Abstract
The ability of nitric oxide (NO)-releasing silica nanoparticles to kill biofilm-based microbial cells is reported. Biofilms of Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, Staphylococcus epidermidis, and Candida albicans were formed in vitro and exposed to NO-releasing silica nanoparticles. Replicative viability experiments revealed that ≥ 99% of cells from each type of biofilm were killed via NO release, with the greatest efficacy (≥ 99.999% killing) against gram-negative P. aeruginosa and E. coli biofilms. Cytotoxicity testing demonstrated that the highest dose of NO-releasing silica nanoparticles inhibited fibroblast proliferation to a lesser extent than clinical concentrations of currently-administered antiseptics (e.g., chlorhexidine) with proven wound-healing benefits. This study demonstrates the promise of employing nanoparticles for delivering an antimicrobial agent to microbial biofilms.
Keywords: Nitric oxide, nanoparticle, biofilm, antimicrobial, cytotoxicity
Introduction
Infections resulting from microbial biofilm formation remain a serious threat to patients worldwide. Particularly problematic are wound infections [1–5], with chronic wounds such as foot, leg, and pressure ulcers being particularly susceptible to biofilm infections [1]. Wound infections are responsible for >80% of the 100,000 limb amputations performed on diabetic patients in the U.S. each year [1]. While most wound infections are polymicrobial (i.e., caused by more than one species of bacteria or fungi) [6], the most common isolated species include gram-positive Staphylococcus aureus and Staphylococcus epidermidis, and gram-negative Pseudomonas aeruginosa [3]. Despite the many effective antimicrobial strategies against planktonic bacteria, most antimicrobials are rarely tested or effective against biofilms [5]. Novel approaches to treat established biofilms are thus urgently needed.
Biofilms are complex communities that form when a group of microorganisms self-secretes a polysaccharide matrix that retains nutrients for the constituent cells and protects them from both the immune response and antimicrobial agents [7]. The biofilm matrix itself may inhibit the penetration of antibiotics and prevent them from reaching embedded cells [8]. It has been shown that killing bacteria in a biofilm may require up to 1000 times the antibiotic dose necessary to achieve the same result in a suspension of cells [9]. While biofilm-embedded microbial cells communicate with each other via quorum sensing, phenotypic variations may occur, exacerbating virulence to the host [7].
Antiseptic wound dressings are currently the most common clinical strategy employed to address wound infections [10]. Although systemic antibiotic administration has shown some efficacy against wound infections [11–13], application of antibiotics directly to wounds is unacceptable clinical practice due to the threat of promoting antibiotic resistance [10]. The threat of bacterial resistance is significantly exacerbated in biofilms where the close proximity of cells allows facile transfer of resistance-encoding DNA [8]. Current clinical protocols call for applying creams, solutions, or wound dressings that contain antiseptics such as silver compounds (e.g., silver sulfadiazine, silver nitrate), iodine (e.g. povidone iodine), or chlorhexidine [10]. Each of these antiseptics has demonstrated broad-spectrum activity against both gram-positive and gram-negative bacterial species. Unfortunately, the efficacy of current antiseptics has been evaluated primarily against planktonic bacteria, not biofilms [5]. Despite their success, Ag+ and iodine wound treatments have some undesirable properties. For example, silver treatment has been reported to result in permanent skin discoloration (argyria) [14]. More problematic, Ag+-resistant bacteria have emerged, raising serious concerns [15–17]. Povidone iodine has been shown to be toxic to fibroblasts in vitro [18] and its efficacy as a safe antimicrobial agent for wound healing questioned [19–24]. Recently, both povidone iodine and chlorhexidine have been shown to be ineffective at treating biofilms of Pseudomonas aeruginosa and Enterococcus faecalis [25, 26]. Alarmingly, a growing number of reports document life-threatening anaphylactic shock in response to chlorhexidine treatment [27–30]. Clearly, new strategies for battling biofilms are warranted.
Recent research has highlighted the antimicrobial properties of nitric oxide (NO) [31, 32], a reactive free radical produced by inflammatory cells (e.g., neutrophils and macrophages) to battle infection. Using small molecule NO donors, Raulli et al. demonstrated that NO possesses broad-spectrum antibacterial properties against both gram-positive and gram-negative bacteria [33]. Ghaffari et al. reported on NO’s effectiveness at killing methicillin-resistant Staphylococcus aureus (MRSA) [34]. The importance of NO in mammalian defense against invading pathogens was demonstrated by MacMicking and coworkers using mice lacking the ability to endogenously generate NO. Such mice were significantly more susceptible to bacterial infection than mice possessing full NO-production capabilities [35]. In terms of biofilms, Barraud noted that NO-releasing small molecules promoted cell dispersal in P. aeruginosa biofilms [36].
As an alternative strategy for delivering NO to pathogenic bacteria, we recently reported on the antibacterial properties of NO-releasing silica nanoparticles [37]. The nanoparticles exhibited enhanced bactericidal efficacy against planktonic P. aeruginosa cells compared to small molecule NO donors [37]. To date, the effectiveness of NO-releasing nanoparticles against established biofilms remains unclear. The rapid diffusion properties of NO may result in enhanced penetration into the biofim matrix and thus improve efficacy against biofilm-embedded bacteria [38]. Moreover, a promising advantage of nanoparticles over small molecules is that their physicochemical properties (e.g., hydrophobicity, charge, size etc.) may be tuned by varying synthetic precursors and procedures [39, 40]. Herein, we present studies aimed at understanding the ability of NO-releasing nanoparticles to kill microbial cells within established biofilms.
Methods and materials
N-Methylaminopropyltrimethoxysilane (MAP3) and N-(6-aminohexyl)aminopropyl-trimethoxysilane (AHAP3) were obtained from Gelest (Morrisville, PA) and stored either under nitrogen or in a desiccator. Tetraethyl orthosilicate (TEOS) was purchased from Fluka (Buchs, Switzerland) and stored in a desiccator. Ethanol (EtOH; absolute), methanol (MeOH), and ammonia solution (NH4OH, 30 wt% in water) were purchased from Fisher Scientific (Fair Lawn, NJ). Tryptic soy broth (TSB), tryptic soy agar (TSA), yeast peptone dextrose broth (YPD), and yeast peptone dextrose agar were purchased from Becton, Dickinson and Company (Sparks, MD). Nitrogen (N2) and argon (Ar) were purchased from National Welders (Raleigh, NC) while nitric oxide (NO, 99.5%) was obtained from Linde (Raleigh, NC). Other solvents and chemicals were analytical-reagent grade and used as received. Pseudomonas aeruginosa (ATCC #19143), Escherichia coli (ATCC #53323), Staphylococcus aureus (ATCC #29213), Staphylococcus epidermidis (ATCC #35983), and Candida albicans (ATCC #90028) were purchased from American Type Culture Collection (Manassas, VA). Class VI medical-grade silicone rubber (SiR) was purchased from McMaster-Carr (Atlanta, GA). Distilled water was purified with a Millipore Milli-Q Gradient A-10 water purification system (Bedford, MA) to a final resistivity of 18.2 MΩ·cm and a total organic content of <6 ppb. The following standard cell-culture products were obtained from Invitrogen (Carlsbad, CA): Eagles minimal essential medium (MEM), fetal bovine serum (FBS), penicillin/streptomycin (P/S), trypsin and 3-(4,5-dimethylthiazol–2-yl)-2,5-diphenyltetrazolium bromide (MTT). L929 mouse fibroblasts (ATCC #CCL-1) were purchased from the University of North Carolina Tissue Culture Facility (Chapel Hill, NC).
Synthesis of NO-releasing silica nanoparticles
The synthesis and characterization of NO-releasing silica nanoparticles have been described previously [41, 42]. Briefly, an aminoalkoxysilane solution was prepared by dissolving either AHAP3 (2.3 mmol) or MAP 3 (6.8 mmol) in 16 mL of EtOH and 4 mL of MeOH in the presence of NaOCH3 (equimolar with either AHAP3 or MAP3). The solution was then placed into 10-mL vials equipped with stir bars. The vials were placed in a Parr bottle, connected to an in-house NO reactor, and flushed with Ar six times to remove O2 in the solution. The reaction bottle was pressurized to 5 atm NO for 3 days with continuous stirring of the silane solution. Prior to removing the diazeniumdiolate-modified silane sample (AHAP3/NO or MAP3/NO), unreacted NO was purged from the chamber with Ar. Silane solutions were prepared by mixing TEOS (2.8 mmol) and AHAP3/NO (2.3 mmol; corresponding to 45 mol%, balance TEOS) or MAP3/NO (6.5 mmol; corresponding to 70 mol%, balance TEOS) in the EtOH/MeOH solution for 2 min. The silane solution was then added into EtOH (22 mL) and ammonia catalyst (6 mL, 30 wt % in water) and mixed vigorously for 30 min at 4 °C. The precipitated nanoparticles were collected by centrifugation (5000 rpm, 5 min), washed with EtOH several times, dried under ambient conditions for 1 h, and stored in a sealed container at −20 °C.
Nitric oxide release measurements
Real-time NO release data were collected using a Sievers 280 chemiluminescent NO analyzer (Boulder, CO). The instrument was calibrated with an atmospheric sample that had been passed through a NO zero filter and a 24.1 ppm NO gas standard (balance N2). Next, a known mass of diazeniumdiolate-modified silica nanoparticles were immersed in deoxygenated PBS (pH 7.4) at 37 °C. Liberated NO was carried from the buffer to the analyzer with a stream of N2 bubbled into the solution at a flow rate of 80 mL/min. In the instrument, NO was detected via chemiluminescent reaction with ozone [43].
Treatment of established biofilms with NO-releasing silica nanoparticles
P. aeruginosa, E. coli, S. aureus, S. epidermidis, and C. albicans were cultured at 37 °C in either tryptic soy broth (TSB; bacteria) or yeast-peptone-dextrose broth (YPD; fungi), pelleted by centrifugation, resuspended in 15% glycerol (v/v in PBS), and stored at −80 °C. Cultures for biofilm studies were grown from a −80 °C stock at 37 °C in TSB overnight. A 1-mL aliquot of overnight culture was inoculated into 100 mL fresh TSB, incubated at 37 °C with rotation, and grown to an optical density (ODλ=650 nm) required to achieve ~108 colony forming units [CFU] per mL, as verified by serial 10-fold dilutions and plating on nutrient agar plates. The bacteria or fungi were pelleted by centrifugation, rinsed with ultrapure water, and resuspended in sterile phosphate buffered saline (PBS; 10 mM, pH 7.4).
Class VI medical-grade silicone rubber (SiR) was sectioned into squares measuring 8 × 6 × 2 mm2. The SiR squares were cleaned with ethanol, dried, and sterilized in an autoclave at 121 °C for 25 min. Under aseptic conditions, the SiR squares were then immobilized onto the end of sterile syringe needles and submerged in sterile TSB or YPD (5 mL) in sterile 10-mL glass vials. The 108 CFU/mL microbial suspension was then diluted to 106 CFU/mL, and 50 μL of the diluted suspension was added to the nutrient broth in each vial containing the SiR squares (final microbial concentration = 104 CFU/mL). The vials containing bacteria, broth and SiR squares were placed in a 37 °C incubator with gentle agitation. After 24 h, the SiR squares were removed from the nutrient broth, rinsed twice in sterile PBS, and individually transferred into new 10-mL glass vials containing a suspension of either 45 mol% AHAP3/TEOS nanoparticles or 70 mol% MAP3/TEOS nanoparticles in PBS. The vials were returned to the 37 °C incubator and gently agitated. After 24 h, the SiR squares were rinsed twice in sterile PBS and aseptically transferred into polypropylene test tubes containing 2 mL of sterile PBS. To remove the biofilm cells from the SiR substrates, each test tube was vortexed for 10 s, sonicated in a 125 W ultrasonic cleaner for 30 min, and vortexed for an additional 10 s. The resulting bacterial suspension was subjected to serial 10-fold dilutions, and 100 μL of appropriate dilutions was plated on either tryptic soy agar (TSA; bacteria) or yeast peptone dextrose agar (fungi). The nutrient agar plates were then incubated at 37 °C. The next day, the colonies that grew on each plate were counted and the number of viable biofilm bacteria removed from each substrate determined. Efficacy is defined as the reduction in viable cells recovered from biofilms treated with nanoparticles compared to control biofilms of the same species not treated with nanoparticles.
Determination of nanoparticle association with biofilms
Biofilms of P. aeruginosa, E. coli, S. aureus, S. epidermidis, and C. albicans were grown on silicone rubber substrates as described above. The biofilms were then incubated with 8 mg/mL MAP3 silica nanoparticles in PBS. After 24 h, the biofilms were removed from the nanoparticle suspension and transferred to 3 mL of PBS in polypropylene test tubes. The test tubes were vortexed for 10 s, sonicated for 30 min, and again vortexed for 10 s. The PBS was then transferred to new vials to which 3 mL of ethanol (absolute) was added to kill any bacteria or fungi. As a measure of the amount of nanoparticles recovered from the biofilms, the solutions were analyzed for Si via direct current optical emission spectrometry (DCP-OES; ARL-Fisons Spectraspan 7; Beverly, MA) calibrated with 0–54 ppm Si standard solutions prepared in 50:50 PBS:absolute ethanol.
In vitro toxicity testing of NO-releasing nanoparticles
L929 mouse fibroblasts were grown to subconfluency in MEM with 10% FBSsupplemented with 0.2% P/S at 37 °C and 5% CO2. Cells were then trypsinized and resuspended in media at a concentration of 2 × 105 cells/mL and plated onto tissue culture treated polystyrene 96-well plates. After incubation for 24 h, the media from each well was discarded. Control and NO-releasing 70 mol% MAP3/TEOS nanoparticles were added at concentrations of 1, 2, 4, and 8 mg/mL (200 μL). After incubation with the nanoparticles for 24 h, a standard MTT viability assay was performed [44]. Briefly, 40 μL of a 1 mg/mL MTT solution in sterile PBS was added to each well and incubated for 3 h, after which all solution was removed from the well and 100 μL DMSO was added to solubilize the crystals. The absorbance measured at 570 nm was proportional to the concentration of viable cells in each well. Fibroblast viability in the presence of control and NO-releasing silica nanoparticles is reported relative to the viability of fibroblasts not exposed to silica nanoparticles.
Results and discussion
While the antimicrobial efficacy of small molecule NO donors has been explored against planktonic bacteria [33] and fungi [45], only one study to date has detailed the effect of NO on established biofilms [36]. To extend the study of NO treatment to other species of biofilm-forming pathogens, the efficacy of NO-releasing silica nanoparticles was herein examined against a broader spectrum of gram-negative, gram-positive, and fungal biofilms. Silica nanoparticles modified to release NO have previously been shown to kill planktonic P. aeruginosa cells more effectively than the small molecule NO donor PROLI/NO [37]. Herein, such studies were expanded and modified to examine the efficacy of NO-releasing nanoparticles against established biofilms of P. aeruginosa, E. coli, S. aureus, S. epidermidis, and C. albicans, all of which are know to cause biofilm-based infections [1, 3, 46, 47]. To closely mimic the established MBEC (minimum biofilm eradication concentration) anti-biofilm assay [48, 49], medical-grade silicone rubber squares were employed as substrates on which biofilms were formed in vitro. The biofilms were then exposed to NO-releasing silica nanoparticles suspended in PBS, and the anti-biofilm efficacy was measured via a replicative viability assay.
Nitric oxide release from silica nanoparticles
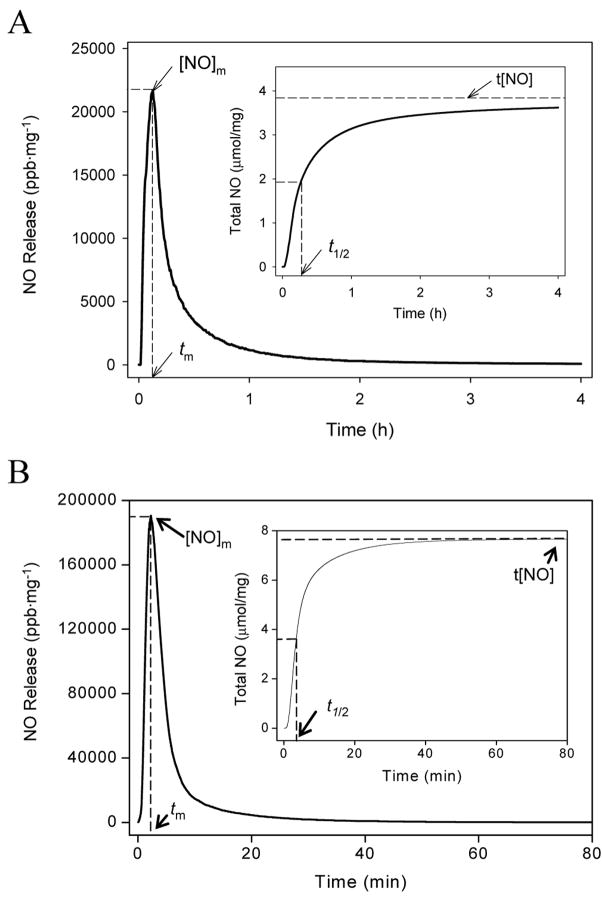
The NO-releasing silica nanoparticles have been characterized and described previously [41, 42, 50]. As shown in Figure 1A, the total amount of NO (t[NO]) released by 45 mol% AHAP3 silica nanoparticles was approximately 3.8 μmol mg−1, with a maximum NO flux ([NO]m) of 21700 ppb mg−1 and a NO release half life (t1/2) of 18 min. In contrast, NO release from 70 mol% MAP3 nanoparticles was much more rapid, with a t1/2 of ~6 min. The amount of NO released from the MAP3 nanoparticles was much greater than from AHAP3 nanoparticles, with t[NO] and [NO]m values of 7.6 μmol mg−1 and 190000 ppb mg−1, respectively, for MAP3 (Fig. 1B). As characterized by atomic force microscopy (AFM), the size of the AHAP3 and MAP3 nanoparticles were 136 ± 15 and 90 ± 10 nm in diameter, respectively (data not shown) [37, 50].
Figure 1.
Nitric oxide release profiles of (A) 45 mol% AHAP3 and (B) 70 mol% MAP3 silica nanoparticles (balance TEOS). Insets represent total NO release. [NO]m = maximum NO flux; tm = time to reach maximum NO flux; t[NO] = total NO released; t1/2 = half life of NO release.
Anti-biofilm efficacy of AHAP3 and MAP3 silica nanoparticles
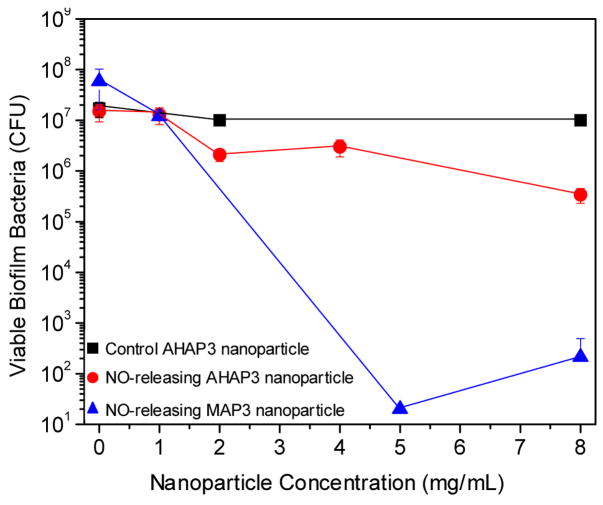
To test the influence of the delivery vehicle (i.e., AHAP3 vs. MAP3 silica nanoparticles) on NO’s ability to kill biofilm-embedded bacterial cells, P. aeruginosa biofilms were exposed to a range of concentrations of both AHAP3 and MAP3 nanoparticles (0 – 8 mg/mL) optimized based on initial efficacy screening and the amount of particles synthesized per batch. To quantitatively determine the number of viable biofilm cells remaining after treatment, the biofilm was dispersed in sterile PBS via vortexing and sonication [48]. Of note, control experiments were performed to confirm that the vortexing and sonication procedure did not influence cell viability (data not shown). As shown in Figure 2, AHAP3 nanoparticles administered at a dose of 8 mg/mL exhibited approximately 2 logs of biofilm killing (i.e., the number of viable cells was reduced from ~2 × 107 to ~4 × 105 CFU), representing ~99% killing of the cells within the biofilm. When administered at an equivalent dose (i.e., 8 mg/mL), MAP3 nanoparticles resulted in >5 logs of killing, reducing the number of viable biofilm bacteria from ~7 × 107 to ~3 × 102, effectively killing >99.999% of the biofilm bacterial cells. Neither AHAP3 nor MAP3 control nanoparticles (i.e., depleted of NO) led to any measurable bacterial killing, indicating that the NO from the particles accounted for the biocidal action.
Figure 2.
Anti-biofilm efficacy of 45 mol% AHAP3 silica nanoparticles (control and NO-releasing) and NO-releasing 70 mol% MAP3 silica nanoparticles against established P. aeruginosa biofilms on medical-grade silicone rubber substrates.
At an equivalent dose (i.e., 8 mg/mL), the MAP3 silica nanoparticles demonstrated ~1000-fold greater efficacy against P. aeruginosa biofilms than the AHAP3 nanoparticles. While MAP3 nanoparticles release more total NO than AHAP3 nanoparticles per mg (7.6 vs. 3.8 μmol mg−1 for MAP3 and AHAP3 nanoparticles, respectively), the 1000-fold increase in killing is not easily accounted for by the 2-fold increase in t[NO] alone. In addition to the greater amount of NO released by MAP3 nanoparticles, it is hypothesized that the more rapid delivery (i.e., shorter t1/2) leads to a greater instantaneous concentration of NO in solution. The higher concentration of NO in solution would thus enhance diffusion into the biofilm matrix. Other explanations for the improved anti-biofilm efficacy of MAP3 nanoparticles include their smaller size potentially allowing them to penetrate the biofilm matrix more effectively, and/or a possible difference in the surface charge of the particles due to the identity and amount of synthetic precursor (i.e., AHAP3 vs. MAP3) employed. Studies to determine the influence of particle size and surface charge on anti-biofilm efficacy are currently underway. Due to their enhanced efficacy over AHAP3 nanoparticles, MAP3 nanoparticles were used as the NO delivery vehicle for the remainder of the study.
Broad-spectrum anti-biofilm efficacy of MAP3 silica nanoparticles
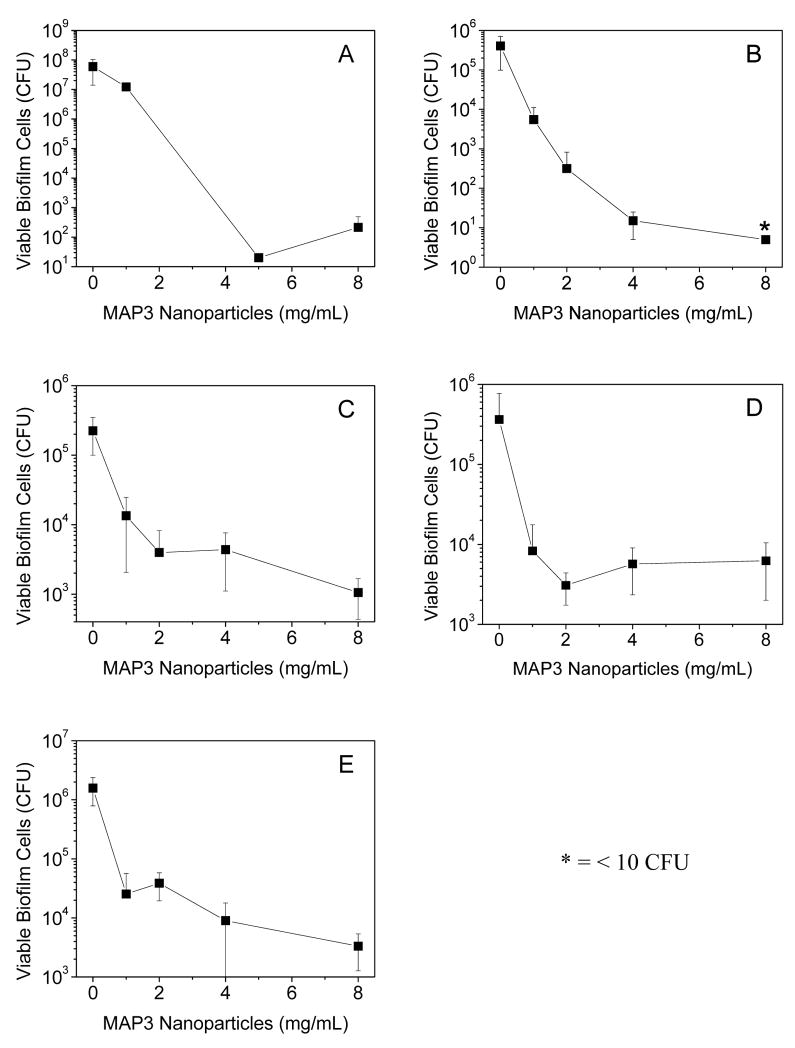
The anti-biofilm properties of MAP3 nanoparticles were tested against a broad spectrum of biofilm-forming pathogens, including gram-negative (P. aeruginosa and E. coli), gram-positive (S. aureus and S. epidermidis), and fungal (C. albicans) species. As shown in Figure 3, nanoparticle-derived NO was effective against biofilms of all species tested. Anti-biofilm efficacy was greatest against the gram-negative species, with ≥5 logs of killing at the highest dose tested (8 mg/mL) for both P. aeruginosa and E. coli. Intermediate efficacy was observed for C. albicans, with 8 mg/mL MAP3 nanoparticles achieving 3 logs of biofilm killing. The NO-releasing particles were least effective against gram-positive S. aureus and S. epidermidis biofilms, with the highest dose of nanoparticles killing ~2 logs of biofilm bacteria. The efficacy of NO-releasing nanoparticles against biofilms of all species tested is summarized in Table 1. As expected, control silica nanoparticles (i.e., depleted of NO) at 8 mg/mL demonstrated negligible anti-biofilm activity against all species tested (Supplementary Materials, Fig. S1), indicating that the NO itself and not the particle scaffold was responsible for the observed anti-biofilm properties.
Figure 3.
Broad spectrum anti-biofilm properties of 70 mol% MAP3 silica nanoparticles (balance TEOS) against (A) P. aeruginosa and (B) E. coli (gram-negative); (C) S. aureus and (D) S. epidermidis (gram-positive); and (E) C. albicans (pathogenic fungus) biofilms.
Table 1.
Log-based and percent reductions in biofilm viability at the highest dose of NO-releasing MAP3 silica nanoparticles tested (8 mg/mL) versus blanks (0 mg/mL nanoparticles).
| Species | Classification | Log Reduction | Percent Reduction |
|---|---|---|---|
| P. aeruginosa | gram-negative | 5 | 99.999% |
| E. coli | gram-negative | 5 | 99.999% |
| S. aureus | gram-positive | 2 | 99% |
| S. epidermidis | gram-positive | 2 | 99% |
| C. albicans | fungus | 3 | 99.9% |
With respect to wound-based biofilms, several aspects of the anti-biofilm properties of NO-releasing silica nanoparticles show promise as a potential therapeutic. Healing is impaired when the bacterial bioburden within a wound is greater than 105 bacterial cells per gram of tissue [51]. For each pathogen studied, the NO-releasing silica nanoparticles reduced the number of viable biofilm cells by at least approximately 2 orders of magnitude (i.e., 99% killing), a desirable characteristic that may lower the bioburden to levels below the 105 threshold, allowing normal healing to progress. Furthermore, the anti-biofilm activity of the nanoparticles was broad-spectrum. While NO-releasing nanoparticles were found to be most effective against gram-negative species, the ability to kill multiple species, including gram-positive and fungal biofilms, is key for treating polymicrobial wound-based infections [6]. Finally, the excellent efficacy against both P. aeruginosa and E. coli (≥99.999% biofilm killing) is promising since gram-negative bacteria are generally more invasive than gram-positive infections, and thus more difficult to treat [6]. Gram-negative pathogens also exhibit certain virulence factors including toxins, proteolytic enzymes, and extracellular polysaccharides [6] that coupled with the increasing antibiotic resistance of gram-negative species [52–54] present a threat for which new treatments are urgently needed.
To determine if the differential efficacy of the NO-releasing silica against gram-negative and gram-positive biofilms was the result of differing extents of nanoparticle association with the biofilm matrices, elemental analysis for Si was performed on dispersed biofilms. Increased nanoparticle association with biofilms would be expected to result in greater NO delivery to the biofilm-based cells due to proximity. As shown in Table 2, the amount of Si recovered from blank experiments (i.e., biofilms not exposed to nanoparticles) ranged from 783 to 890 pmol Si for all pathogens. These Si levels may be attributed to the silicone rubber substrates on which the biofilms were formed and/or any Si that leached from the glass flask employed during the sterilization of the PBS. In contrast, the Si levels from biofilms exposed to 8 mg/mL MAP3 silica nanoparticles were significantly larger, ranging from 1246 to 1709 pmol of Si recovered, indicating that the nanoparticles associated with each species tested. After accounting for the Si measured from blanks not exposed to nanoparticles, the greatest amount of Si (890 pmol) was recovered from the gram-negative E. coli biofilm, which was also characterized by the greatest amount of biofilm killing (5 logs of killing, Table 1). Less Si was measured from the C. albicans biofilm (606 pmol), with even less Si recovered from the gram-positive biofilms (427 and 534 pmol for S. epidermidis and S. aureus, respectively). This might be anticipated as the C. albicans and S. epidermidis/S. aureus biofilms were characterized by less biofilm killing (3 and 2 logs, respectively) compared to E. coli. Indeed, the greatest amount of Si was recovered from the biofilm of the species most susceptible to NO-releasing silica nanoparticles (i.e., E. coli). The different nanoparticle/biofilm association as a function of pathogen type may be due to differences in the electrostatic properties of each type of biofilm [55]. Indeed, while many bacterial species are characterized by a negatively-charged biofilm matrix, certain strains of S. epidermidis biofilms have been found to generate polycationic biofilms [55]. Research in our laboratory is currently focused on tuning the electrostatic properties of NO-releasing nanoparticles to maximize their association with particular biofilms to enhance NO delivery to biofilm-based microbes. Surprisingly, the elemental analysis data indicate that comparatively little Si was recovered the P. aeruginosa biofilm (284 pmol), despite the fact that the P. aeruginosa biofilms had the greatest cell density. Such little Si recovery was unexpected due to the extreme efficacy of the nanoparticles against P. aeruginosa biofilms (5 logs of killing). It is possible that instead of only killing the P. aeruginosa cells, the NO also initiated cell dispersion from the biofilm [36]. Biofilm dispersion would not be distinguished from cell killing via the assay employed herein. We are currently seeking to determine if NO-releasing silica nanoparticles kill P. aeruginosa cells within a biofilm and/or preferentially encourage their dispersal. Nevertheless, the ability to disperse bacteria from a biofilm is promising in that once bacterial cells have re-entered the planktonic state, they are more susceptible to antimicrobial agents than in their biofilm state [9]. Previous studies from our laboratory have demonstrated enhanced bactericidal efficacy of NO-releasing silica nanoparticles against planktonic P. aeruginosa cells [37].
Table 2.
Amount of Si recovered from biofilms after 24 h incubation with 8 mg/mL MAP3 silica nanoparticles.
| Amount of Si Recovered (pmol)a |
||||
|---|---|---|---|---|
| Sample |
Without Nanoparticles |
With Nanoparticlesb |
Total Si Recovered from Biofilm (pmol) |
Si Attributable to Nanoparticlesc |
| P. aeruginosa | 855 ± 36 | 1139 ± 107 | 284 | 25% |
| E. coli | 819 ± 18 | 1709 ± 499 | 890 | 52% |
| S. aureus | 890 ± 36 | 1424 ± 178 | 534 | 38% |
| S. epidermidis | 819 ± 16 | 1246 ± 321 | 427 | 34% |
| C. albicans | 783 ± 4 | 1389 ± 285 | 606 | 44% |
Measured via direct current plasma optical emission spectrometry
8 mg/mL MAP3 nanoparticles, 24-h incubation in PBS
Amount of total Si recovered from sample that is attributable to nanoparticles associated with the biofilm
Experiments are currently underway to examine possible synergistic enhancement of NO treatment with other antibacterial agents, including silver ion and traditional antibiotics. It will also become important to understand the role of nanoparticle size on anti-biofilm efficacy. Size and other parameters (i.e., surface charge, hydrophobicity, etc.) are easily tuned during synthesis by varying the solvent and/or precursor types and concentrations [42]. A primary advantage of nanoparticle-based drug delivery involves the ability to graft different ligands to the nanoparticle surface to target the particles to particular cells. Ligands useful for promoting the antimicrobial efficacy of NO-releasing silica nanoparticles may include antibodies and/or sugars, both of which may promote nanoparticle association with and/or uptake by bacterial cells. With respect to wounds, NO release is expected to exert beneficial secondary effects on the healing process. We have demonstrated previously that NO modulates inflammation, angiogenesis, and tissue remodeling [62]. Since wounds are known to be deficient in NO [63], application of NO-releasing silica nanoparticles may speed healing by killing bacteria and overcoming the general NO deficiency. To understand the dose of NO necessary to promote wound healing and aid the host response to infection, in vivo wound-healing experiments both with and without bacterial challenge are necessary.
Cytotoxicity of MAP3 nanoparticles to mammalian fibroblasts
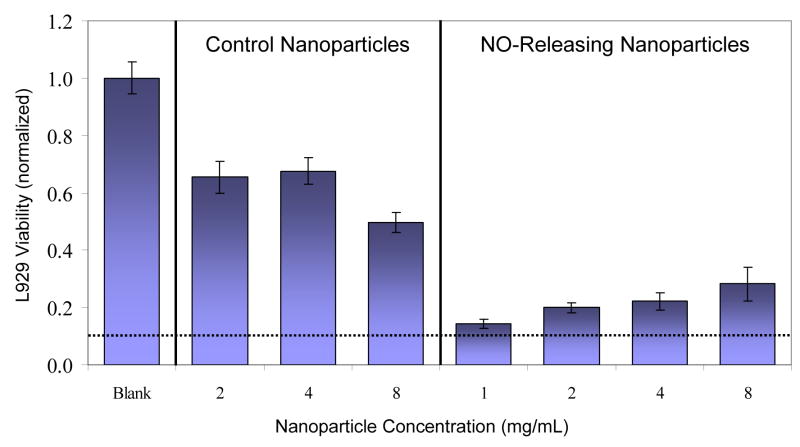
The toxicity of the nanoparticles to healthy mammalian cells is critical to their potential as a future therapeutic against biofilms. We thus evaluated the toxicity of the particles against fibroblast cells. Such cells are an excellent model as they are instrumental in wound healing and maintaining the extracellular matrix [56, 57]. Previous studies have shown that NO-releasing 45 mol% AHAP3 silica nanoparticles exhibited minimal (<10%) acute toxicity to fibroblasts at concentrations up to 1 mg/mL [37]. In the present study, the concentration range was expanded to 8 mg/mL to include the highest dose tested against microbial biofilms. As shown in Figure 4, a standard MTT viability assay [44] revealed that both control and NO-releasing silica nanoparticles reduced the proliferation of L929 fibroblasts. At 8 mg/mL, control and NO-releasing nanoparticles inhibited fibroblast proliferation by approximately 50% and 70%, respectively. Surprisingly, the marginal effect of administering greater amounts of NO did not result in greater toxicity to the fibroblasts. In fact, increasing the dose of NO-releasing silica nanoparticles from 1 to 8 mg/mL resulted in greater fibroblast viability. It is unclear to what extent the decreased fibroblast proliferation was the result of NO-induced cell death or NO-mediated signaling. Other reports have documented reduced proliferation of fibroblasts and other cell types (e.g., keratinocytes) as a result of NO treatment [58–60]. Inhibition of fibroblast proliferation may thus be the result of NO-mediated cell signaling and not necessarily NO-induced cell death. Others have suggested that reduced fibroblast proliferation after treatment with NO donors likely occurs via a cGMP-independent mechanism [58].
Figure 4.
Viability of L929 mouse fibroblasts exposed to control and NO-releasing 70 mol% MAP3 silica nanoparticles (balance TEOS) at various concentrations. Viability was measured via MTT reduction by metabolically-active (viable) fibroblast cells and is expressed normalized to untreated fibroblasts. Dotted line represents fibroblast viability after treatment with clinical concentration of povidone iodine. Treatment with clinical concentration of chlorhexidine results in 0% viability. Povidone iodine and chlorhexidine data adapted from Ref. [61].
Notwithstanding, the toxicity of silica nanoparticles to fibroblasts is minimal compared to other commonly-applied topical antiseptics [18, 61]. For example, Pyo et al. found that administering clinical concentrations of povidone iodine and chlorhexidine to human fibroblasts reduced cell viability by 89 ± 4% and 100 ± 4%, respectively [61]. Despite considerable in vitro toxicity to fibroblasts, clinical application of both povidone iodine and chlorhexidine has been demonstrated to enhance wound healing by killing wound-based microbes [10]. Thus, the in vitro toxicity of NO-releasing silica nanoparticles to fibroblasts may not represent a significant detriment to their continued study as potential treatments for wound-based biofilms.
Conclusions
Nitric oxide-releasing silica nanoparticles are effective at killing biofilm-based microbes and may represent a new paradigm for addressing biofilms. When compared to AHAP3 nanoparticles, MAP3 nanoparticles exhibited a 1000-fold improvement in efficacy, suggesting that rapid delivery of NO may be more effective at biofilm killing than slow/prolonged NO delivery. The MAP3 silica nanoparticles demonstrated anti-biofilm activity against a range of pathogens with the greatest efficacy (≥99.999% killing) against the species most problematic for wound infections (i.e., gram-negative bacteria). The toxicity of the nanoparticles to fibroblasts was also examined and found to be comparable to or less than currently-applied antiseptics with proven wound-healing benefits.
Acknowledgments
This research was supported by the National Institutes of Health (NIH EB000708). E.M.H. gratefully acknowledges financial support from the Pfizer Analytical Chemistry Graduate Research Fellowship and Novan, Inc. H.S.P. acknowledges support from the Department of Education’s Graduate Assistance in Areas of National Need (GAANN) Fellowship. The authors acknowledge Dr. Channing Der of the Lineberger Comprehensive Cancer Center at UNC for the use of his laboratory’s cell culture resources.
Appendix A. Supplementary Materials
Supplementary data associated with this article can be found in the online version at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.James GA, Swogger E, Wolcott R, deLancey Pulcini E, Secor P, Sestrich J, et al. Biofilms in chronic wounds. Wound Rep Reg. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 2.Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Rep Reg. 2008;16:23–9. doi: 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, Madsen KG, Phipps R, Krogfelt K, et al. Why chronic wounds will not heal: A novel hypothesis. Wound Rep Reg. 2008;16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 4.Percival SL, Bowler P. Biofilms and their potential role in wound healing. Wounds. 2004;16:234–40. [Google Scholar]
- 5.Percival SL, Bowler P, Woods EJ. Assessing the effect of an antimicrobial wound dressing on biofilms. Wound Rep Reg. 2008;16:52–7. doi: 10.1111/j.1524-475X.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- 6.Pruitt BA, McManus AT, Kim SH, Goodwin CW. Burn wound infections: Current status. World J Surg. 1998;22:135–45. doi: 10.1007/s002689900361. [DOI] [PubMed] [Google Scholar]
- 7.DeQueiroz GA, Day DF. Antimicrobial activity and effectiveness of a combination of sodium hypochlorite and hydrogen peroxide in killing and removing Pseudomonas aeruginosa biofilms from surfaces. J Appl Microbiol. 2007;103:794–802. doi: 10.1111/j.1365-2672.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- 8.Davis SC, Martinez L, Kirsner R. The diabetic foot: The importance of biofilms and wound bed preparation. Curr Diabet Rep. 2006;6:439–45. doi: 10.1007/s11892-006-0076-x. [DOI] [PubMed] [Google Scholar]
- 9.Smith AW. Biofilms and antibiotic therapy: Is there a role for combating bacterial resistance by the use of novel drug delivery systems? Adv Drug Delivery Rev. 2005;57:1539–50. doi: 10.1016/j.addr.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 10.White RJ, Cutting K, Kingsley A. Topical antimicrobials in the control of wound bioburden. Ostomy/Wound Management. 2006;52:26–58. [PubMed] [Google Scholar]
- 11.Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14:244–69. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebright JR. Microbiology of chronic leg and pressure ulcers: Clinical significance and implications for treatment. Nurs Clin North Am. 2005;40:207–16. doi: 10.1016/j.cnur.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Eron LJ, Lipsky BA, Low DE, Nathwani D, Tice AD, Volturo GA. Managing skin and soft tissue infections: Expert panel recommendations on key decision points. J Antimicrob Chemother. 2003;52(Suppl S1):i3–i17. doi: 10.1093/jac/dkg466. [DOI] [PubMed] [Google Scholar]
- 14.Wright JB, Lam K, Burrell RE. Wound management in an era of increasing bacterial antibiotic resistance: A role for topical silver treatment. Am J Infect Control. 1998;26:572–7. doi: 10.1053/ic.1998.v26.a93527. [DOI] [PubMed] [Google Scholar]
- 15.Bad bugs, no drugs: As antibiotic discovery stagnates, a public health crisis brews. Infectious Diseases Society of America. 2004:1–35. [Google Scholar]
- 16.Silver S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev. 2003;27:341–53. doi: 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 17.Li XZ, Nikaido H, Williams KE. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J Bacteriol. 1997;179:6127–32. doi: 10.1128/jb.179.19.6127-6132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balin AK, Pratt L. Dilute povidone-iodine solutions inhibit human skin fibroblast growth. Dermatol Surg. 2002;28:210–4. doi: 10.1046/j.1524-4725.2002.01161.x. [DOI] [PubMed] [Google Scholar]
- 19.Rodeheaver G. Controversies in topical wound management. WOUNDS. 1989;1:19–27. [Google Scholar]
- 20.Lawrence JC. The use of iodine as an antiseptic agent. J Wound Care. 1998;7:421–5. doi: 10.12968/jowc.1998.7.8.421. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence JC. A povidone-iodine medicated dressing. J Wound Care. 1998;7:332–6. doi: 10.12968/jowc.1998.7.7.332. [DOI] [PubMed] [Google Scholar]
- 22.Thomas C. Nursing alert - wound healing halted with the use of povidone iodine. Ostomy/Wound Management. 1988;18:30–3. [Google Scholar]
- 23.Goldheim PD. An Appraisal of povidone-iodine and wound healing. Postgrad Med J. 1993;69 (Suppl):S97–S105. [PubMed] [Google Scholar]
- 24.Gulliver G. Arguments over iodine. Nurs Times. 1999;95:68–70. [PubMed] [Google Scholar]
- 25.Stickler D, Hewett P. Activity of antiseptics against biofilms of mixed bacterial species growing on silicone surfaces. Eur J Clin Microbiol Infect Dis. 1991;10:157–62. doi: 10.1007/BF01964448. [DOI] [PubMed] [Google Scholar]
- 26.Presterl E, Suchomel M, Eder M, Reichmann S, Lassnigg A, Graninger W, et al. Effects of alcohols, povidone-iodine and hydrogen peroxide on biofilms of Staphylococcus epidermidis. J Antimicrob Chemother. 2007;60:417–20. doi: 10.1093/jac/dkm221. [DOI] [PubMed] [Google Scholar]
- 27.Torricelli R, Wuthrich B. Life-threatening anaphylactic shock due to skin application of chlorhexidine. Clin Experiment Allergy. 1996;26:112. doi: 10.1111/j.1365-2222.1996.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 28.Autegarden JE, Pecquet C, Huet S, Bayrou O, Leynadier F. Anaphylactic shock after application of chlorhexidine to unbroken skin. Contact Dermatitis. 1999;40:215. doi: 10.1111/j.1600-0536.1999.tb06039.x. [DOI] [PubMed] [Google Scholar]
- 29.Krautheim AB, Jermann THM, Bircher AJ. Chlorhexidine anaphylaxis: Case report and review of the literature. Contact Dermatitis. 2004;50:113–6. doi: 10.1111/j.0105-1873.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 30.Meiller TF, Kelley JI, Jabra-Rizk MA, DePaola LG, Baqui AAMA, Falkler WA. In vitro studies of the efficacy of antimicrobials against fungi. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91:663–70. doi: 10.1067/moe.2001.113550. [DOI] [PubMed] [Google Scholar]
- 31.Fang FC. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2818–25. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang fC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2:820–32. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 33.Raulli R, McElhaney-Feser G, Hrabie JA, Cihlar RL. Antimicrobial properties of nitric oxide using diazeniumdiolates as the nitric oxide donor. Rec Res Devel Microbiol. 2002;6:177–83. [Google Scholar]
- 34.Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide. 2006;14:21–9. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 35.MacMicking J, Xie Q, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–50. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 36.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–53. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hetrick EM, Shin JH, Stasko NA, Johnson CB, Wespe DA, Holmuhamedov E, et al. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2008;2:235–46. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster JR. A tutorial on the diffusability and reactivity of free nitric oxide. Nitric Oxide: Biology and Chemistry. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 39.Bagwe RP, Hilliard LR, Tan W. Surface modification of silica nanoparticles to reduce aggregation and nonspecific binding. Langmuir. 2006;22:4357–62. doi: 10.1021/la052797j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Y, Lohstreter S, Pierce DT, Parisien J, Wu M, Hall C, et al. Silica nanoparticles with continuously tunable sizes: Synthesis and size effects on cellular contrast imaging. Chem Mater. 2008;20:4411–9. [Google Scholar]
- 41.Shin JH, Metzger SK, Schoenfisch MH. Synthesis of nitric oxide-releasing silica nanoparticles. J Am Chem Soc. 2007;129:4612–9. doi: 10.1021/ja0674338. [DOI] [PubMed] [Google Scholar]
- 42.Shin JH, Schoenfisch MH. Inorganic/organic hybrid silica nanoparticles as a nitric oxide delivery scaffold. Chem Mater. 2008;20:239–49. doi: 10.1021/cm702526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beckman JS, Conger KA. Direct measurement of dilute nitric oxide in solution with an ozone chemiluminescent detector. Methods in Enzymology. 1995;7:35–9. [Google Scholar]
- 44.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 45.McElhaney-Feser G, Raulli RE, Cihlar RL. Synergy of nitric oxide and azoles against Candida species in vitro. Antimicrob Agents Chemother. 1998;42:2342–6. doi: 10.1128/aac.42.9.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hancock V, Ferrieres L, Klemm P. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol Lett. 2007;267:30–7. doi: 10.1111/j.1574-6968.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 47.Schinabeck MK, Long LA, Hossain MA, Chandra J, Mukherjee PK, Mohamed S, et al. Rabbit model of Candida albicans biofilm infection: Liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Chemother. 2004;48:1727–32. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceri H, Olson M, Morck D, Storey D, Read R, Buret A, et al. The MBEC assay system: Multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol. 2001;337:377–85. doi: 10.1016/s0076-6879(01)37026-x. [DOI] [PubMed] [Google Scholar]
- 49.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–6. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin JH, Stevens EV, Der C, Schoenfisch MH. Nitric oxide-releasing silica nanoparticles: Synthesis, characterization, and efficacy against ovarian cancer cells. PMSE Preprints. 2006;95:685–6. [Google Scholar]
- 51.Ryan TJ. Infection following soft tissue injury: Its role in wound healing. Curr Opin Infect Dis. 2007;20:124–8. doi: 10.1097/QCO.0b013e32801a3e7c. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz C, McMurry LM, Levy SB. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J Bacteriol. 2008;190:1290–7. doi: 10.1128/JB.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microb Infect. 2000;2:1051–60. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 54.Hsueh PR, Teng LJ, Yang PC, Chen YC, Ho SW, Luh KT. Persistence of a multidrug-resistant Pseudomonas aeruginosa clone in an invasive care burn unit. J Clin Microbiol. 1998;36:1347–51. doi: 10.1128/jcm.36.5.1347-1351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutherland IW. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 56.Wong T, McGrath JA, Navsaria H. The role of fibroblasts in tissue engineering and regeneration. British J Dermatol. 2007;156:1149–55. doi: 10.1111/j.1365-2133.2007.07914.x. [DOI] [PubMed] [Google Scholar]
- 57.Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Zhu YK, Wang H, Kohyama T, Wen F, Rennard SI. Effect of nitric oxide donors on human lung fibroblast proliferation in vitro. Chest. 2001;120:13–4. [Google Scholar]
- 59.Witte MB, Thornton FJ, Efron DT, Barbul A. Enhancement of fibroblast collagen synthesis by nitric oxide. Nitric Oxide. 2000;4:572–82. doi: 10.1006/niox.2000.0307. [DOI] [PubMed] [Google Scholar]
- 60.Schwentker A, Billiar TR. Nitric oxide and wound repair. Surg Clin N Am. 2003;83:521–30. doi: 10.1016/S0039-6109(02)00207-4. [DOI] [PubMed] [Google Scholar]
- 61.Pyo HC, Kim YK, Whang KU, Park YL, Eun HC. A comparative study of cytotoxicity of topical antimicrobials to cultured human keratinocytes and fibroblasts. Korean J Dermatol. 1995;33:895–906. [Google Scholar]
- 62.Hetrick EM, Prichard HL, Klitzman B, Schoenfisch MH. Reduced foreign body response at nitric oxide-releasing subcutaneous implants. Biomaterials. 2007;28:4571–80. doi: 10.1016/j.biomaterials.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isenberg JS, Ridnour LA, Espey MG, Wink DA, Roberts DD. Nitric oxide in wound-healing. Microsurg. 2005;25:442–51. doi: 10.1002/micr.20168. [DOI] [PubMed] [Google Scholar]