SUMMARY
The nucleotide excision repair pathway corrects many structurally unrelated DNA lesions. Damage recognition in bacteria is performed by UvrA, a member of the ABC ATPase superfamily whose functional form is a dimer with four nucleotide-binding domains (NBD), two per protomer. In the 3.2-Å structure of UvrA from Bacillus stearothermophilus, we observe that the nucleotide-binding sites are formed in an intramolecular fashion and are not at the dimer interface as is typically found in other ABC ATPases. UvrA also harbors two unique domains; we show that one of these is required for interaction with UvrB, its partner in lesion recognition. In addition, UvrA contains three zinc modules, the number and ligand sphere of which differ from previously published models. Structural analysis, biochemical experiments, surface electrostatics and sequence conservation form the basis for models of ATP-modulated dimerization, UvrA-UvrB interaction and DNA binding during the search for lesions.
INTRODUCTION
Among the various DNA repair mechanisms, nucleotide excision repair (NER) is unique in its ability to remove a broad range of structurally unrelated DNA lesions (Goosen and Moolenaar, 2001; Truglio et al., 2006a). NER is a multi-step, ATP-dependent process that involves damage recognition, incisions on the 3′ and 5′ sides of the lesion, repair synthesis, and ligation. The mechanism of NER is essentially conserved throughout evolution. In bacteria, the damage recognition and incision steps are carried out by three proteins, UvrA, UvrB, and UvrC.
The UvrAB complex operates as a damage sensor by monitoring DNA and recognizing sites of damage. After a lesion is encountered, UvrA dissociates from the complex and leaves UvrB stably bound in the so-called pre-incision complex (Orren and Sancar, 1990). Damage searching, dissociation of UvrA, and formation of the pre-incision complex are regulated by ATP binding and hydrolysis (Goosen and Moolenaar, 2001; Mazur and Grossman, 1991; Moolenaar et al., 2000a; Oh et al., 1989). The UvrB·DNA pre-incision complex recruits the endonuclease UvrC to the site of damage. UvrC makes the first incision at the 4th or 5th phosphodiester bond 3′ to the lesion, followed by the second incision at the 8th phosphodiester bond 5′ to the lesion (Verhoeven et al., 2000). Additional processing reactions lead to removal of the damage-containing oligonucleotide and restoration of the original DNA sequence (Orren et al., 1992).
UvrA plays a key role in DNA damage recognition, since it preferentially binds to damaged DNA in the absence of other NER components, and is required for the loading of UvrB to form the pre-incision complex at site of a DNA lesion (Truglio et al., 2006a). At physiological concentrations, UvrA is a dimer and this is believed to represent the functional form of the protein.
UvrA belongs to the ATP-binding cassette (ABC) superfamily of ATPases. Proteins of this superfamily couple ATP hydrolysis to diverse cellular functions that include transport of solutes through membranes, ribosome biogenesis, chromosome condensation, and DNA repair (Hopfner and Tainer, 2003; Junop et al., 2001; Lebbink and Sixma, 2005; Locher, 2004; Sixma, 2001). ABC ATPases share a number of sequence and structural features. First, ABC ATPases contain a 200–250 residue nucleotide-binding domain (NBD), which harbors several conserved functional regions termed Walker A/P-loop, Q-loop, ABC signature, Walker B, D-loop, and H-loop/switch (Linton, 2007). These elements surround the bound nucleotide. Second, ABC ATPases exhibit composite nucleotide-binding sites. One NBD (NBD-I) provides the Walker A, Walker B, Q-loop, and H-loop substructures, while the ABC signature motif and D-loop are donated by the second NBD (NBD-II). The second nucleotide is bound in a similar manner except that the NBDs that provide the contacts are switched. In some ABC ATPases, the two NBDs involved in the formation of the composite sites lie on different protein chains that come together across the intermolecular interface upon dimerization. In other ABC ATPases whose functional form is a monomer, the two NBDs lie within the same chain and the composite sites are formed in an intramolecular fashion. UvrA presents a unique case as it harbors two ABC modules (Doolittle et al., 1986; Gorbalenya and Koonin, 1990), but also functions as a dimer. Thus, the NBDs of dimeric UvrA form four composite nucleotide-binding sites. In principle, these could be arranged in either an intra- or intermolecular manner, which could not readily be distinguished absent structural information.
In addition to the nucleotide-binding substructures, UvrA has a number of specific structural elements that are not found in other ABC ATPases. For example, the region between the Walker A and ABC signature motifs in the N-terminal NBD (NBD-I) of UvrA contains a large inserted segment whose functions are unknown. Also, a Zn structural module was predicted to lie between the Walker A and ABC signature motifs in each NBD; the roles of these Zn modules have been studied experimentally (Croteau et al., 2006; Moolenaar et al., 2000b; Navaratnam et al., 1989; Visse et al., 1993; Wang et al., 1994).
Understanding the molecular mechanisms of DNA damage recognition and repair requires detailed structural information on the NER proteins and the complexes they form with DNA substrates. To date, the structures of UvrB (alone and in complex with short oligonucleotides) (Alexandrovich et al., 2001; Eryilmaz et al., 2006; Machius et al., 1999; Nakagawa et al., 1999; Sohi et al., 2000; Theis et al., 1999; Truglio et al., 2004; Truglio et al., 2006b; Waters et al., 2006) and fragments of UvrC (Karakas et al., 2007; Singh et al., 2002; Truglio et al., 2005) have been described. While these studies have expanded our understanding of the structural basis of NER, no structural information on UvrA is currently available and the precise mechanism of lesion recognition remains to be elucidated.
In order to better understand the structure and function of UvrA, we have determined the crystal structure of Bacillus stearothermophilus UvrA bound to ADP at 3.2-Å resolution. The structure of the UvrA dimer reveals unique features of nucleotide-modulated dimerization not seen in other ABC ATPases. An additional unexpected finding concerns the number and coordination scheme of the three structural Zn atoms in each UvrA protomer, which differ from that predicted. An especially prominent feature of the overall structure is the presence of two inserted domains in the N-terminal nucleotide-binding domain (NBD-I) of UvrA, which are not found in other ABC ATPases. On the basis of biochemical experiments, we show here that one of these domains is necessary for interaction with UvrB; this is the first mapping of UvrB-interaction surface on UvrA. We have used the crystallographic model, biochemical experiments, and calculations of surface electrostatics and sequence conservation to derive models for the regulation of UvrA dimerization by ATP, UvrA-UvrB interaction and DNA binding.
RESULTS AND DISCUSSION
Description of the Structure
The structure of Bacillus stearothermophilus UvrA was solved at 3.2-Å resolution by multiwavelength anomalous diffraction (MAD) using selenomethionine-substituted protein. The final model consists of a UvrA dimer (residues 1–153, 200–309, and 314–949 of protomer A; residues 1–151, 205–229, and 247–949 of protomer B), 4 ADP molecules, 6 Zn atoms, and 4 water molecules, with a crystallographic R factor of 25.3 % and Rfree of 29.2 % (Table 1). The electron density for residues 89–92 and 355–369 of protomer A, and residues 142–143 of protomer B, was not satisfactorily resolved; therefore, these regions of our model should be considered tentative.
Table 1.
Data collection, phasing, and refinement statistics.
| BstUvrA (Space group: P21, Cell parameters: a = 102.71 Å, b = 94.72 Å, c = 130.48 Å, α = γ = 90.0°, and β = 108.8°) | ||||
|---|---|---|---|---|
| Data collectiona | SeMet λ1 | SeMet λ2 | SeMet λ3 | Zn λ1 |
| Source | APS 24ID–C | APS 24ID–C | APS 24ID–C | APS 24ID–C |
| Wavelength (Å) | 0.97925 | 0.97941 | 0.96403 | 1.28281 |
| Resolution (Å) | 50–3.20 (3.31–3.20) | 50–3.25 (3.37–3.25) | 50–3.25 (3.37–3.25) | 50–3.80 (3.94–3.80) |
| Rsym (%)b | 7.9 (30.5) | 7.8 (35.2) | 7.4 (33.7) | 7.6 (17.7) |
| Total no. of obs. | 266945 | 250620 | 253614 | 123992 |
| No. of unique obs. | 38657 | 36928 | 36934 | 22212 |
| Completeness (%) | 98.3 (85.9) | 97.6 (80.6) | 98.3 (86.0) | 93.2 (82.6) |
| <I>/σ<I> | 18.4 (4.5) | 19.0 (3.6) | 21.7 (3.7) | 18.8 (7.2) |
|
| ||||
| Phasing | ||||
| Overall figure of meritc | 0.58778 | |||
|
| ||||
| Refinement | ||||
| Resolution (Å) | 33–3.20 | |||
| No. of non-hydrogen atoms | 13988 | |||
| Rwork (%)d | 25.30 | |||
| Rfree (%)e | 29.21 | |||
| Mean B value (Å2)f | 68.793 | |||
| Rmsd bond (Å)f | 0.007 | |||
| Rmsd angle (°)f | 1.247 | |||
| Ramachandran plot analysisg (% most favored, additional allowed, generously allowed, disallowed) | 85.9, 13.1, 1.0, 0.0 | |||
All SeMet data were measured from a single crystal and Zn data were from a different crystal. Values in parentheses are for the highest resolution shell.
Rsym = Σ|I-<I>|/ΣI where I is the integrated intensity of a given reflection.
Figure of merit = <|ΣP(α)eia/Σ|P(α)|>, where a is the phase and P(α) is the phase probability distribution.
Rwork = Σ|F(obs)-F(calc)|/ΣF(obs).
Rfree = Σ|F(obs)-F(calc)|/ΣF(obs), calculated using 9.9% of the data.
From REFMAC.
From PROCHECK.
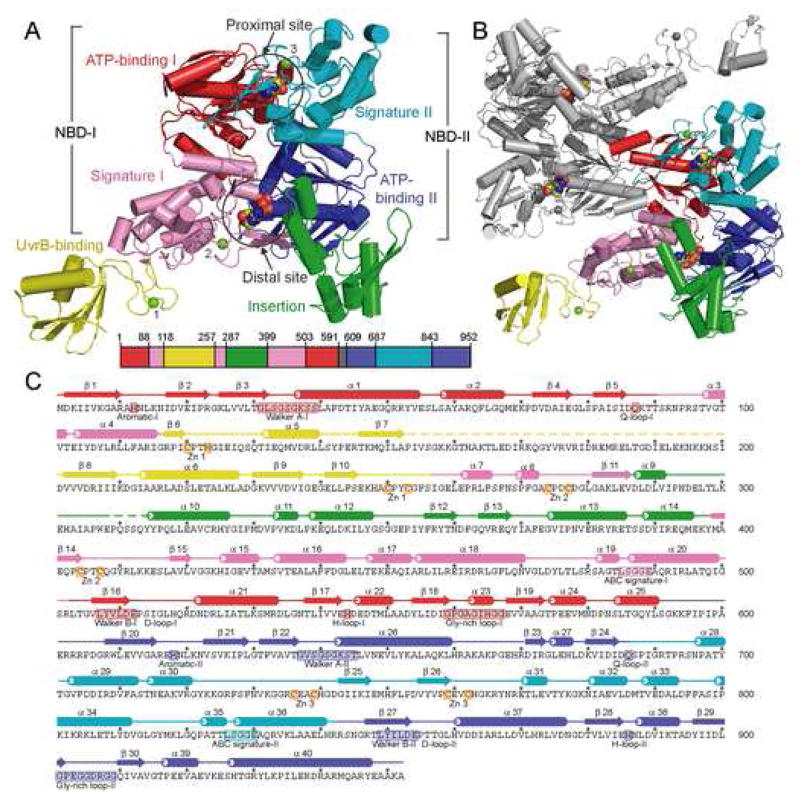
Each UvrA protomer contains two ABC ATPase structural modules, which we have designated nucleotide binding domain I (NBD-I) and nucleotide-binding domain II (NBD-II), connected by a flexible linker (residues 591–608). NBD-I can be further divided into ATP-binding domain I (residues 1–87 and 503–590) and signature domain I (residues 88–117, 257–286, and 399–502), while NBD-II can be divided into ATP-binding domain II (residues 609–686 and 843–952) and signature domain II (residues 687–842). The ATP-binding domain and the signature domain together comprise the nucleotide-binding domain (NBD) as observed in other ABC ATPases. Inserted in NBD-I are two domains not found in other ABC ATPases, namely the UvrB-binding domain (residues 118–256, see below) and the insertion domain (residues 287–398); these are connected to the signature domain I through Zn modules (Figure 1). The two NBDs of UvrA are structurally related and can be superimposed with an RMSD of 2.1 Å over 296 Ca atoms (Figure S1). The similarity between the N- and C-terminal NBDs is consistent with the proposal that UvrA is a tandem ABC ATPase resulting from gene duplication (Doolittle et al., 1986).
Figure 1.
Structure of Bacillus stearothermophilus UvrA. (A) Overall structure of the UvrA monomer, with a-helices depicted as cylinders and β-strands as arrows. The protein is colored by domains, ATP-binding I (1–87,503–590), red; signature I (88–117, 257–286, 399–502), pink; ATP-binding II (609–686, 843–952), blue; signature II (687–842), cyan; UvrB-binding (118–256), yellow; insertion (287–398), green; and linker (591–608), gray; with the Zn atoms, numbered by module, in light green. The bound ADP molecules are shown as space-filling models. The location of each domain, colored as above, is projected onto the primary sequence of BstUvrA, shown as a bar. (B) Overall structure of the UvrA dimer as observed in the crystal asymmetric unit. One protomer is colored as in (A), and the second protomer in gray. (C) Secondary structure assignment of BstUvrA, colored by domains as above. Disordered regions are depicted as dashed lines. Locations of the conserved ABC ATPase motifs, glycine-rich loops, and Zn-coordinating residues are depicted on the amino acid sequence.
The two ATP-binding domains each consists of two β-sheets and six a-helices. These domains form nucleotide binding sites that are common in all ABC-type ATPases and contain the Walker A and Walker B motifs, as well as the Q-, D-, and H-loops. The locations of the ATPase motifs in the primary sequence of UvrA are shown in Figure 1C. The signature domains contain the ABC signature motif and are positionally equivalent to the helical domain in the NBD of ABC transporters (Gaudet and Wiley, 2001; Schmitt et al., 2003). The UvrA signature domains are, however, significantly larger than the helical domains of transporters, comprising roughly twice as many α-helices and a unique, highly conserved Zn module (Figures 1, S1). The signature domains are joined to the ATP-binding domains through the Q-loop, which has been proposed to be the site of conformational changes that couple ATP hydrolysis to function in ABC proteins (Hopfner and Tainer, 2003; Linton, 2007).
Nucleotide-Binding Sites
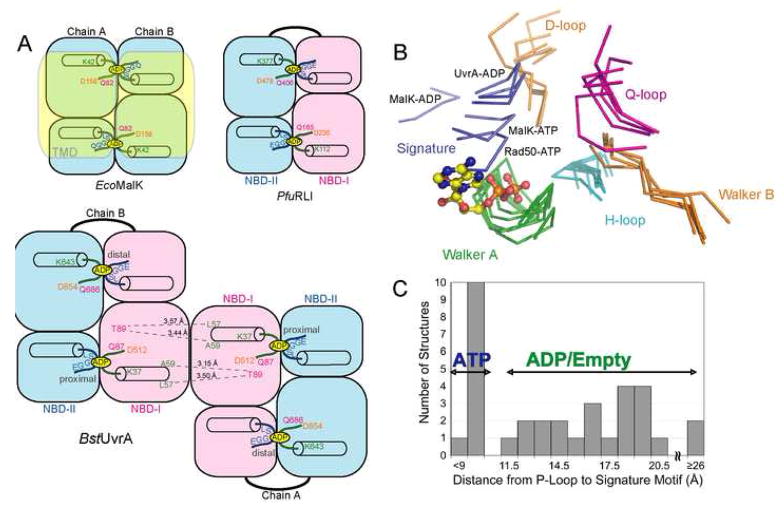
The UvrA protomer is a tandem ABC ATPase, possessing two composite nucleotide-binding sites that we have designated the proximal and the distal sites, respectively (Figures 1A, 1B, 2A). The proximal site is located at the interface between ATP-binding domain I and signature domain II, closest to the dimer interface. The distal site is located further away from the dimer contacts at the interface between ATP-binding domain II and signature domain I (Figures 1B, 2A). For each composite site, the ATP-binding domain provides the Walker A and Walker B motifs, as well as the Q- and H-loop, whereas the ABC signature motif and D-loop are donated by the signature domain. The composite sites in UvrA are formed by the NBDs present within the same polypeptide chain; this arrangement resembles that in the tandem ABC protein RNaseL inhibitor (RLI) (Karcher et al., 2005). Unlike other tandem ABC ATPases, however, the functional form of UvrA is a dimer. Thus, there are four sites that can be occupied by nucleotides (Figures 1B, 2A).
Figure 2.
Structural comparison of the NBDs of BstUvrA and other ABC ATPases. The conserved ATPase motifs are colored as follows: Walker A/P-loop, green; Walker B and D-loop, orange; ABC signature motif, blue; Q-loop, magenta; and H-loop/switch, cyan. (A) Arrangement of the NBDs in E. coli MalK, Pyrococcus furiosus RNaseL inhibitor (RLI), and BstUvrA. Key residues important for nucleotide binding and interactions across the dimer interface are shown. The transmembrane portion of MalK is depicted in transparent yellow. (B) NBDs of UvrA were superimposed with the NBDs of the maltose transporter MalK (ADP-bound, PDB code 2AWO; ATP-bound, PDB code 1Q12), and Rad50 (ATP-bound, PDB code 1F2U) using their respective ATP-binding domains. All the motifs are from the same NBD except for the ABC signature motif and the D-loop, which are part of the opposing NBD. The bound ADP is from the proximal site of UvrA protomer A and is represented as ball-and-stick. (C) Histogram showing the distance between the Cα atoms of the conserved Lys residue in the Walker A motif and Ser residue in the ABC signature motif in the structures of ABC ATPases solved in the dimeric state (PDB codes 2R6F, 2AWO, 1Q1B, 1Q1E, 2AWN, 1YQT, 1L7V, 2ONK, 1Q12, 1F2U, 1XEF, 1XEX). Asterisk denotes the average distance for the four nucleotide-binding sites in UvrA.
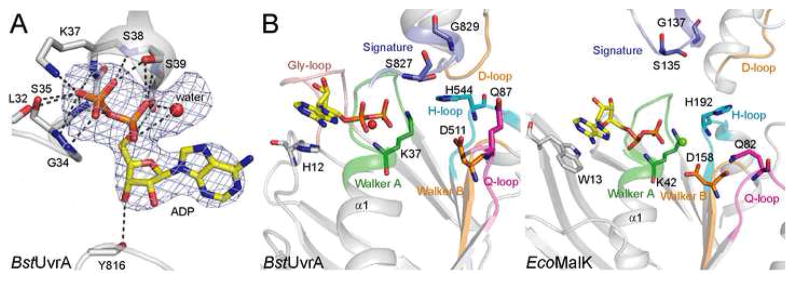
In the structure, all four nucleotide-binding sites in the UvrA dimer are occupied by ADP, which was present in the crystallization buffer. The adenine ring of ADP stacks against H12 of the proximal site (Figure 3B) and H618 of the distal site. These conserved histidine residues in UvrA align with the ABC aromatic residue motif and interact with the nucleotide in much the same manner as that seen in the NBDs of ABC transporters (Kim et al., 2006; Linton, 2007). The phosphate moiety of ADP is recognized by H-bonding to the residues of the Walker A motif (Figure 3A), similar to what is observed in the structures of other ABC ATPases. Although MgCl2 was included in the crystallization buffer, no electron density was observed at the position adjacent to the β-phosphate normally occupied by Mg2+ in the structures of ABC ATPases bound to Mg-ADP. However, we note a peak of positive electron density close to the a-phosphate. As this is an unusual location for Mg2+, we have modeled this peak as water only for the purpose of crystallographic refinement (Figure 3A). The identity of this peak should be considered tentative.
Figure 3.
Nucleotide binding by BstUvrA. (A) Binding of ADP by the Walker A motif. ADP and the interacting protein residues are shown as sticks, with the unknown solvent component, modeled as a water molecule, depicted as a red sphere. Hydrogen bonds are drawn as dashed lines. The difference electron density calculated with the nucleotide and water molecule omitted from the model is shown at 3σ. (B) Comparison of the nucleotide-binding sites from UvrA (left) and MalK (right, PDB code 2AWO). The ATPase motifs are colored as in Figure 2. Important conserved residues are shown as sticks. The conserved glycine-rich loop found in UvrA is peach. All four nucleotide-binding sites in the UvrA dimer are structurally very similar, thus only the proximal site of protomer A is shown.
In addition to the motifs common in all ABC ATPases, UvrA contains a unique glycine-rich loop that forms part of the nucleotide-binding site (peach in Figure 3B). This loop corresponds to residues 559–567 of the proximal site and residues 901–909 of the distal site. Each of these regions contains 5 glycine residues, four of which are essentially invariant in all UvrA orthologs. Deletion mutagenesis studies have established the importance of this loop for the NER activity of UvrA (Claassen and Grossman, 1991; Kulkarni et al., 2006), but its precise function is not known. The structure reveals that the Gly-rich loop caps off the nucleotide-binding site towards the side of the ribose ring, and although residues of the loop do not contact the nucleotide directly, they appear to buttress nucleotide-interacting residues. It is possible that additional roles for the Gly-rich loop will be evident when structures in different nucleotide states become available.
When the structures of the NBDs from UvrA are compared to other ABC ATPases, we observe that the signature motifs, which are not included in the superposition, align well. However, the unique elements of the NBDs are quite divergent (Figure S1). Closer examination of the active site loops forming each nucleotide-binding site reveals that the distance between the Walker A and the ABC signature motifs from opposing NBDs of UvrA (an average of 12.6 Å for the 4 sites) is shorter than that in most NBD structures solved with bound ADP or without nucleotide, but longer than in the ATP-bound structures (Figures 2B, C). This is illustrated in the comparison between the nucleotide-binding sites of UvrA and E. coli MalK (PDB code 2AWO) (Figure 3B), which were both determined in the presence of ADP. We note that while the overall structures are very similar, the positions of the signature motif relative to the Walker A motif, and the conformations of some of the active site loops in UvrA and MalK are different. Therefore, it appears that in the absence of the -phosphate, the active site loops of the ATPases can assume different conformations, and only upon binding of ATP do they become properly positioned for hydrolysis (Chen et al., 2003; Karpowich et al., 2001).
Dimerization of UvrA
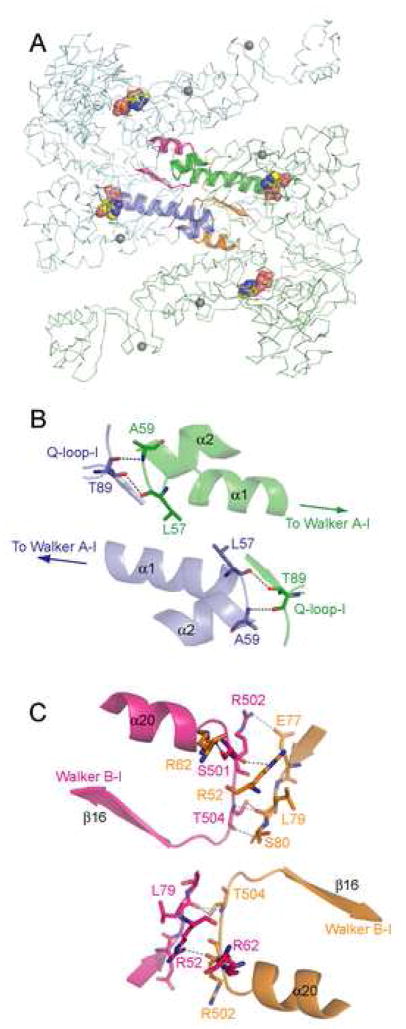
The dimer of UvrA observed in the crystal asymmetric unit displays an unexpected dimerization mode. Unlike other structures of dimeric ABC ATPases, the UvrA dimer interface does not contain bound nucleotides, and comprises regions of the NBDs not seen to participate in dimerization in other ABC proteins (Figures 1B, 2A). Residues that form the dimer interface are well conserved and are mostly borne on ATP-binding domain I and signature domain I, with the exception of H750, F751, and L752, which are part of the β-substructure in signature domain II. The interface buries ~4300 Å2 of accessible surface area, involving a large number of hydrophobic interactions and an extensive hydrogen-bonding network. The high degree of conservation and a large buried surface area suggest that the dimer interface observed in the asymmetric unit is not due to crystal packing. This extensive interaction might explain why dimerization of UvrA is not strictly dependent on the presence of nucleotide, and why the Kd of dimer formation by UvrA (~10 nM) (Myles et al., 1991) is much lower than those of other ABC ATPases, which are in the micromolar range (Zaitseva et al., 2005).
We observe a second potential dimer interface between the insertion domain (residues 287–398) of two UvrA monomers in the crystal. However, a UvrA mutant lacking this domain is still dimeric. Further, this interface buries only ~2600 Å2 and is formed by residues that are poorly conserved. On this basis, we considered this interface a likely artifact of crystal packing.
The precise role for ATP in UvrA dimerization and the interaction between UvrA, damage DNA, and other NER components is not well understood. Analysis of the UvrA dimer interface provides clues that could serve as the basis for future experiments. T89, which belongs to the Q-loop-I of one protomer, participates in the dimer interface by forming H-bonds with the main chain atoms of L57 and A59 of the opposing protomer (Figure 4A, B). The Q-loop has been implicated in the coupling of ATP hydrolysis to conformational changes that are crucial for function of ABC proteins. In ABC transporters, the Q-loop interacts with a cytoplasmic loop of the transmembrane domain (Dalmas et al., 2005; Locher et al., 2002), while in Rad50 and SMC proteins, it is connected to the coiled-coil region that has been implicated in their DNA tethering function (Hopfner and Tainer, 2003). Therefore, the interaction observed at the UvrA dimer interface might be important in the regulation of UvrA dimerization state by ATP binding and hydrolysis.
Figure 4.
BstUvrA dimer interface. Cα trace of protomers A and B are shown in pale green and pale blue, respectively, with the Zn atoms in gray. The regions involved in polar contacts across the dimer interface are shown using ribbon diagram (green and orange, protomer A; blue and magenta, protomer B). Hydrogen bonds are depicted as dashed lines and the bound ADP molecules as space-filling models. Illustrations in panels A–C are shown in the same orientation as in Figure 1B. (A) UvrA dimer. (B) Interactions between the Q-loop-I and the loop following α-helix 1, which contains the Walker A–I motif at its N-terminus. (C) Interactions between the loop preceding Walker B-I and residues of the ATP-binding domain I of the opposing monomer.
Intriguingly, the C-terminus of a-helix 1, which contains the Walker A-I motif at its N-terminus, contributes directly to the dimer interface via residue L57. Additional interactions are also found between the loop (residues 502-504) preceding the Walker B-I motif, and residues 52, 62, and 77-79 of the opposing monomer (Figure 4A, C). These interactions could account for the loss of cooperativity in ATP hydrolysis upon mutation of the proximal site (Myles et al., 1991).
UvrA-Specific Structural Features: Identification of the UvrB-Binding Domain
Little is known about how UvrA contacts UvrB in the UvrAB complex. Previous studies suggest that elements within the first 230 residues of UvrA are important for contacts to UvrB (Claassen and Grossman, 1991). In the structure of UvrA, NBD-I contains two inserted domains (residues 118–257 and 287–399, respectively) whose functions are unknown. Database searches (Holm and Sander, 1993) failed to find significant similarity to any structure in the Protein Data Bank.
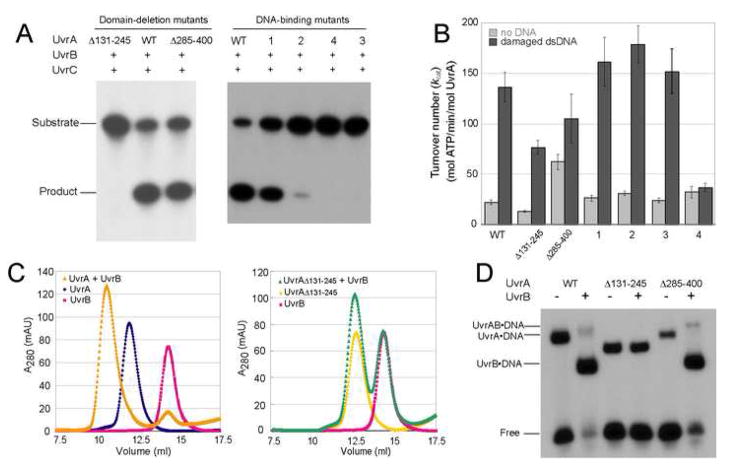
Deletion mutagenesis and biochemical analyses were performed to study the roles of these domains. Two mutant proteins were made; UvrAΔ131–245 has residues 131-245 deleted and replaced with a GT dipeptide, while UvrAΔ285–400 has residues 285-400 deleted and replaced in the same manner. Both deletion mutant proteins eluted as dimers upon size exclusion chromatography and exhibited DNA-stimulated ATPase activity (Figure 5B). These observations suggest that they are properly folded and further indicate that neither of the inserted domains contributes to the dimer interface, consistent with our structure.
Figure 5.
Biochemical characterizations of BstUvrA mutants. The deleted residues are indicated and the mutated regions in DNA-binding mutants 1–4 are defined in Figure 6C and 6D. (A) Incision of the 5′ end-labeled 50-bp duplex containing the N3-menthol lesion to yield a 19-bp product. (B) ATP hydrolysis as monitored by the coupled enzyme system (see supplemental experimental procedures), reported as mean turnover number (kcat) (mol ATP/min/mol UvrA) ± standard error of the mean (n = 8). (C) UvrA-UvrB interaction as assessed by size exclusion chromatography. (D) Binding of UvrA and UvrB to the 50-bp duplex containing the N3-menthol lesion. The position of the UvrA·DNA, UvrAB·DNA, and UvrB·DNA complexes are indicated.
An incision assay (Verhoeven et al., 2002) revealed that UvrAΔ131–245 was completely inactive, pointing to an important role for this region in incision activity. On the other hand, UvrAΔ285–400 exhibited near wild-type incision activity (Figure 5A), suggesting that residues 285-400 are not critical for UvrA function in vitro.
To examine whether the loss of function in UvrAΔ131–245 was due to a defect in UvrB interaction, the mixture of wild-type UvrA or UvrAΔ131–245 and UvrB was analyzed by size exclusion chromatography in the presence of Mg-ATP (Figure 5C, S2). Wild-type UvrA and UvrB eluted together as a complex, while UvrAΔ131–245 failed to associate stably with UvrB. This defect was confirmed by an electrophoretic mobility shift assay (Figure 5D), in which no loading of UvrB onto the DNA was observed when UvrAΔ131–245 was used. UvrAΔ285–400, on the other hand, retained the ability to load UvrB onto DNA in a damage-specific manner. The isolated UvrA domain construct (residues 131-245) was also found to interact with isolated domain 2 of UvrB (residues 149-250) (data not shown), which has been identified as the UvrA-binding surface on UvrB (Truglio et al., 2004). Taken together, these analyses clearly demonstrate that the inserted domain containing residues 131-245 is required for the interaction between UvrA and UvrB and is important for NER function. On this basis, we named this element of the UvrA structure, the UvrB-binding domain.
Previous studies indicate that electrostatic forces contribute primarily to the UvrA-UvrB interaction (Truglio et al., 2004). Consistent with these results, analysis of UvrA sequence conservation reveals that most of the conserved charged residues in the UvrB-binding domain are located on the one side that is solvent-exposed. These conserved residues include D205, R206, R216 and E222, the first three of which form a highly conserved patch on the surface of the domain that could play a key role in the UvrA-UvrB interaction. We further note the presence of numerous conserved hydrophobic residues on the same side of the domain, namely I150, L151, A152, P153, I154, V204, L217, A224, and L225. These might also contribute to the interaction with UvrB.
Zn-Binding Modules in UvrA
The presence of one Zn structural module in each NBD was predicted on the basis of sequence analysis and biochemical studies (Doolittle et al., 1986; Navaratnam et al., 1989; Visse et al., 1993; Wang et al., 1994). However, analysis of Zn anomalous diffraction as part of the UvrA structure determination revealed the presence of three Zn atoms bound to each UvrA monomer (Figure 1A). These Zn modules do not adopt the classical TFIIIA-type Zn finger structure and most likely play structural roles. The first Zn atom is coordinated by C120, H123, C250, and C253 (Zn module 1), and is located between the signature domain I and the UvrB-binding domain. Zn module 1 in UvrA most resembles the Zn module in the protein YfgJ (PDB code 2JNE) (Figure S2). The second Zn atom is coordinated by C274, C277, C404, and C407 (Zn module 2) and is located between the signature domain I and the insertion domain. The third Zn atom is coordinated by C736, C739, C759, and C762 (Zn module 3) and connects the helical region of the signature domain II to the dimer interface. Only the coordination scheme for Zn module 3 agrees with the previously published model (Doolittle et al., 1986). Zn modules 2 and 3 of UvrA resemble the ‘Zn ribbon’ found in the cysteine-rich (CR) domain of the chaperone DnaJ (PDB code 1EXK) (Figure S2). The conserved motif found in the DnaJ Zn ribbon is CxxCxGxG repeated twice for each Zn site. For Zn modules 2 and 3 in UvrA, the motif is CxxCxGxG-(x)n-CxxCxGxR. Substitution of Arg or Lys for the second conserved Gly residue was also found in some DnaJ homologs, in which the positively charged side chain is surrounded by exposed carbonyl groups (Martinez-Yamout et al., 2000). Similar interactions between the Arg side chain and several carbonyl groups are observed in UvrA and might contribute to the local structural stability.
Mutational analyses of the Zn-coordinating residues in modules 1 and 3, and the nearby protein residues, have demonstrated their importance in DNA repair and replication (Croteau et al., 2006; Moolenaar et al., 2000b; Navaratnam et al., 1989; Visse et al., 1993; Wang et al., 1994). The coordination schemes of the three Zn modules, revealed by the UvrA structure, will enable experiments that more precisely define their specific roles.
Mutational studies and analysis of the surface properties of UvrA reveal the DNA binding surface
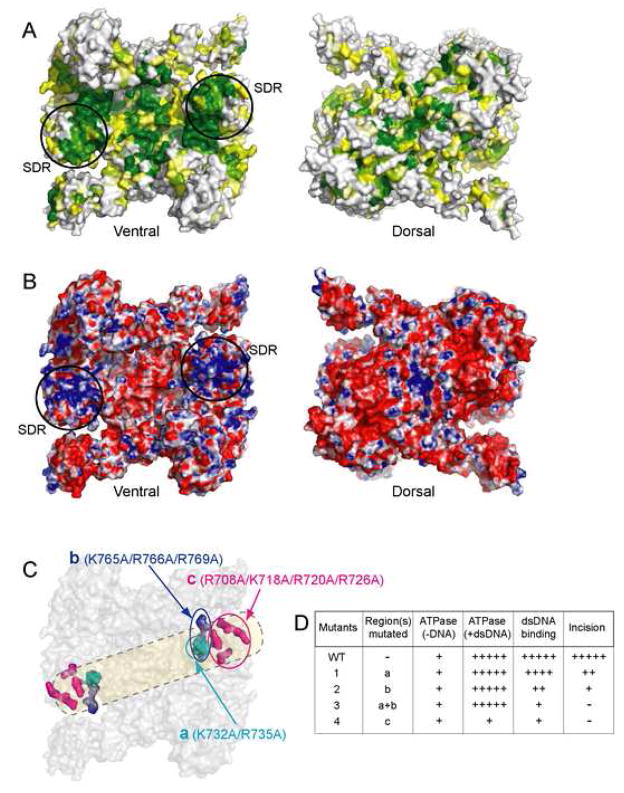
One of the main unanswered questions in the mechanism of NER centers around how damaged DNA is recognized. However, the regions of UvrA that are involved in DNA binding are not known. In order to gain insight into how UvrA might interact with DNA, we inspected the surface for conserved amino acids and the distribution of charged residues (Figures 6A, B). The ventral surface of UvrA is strikingly concave, whereas its dorsal surface is quite flat. Mapping sequence conservation (Figure S4) to the surface of UvrA reveals several regions of interest on the ventral surface, while the dorsal surface is less conserved. These are 1) around the ATPase active sites, 2) at the dimer interface, and 3) in the concave cleft of the UvrA dimer (Figure 6A). Further, the highly conserved regions in the concave cleft of the UvrA dimer coincide with regions of positive electrostatic potential (Figure 6B). The importance of these regions is indicated by their alignment with the structurally diverse region (SDR), a functionally significant region in the ABC superfamily of proteins. These generally perform functions unique to each family member (Hopfner et al., 2001; Schmitt et al., 2003).
Figure 6.
A DNA-binding model for UvrA as inferred from sequence conservation and electrostatic surface potential. The ventral surface of the UvrA dimer is shown in the same view as in Figure 1B, while the dorsal surface is rotated 180° about a vertical axis. (A) Sequence conservation index was calculated and projected onto BstUvrA molecular surface. The color ranges from yellow (60% conservation) to dark green (≥96% conservation). Residues with <60% conservation are colored white. (B) Electrostatic potential for BstUvrA. The colors range from red (corresponding to an electrostatic potential energy of −10 kBT, where kB is the Boltzmann’s constant and T is the temperature) to blue (+10 kBT). In (A) and (B), the structurally diverse region (SDR), which is part of the signature domain II, is circled. (C) Sequence conservation and surface electrostatics in combination with biochemical analysis permit the approximate path for DNA to be defined. Biochemical studies were performed on wild-type and mutant UvrA containing K→A/R→A substitutions at the positions depicted in colors; the rest of the protein is shown in gray. The path of the DNA is drawn in transparent yellow. (D) Summary of biochemical analyses of the mutants.
Towards building a working model of the interaction between UvrA and damaged DNA, we have targeted several conserved positively charged residues for mutation and subsequent biochemical analyses (Figure 6C, 6D, 5A, 5B, S5, and S6). The mutations were made in three regions (a: K732/R735, b: K765/R766/R769, and c: R708/K718/R720/R726, Figure 6C). Four mutant proteins were constructed with Lys and Arg residues mutated to Ala. Mutants 1, 2, 3 and 4 have changes in regions a, b, a+b, and c, respectively. The mutant proteins migrated on a gel filtration column as dimers, indicating that the substitutions did not disrupt dimerization. Also, both protomers in the UvrA dimer contain the mutations. An incision assay revealed that only mutant 1 could support a significant level of incision (~70% of wild-type incision activity). Mutant 2 gave little incision product, whereas mutants 3 and 4 were inactive in this assay (Figure 5A). Probing further, we observed that the interaction with damaged DNA, as judged by electrophoretic mobility shift, was impaired in all mutant proteins. The apparent affinity of the UvrA mutants towards damaged DNA (1>2>3>4) is consistent with their incision activity (Figure 5A, S5, S6).
The four mutant proteins displayed wild-type basal ATPase activity, confirming that they are properly folded (Figure 5B). Further, the ATPase activity of mutants 1, 2, and 3 was stimulated by the presence of damaged DNA, suggesting that even a weak interaction with DNA can result in stimulation. This stimulation was not observed in mutant 4, which is the most impaired for DNA binding (Figure 5B).
On the basis of biochemical studies of the mutant proteins in combination with analysis of the surface properties of UvrA, we have identified that the DNA binding surface lies on the ventral side of UvrA dimer (Figure 6C). The positions of the mutated residues define the likely path for DNA binding and imply that UvrA could interact with approximately 30 bp of B-form DNA. This finding is in good agreement with DNaseI footprinting of the complex which suggests protection of 33 bp of DNA (Van Houten et al., 1987). Electrostatic interactions have also been implicated in the detection of helical deformations in DNA by the eukaryotic NER protein XPA (Camenisch et al., 2006; Camenisch et al., 2007).
We expect that future structural and biochemical studies will support and further refine our model of the UvrA surfaces involved in DNA binding and interaction with UvrB. The structure of the UvrA dimer provides a much needed starting point for future experiments in the NER field for addressing the mechanism of initial lesion recognition by UvrA and UvrB.
EXPERIMENTAL PROCEDURES
Expression and Purification of Bacillus stearothermophilus UvrA, UvrB, and UvrC
The genes for Bacillus stearothermophilus UvrA, UvrB, and UvrC were identified by searching with the Escherichia coli sequences against the genome of B. stearothermophilus strain 10 (The University of Oklahoma Advance Center for Genome Technology). The full-length genes were cloned into pET-28a (+) (Novagen) (Table S1) and their sequences were verified.
UvrA, UvrB, and UvrC proteins were expressed in E. coli using standard techniques. Selenomethionine-substituted UvrA was expressed in E. coli as described (Van Duyne et al., 1993). UvrA and UvrC were purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen), heparin agarose, and size-exclusion chromatography (GE Healthcare). UvrB was purified using Ni-NTA agarose, MonoQ, and size-exclusion chromatography (GE Healthcare).
For biochemical experiments, proteins were dialyzed into 25 mM Tris-HCl pH 7.4, 250 mM NaCl, 20% (v/v) glycerol, 5 mM β-ME, flash frozen in liquid nitrogen and stored at −80°C.
Crystallization of BstUvrA
Selenomethionine-substituted BstUvrA was crystallized using hanging drop vapor diffusion, with the drop consisting of a 1.25 to 1 ratio of protein solution (7.5 mg/ml BstUvrA in 25 mM Tris-HCl pH 7.4, 400 mM NaCl, 5 mM β-ME, 5 mM MgCl2, 5 mM ADP) and reservoir solution (13% PEG2000 MME, 50 mM HEPES pH 7.0, 500 mM NaCl). The crystals grew to approximately 175 × 50 × 50 μm in one week, and were transferred to a 10-μl drop of crystallization buffer containing 15% (±) 1, 2-propanediol for cryoprotection and X-ray diffraction.
Structure Determination
Experimental crystallographic phases were calculated using the multi-wavelength anomalous diffraction (MAD) data from a selenomethionine-substituted UvrA crystal collected on the NE-CAT beamline 24ID at the Advanced Photon Source (APS), Argonne National Laboratory. The processed data (HKL2000) (Otwinowski and Minor, 1997) revealed that the crystal belonged to the monoclinic space group P21 with cell parameters a = 102.71 Å, b = 94.72 Å, c = 130.48 Å, α = γ = 90.0°, and β = 108.8°, and contained two molecules in the asymmetric unit. Seventeen of the 34 Se positions, located using direct methods (SnB) (Hauptman, 1997), were used to calculate the initial phases, which were then used to identify an additional 10 Se sites (SHARP) (de La Fortelle and Bricogne, 1997). The Se positions were used to calculate a MAD-phased electron density map to 3.2 Å. Density modification (Solomon, DM) (Collaborative Computational Project, 1994) improved the quality of the map and allowed a polyalanine model corresponding to ~70% of UvrA to be built using O (Jones et al., 1991). The Se positions in combination with 6 Zn sites, which were identified with diffraction data collected around the Zn absorption peak (λ = 1.28281 Å), allowed us to reliably assign the amino acid sequence to the electron density. Rounds of model building in Coot (Emsley and Cowtan, 2004) interspersed with crystallographic refinement with CNS (Brunger et al., 1998) resulted in a model with > 90% of the protein residues. Refinement was completed in REFMAC since we found that the application of TLS (translation/libration/screw) tensors permitted improvement of Rfree (Howlin et al., 1993; Murshudov et al., 1997). The model shows no outliers in the Ramachandran plot (PROCHECK) (Laskowski et al., 1993). Data collection, phasing, and refinement statistics are in Table 1. The coordinates and structure factors have been deposited to the Protein Data Bank with the accession code 2R6F.
Structural Analysis
Structural analysis and the preparation of illustrations were carried out in PyMOL (DeLano, 2002). Protein structure database comparisons were performed with DALI (Holm and Sander, 1993) using the coordinates of the BstUvrA monomer or the protein domains as the search models. Evolutionarily conserved protein surfaces were identified by mapping a sequence conservation index, calculated at each position of an alignment of 135 UvrA orthologs using ClustalW (Thompson et al., 1994) and the BLOSUM62 matrix (Henikoff and Henikoff, 1992), to the surface of UvrA. APBS (Baker et al., 2001) was used to calculate the surface electrostatic potential.
Construction of BstUvrA Mutants
Deletion and point mutants of BstUvrA were constructed using QuikChange® II XL site-directed mutagenesis kit (Stratagene) and mutations confirmed by sequencing (Table S1). The mutant proteins were purified using the wild-type protocol with slight modifications.
Biochemical characterization of BstUvrA Mutants
Interaction between UvrA and UvrB (10 nmol each) was analyzed by size exclusion chromatography (Superdex200, GE Healthcare) at 4°C in UvrAB Complex Buffer (50 mM Tris-HCl pH 7.5, 150 mM KCl, 5% (v/v) glycerol, 5 mM β-ME, 10 mM MgCl2, 2 mM ATP). The presence of proteins in each fraction was determined by SDS-PAGE. Incision and electrophoretic mobility shift assays were carried out as described previously (Verhoeven et al., 2002; Visse et al., 1992). ATPase activity was measured using a coupled enzyme assay system (Kiianitsa et al., 2003). The effect of DNA on ATP hydrolysis was studied by adding 0.2 μM 50-mer dsDNA with fluorescein at the central position to the assay mixture.
Supplementary Material
Acknowledgments
We thank the entire staff at the NE-CAT beamline 24ID, the Advance Photon Source, especially K. Rajashankar for assistance during data collection; Rachelle Gaudet, Alexander Ruthenburg, Wilhelm Weihofen, and members of the Verdine laboratory for useful discussions. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was supported by a grant from the National Institutes of Health (CA100742 to G.L.V). D.P. is supported by the Anandamahidol Foundation under the Royal Patronage of His Majesty the King of Thailand. B.R.B. is the recipient of an NRSA grant from the NIH (F32 GM077935-01).
Footnotes
Supplemental data includes detailed experimental procedures, additional references, six figures and one table and can be found with this article online at XXXXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandrovich A, Czisch M, Frenkiel TA, Kelly GP, Goosen N, Moolenaar GF, Chowdhry BZ, Sanderson MR, Lane AN. Solution structure, hydrodynamics and thermodynamics of the UvrB C-terminal domain. J Biomol Struct Dyn. 2001;19:219–236. doi: 10.1080/07391102.2001.10506734. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol. 2006;13:278–284. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]
- Camenisch U, Dip R, Vitanescu M, Naegeli H. Xeroderma pigmentosum complementation group A protein is driven to nucleotide excision repair sites by the electrostatic potential of distorted DNA. DNA Repair. 2007 doi: 10.1016/j.dnarep.2007.07.011. in press. [DOI] [PubMed] [Google Scholar]
- Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Claassen LA, Grossman L. Deletion mutagenesis of the Escherichia coli UvrA protein localizes domains for DNA binding, damage recognition, and protein-protein interactions. J Biol Chem. 1991;266:11388–11394. [PubMed] [Google Scholar]
- Collaborative Computational Project n. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J Biol Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmas O, Orelle C, Foucher AE, Geourjon C, Crouzy S, Di Pietro A, Jault JM. The Q-loop disengages from the first intracellular loop during the catalytic cycle of the multidrug ABC transporter BmrA. J Biol Chem. 2005;280:36857–36864. doi: 10.1074/jbc.M503266200. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Doolittle RF, Johnson MS, Husain I, Van Houten B, Thomas DC, Sancar A. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature. 1986;323:451–453. doi: 10.1038/323451a0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eryilmaz J, Ceschini S, Ryan J, Geddes S, Waters TR, Barrett TE. Structural insights into the cryptic DNA-dependent ATPase activity of UvrB. J Mol Biol. 2006;357:62–72. doi: 10.1016/j.jmb.2005.12.059. [DOI] [PubMed] [Google Scholar]
- Gaudet R, Wiley DC. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. Embo J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N, Moolenaar GF. Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res Microbiol. 2001;152:401–409. doi: 10.1016/s0923-2508(01)01211-6. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J Mol Biol. 1990;213:583–591. doi: 10.1016/S0022-2836(05)80243-8. [DOI] [PubMed] [Google Scholar]
- Hauptman HA. Shake-and-bake: an algorithm for automatic solution ab initio of crystal structures. Methods Enzymol. 1997;277:3–13. doi: 10.1016/s0076-6879(97)77003-4. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- Howlin B, Butler SA, Moss DS, Harris GW, Driessen HPC. TLSANL: TLS parameter analysis program for segmented anisotropic refinement of macromolecular structures. J Appl Cryst. 1993;26:622–624. [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. Embo J. 2007;26:613–622. doi: 10.1038/sj.emboj.7601497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher A, Buttner K, Martens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure. 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Kim IW, Peng XH, Sauna ZE, FitzGerald PC, Xia D, Muller M, Nandigama K, Ambudkar SV. The conserved tyrosine residues 401 and 1044 in ATP sites of human P-glycoprotein are critical for ATP binding and hydrolysis: evidence for a conserved subdomain, the A-loop in the ATP-binding cassette. Biochemistry. 2006;45:7605–7616. doi: 10.1021/bi060308o. [DOI] [PubMed] [Google Scholar]
- Kulkarni AS, Khalap N, Joshi VP. Effect of C-terminal 44 amino acids deletion on activity of Haemophilus influenzae UvrA protein. Indian J Exp Biol. 2006;44:7–13. [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lebbink JH, Sixma TK. Variations on the ABC. Structure. 2005;13:498–500. doi: 10.1016/j.str.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Linton KJ. Structure and function of ABC transporters. Physiology (Bethesda) 2007;22:122–130. doi: 10.1152/physiol.00046.2006. [DOI] [PubMed] [Google Scholar]
- Locher KP. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2004;14:426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad Sci U S A. 1999;96:11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Yamout M, Legge GB, Zhang O, Wright PE, Dyson HJ. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J Mol Biol. 2000;300:805–818. doi: 10.1006/jmbi.2000.3923. [DOI] [PubMed] [Google Scholar]
- Mazur SJ, Grossman L. Dimerization of Escherichia coli UvrA and its binding to undamaged and ultraviolet light damaged DNA. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, Herron MF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N. The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J Biol Chem. 2000a;275:8044–8050. doi: 10.1074/jbc.275.11.8044. [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, Moorman C, Goosen N. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J Bacteriol. 2000b;182:5706–5714. doi: 10.1128/jb.182.20.5706-5714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Myles GM, Hearst JE, Sancar A. Site-specific mutagenesis of conserved residues within Walker A and B sequences of Escherichia coli UvrA protein. Biochemistry. 1991;30:3824–3834. doi: 10.1021/bi00230a004. [DOI] [PubMed] [Google Scholar]
- Nakagawa N, Sugahara M, Masui R, Kato R, Fukuyama K, Kuramitsu S. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J Biochem (Tokyo) 1999;126:986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- Navaratnam S, Myles GM, Strange RW, Sancar A. Evidence from extended X-ray absorption fine structure and site-specific mutagenesis for zinc fingers in UvrA protein of Escherichia coli. J Biol Chem. 1989;264:16067–16071. [PubMed] [Google Scholar]
- Oh EY, Claassen L, Thiagalingam S, Mazur S, Grossman L. ATPase activity of the UvrA and UvrAB protein complexes of the Escherichia coli UvrABC endonuclease. Nucleic Acids Res. 1989;17:4145–4159. doi: 10.1093/nar/17.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Sancar A. Formation and enzymatic properties of the UvrB.DNA complex. J Biol Chem. 1990;265:15796–15803. [PubMed] [Google Scholar]
- Orren DK, Selby CP, Hearst JE, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J Biol Chem. 1992;267:780–788. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schmitt L, Benabdelhak H, Blight MA, Holland IB, Stubbs MT. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J Mol Biol. 2003;330:333–342. doi: 10.1016/s0022-2836(03)00592-8. [DOI] [PubMed] [Google Scholar]
- Singh S, Folkers GE, Bonvin AM, Boelens R, Wechselberger R, Niztayev A, Kaptein R. Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli. Embo J. 2002;21:6257–6266. doi: 10.1093/emboj/cdf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma TK. DNA mismatch repair: MutS structures bound to mismatches. Curr Opin Struct Biol. 2001;11:47–52. doi: 10.1016/s0959-440x(00)00169-x. [DOI] [PubMed] [Google Scholar]
- Sohi M, Alexandrovich A, Moolenaar G, Visse R, Goosen N, Vernede X, Fontecilla-Camps JC, Champness J, Sanderson MR. Crystal structure of Escherichia coli UvrB C-terminal domain, and a model for UvrB-uvrC interaction. FEBS Lett. 2000;465:161–164. doi: 10.1016/s0014-5793(99)01690-7. [DOI] [PubMed] [Google Scholar]
- Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. Embo J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. Interactions between UvrA and UvrB: the role of UvrB’s domain 2 in nucleotide excision repair. Embo J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006a;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol. 2006b;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. Embo J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Gamper H, Sancar A, Hearst JE. DNase I footprint of ABC excinuclease. J Biol Chem. 1987;262:13180–13187. [PubMed] [Google Scholar]
- Verhoeven EE, van Kesteren M, Moolenaar GF, Visse R, Goosen N. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J Biol Chem. 2000;275:5120–5123. doi: 10.1074/jbc.275.7.5120. [DOI] [PubMed] [Google Scholar]
- Verhoeven EE, van Kesteren M, Turner JJ, van der Marel GA, van Boom JH, Moolenaar GF, Goosen N. The C-terminal region of Escherichia coli UvrC contributes to the flexibility of the UvrABC nucleotide excision repair system. Nucleic Acids Res. 2002;30:2492–2500. doi: 10.1093/nar/30.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse R, de Ruijter M, Moolenaar GF, van de Putte P. Analysis of UvrABC endonuclease reaction intermediates on cisplatin-damaged DNA using mobility shift gel electrophoresis. J Biol Chem. 1992;267:6736–6742. [PubMed] [Google Scholar]
- Visse R, de Ruijter M, Ubbink M, Brandsma JA, van de Putte P. The first zinc-binding domain of UvrA is not essential for UvrABC-mediated DNA excision repair. Mutat Res. 1993;294:263–274. doi: 10.1016/0921-8777(93)90009-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Mueller KL, Grossman L. A mutational study of the C-terminal zinc-finger motif of the Escherichia coli UvrA protein. J Biol Chem. 1994;269:10771–10775. [PubMed] [Google Scholar]
- Waters TR, Eryilmaz J, Geddes S, Barrett TE. Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett. 2006;580:6423–6427. doi: 10.1016/j.febslet.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Zaitseva J, Jenewein S, Wiedenmann A, Benabdelhak H, Holland IB, Schmitt L. Functional characterization and ATP-induced dimerization of the isolated ABC-domain of the haemolysin B transporter. Biochemistry. 2005;44:9680–9690. doi: 10.1021/bi0506122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.