMPIISC, was synthesized, coupled to KLH, and used to generate polyclonal antibodies in two rabbits (Sigma Genosys; The Woodlands, TX). Additionally, a fragment representing the DEAD-box of S. purpuratus vasa (amino acids 300 to 583, submitted to Genbank) was used to produce a 6-histidine tagged protein fusion and the resulting antiserum was affinity-purified on a column containing the protein immunogen. Additional antibodies used in this study included an anti pan-insect vasa, called For2 (Chang et al., 2002), and an anti-zebrafish vasa antibody obtained from Dr. Zivkovic at the Hubrecht Laboratory, Netherlands Institute for Developmental Biology (Braat et al., 2000). Immunofluorescence localization was performed as described (Laidlaw and Wessel, 1994; Voronina et al., 2003).
Protein samples of S. purpuratus eggs and several embryonic stages were prepared, subjected to SDS-PAGE and immunoblot analysis as described (Voronina et al., 2003). For antibody competition experiments, the diluted anti-fragment antiserum was incubated for 1 hour with 100 μg/ml purified antigenic protein and then pelleted at 10,000 g for 15 minutes. The signal intensities on the resulting blots were quantified using Metamorph software (Universal Imaging Corporation, Downingtown, PA), and the values were normalized per intensity of duplicate Coomassie-stained gel or per intensity of a loading control, yolk protein YP30 band.
Whole-mount in situ RNA hybridizations were performed as previously described (Minokawa et al., 2004).
Results
Sea urchin vasa homologs
Full length vasa cDNA homologs were isolated from two sea urchin species, Strongylocentrotus purpuratus and Lytechinus vareigatus. These proteins are 762 and 796 amino acid residues, respectively, and were concluded to be vasa based on sequence similarity to known vasa proteins and the characteristic domain architecture (Fig. 1). These included: Zn-fingers, glycine-rich regions, and a C-terminal DEAD-box motif. Despite these conserved structures, the N-terminal halves of vasa orthologs have considerable amino acid divergence (Fig. 1). Computational searches failed to identify additional vasa isoforms in the S. purpuratus genome, however we did detect two vasa splice forms in each species. These isoforms differ in the N-terminus of the transcript, and produce protein isoforms with varying number of Zn finger domains (Fig. 1 and data not shown).
Figure 1.
Functional conservation of protein domains across vasa subfamily of DEAD box helicses. Pictured domain structures of the proteins were predicted by SMART (Letunic et al., 2006). Cloned sea urchin vasa homologs are identified with species name in red. Major splice variants are pictured; exons missing in minor splice variants are indicated with dotted lines. a – ecdysozoa, b – lophotrochozoa, RGG – glycine-rich region containing RGG repeats, ZnF – zinc finger motif, RRM – RNA-binding domain, DEXDc/HELICc – DEAD box RNA helicase domain.
Vasa mRNA analysis
As previously shown (Juliano et al., 2006), in situ RNA hybridization of vasa in S. purpuratus demonstrates that embryos developing through blastula stage have uniform vasa mRNA accumulation (Fig. 2A, B), which becomes restricted to the vegetal plate of mesenchyme blastulae (Fig. 2C), and then to a small population of about 6–8 cells at the tip of the archenteron in gastrulae (Fig. 2D). In the larvae, vasa mRNA is detected in a subset of cells initially in both coelomic pouches, and subsequently only in the left pouch (Fig. 2E). This is significant since the embryonic rudiment is largely derived from this same region (see e.g. Pearse and Cameron, 1991). Similar vasa mRNA accumulation was detected in L. variegatus, and were supported by qPCR results (data not shown, and Juliano et al., 2006).
Figure 2.

Vasa mRNA accumulates uniformly in early embryos. In situ RNA hybridization detects uniform vasa mRNA accumulation in early embryonic stages (A, four cells; B, 16 cells, with arrow indicating the micromeres) becoming restricted to the vegetal region in mesenchyme blastula (C) and during gastrulation (D) the mRNA is restricted to small micromere descendents that in larvae (E) accumulate selectively in the left coelomic pouch (E, arrow indicates the small micromere descendents in the left coelomic pouch). Scale bar = 20 microns.
Vasa protein analysis
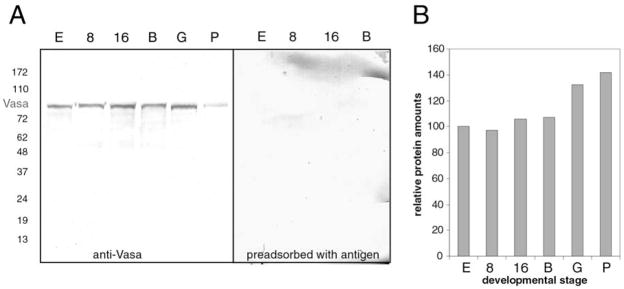
To analyze the distribution of vasa protein throughout embryogenesis, we made and obtained several different anti-vasa antibodies. We generated antibodies to an internal 20 amino acid peptide (aa 330–344) and to the C-terminal RNA helicase fragment, both from the S. purpuratus sequence. We also obtained antibodies against zebrafish vasa (Braat et al., 2000) and anti-pan-vasa antibody developed originally against grasshopper vasa (Chang et al., 2002). Each of these antibodies yielded similar results in immunolocalizations (see below), and western blots show that the approximately 80 kDa vasa band does not change in abundance substantially during embryogenesis (Fig. 3).
Figure 3.
Vasa protein is expressed throughout embryonic development. A. 120 micrograms of protein lysates were loaded per lane and probed with anti-DEAD box domain affinity-purified antiserum. Vasa-specific bands are competed away by preincubation of the antibody with the antigen. B. Quantification of the western blot signal in (A) normalized per Coomassie staining intensity of the duplicate gel (levels of vasa in the egg are set to 100%). Developmental stages used: E, egg; 8, 8-cell embryos; 16, 16-cell embryos; B, blastula; G, gastrula; P, pluteus.
Accumulation of vasa protein during embryogenesis and larval development
Vasa protein is present in unfertilized eggs, consistent with mRNA accumulation in oocytes (Juliano et al., 2006), and is enriched along the periphery of the cell (Fig. 4A). This pattern of labeling is not due to limited antibody permeability, as it is similar on immunolabeled sections of eggs (data not shown). The immunolabeling during early cleavage is homogenous through the 8-cell stage (Fig. 4B–C), similar to the mRNA distribution. After the first asymmetric division in the embryo at 16 cells, in contrast to the distribution of the vasa mRNA (Fig. 2B), vasa protein is selectively enriched in the micromeres (Fig. 4D). The fifth cell division, generating small micromeres and large micromeres (32 cell stage) further restricts vasa to just the small micromeres; the large micromeres lose vasa signal and the small micromeres remain vasa-positive through gastrulation (Fig. 4F–I). Since the overall amount of protein appears to stay relatively constant through early development (Figure 3), we conclude that translation of new vasa protein likely continues in the small micromere lineage, accompanied by protein turnover in the non-small micromere lineages. In gastrulae, the eight vasa-positive cells (four small micromeres after a single round of cell division) are located at the tip of the invaginating archenteron and in early larval stages they are associated with the developing coelomic pouches (Fig. 4I–K). In more advanced larvae (approximately 5 days in S. purpuratus, 3 days in Lytechinus variegatus), the vasa positive cells are restricted to the left coelomic sac (Fig. 4L). We do not know if this change in the location of vasa-positive cells reflects a turnover of vasa in cells of the right coelomic pouch, migration of vasa positive cells from the right to the left pouch, selective death of the vasa positive cells in the right coelomic pouch, or a combination thereof. The number of vasa-positive cells subsequently increases significantly in the left coelomic pouch of larvae and following metamorphosis, vasa is expressed in the germ cells of the developing juvenile gonads (data not shown). This vasa immunolabeling pattern was similar in other sea urchins, e.g. L. variegatus (and data not shown), and with other anti-vasa antibodies (Fig. 4N, O, and data not shown), suggesting a stereotypic and specific vasa distribution common to sea urchins.
Figure 4.
Uniform distribution of vasa protein in the early embryonic stages is followed by restriction of vasa to micromeres at 16-cell stage, the small micromeres at 32-cell stage, and remains associated with the small micromeres throughout embryonic development. Indicated embryonic stages were fixed and immunolabeled with anti-vasa DEAD-box antibody (red in A–M), or anti-pan insect vasa For2 (green in N, O). DNA was counterstained with Hoechst (blue). Corresponding DIC images of the embryos are shown at right. A–L: vasa localizations in S. purpuratus. M: 16 cell control, primary antibody following preadsorption with the antigenic protein does not exhibit specific staining pattern in the micromeres. N, O: For2 antibody (Chang et al., 2002) labels small micromeres in L. variegates blastula.
Following gastrulation, we find a strict coincidence of vasa RNA and protein expression in each species. In contrast, early in development the vasa mRNA is distributed uniformly throughout the embryo, as the vasa protein becomes restricted to the small micromeres. Progressive restriction of vasa-protein expressing cells begins at the 16 cell stage, and we focused our attention in the regulation of this mechanism.
Vasa positive cells of the embryo are exclusively the small micromere lineage
Based on morphological criteria, the vasa-positive cells in the sea urchin embryo appear to be small micromeres. This is a reliable determination up to ~128 cell stage, but recognition is lost later in development. To test this premise using molecular markers, we employed BrdU pulse-chase labeling of the sea urchin embryos, as described previously (Tanaka and Dan, 1990). BrdU is used to label replicating DNA during the first embryonic cell cycle that is diluted out with subsequent cell divisions. By virtue of the slow cell cycle in small micromeres, these cells preferentially retain BrdU relative to the other, more rapidly dividing cells. Embryos were pulsed with BrdU following fertilization and then cultured until late gastrula stage when they were processed for both BrdU and vasa immunolabeling. The results show a perfect correlation of BrdU retention and vasa-protein (Fig. 5). Thus, by using both morphological and molecular criteria, we conclude that the vasa-positive cells in the embryo and larvae are the small micromeres and their immediate descendents.
Figure 5.
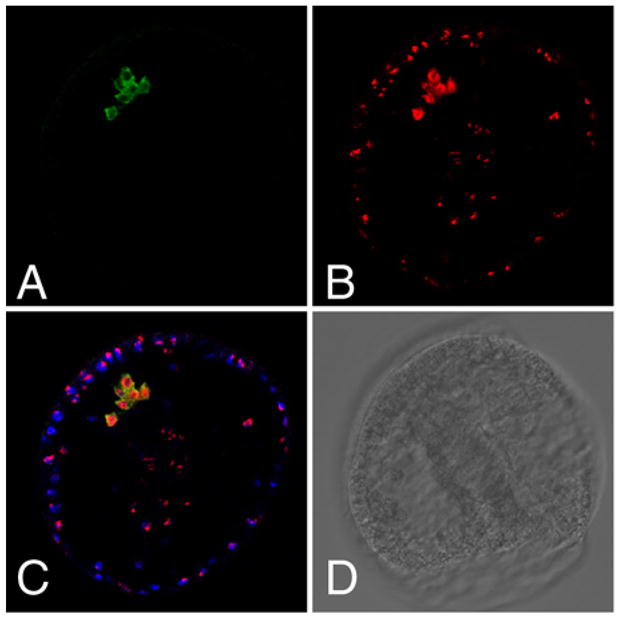
BrdU incorporation marks small micromeres that are vasa-positive during gastrulation. A. Anti-Vasa antibody staining (green). B. Anti-BrdU antibody staining, red. C. Red and green channels overlay; blue is nuclear staining of the embryo (Hoechst). D. Brightfield view of the embryo.
Inherent vasa expression patterns
To test the lineage restrictions of vasa we cultured blastomeres isolated from 2, 4 or 8 cell stages as pairs or as individuals, until untreated siblings reached the early blastula stage. (Fig. 6). This end point was chosen as an easily recorded and identifiable 4-vasa-cell stage (the 4 small micromeres). The results show that the number of vasa – positive cells is imperturbable: a blastomere isolated from a two cell stage had precisely half the number of small micromeres, two, as normal embryos (Fig. 6B); and a blastomere from a four cell embryo developed into a yet even smaller embryo and had one vasa positive cell (Fig. 6C). Tiny blastulae resulting from an eight cell stage blastomere had either none or one vasa-positive cell, reflecting the equatorial cleavage that occurred at this time resulting in an animal and vegetal tier of blastomeres (Fig. 6D, E). No changes were detected in vasa accumulation in other cells of the embryoids. Furthermore, dissociation of 2-cell embryos followed by reaggregation into 4-cell embryos results in a blastula that contains two clusters of 4 vasa-positive cells (Fig. 6F).
Figure 6.
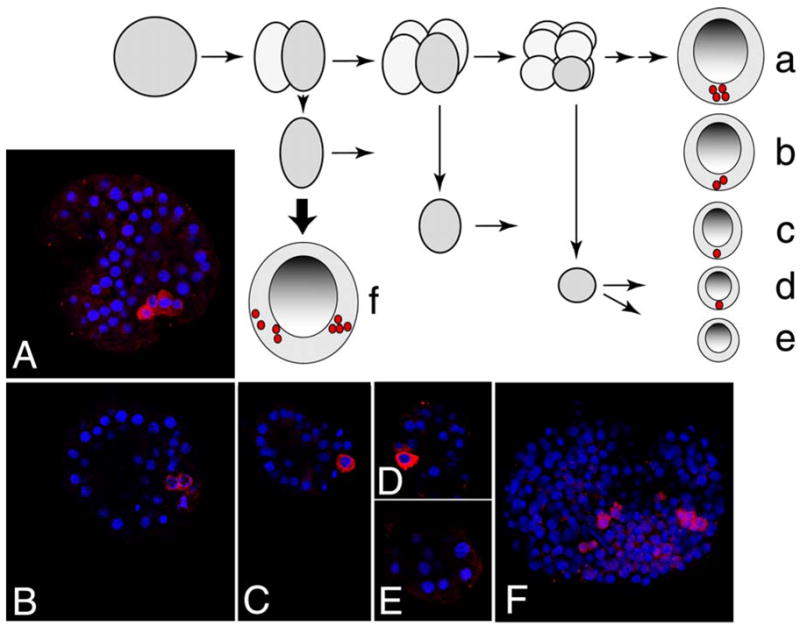
Vasa expression following blastomere dissociation. Schematic of the experiments in A–E is shown on top. Fertilized eggs were dissociated at 2 cell, 4 cell or 8 cell stage, or left untreated. At 20 hours of development, embryos arising from blastomeres or controls were fixed and labeled with anti-Vasa antibodies (red); DNA is stained with Hoechst (blue). Representative embryos are shown for each treatment group. A. Control embryo, 4 vasa-positive cells. B. Progeny of blastomere dissociated at 2 cell stage; 2 vasa-positive cells (100%). C. Progeny of blastomere dissociated at 4 cell stage; 1 vasa-positive cell (100%). D–E. Progeny of blastomeres dissociated at 8 cell stage; 1 vasa-positive cell (50%) or no vasa-positive cells (50%). F. After dissociation at 2-cell stage, 4 blastomeres (2 embryos worth) were induced to aggregate and cultured for 20 hours, resulting in 2 clusters of 4 vasa-positive cells.
Regulation of vasa protein expression
To understand the mechanism of vasa protein localization selectively in small micromeres, we tested disruptions of embryonic patterning on vasa protein expression. A widely used experimental perturbation to test mechanisms of embryonic pattern formation is LiCl exposure. This treatment uniformly activates the wnt/β-catenin signaling pathways by blocking GSK3β activity, resulting in a vegetalized, embryonic phenotype as a result of nuclearizing β-catenin (Logan et al., 1999). Nuclear β-catenin is first detected in the nuclei of both large and small micromeres after 5th cleavage, and persists until gastrulation, driving expression of the micromere gene program, starting with a paired homeodomain family transcription factor Pmar1 (Logan et al., 1999; Oliveri et al., 2002). This pattern could implicate nuclear β-catenin in the upkeep of high levels of vasa protein expression.
To test the mechanism of selective vasa protein expression, embryos first were treated with 30 mM LiCl from the 2-cell stage to the late gastrula stage of the control group (48 hours), and several time points were taken along this interval and the resulting vasa protein patterns were compared with those in untreated embryos (Fig. 7A–D). That the embryos were indeed vegetalized was determined by morphological and molecular criteria including a preponderance of exogastrulae, and aberrant Endo 1 expression (data not shown). The results show that LiCl treatment did not disrupt the pattern of vasa expression: protein was found enriched in the four small micromeres in both control and the treated group. However, LiCl treatment did induce more vasa protein in the embryos overall (Fig. 7A–D). This was also apparent by immunoblot analysis of treated and control samples indicating a 50% increase in overall vasa protein levels in the treated sample (data not shown). We do not, however, know if the detected increase in vasa expression represents an overall increase in synthesis from the ubiquitous transcript, or a lack of protein turnover normally seen in non-small micromeres.
Figure 7.
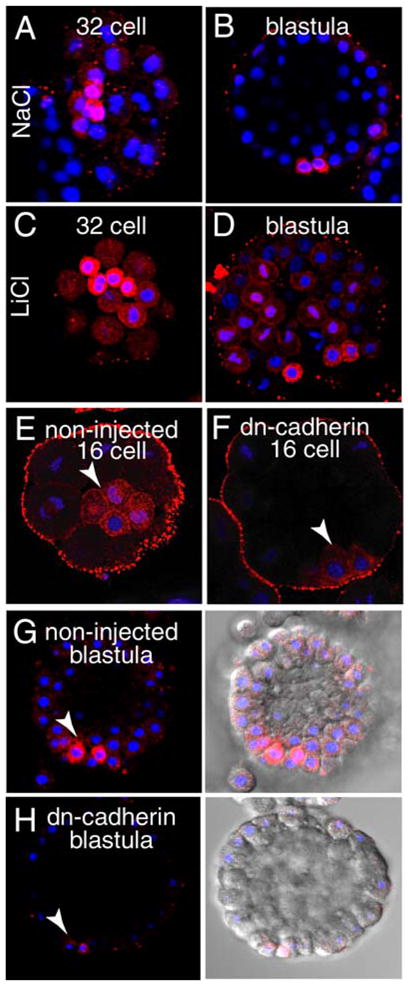
Regulation of vasa protein expression. LiCl induces vasa protein accumulation. Embryonic cultures were treated with 30 mM LiCl or NaCl (as a control) in sea water from 2-cell stage (1.5 hours) until blastula (24 hours). Samples of the treated embryos were removed, fixed at several stages, and the vasa protein expression pattern was detected by immunolocalization (red). DNA is counterstained with Hoechst (blue). In contrast to control (NaCl), LiCl-treated embryos show higher general expression of vasa protein (A, C). By blastula, increased vasa protein level outside of the small micromere population is still apparent (B, D). Conversely, repression of the β-catenin signaling pathway by over-expression of a dominant-negative cadherin fragment, resulted in significantly less vasa protein, particularly in the micromeres in 16-cell embryos (E–F), and in the small micromeres of the blastula (G–H). Micromeres and small micromeres are indicated with an arrowhead in each panel.
To further examine this vasa inducibility, we then microinjected into embryos mRNA encoding a dominant-negative cadherin that inhibits nuclear beta-catenin localization (Logan et al., 1999). Down regulation of β-catenin by DN-cadherin decreased expression of vasa protein at the 16-cell and 32-cell stage (5 hours) to the blastula stage (9 hours) by about 30%, both in the micromeres, and overall throughout the embryo (Fig. 7E–H). These changes were apparent both by in situ immunolabeling as well as by immunoblots (data not shown). Further, the homeodomain containing protein pmar, a downstream target of β-catenin and a skeletogenic determinant by virtue of its repression of HesC (Revilla-i-Domingo et al., 2007), appears to have a transient stimulus for vasa protein expression (data not shown). Overall, it appears that interference with the function of β-catenin signaling changes the total levels of vasa protein expression throughout the embryo without disturbing the selective vasa expression in the micromeres. In particular, nuclear β-catenin appears essential for significant vasa translation in the micromeres and small micromeres. This result argues for a β-catenin-sensitive gene transcribed in the micromeres and the small micromeres that directs vasa translation. Several such candidates have been identified (Ransick et al., 2002). Again, we do not know if the altered vasa protein accumulation represents strictly an overall increase in synthesis from the ubiquitous transcript, and/or a lack of protein turnover normally seen in non-small micromeres. It is clear that even though these manipulations impinge on the accumulation of vasa protein throughout the embryo, the small micromeres remain distinguishable from all other cells by their higher level of vasa protein accumulation.
Vasa regulation – micromere removal causes ectopic vasa translation
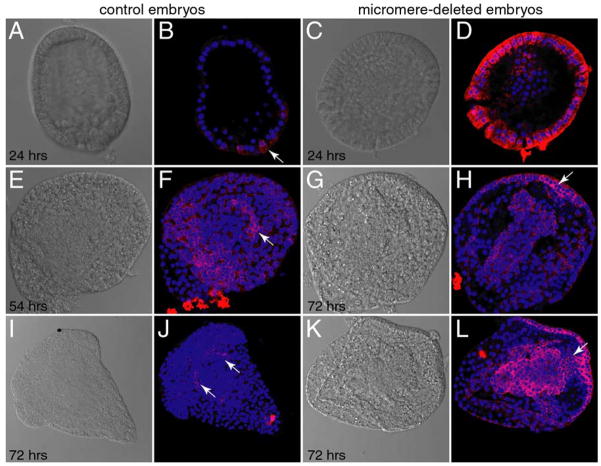
Ransick et al. (1996) surgically removed the micromeres (the vasa positive cell lineage) from 16-cell embryos, yet the adults resulting from these embryos made gametes normally, a finding inconsistent with the vasa-positive, small micromeres being the obligate primordial germ cells. We assessed the fate of vasa protein in micromere depleted embryos cultured to early blastula, gastrula, or to larval stages. When blastula resulting from micromere removal were examined for vasa expression we found that vasa protein expression dramatically increased throughout the embryo (Fig. 8D) without a detectable change of vasa mRNA levels compared to controls (data not shown). When gastrulae resulting from micromere depletion were examined, we found that vasa signal overall was still increased, but was now enriched within the gut and oral ectoderm (Fig. 8H). Finally, when larvae resulting from micromere depletion were analyzed, vasa expression was again restricted to cells of the coelomic region, the endoderm, and to the oral ectoderm. Thus, the micromeres must in some way repress the translation and accumulation of vasa throughout the embryo and their removal enables a compensatory induction leading to vasa-expression (Figure 9).
Figure 8.
Compensatory vasa expression upon micromere deletions. Upon surgical removal of micromeres, vasa protein is expressed broadly in the embryo, and subsequently is restricted to a smaller domain of expression. Here, manipulated embryos were fixed and immunostained with anti-Vasa (red); DNA is stained with Hoechst (blue). Arrows mark vasa expression domains. A, B: following 24 hours of culture, control embryos reach mesenchyme blastula stage (vasa only in small micromeres). C, D: deleted embryos form blastula at 24 hrs (broad vasa expression). E, F: control embryos at 54 hours, gastrula (vasa at the tip of archenteron) G, H: deleted embryos gastrulate following 72 hours (vasa in oral ectoderm). I, J: control embryos form plutei by 72 hours (vasa in coelomic pouches). K, L: late gastrula/prism stage deleted embryos at 72 hours (vasa at junction of gut/oral ectoderm).
Figure 9.

Summary diagram of vasa regulation during early embryonic development. Blue shading represents mRNA accumulation, whereas red shading indicates vasa protein.
Discussion
Germ line determinants are conserved between animals, but how cells of the embryo acquire such determinants, and their function in cell fate are very different. Some animals localize determinants, proteins and/or mRNAs, in distinct regions of the egg or early embryo, which then dictates a germ cell lineage to the cells that acquire this cytoplasm (Seydoux and Braun, 2006). Other animals employ inductive interactions and synthesize these same determinants in a particular cell lineage that then forms the primordial germ cells. It is becoming apparent that in animals using inductive interactions for germ cell formation that a progression occurs from a general stem cell, to a diversification of both somatic cells and stem cells, some of which are restricted to form the germ line. This progression was demonstrated both in polycheates and in mice (Hayashi et al., 2007; Rebscher et al., 2007). The results presented here, and by others, support the contention that the small micromeres of the sea urchin are an early stem cell of the larval rudiment. These cells contain germ line determinants, do not divide frequently early in development, but later in larval growth they begin to proliferate and diversify, likely into both somatic cells of the adult rudiment, and into primordial germ cells. Surprisingly, the micromeres (shown here), and presumably the small micromeres somehow repress vasa translational up-regulation in other cells of the embryo (Kurihara and Amemiya, 2005). Vasa up-regulation in the absence of micromeres appears to be largely a result of translational regulation since we do not see any increased vasa mRNA accumulation to otherwise explain the massive increase in vasa. As far as we know, this is the only known translational response from micromere removal. By virtue of vasa function in translational regulation, we hypothesize that the micromere-null embryos increase translation of ubiquitous mRNAs, the products of which may regulate additional reprogramming of developmental fate decisions. It is hard to imagine that the entire embryo becomes stem-cell-like but perhaps the increase in vasa protein allows for retention of plasticity in the embryonic cells for lack of the original signaling center. Vasa restriction to a small percentage of cells then occurs during gastrulation and may indicate committed fate decisions, except for the vasa-positive cells. Remaining vasa-positive cells presumably have taken over the stem cell function and will contribute to the adult rudiment.
Vasa protein accumulation in the small micromeres appears to be the result of two selective activities. First, an increase in vasa translation likely occurs specifically in the small micromeres from the maternal mRNA; the mRNA levels in the embryo are uniform whereas vasa protein accumulates only in the small micromeres. Future studies will address the mechanism of this selective translational activation with the hypothesis that 3′ UTR control elements are responsible for this selection. Second, vasa protein appears to selectively turnover in non-small micromere cells. Vasa is present uniformly in early embryos, and beginning after the 16 cell stage, vasa protein is lost in all cells except the small micromeres. This turnover mechanism is not known, though it should be pointed out that in Drosophila, a well-conserved SOCs-box protein, gustavus, associates with vasa and may be involved in vasa protein longevity (Styhler et al., 2002). The sea urchin embryo also contains this putative ubiquitin ligase, and intriguingly, its mRNA accumulates throughout the early embryo, and then encircles the vasa-positive, small micromeres (data not shown). Perhaps then vasa protein is selectively degraded in non-small micromeres by a gustavus-dependent ubiqutination pathway.
It is not yet known whether the small micromeres have a repressive function on vasa expression on their own, or whether the micromeres (including the large micromeres) have this sole function. One possibility is that the large micromeres repress trans-fating of the skeletal lineage later in development (mesenchyme blastula), and that the small micromeres repress only the alternative vasa-expression pathway. A series of transplants using only large or small micromeres followed by vasa immunolabeling may help resolve this functional difference.
It is noteworthy that only one other animal, the larva of Ascidians, has been shown to be able to rescue vasa-positive cells upon their removal (Takamura et al., 2002). In all other animals reported, removal of the vasa-positive cells results in adult sterility. For example, removal of pole cells in Drosophila, or a small piece at the posterior end of the primitive streak of a 7-day old mouse embryo, the remainder of the embryo loses the ability to make germ cells (McLaren, 1981). The “rescue” revealed in this sea urchin by micromere removal may reflect an ancestral mechanism of vasa expression and stem cell formation retained by the typically developing sea urchin.
Primordial germ cells become highly proliferative when they reach the gonad of the juvenile, giving rise to a large number of germ cells that differentiate into eggs and/or sperm. Thus, they are true stem cells, but are restricted to a single, germ line fate. Recent evidence points to vasa association with stem cells that give rise to the primordial germ cells and somatic stem cells (e.g. Agata et al., 2006; Rebscher et al., 2007). In the polychaete, Platynereis dumerilii, vasa is present in a population of mesodermal stem cells that proliferate in the posterior growth zone of annelids, the so called MPGZ cells. These cells are highly proliferative, give rise to several different cells fates - including primordial germ cells - and express vasa. Vasa expression is limited to the unspecified MPGZ cells, and the primordial germ cells that emanate from them. Of additional interest is that these cells also express other germ cell markers, including nanos, piwi, and the stem cell marker, PL10. Therefore, primordial germ cells may have a common origin with certain somatic stem cells such as those found in Planaria and Cnidaria (e.g. Extavour and Akam, 2003; Mochizuki et al., 2001) and thus may reflect an ancestral mode of germ cell specification. The sea urchin is a basal deuterostome and perhaps shares this ancestral mechanism in germ cell and somatic stem cell function.
Acknowledgments
We thank Dr. D. Zivkovic for a gift of anti-zebrafish vasa antibodies, Dr. Andrew Ransick for participating in micromere deletion experiments, and Dr. Eric Davidson and other members of his lab for helpful feedback. This work was supported by grants from the NIH and the NSF (GMW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agata K, Nakajima E, Funayama N, Shibata N, Saito Y, Umesono Y. Two different evolutionary origins of stem cell systems and their molecular basis. Semin Cell Dev Biol. 2006;17:503–9. doi: 10.1016/j.semcdb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Braat AK, van de Water S, Goos H, Bogerd J, Zivkovic D. Vasa protein expression and localization in the zebrafish. Mech Dev. 2000;95:271–4. doi: 10.1016/s0925-4773(00)00344-0. [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Hentze MW, Ephrussi A, Gebauer F. Control of oskar mRNA translation by Bruno in a novel cell-free system from Drosophila ovaries. Development. 2000;127:1063–8. doi: 10.1242/dev.127.5.1063. [DOI] [PubMed] [Google Scholar]
- Chang CC, Dearden P, Akam M. Germ line development in the grasshopper Schistocerca gregaria: vasa as a marker. Dev Biol. 2002;252:100–18. doi: 10.1006/dbio.2002.0840. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–84. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Fujii T, Mitsunaga-Nakatsubo K, Saito I, Iida H, Sakamoto N, Akasaka K, Yamamoto T. Developmental expression of HpNanos, the Hemicentrotus pulcherrimus homologue of nanos. Gene Expr Patterns. 2006 doi: 10.1016/j.modgep.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Takamura K. Characterization of an ascidian DEAD-box gene, Ci-DEAD1: specific expression in the germ cells and its mRNA localization in the posterior-most blastomeres in early embryos. Dev Genes Evol. 2000;210:64–72. doi: 10.1007/s004270050012. [DOI] [PubMed] [Google Scholar]
- George SB, Lawrence JM, Lawrence AL. Complete larval development of the sea urchin Lytechinus variegatus fed an artificial feed. Aquaculture. 2004;242:217–228. [Google Scholar]
- Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–6. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Crother B, White ME, Patient R, Bachvarova RF, Drum M, Masi T. Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Philos Trans R Soc Lond B Biol Sci. 2003;358:1371–9. doi: 10.1098/rstb.2003.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–78. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–15. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–12. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C. An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol. 2002;12:454–66. doi: 10.1016/s0960-9822(02)00723-6. [DOI] [PubMed] [Google Scholar]
- Komiya T, Itoh K, Ikenishi K, Furusawa M. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162:354–63. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- Kurihara H, Amemiya S. Developmental potential of small micromeres in sea urchin embryos. Zoolog Sci. 2005;22:845–52. doi: 10.2108/zsj.22.845. [DOI] [PubMed] [Google Scholar]
- Laidlaw M, Wessel GM. Cortical granule biogenesis is active throughout oogenesis in sea urchins. Development. 1994;120:1325–33. doi: 10.1242/dev.120.5.1325. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–7. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Lasko P. Bent out of shape: RNA unwinding by the DEAD-box helicase Vasa. Cell. 2006;125:219–21. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–57. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- MacArthur H, Houston DW, Bubunenko M, Mosquera L, King ML. DEADSouth is a germ plasm specific DEAD-box RNA helicase in Xenopus related to eIF4A. Mech Dev. 2000;95:291–5. doi: 10.1016/s0925-4773(00)00357-9. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- McLaren A. Germ Cells and Soma: A New Look at an Old Problem. Yale University Press; New Haven, CT: 1981. [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–56. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T. Expression and evolutionary conservation of nanos-related genes in Hydra. Dev Genes Evol. 2000;210:591–602. doi: 10.1007/s004270000105. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–28. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- Pearse JS, Cameron AR. Echinodermata: Echinoidea. In: Giese AC, Pearse JS, Pearse VB, editors. Reproduction of Marine Invertebrates. VI. Boxwood Press; Pacific Grove, CA: 1991. pp. 514–664. [Google Scholar]
- Pehrson JR, Cohen LH. The fate of the small micromeres in sea urchin development. Dev Biol. 1986;113:522–6. doi: 10.1016/0012-1606(86)90188-0. [DOI] [PubMed] [Google Scholar]
- Ransick A, Cameron RA, Davidson EH. Postembryonic segregation of the germ line in sea urchins in relation to indirect development. Proc Natl Acad Sci U S A. 1996;93:6759–63. doi: 10.1073/pnas.93.13.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol. 2002;246:132–47. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306:599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci U S A. 2007;104:12383–8. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Genet Dev. 2001;11:374–83. doi: 10.1016/s0959-437x(00)00207-0. [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–76. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–78. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Takamura K, Fujimura M, Yamaguchi Y. Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev Genes Evol. 2002;212:11–8. doi: 10.1007/s00427-001-0204-1. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–32. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Dan K. Study of the lineage and cell cycle of small micromeres in embryos of the sea urchin, Hemicentrotus pulcherrimus. Develop Growth & Differ. 1990;32:145–156. doi: 10.1111/j.1440-169X.1990.00145.x. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93:139–49. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- Voronina E, Marzluff WF, Wessel GM. Cyclin B synthesis is required for sea urchin oocyte maturation. Dev Biol. 2003;256:258–75. doi: 10.1016/s0012-1606(02)00134-3. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 1997;11:2510–21. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Berg L, Adelson DL, Cannon G, McClay DR. A molecular analysis of hyalin--a substrate for cell adhesion in the hyaline layer of the sea urchin embryo. Dev Biol. 1998;193:115–26. doi: 10.1006/dbio.1997.8793. [DOI] [PubMed] [Google Scholar]
- Yan N, Macdonald PM. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics. 2004;168:1433–42. doi: 10.1534/genetics.104.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–65. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]