Abstract
Carbonylation is a highly prevalent protein modification in skeletal muscle mitochondria, possibly contributing to its functional decline with age. Using quantitative proteomics, we identified mitochondrial proteins susceptible to carbonylation in a muscle type (slow- vs fast-twitch)-dependent and age-dependent manner from Fischer 344 rat skeletal muscle. Fast-twitch muscle contained twice as many carbonylated mitochondrial proteins than did slow-twitch muscle, with 22 proteins showing significant changes in carbonylation state with age, the majority of these increasing in their amount of carbonylation. Ingenuity pathway analysis revealed that these proteins belong to functional classes and pathways known to be impaired in muscle aging, including cellular function and maintenance, fatty acid metabolism, and citrate cycle. Although our studies do not conclusively link protein carbonylation to these functional changes in aging muscle, they provide a unique catalogue of promising protein targets deserving further investigation because of their potential role in aging muscle decline.
Keywords: Carbonylation, Muscle, Aging, Mitochondria, Quantitative proteomics, Mass spectrometry, Ingenuity pathway analysis
Oxidative damage occurs in lipids, nucleic acids, and proteins (1,2). It is known that oxidatively modified forms of proteins accumulate under conditions of oxidative stress associated with aging, and in some age-related diseases. Introduction of reactive carbonyls into proteins, known as protein carbonylation, is a prominent marker of oxidative damage in aged tissue (3,4), including human skeletal muscle (5,6), as well as other tissues and cells (7–11). Because of the production of reactive oxygen species (ROS) in skeletal muscle mitochondria (12), which increases with age and drives these protein modifications, mitochondrial muscle proteins have been shown to be particularly susceptible to carbonylation (13). However, the dependence of these modifications on muscle type (fast or slow twitch) and age has not been studied. Such information is needed to better understand the possible role of protein carbonylation in altering protein function in aging muscle.
There are two major mechanisms leading to protein carbonylation: (a) metal catalyzed oxidation (MCO) and (b) reaction of nucleophilic amino acid side chains with lipid oxidation products such as 4-hydroxyl-2-nonenal (HNE). In the former mechanism, metals such as copper and iron catalyze the formation of highly reactive, short-lived hydroxyl radicals that modify nearby amino acids, like proline, arginine, lysine, and threonine (3,14–16). In the latter mechanism, lipid peroxidation leads to the generation of aldehyde-containing byproducts, which covalently modify nucleophilic amino acid side chains on proteins, such as cysteine, histidine, and lysine (3,15).
Carbonylated proteins have been identified in several ways from cells and tissues. Immunoassays for carbonylated proteins have been based on derivatization with 2,4-dinitrophenyhydrazine followed by treatment with anti-2,4-dinitrophenol antibodies and secondary peroxidase-labeled antibodies (17,18). Several reports have used biotin hydrazide for derivatization of proteins with carbonyl groups followed by: (a) two-dimensional gel separation and detection with fluorescently labeled avidin (19); (b) affinity enrichment with biotin–streptavidin liquid chromatography tandem mass spectrometric (LC-MS/MS) analysis (20); (c) enrichment using avidin affinity chromatography, followed by LC-MS/MS (21); and (d) enrichment using avidin affinity chromatography followed by iTRAQ-based quantitative proteomics, as described by our group (13). In this work, we identified carbonylated proteins in the mitochondria of mixed skeletal muscle types from young rats, and demonstrated the potential of this method for quantitative studies of protein carbonylation.
Here we extend our proteomic method to investigate differences in mitochondrial protein carbonylation attributed to muscle type (fast vs slow twitch) and age. Our findings showed that fast-twitch muscle had about two times more proteins susceptible to carbonylation, with more than 20 of these proteins showing significant increases in carbonylation with age in fast-twitch muscle. Using ingenuity pathway analysis (IPA) (22–24), we found that these carbonylated proteins belong to pathways and functional classes already known to be impaired in aging skeletal muscle. Our findings provide novel targets for studies investigating the possible contribution of carbonylation to protein functional changes playing a role in muscle decline with age.
Methods
Materials
Biotin hydrazide, monomeric avidin columns, Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl), and bicinchoninic acid (BCA) reagents were purchased from Pierce (Rockford, IL). Oasis mixed-mode MCX cartridges were purchased from Waters (Milford, MA). Sequencing grade trypsin was purchased from Promega (Madison, WI). Methanol, sucrose, potassium chloride (KCl), Tris-HCl, sodium acetate, potassium phosphate monobasic (KH2PO4), HEPES buffer (1 M), sodium cyanoborohydride (NaBH3CN), nagarse, EGTA, and sodium dodecyl sulfate (SDS) were purchased from Sigma (St. Louis, MO). Phosphate-buffered saline (PBS; 10X) was obtained from Invitrogen (Grand Island, NY). iTRAQ reagents were obtained from Applied Biosystems (Foster City, CA).
Animal and Tissue Preparation
Fisher 344 rats used in this study were purchased from the Minneapolis Veterans Administration Aged Rodent Colony, maintained by the University of Minnesota and fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The University of Minnesota Institutional Animal Care and Use Committee approved the study. The slow-twitch muscles (soleus) and the fast-twitch muscles (semimem-branosus, plantaris, extensor digitorum longus, and tibialis anterior) were isolated from 16 Fisher 344 female rats. The rats were divided into two groups based on their age: 8 young adult rats (12 months old, 100% survival rate) and 8 old rats (26 months old, ~25% survival rate).
Protein Preparation
We described previously the basic methodology used here for identifying and quantifying carbonylated proteins from skeletal rat muscle mitochondria (13). A flow chart for such methodology is shown in Figure 2 and briefly described in the Results section. To demonstrate reproducibility of the mitochondrial enrichment procedure, we conducted the tissue homogenization and differential centrifugation steps in triplicate. The isolated mitochondrial fractions were then separated in a 4%–15% gradient SDS–polyacrylamide gel electrophoresis (PAGE) gel and silver stained (Bio-Rad silver stain kit; Hercules, CA).
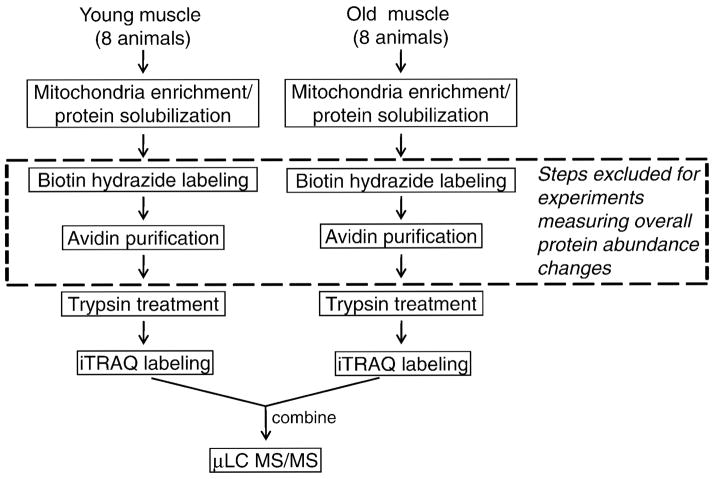
Figure 2.
Quantitative proteomic strategy. All the steps detailed were used in determining changes in carbonylation from both slow- and fast-twitch muscle. The steps within the dashed box are excluded when determining changes in overall protein abundance in fast-twitch muscle. μLC-MS/MS = microcapillary liquid chromatography tandem mass spectrometry.
For a given muscle type, mitochondria were isolated from pooled muscles taken from several animals from the same age group; the isolation procedure was done in parallel for the two age groups (Figure 1). When investigating carbonylation, 2.5 mg of solubilized proteins were treated with biotin hydrazide to derivatize carbonyls and then were subjected to avidin affinity enrichment, as previously described (13). This step was omitted when quantifying age-related changes in protein abundance of the fast-twitch muscles. For these studies, protein mixtures of 90 μg per each portion from mitochondria of fast-twitch muscle from young and old rats were used in protein abundance profiling.
Figure 1.
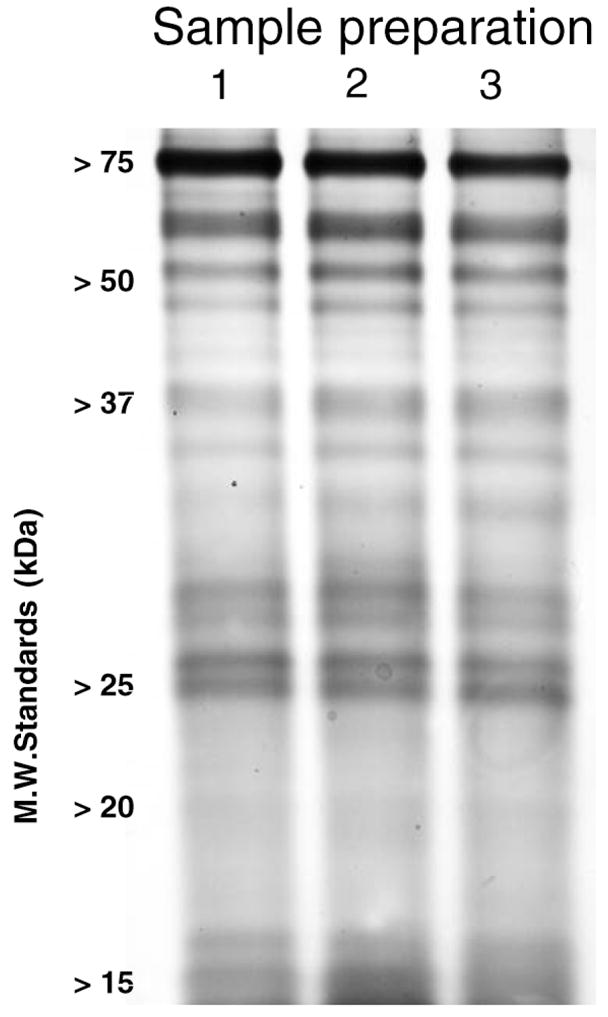
Reproducibility of mitochondrial fraction preparation from skeletal muscle. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and silver staining of independent mitochondrial protein preparations.
For all samples (either the enriched, carbonylated proteins or the proteins solubilized directly from mitochondria), the proteins were then trypsin digested and the resulting peptides were purified and desalted in a mixed-mode cation exchange column (MCX; Waters) followed by Sep-Pak reverse-phase extraction. In comparing carbonylation, fast-twitch muscle samples from the young and old animals were labeled with iTRAQ 114 and 116 labels, respectively. Similarly, slow-twitch muscle samples from the young and old animals were labeled with iTRAQ 115 and 117 labels, respectively. When investigating age-related changes in protein abundance of fast-twitch muscles, two experiments were carried out: In the first one, samples from the young and old animals were labeled with iTRAQ 114 and 116 labels, respectively. In the second experiment, the iTRAQ 115 and 117 labels were used for young and old animals, respectively.
For all samples, the combined, iTRAQ reagent-labeled peptide mixtures were further purified with a mixed-mode cation exchange column prior to LC-electrospray ionization (ESI) MS/MS analysis. Between purification and analysis of the samples in the protein abundance study only, the additional step of strong cation exchange (SCX) high-performance liquid chromatography (HPLC) was performed to increase coverage of proteins from mitochondrial protein isolate. In this step, fractionation of iTRAQ reagent-labeled peptide mixtures was performed using a PolySULFOETHYL SCX guard column (Javelin guard column, 1.0 mm i.d. × 10 mm, 5 μm, 300 Ǻ; PolyLC, Inc., Columbia, MD) using an automated syringe pump capable of highly accurate sub-microliter per minute flow rates (Harvard Apparatus Inc., Holliston, MA). The peptides were redissolved in 200 μL of SCX loading buffer (10 mM KH2PO3 containing 20% acetonitrile, pH 3.0) and loaded onto a preconditioned SCX column at a flow rate of 50 μL/min. After washing with loading buffer, the column was washed with buffer containing 15 mM KCl, to remove loosely retained contaminants, and then peptides were eluted using a step-gradient chromatography, using steps with increasing KCl concentration of 20, 25, 50, and 200 mM, at a flow rate of 50 μL/min collecting elution fractions 200 μL in volume. Each collected fraction was concentrated by vacuum centrifugation and reconstituted in 30 μL of HPLC load buffer prior to microcapillary LC-ESI MS/MS (μLC-ESI MS/MS) analysis.
μLC-ESI MS/MS Analysis
For identifying carbonylated proteins, the sample was divided into three equal portions and analyzed three times using the instrumental conditions described previously (13), to improve protein coverage. Peptide mixtures were first separated on an automated Paradigm MS4 LC system (Michrom Bioresources, Auburn, CA), and identified via μLC-ESI MS/MS on a linear ion trap mass spectrometer (LTQ; Thermo Electron, San Jose, CA) operating in pulsed Q dissociation (PQD) mode, facilitating the analysis of iTRAQ reagent-labeled peptide mixtures. We have described the effectiveness of PQD-LTQ for the analysis of iTRAQ reagent-labeled peptide mixtures (25). Peptides were selected for MS/MS using PQD operating mode with a normalized collision energy setting of 42% optimized for iTRAQ reagent-labeled peptides as described (25) and a data-dependent procedure that alternated between one MS scan followed by four MS/MS scans for the four most abundant precursor ions in the MS survey scan.
Sequence Database Searching and Data Analysis
MS/MS spectra were searched using SEQUEST (26) (Thermo Electron) against a nonredundant rat proteome sequence database from the European Bioinformatics Institute (version 3.18, containing approximately 42,000 sequence entries) using a seven-node (14 processors) cluster maintained at the University of Minnesota Supercomputing Institute. The sequence database contained reversed sequences appended to the forward sequences to estimate false-positive rates for peptide matches (27). Search parameters included differential amino acid mass shifts for oxidized methionine (+ 16 Da), and biotin hydrazide-labeled carbonylated residues, with mass shifts appropriate for commonly known carbonyl groups (e.g., produced via MCO or lipid peroxidation products such as HNE). All search results were validated using the publicly available peptide validation program PeptideProphet (28), which assigns a comprehensive probability (P) score from 0 to 1 to each peptide sequence match based on its SEQUEST scores (Xcorr, dCn, Sp, RSp) and additional information, including the mass difference between the precursor ion and the assigned peptide, and the number of tryptic termini. The peptide sequence-match results were organized and interpreted using the software tool Interact (29). Only the peptide sequence matches at least partially tryptic termini. The P score cutoff value was set as 0.2 for multiple hits (two or more unique peptides), and the false-positive rate for the peptide sequence matches using this criterion was estimated to be 0.8% via reverse database searching, consistent with prior studies using the same filtering strategy (30). For single hits, the P score threshold was set at 0.80, and the false-positive rate for the obtained sequence matches was 2%. Single peptide matches were accepted only after manual inspection, validating that major detected peaks matched expected b and y ions.
The intensities of iTRAQ reporter ions 114, 115, 116, and 117 were extracted from the raw data files using a perl script developed in-house, as described (25). The ion ratios (e.g., 116:114 for fast-twitch old/young) of an identified protein were calculated using the sum of intensities for each iTRAQ reporter ion from all peptide sequence matches that satisfied the criteria mentioned above. Only proteins identified by at least two MS/MS were considered for quantitative analysis, to ensure more accurate quantitative results (25).
As a quantitative internal control, in vitro carbonylated bovine serum albumin (BSA) protein, modified using MCO (31), was included at an equal amount in each separate biotin hydrazide-labeled mitochondrial protein preparation from young adult and old rats. The summed iTRAQ reporter intensities across all the MS/MS acquired for carbonylated BSA resulted in a 116:114 (for fast-twitch experiment) ratio of 0.83 and a 117:115 (for slow-twitch experiment) ratio of 1.26. Given this observation, we normalized the calculated ratios for each protein in fast twitch and slow twitch by dividing their iTRAQ ratios by 0.83 and 1.26, respectively. To find proteins whose carbonylation levels are significantly changed with age, we calculated that the 90% interval range of iTRAQ ratios lies between 0.1 and 9.4. Any proteins outside of this range are considered to have significant change of carbonylation.
Regarding the iTRAQ ratios for measuring overall protein abundance in the fast-twitch muscle, the ratio of old to young was 0.91 ± 0.42 (average ± interval of confidence, at p = .05), indicating that the mean value of protein abundance does not differ much from the expected ratio of 1.0. Given this observation, we normalized the calculated ratios for each protein in the abundance profiling data set by dividing each iTRAQ ratio (old/young) by 0.91. To determine whether the selected proteins change in total abundance, we calculated that a 90% interval range of iTRAQ ratios lies between 0.5 and 1.7. For the quantitative carbonylation data from fast-twitch muscle, our measured value for oxidized BSA as a control was 0.83 for fast-twitch muscle. We conservatively estimated that the experimental error is small (about 20%) for the carbonylation data, so setting threshold values of 0.5/1.7 is outside of our measured error. Any proteins outside of this interval from carbonylation profiling, although inside of this interval from protein abundance profiling, are considered significantly changing.
IPA Analysis
We used IPA (Ingenuity Systems, www.ingenuity.com) to identify functions and canonical pathways significantly represented by the set of carbonylated proteins. IPA is a knowledge database generated from peer-reviewed scientific publications that enables discovery of highly represented functions and pathways (p < .001) from large, quantitative data sets (22–24). Fisher’s Exact test was used to calculate a p value determining the probability that each biological function or canonical pathway assigned to that data set is due to chance alone. The smaller the p value, the less likely that the association is random, and the more significant is the association.
Results
Carbonylated Mitochondrial Proteins in Fast-Twitch and Slow-Twitch Muscle
Validation of the method to measure changes in protein carbonylation in mitochondrial fractions of skeletal muscle proteins requires investigating the reproducibility of the mitochondrial preparation as well as the proteomic method, beginning with the affinity enrichment of carbonylated proteins. Although we have previously described the reproducibility of the latter (13), the reproducibility of our mitochondrial preparation method needed to be demonstrated. For this reason, we prepared three separate preparations of crudely enriched mitochondria from rat skeletal muscle, solubilized the proteins, and detected the isolated proteins by PAGE and silver staining. As shown in Figure 1, the three separate preparations showed equal staining intensities and patterns, confirming the reproducibility our mitochondrial preparation protocol.
Next, we used the procedure outlined in Figure 2 to quantify age-related changes in the carbonylation of fast- and slow-twitch mitochondrial muscle proteins isolated from young (12 months) and old (26 months) rat tissues. After digestion, the peptide mixtures were separately labeled with the 114 iTRAQ reagent (young) and the 116 iTRAQ reagent (old), and the samples were combined. Using μLC-ESI MS/MS, a total of 78 and 38 carbonylated proteins in the fast-twitch and slow-twitch muscle, respectively, were identified, and their changes in relative abundance with age were quantified (Table 1). Among the proteins identified, 29 were common to both muscle types (see Table 1 and Figure 3).
Table 1.
Carbonylated Mitochondrial Proteins From Fast-Twitch and Slow-Twitch Skeletal Muscle
| No. | Accession* | Proteins | Unique Peptides and Total MS/MS (Fast-Twitch)† | Ratio (Fast-Twitch) Old:Young‡ | Unique Peptides and Total MS/MS (Slow-Twitch)† | Ratio (Slow-Twitch) Old:Young§ |
|---|---|---|---|---|---|---|
| Found in Both Muscle Types | ||||||
| 1 | Q5M7W0 | Electron-transferring flavoprotein, α polypeptide | 2,3 | 40.2 | 1,8 | 13.8 |
| 2 | P08461 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex (fragment) | 4,13 | 15.1 | 1,2 | 0.5 |
| 3 | Q64428 | Trifunctional enzyme subunit α | 3,13 | 9.9 | 1,1 | 0.5 |
| 4 | P35434 | ATP synthase Δ chain | 1,4 | 9.5 | 1,4 | 5.3 |
| 5 | Q05962 | ADP/ATP translocase 1 | 1,4 | 8.7 | 2,4 | 1 |
| 6 | Q01205 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | 3,3 | 6.2 | 4,12 | 0.2 |
| 7 | P13437 | Acetyl-CoA acyltransferase | 1,4 | 4.7 | 1,4 | 0.1 |
| 8 | Q5XIH3 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 1,7 | 3.3 | 2,19 | 7.9 |
| 9 | IPI00390435.1 | 76-kd protein | 10,32 | 3.1 | 3,13 | 0.5 |
| 10 | P45953 | Very-long-chain specific acyl-CoA dehydrogenase | 2,5 | 2.7 | 1,2 | 1.9 |
| 11 | P20788 | Ubiquinol–cytochrome c reductase iron–sulfur subunit | 4,6 | 2.4 | 1,6 | 0.4 |
| 12 | Q68FY0 | Ubiquinol–cytochrome c reductase core protein 1 | 1,14 | 2.3 | 1,9 | 7 |
| 13 | Q641Y2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | 1,1 | 2.1 | 1,2 | 0.7 |
| 14 | IPI00360013.2 | Voltage-dependent anion channel | 3,21 | 1.8 | 2,11 | 2.9 |
| 15 | P15999 | ATP synthase subunit α | 5,14 | 1.8 | 8,22 | 2.2 |
| 16 | IPI00207891.2 | 31-kd protein | 1,2 | 1.7 | 2,2 | 1.5 |
| 17 | P10719 | ATP synthase subunit β | 27,127 | 1.5 | 14,98 | 1.8 |
| 18 | P49432 | Pyruvate dehydrogenase E1 component subunit β | 12,34 | 1.4 | 1,5 | 0.4 |
| 19 | Q68FX0 | Isocitrate dehydrogenase (NAD) subunit β | 2,4 | 1.4 | 1,5 | 0.3 |
| 20 | P04636 | Malate dehydrogenase | 18,63 | 1.3 | 6,37 | 1.5 |
| 21 | Q5BJZ3 | Nicotinamide nucleotide transhydrogenase | 4,5 | 1.3 | 3,18 | 1.1 |
| 22 | IPI00366416.1 | Cytochrome c-1 | 3,23 | 1.3 | 2,2 | 0.6 |
| 23 | IPI00371236.2 | TU translation elongation factor | 1,2 | 1.2 | 1,1 | 0.1 |
| 24 | P21571 | ATP synthase coupling factor 6 | 6,14 | 1.1 | 1,4 | 0.7 |
| 25 | P19234 | NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial (precursor) | 5,14 | 1.0 | 3,13 | 0.4 |
| 26 | Q9WVK7 | Short chain 3-hydroxyacyl-CoA dehydrogenase | 4,8 | 1.0 | 2,6 | 1.3 |
| 27 | Q9ER34 | Aconitate hydratase | 2,3 | 0.9 | 4,11 | 0.8 |
| 28 | P10860 | Glutamate dehydrogenase 1 | 2,3 | 0.7 | 4,8 | 0.5 |
| 29 | IPI00359981.1 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 13 | 1,5 | 0.2 | 1,19 | 1.1 |
| Found in Fast-Twitch Muscle Only | ||||||
| 30 | Q6GSM4 | Malate dehydrogenase | 1,3 | 193.2 | ||
| 31 | P15650 | Long-chain specific acyl-CoA dehydrogenase | 1,2 | 134.9 | ||
| 32 | IPI00365813.2 | Voltage-dependent anion-selective channel protein 1 | 1,2 | 27.9 | ||
| 33 | P14604 | Enoyl-CoA hydratase | 2,4 | 10.0 | ||
| 34 | P81155 | Voltage-dependent anion-selective channel protein 2 | 1,5 | 7.3 | ||
| 35 | Q62651 | Delta3,5-delta2,4-dienoyl-CoA isomerase | 1,2 | 6.7 | ||
| 36 | P48721 | Stress-70 protein | 5,29 | 6.4 | ||
| 37 | P35435 | ATP synthase γ chain | 6,30 | 5.3 | ||
| 38 | P63039 | 60-kd heat shock protein | 6,9 | 4.8 | ||
| 39 | Q5RJN0 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | 3,9 | 4.8 | ||
| 40 | Q5BK63 | Ndufa9 protein (fragment) | 2,4 | 4.5 | ||
| 41 | P15651 | Short-chain specific acyl-CoA dehydrogenase | 1,5 | 4.3 | ||
| 42 | Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | 1,1 | 4.1 | ||
| 43 | P32551 | Ubiquinol-cytochrome-c reductase complex core protein 2 | 11,38 | 3.3 | ||
| 44 | Q9Z0J5-1 | Thioredoxin reductase 2 | 1,1 | 2.9 | ||
| 45 | P17764 | Acetyl-CoA acetyltransferase | 15,40 | 3.0 | ||
| 46 | P97615 | Thioredoxin | 2,3 | 3.0 | ||
| 47 | P18886 | Carnitine O-palmitoyltransferase 2 | 1,4 | 2.6 | ||
| 48 | Q80WE0 | NADH dehydrogenase 1 α subcomplex 10-like protein | 1,1 | 2.6 | ||
| 49 | IPI00366206.2 | 13-kd differentiation-associated protein | 2,6 | 2.5 | ||
| 50 | Q6DGF1 | NADP+-specific isocitrate dehydrogenase | 6,10 | 2.3 | ||
| 51 | O96000 | NADH–ubiquinone oxidoreductase PDSW subunit (complex I-PDSW) (CI-PDSW) isoform 1 | 1,1 | 2.3 | ||
| 52 | IPI00202616.1 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | 2,14 | 2.2 | ||
| 53 | Q99NA5 | Isocitrate dehydrogenase (NAD) subunit α | 1,1 | 2.1 | ||
| 54 | IPI00204641.1 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 9 | 1,1 | 2.0 | ||
| 55 | Q5XI78 | Similar to oxoglutarate dehydrogenase | 3,6 | 1.9 | ||
| 56 | P11608 | ATP synthase protein 8 | 1,1 | 1.9 | ||
| 57 | IPI00372007.2 | Mitochondrial ribosomal protein S36 | 1,1 | 1.9 | ||
| 58 | Q5UAJ8 | NADH dehydrogenase subunit 2 | 1,6 | 1.8 | ||
| 59 | IPI00191103.1 | Mitochondrial ribosome | 2,5 | 1.7 | ||
| 60 | Q60587 | Trifunctional enzyme subunit β | 2,5 | 1.7 | ||
| 61 | P11884 | Aldehyde dehydrogenase | 2,4 | 1.4 | ||
| 62 | P09605 | Creatine kinase | 1,1 | 1.4 | ||
| 63 | Q9R063-1 | Peroxiredoxin-5 | 1,2 | 1.3 | ||
| 64 | P21913 | Enoyl-CoA hydratase | 1,1 | 1.3 | ||
| 65 | Q91ZW1-1 | Transcription factor A | 1,1 | 1.3 | ||
| 66 | Q920L2 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit | 7,20 | 1.2 | ||
| 67 | Q63704 | Carnitine O-palmitoyltransferase I | 2,2 | 1.0 | ||
| 68 | P11662 | NADH-ubiquinone oxidoreductase chain 2 | 1,1 | 1.0 | ||
| 69 | IPI00364431.2 | Succinate-CoA ligase, ADP-forming, β subunit | 2,5 | 0.9 | ||
| 70 | P13803 | Electron transfer flavoprotein subunit α | 1,3 | 0.9 | ||
| 71 | Q6UPE1 | Electron transfer flavoprotein-ubiquinone oxidoreductase | 1,1 | 0.9 | ||
| 72 | Q9Z2L0 | Voltage-dependent anion-selective channel protein 1 | 6,11 | 0.8 | ||
| 73 | P00507 | Aspartate aminotransferase | 6,17 | 0.7 | ||
| 74 | P26772 | 10-kd heat shock protein | 3,10 | 0.7 | ||
| 75 | Q2XTA8 | NADH dehydrogenase 1 β 4 (fragment) | 2,6 | 0.7 | ||
| 76 | IPI00208203.2 | Peroxisomal Δ3, Δ2-enoyl-CoA isomerase | 1,1 | 0.7 | ||
| 77 | P26284 | Pyruvate dehydrogenase E1 component α subunit | 2,2 | 0.4 | ||
| 78 | P31399 | ATP synthase D chain | 1,2 | 0.1 | ||
| Found in Slow-Twitch Muscle Only | ||||||
| 79 | Q6AYK4 | Apoptosis facilitator Bcl-2-like protein 14 | 1,2 | 9.0 | ||
| 80 | P11240 | Cytochrome c oxidase subunit 5A | 2,7 | 6.6 | ||
| 81 | Q7TQ85 | Ac1164 | 1,5 | 0.9 | ||
| 82 | IPI00365505.2 | Cytochrome c oxidase subunit VIIA polypeptide 2-like | 1,4 | 0.9 | ||
| 83 | P41565 | Isocitrate dehydrogenase (NAD) subunit γ | 1,3 | 0.9 | ||
| 84 | P09605 | Creatine kinase | 1,10 | 0.8 | ||
| 85 | IPI00370372.2 | NADH dehydrogenase (ubiquinone) 1 β subcomplex 8 | 1,6 | 0.8 | ||
| 86 | O55171 | Acyl-CoA thioesterase 2 | 1,4 | 0.5 | ||
| 87 | Q6P6R2 | Dihydrolipoyl dehydrogenase | 1,2 | 0.3 |
Notes: Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
The number of unique peptides and total number of tandem mass spectrometry (MS/MS) spectra matched to each protein via sequence database searching are given.
Ratio 116:114 indicates protein carbonylation ratio between old and young in fast-twitch muscle.
Ratio 117:115 indicates protein carbonylation ratio between old and young in slow-twitch muscle.
MS/MS = tandem mass spectrometry; ATP = adenosine-5′-triphosphate; ADP = adenosine diphosphate; CoA = coenzyme A; NADH = reduced nicotinamide adenine dinucleotide; NAD = nicotinamide adenine dinucleotide; NADP = nicotinamide adenine dinucleotide phosphate.
Figure 3.
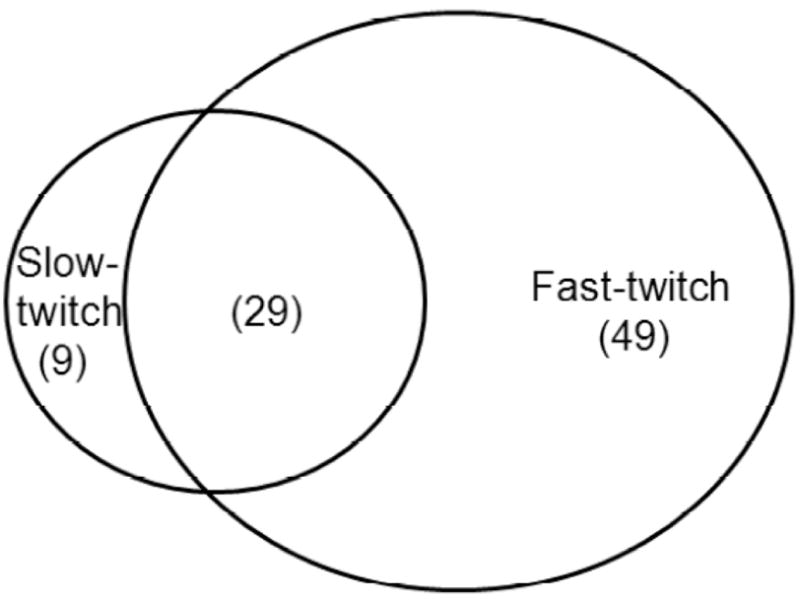
Venn diagram of carbonylated proteins identified from slow- and fast-twitch skeletal muscle mitochondria.
Pathways Represented by Carbonylated Proteins From Fast-Twitch and Slow-Twitch Muscle
Using IPA, we mapped the carbonylated proteins found in fast- and slow-twitch muscle to the curated biochemical pathways and biological functions of the IPA environment. The significant (p < .001) canonical pathways and functions represented in our data set are included in Table 2. Common to both muscle types were eight canonical pathways (e.g., oxidative phosphorylation) and six biological functions (e.g., carbohydrate metabolism). No significant functions or pathways were assigned to the carbonylated proteins identified only in the slow-twitch muscle. In contrast, the carbonylated proteins unique to fast-twitch muscle mapped to two distinct pathways (cellular function/maintenance and cell death) and two distinct functions (tryptophan metabolism and synthesis/degradation of ketone bodies) in the IPA environment. The carbonylated proteins matching to these categories are also included in Table 2.
Table 2.
Ingenuity Pathway Analysis (IPA) Pathways and Functions Significantly Represented by Carbonylated Proteins
| Significant Pathways and Function in Both Muscle Types | ||
| Canonical Pathway | Function | |
| Oxidative phosphorylation | Carbohydrate metabolism | |
| Mitochondrial dysfunction | Cell signaling | |
| Butanoate metabolism | Energy production | |
| Fatty acid metabolism | Amino acid metabolism | |
| Valine, leucine, and isoleucine degradation | Lipid metabolism | |
| Citric cycle | Small molecule biochemistry | |
| Fatty acid elongation in mitochondria | ||
| Pyruvate metabolism | ||
| Significant Functions Unique Only to Fast-Twitch Muscle* | ||
| Accession†,‡ | Protein Name | |
| Cellular function and maintenance | P63039 | Heat shock 60 kd protein 1 (chaperonin) |
| p = 2.5 × 10−18 (fast-twitch) | P32551 | Ubiquinol–cytochrome c reductase core protein II |
| p = 1.6 × 10−2 (slow-twitch) | Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| Q9Z2L0 | Voltage-dependent anion channel 1 | |
| Q68FY0 | Ubiquinol-cytochrome c reductase core protein I | |
| Cell death | P11884 | Aldehyde dehydrogenase 2 |
| p = 2.0 × 10−5 (fast-twitch) | P10860 | Glutamate dehydrogenase 1 |
| p = 3.4 × 10−3 (slow-twitch) | Q64428 | Trifunctional protein, α subunit |
| P48721 | Heat shock 70 kd protein 9 | |
| P63039 | Heat shock 60 kd protein 1 | |
| P26772 | Heat shock 10 kd protein 1 | |
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | |
| Q920L2 | Succinate dehydrogenase complex, subunit A, flavoprotein | |
| P21913 | Succinate dehydrogenase complex, subunit B, iron sulfur | |
| Q05962 | Adenine nucleotide translocator | |
| Q9Z2L0 | Voltage-dependent anion channel 1 | |
| P81155 | Voltage-dependent anion channel 2 | |
| Significant Canonical Pathways Unique Only to Fast-Twitch Muscle* | ||
| Accession†,‡ | Protein Name | |
| Tryptophan metabolism | P17764 | Acetyl-CoA acetyltransferase 1 |
| p = 1.2 × 10−12 (fast-twitch) | P11884 | Aldehyde dehydrogenase 2 |
| p = 2.5 × 10−3 (slow-twitch) | Q62651 | Enoyl CoA hydratase 1 |
| P14604 | Enoyl CoA hydratase, short chain | |
| Q60587 | Trifunctional protein, β subunit | |
| Q5XI78 | Oxoglutarate dehydrogenase | |
| Synthesis and degradation of ketone bodies | ||
| p = 2.4 × 10−7 (fast-twitch) | P17764 | Acetyl-CoA acetyltransferase 1 |
| p = 6.8 × 10−3 (slow-twitch) | Q60587 | Trifunctional protein, β subunit |
Notes: The canonical pathways and functions are significant (p < .001) in fast-twitch muscle but not significant (p > .001) in slow-twitch muscle.
Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
Only proteins identified in fast-twitch muscle are listed.
NADH = reduced nicotinamide adenine dinucleotide; CoA = coenzyme A.
Carbonylated Proteins Showing Age-Dependent Changes in Fast-Twitch Muscles
Although we measured changes in protein carbonylation with age in both muscle types, we decided to focus on the age-related changes in carbonylated proteins of fast-twitch muscle, for two reasons: (a) we identified more carbonylated proteins from this muscle type, and (b) this muscle type is known to show more susceptibility to oxidative damage and age-related decline (32), suggesting a possible connection between protein carbonylation and muscle decline with age. Because it is not possible to discern from the data on affinity-enriched carbonylated proteins if changes between young adult and old animals are due to changes in carbonyl modification state or simply due to overall abundance changes from expression or turnover, we profiled changes in overall protein abundance in a separate proteomic experiment. We used the same fast-twitch tissue used for quantification of total carbonylation, but with the alternate procedure represented in Figure 2 by the exclusion of the biotin hydrazide labeling and avidin chromatography (shown in a dashed box in Figure 2).
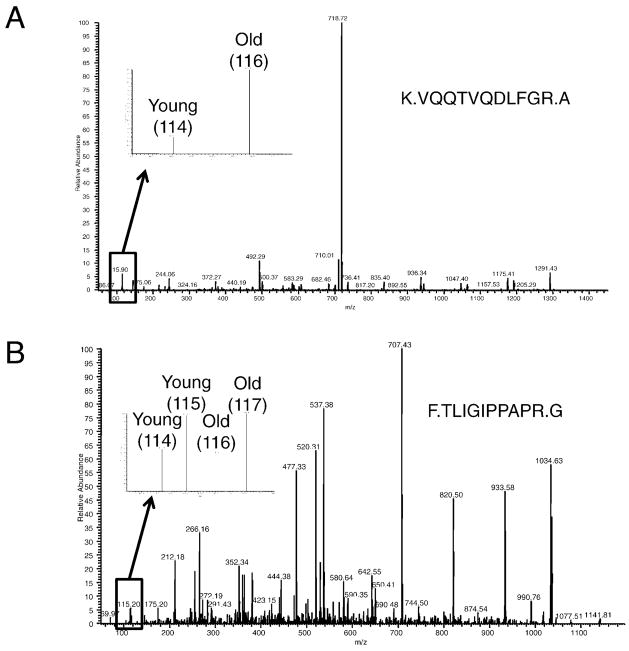
The relative quantification of carbonylated proteins was based on the intensity ratios of the iTRAQ reporter ion masses detected in the MS/MS spectra, using a method we have described previously (25). For example, Figure 4A shows a representative MS/MS spectrum of a peptide corresponding to the mitochondrial Stress-70 protein (also known as mortalin), found in the mitochondrial preparation of fast-twitch muscle (entry 36 in Table 1). This spectrum shows the conspicuous difference in intensity of iTRAQ reagent 114 and 116 reporter ions, indicating an increased abundance of the carbonylated form of this protein in old rats compared to that in young adult rats.
Figure 4.
Representative tandem mass spectrometry (MS/MS) spectra of Stress 70-protein peptides. A, Example MS/MS spectra used for identification of peptides and quantification of protein carbonylation changes. B, Example MS/MS spectra used for peptide identification and protein abundance. Replicate experiments were analyzed in the same MS/MS analysis; one replicate was labeled with iTRAQ reagents 114 and 116, the other with 115 and 117. Therefore, the intensities of 114:116 and 115:117 reporter ion pairs should be compared to each other in the figure.
Figure 4B shows an example of data obtained from experiments measuring overall abundance changes in aging muscle mitochondria. A peptide identified belonging to the Stress-70 protein is shown, which was also identified in the experiments identifying carbonylated proteins and their abundance changes with age (Figure 4A). For the experiment measuring overall abundance changes, young and old muscle mitochondria samples were prepared in duplicate, labeling one pair of young and old samples with iTRAQ reagents 114 and 116, and the other pair of samples with reagents 115 and 117, and combining these four different samples prior to MS/MS analysis. Therefore, reporter ions’ intensities at mass-to-charge values of 114 and 116 were compared for one of the replicates, and 115 and 117 were compared for the other replicate in Figure 4B. This comparison shows that, in the replicate experiments, the overall abundance for Stress-70 protein did not change with age, in stark contrast to the large increase observed for the carbonylated form of this protein (Figure 4A).
From the 200 proteins that were identified in the overall abundance-profiling experiment (false-positive rate of 1%), 28 were selected because they were also identified as being carbonylated (these proteins are shown in Supplemental Table 1). As described in the “Method” section, we used a 90% confidence interval (0.5, 1.7) to determine whether the selected 28 proteins changed in their overall abundance, carbonylation abundance, or both. Using this conservative threshold to assign significance to measured quantitative changes, a total of 15 proteins showed change in their carbonylation state, with no similar change in their overall abundance (Table 3, entries 1–15).
Table 3.
Age-Related Changes in Carbonyl Abundance and Protein Abundance From Fast-Twitch Muscle
| No. | Accession* | Proteins Showing Change in Carbonylation with Correction of Overall Abundance† | Ratio‡ (Overall Abundance) 117:115 | Ratio§ (Carbonylation) 116:114 | Corrected Ratio Carbonylation† |
|---|---|---|---|---|---|
| 1 | Q60587 | Trifunctional enzyme subunit β | 0.9 | 1.7 | 1.9 |
| 2 | Q64428 | Trifunctional enzyme subunit α | 1.0 | 9.9 | 9.9 |
| 3 | P45953 | Very-long-chain specific acyl-CoA dehydrogenase | 0.9 | 2.7 | 3.0 |
| 4 | Q05962 | ADP/ATP translocase 1 | 1.3 | 8.7 | 6.7 |
| 5 | P17764 | Acetyl-CoA acetyltransferase | 0.7 | 3.0 | 4.3 |
| 6 | P63039 | 60 kd heat shock protein | 0.6 | 4.8 | 8.0 |
| 7 | P20788 | Ubiquinol–cytochrome c reductase iron-sulfur subunit | 0.8 | 2.4 | 3.0 |
| 8 | P48721 | Stress-70 protein | 0.9 | 6.4 | 7.1 |
| 9 | Q01205 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | 1.0 | 6.2 | 6.2 |
| 10 | Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | 0.9 | 4.1 | 4.6 |
| 11 | Q99NA5 | Isocitrate dehydrogenase (NAD) subunit α | 0.7 | 2.1 | 3.0 |
| 12 | IPI00390435.1 | 76 kd protein | 1.6 | 3.1 | 1.9 |
| 13 | P15999 | ATP synthase subunit α | 0.5 | 1.8 | 3.6 |
| 14 | Q6DGF1 | NADP+-specific isocitrate dehydrogenase | 0.8 | 2.3 | 2.9 |
| 15 | P15650 | Long-chain specific acyl-CoA dehydrogenase | 0.4 | 134.9 | 337.3 |
| 16 | Q6GSM4 | Malate dehydrogenase | 193.2 | ||
| 17 | Q5M7W0 | Electron transferring flavoprotein | 40.2 | ||
| 18 | IPI00365813.2 | Voltage-dependent anion-selective channel protein | 27.9 | ||
| 19 | P08461 | Dihydrolipoamide S-acetyltransferase | 15.1 | ||
| 20 | P31399 | ATP synthase D chain | 0.1 | ||
| 21 | IPI00371236.2 | TU translation elongation factor | 0.1 | ||
| 22 | P13437 | Acetyl-CoA acyltransferase | 0.1 |
Notes: Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
Corrected ratio carbonylation is calculated from ratios for carbonylated protein divided by those for protein abundance. These proteins are those with a carbonylation ratio outside the 90% confidence interval (0.5, 1.7), but with an overall abundance ratio within this interval. Proteins numbered 16–22 are proteins identified as having significant changes in carbonylation, but were not identified in the proteomic experiments measuring overall abundance changes with age.
Ratios of protein abundance compared between old and young animals are given from quantitative proteomic experiments measuring the overall abundance of enriched mitochondrial proteins.
Ratios of protein abundance compared between old and young animals are given from experiments identifying enriched, carbonylated mitochondrial proteins.
CoA =coenzyme A; ATP = adenosine-5′-triphosphate; ADP =adenosine diphosphate; NADH = reduced nicotinamide adenine dinucleotide; NAD =nicotinamide adenine dinucleotide; NADP = nicotinamide adenine dinucleotide phosphate.
Pathways and Functions Represented by Proteins Showing Increased Carbonylation With Age in Fast-Twitch Muscle
Using IPA, we mapped selected proteins with a significant age-dependent change in carbonylation to the curated biochemical pathways and biological functions of the IPA environment. The significant (p < .001) canonical pathways and functions are included in Tables 4 and 5, respectively. The selected set of proteins included proteins with (a) change in carbonylation corrected for change in overall abundance (Table 3, entries 1–15) and (b) significant changes in overall carbonylation that were not identified in the overall abundance-profiling experiments (bottom of Table 3, entries 16–22). A total of 13 canonical pathways (Table 4) and 7 biological functions (Table 5) mapped to the proteins shown in Table 3. The relatively high number of pathway and function hits (20) compared with the number of proteins in the set (22) reflects the fact that these pathways and functions are highly inter-related, with many of the proteins identified participating in multiple pathways.
Table 4.
Significant Canonical Pathways Mapped to Protein Showing Age-Dependent Quantitative Changes by IPA*
| No. | Canonical Pathway† | p | Accession‡,§ | Protein Name |
|---|---|---|---|---|
| 1 | Valine, leucine, and isoleucine degradation | 5.3 × 10−14 | P13437 ↓ | Acetyl-coenzyme A acyltransferase |
| P15650 | Acyl-coenzyme A dehydrogenase | |||
| P45953 | Very-long-chain acyl-coenzyme A dehydrogenase | |||
| P17764 | Acetyl-coenzyme A acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional enzyme β subunit | |||
| 2 | Fatty acid metabolism | 3.6 × 10−12 | P13437 ↓ | Acetyl-coenzyme A acyltransferase |
| P15650 | Acyl-coenzyme A dehydrogenase | |||
| P45953 | Very-long-chain acyl-coenzyme A dehydrogenase | |||
| P17764 | Acetyl-coenzyme A acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional enzyme β subunit | |||
| 3 | Propanoate metabolism | 1.7 × 10−11 | P15650 | Acyl-coenzyme A dehydrogenase |
| P45953 | Very-long-chain acyl-coenzyme A dehydrogenase | |||
| P17764 | Acetyl-coenzyme A acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional enzyme β subunit | |||
| 4 | Pyruvate metabolism | 5.2 × 10−11 | P17764 | Acetyl-coenzyme A acetyltransferase 1 |
| P08461 | Dihydrolipoamide S-acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| Q6GSM4 | Malate dehydrogenase 2 | |||
| 5 | β-alanine metabolism | 3.0 × 10−9 | P15650 | Acyl-coenzyme A dehydrogenase, long chain |
| P45953 | Acyl-coenzyme A dehydrogenase, very long chain | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| 6 | Synthesis and degradation of ketone bodies | 8.2 × 10−9 | P17764 | Acetyl-coenzyme A acetyltransferase 1 |
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| 7 | Fatty acid elongation | 2.5 × 10−8 | P13437 ↓ | Acetyl-coenzyme A acyltransferase 2 |
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| 8 | Citrate cycle | 1.7 × 10−7 | Q6DGF1 | Isocitrate dehydrogenase 2 (NADP+) |
| Q99NA5 | Isocitrate dehydrogenase 3 (NAD+) α | |||
| Q6GSM4 | Malate dehydrogenase 2 | |||
| 9 | Oxidative phosphorylation | 1.8 × 10−8 | P15999 | ATP synthase, H+ transporting, mitochondrial F1 complex |
| P31399 ↓ | ATP synthase, H+ transporting, mitochondrial F0 complex | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | |||
| P20788 | Ubiquinol–cytochrome c reductase, iron-sulfur polypeptide | |||
| 10 | Lysine degradation | 1.7 × 10−6 | P17764 | Acetyl-coenzyme A acetyltransferase 1 |
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| 11 | Tryptophan metabolism | 1.3 × 10−5 | P17764 | Acetyl-coenzyme A acetyltransferase 1 |
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein β subunit | |||
| 12 | Glutathione metabolism | 2.8 × 10−4 | Q6DGF1 | Isocitrate dehydrogenase 2 (NADP+), |
| Q99NA5 | Isocitrate dehydrogenase 3 (NAD+) α | |||
| 13 | Mitochondrial dysfunction | 9.0 × 10−4 | Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, |
| P20788 | Ubiquinol-cytochrome c reductase, iron-sulfur polypeptide 1 |
Notes: The carbonylated protein used in IPA analysis are those from Table 3.
The canonical pathways are in the order of significance represented by p value. In IPA, p value is a measurement of the likelihood that the pathway is associated with the data set by random chance.
Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
The symbol “↓” after an accession number indicates that the protein has decreased carbonylation with aging. All other proteins showed an increase in carbonylation with age.
IPA = Ingenuity pathway analysis; NADP = nicotinamide adenine dinucleotide phosphate; NAD = nicotinamide adenine dinucleotide; ATP = adenosine-5′-triphosphate; NADH = reduced nicotinamide adenine dinucleotide.
Table 5.
Significant Biological Functions Mapped by Carbonylated Proteins*
| Functions† | p | Accession‡,§ | Protein Name | |
|---|---|---|---|---|
| 1 | Carbohydrate metabolism | 1.7 × 10−15 | P15650 | Acyl-coenzyme A dehydrogenase, long chain |
| P45953 | Acyl-coenzyme A dehydrogenase, very long chain | |||
| P17764 | Acetyl-coenzyme A acetyltransferase 1 | |||
| P15999 | ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1 | |||
| P31399 ↓ | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | |||
| P08461 | Dihydrolipoamide S-acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein, β subunit | |||
| Q6DGF1 | Isocitrate dehydrogenase 2 (NADP+) | |||
| Q99NA5 | Isocitrate dehydrogenase 3 (NAD+) α | |||
| Q6GSM4 | Malate dehydrogenase 2, NAD | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein | |||
| P20788 | Ubiquinol-cytochrome c reductase, iron-sulfur polypeptide 1 | |||
| 2 | Amino acid metabolism | 5.3 × 10−14 | P13437 ↓ | Acetyl-coenzyme A acyltransferase 2 |
| P15650 | Acyl-coenzyme A dehydrogenase, long chain | |||
| P45953 | Acyl-coenzyme A dehydrogenase, very long chain | |||
| P17764 | Acetyl-coenzyme A acetyltransferase 1 | |||
| P08461 | Dihydrolipoamide S-acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein, β subunit | |||
| Q6DGF1 | Isocitrate dehydrogenase 2 (NADP+) | |||
| Q99NA5 | Isocitrate dehydrogenase 3 (NAD+) α | |||
| 3 | Lipid metabolism | 3.6 × 10−12 | P13437 ↓ | Acetyl-coenzyme A acyltransferase 2 |
| P15650 | Acyl-coenzyme A dehydrogenase, long chain | |||
| P45953 | Acyl-coenzyme A dehydrogenase, very long chain | |||
| P17764 | Acetyl-coenzyme A acetyltransferase 1 | |||
| P08461 | Dihydrolipoamide S-acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein, β subunit | |||
| 4 | Small molecule biochemistry | 6.5 × 10−8 | P15650 | Acyl-coenzyme A dehydrogenase, long chain |
| P45953 | Acyl-coenzyme A dehydrogenase, very long chain | |||
| P17764 | Acetyl-coenzyme A acetyltransferase 1 | |||
| P08461 | Dihydrolipoamide S-acetyltransferase | |||
| Q64428 | Trifunctional protein, α subunit | |||
| Q60587 | Trifunctional protein, β subunit | |||
| P63039 | Heat shock 60-kd protein 1 (chaperonin) | |||
| Q01205 | Myeloid/lymphoid or mixed-lineage leukemia | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | |||
| Q05962 | Adenine nucleotide translocator | |||
| 5 | Cell signaling | 1.8 × 10−7 | P15650 | Acyl-coenzyme A dehydrogenase, long chain |
| P15999 | ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1 | |||
| P31399 ↓ | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | |||
| P48721 | Heat shock 70-kd protein 9 | |||
| P63039 | Heat shock 60-kd protein 1 | |||
| Q6DGF1 | Isocitrate dehydrogenase 2 (NADP+) | |||
| Q99NA5 | Isocitrate dehydrogenase 3 (NAD+) α | |||
| Q6GSM4 | Malate dehydrogenase 2, NAD | |||
| Q01205 | Myeloid/lymphoid or mixed-lineage leukemia | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | |||
| P20788 | Ubiquinol–cytochrome c reductase, iron-sulfur polypeptide 1 | |||
| 6 | Energy production | 1.8 × 10−7 | P45953 | Acyl-coenzyme A dehydrogenase, very long chain |
| P15999 | ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1 | |||
| P31399 ↓ | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | |||
| P63039 | Heat shock 60-kd protein 1 | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | |||
| Q05962 | Adenine nucleotide translocator | |||
| P20788 | Ubiquinol–cytochrome c reductase, iron-sulfur polypeptide 1 | |||
| 7 | Cell death | 3.3 × 10−4 | Q64428 | Trifunctional protein, α subunit |
| P48721 | Heat shock 70-kd protein | |||
| P63039 | Heat shock 60-kd protein | |||
| Q66HF1 | NADH dehydrogenase (ubiquinone) Fe-S protein | |||
| Q05962 | Adenine nucleotide translocator |
Notes: The carbonylated protein used in Ingenuity pathway analysis (IPA) are those from Table 3.
The canonical pathways are in the order of significance represented by p value. In IPA, p value is a measurement of the likelihood that the pathway is associated with the data set by random chance.
Swiss-Prot accession numbers are given for each protein. For those proteins without a Swiss-Prot accession number, IPI accession numbers are given instead.
The symbol “↓” after an accession number indicates that the protein has decreased carbonylation with aging. All other proteins showed an increase in carbonylation with age.
ATP = adenosine-5′-triphosphate; NADP = nicotinamide adenine dinucleotide phosphate; NAD = nicotinamide adenine dinucleotide; NADH = reduced nicotinamide adenine dinucleotide.
Discussion
Proteomic Analysis of Slow- and Fast-Twitch Muscle Reveals Muscle-Type Dependence of Protein Carbonylation
Our findings provide novel data on the muscle type dependence of protein carbonylation in skeletal muscle mitochondria. Although carbonyl formation in proteins in skeletal muscle mitochondria has been reported by our group and others (5,13,33), our present study is the first using advanced proteomic tools to identify the differences in proteins susceptible to modification between slow- and fast-twitch muscle. Despite using equal amounts of tissue in our studies, approximately twice as many proteins were detected as carbonylated in the fast-twitch muscle compared to slow-twitch (78 and 38 proteins, respectively), with 29 of these proteins being identified in both muscle types (Figure 3).
Our findings lead to a basic question: Why would fast-twitch muscle produce more carbonylated mitochondrial proteins than slow twitch-muscle? On one hand, our findings are consistent with past reports showing more severe oxidative damage to fast-twitch muscle (32,34,35). On the other hand, our findings are somewhat surprising, given that fast-twitch muscle has less mitochondrial content than slow-twitch muscle (36), and it relies primarily on cytoplasmic glycolysis for adenosine-5′-triphosphate (ATP) production, rather than mitochondrial oxidative phosphorylation like in slow-twitch muscle. Based on this understanding, one might hypothesize that fast-twitch muscle would show less carbonyl formation in mitochondrial proteins than slow-twitch muscle would. Although oxidative damage originating from ROS in the cytoplasm of fast-twitch muscle fibers (e.g., cytoplasmic xanthine oxidase or nitric oxide synthase) cannot be ruled out (37,38), one explanation for our findings may be the lower superoxide scavenging capacity in fast-twitch muscle compared to slow-twitch muscle (36). This indeed could lead to carbonylation of more mitochondrial proteins in fast-twitch muscle, such as oxidative phosphorylation components, which were prominently represented in the carbonylated proteins identified from fast-twitch muscle.
Beyond a simple comparison of the number of proteins identified from different muscle types, advanced bioinformatic analysis of our proteomic data using the IPA tool provided a unique look at cellular pathways and functions potentially affected by carbonylation in the different muscle types. Interestingly, proteins susceptible to carbonylation having a role in two functions, cellular function/maintenance and cell death, were significantly represented in the fast-twitch muscle, but not in the slow-twitch muscle (Table 2). This finding is potentially significant, given the fact that fast-twitch muscle is known to show more rapid decline with age than slow-twitch muscle does (32). Our results provide the identities of specific protein targets of carbonylation and their cellular functions, which could potentially contribute to this observed functional decline in aging fast-twitch muscle.
An Advanced, Two-Pronged Quantitative Proteomic Strategy Reveals Age-Dependent Protein Carbonylation Changes in Fast-Twitch Muscle
Our two-pronged proteomic strategy enabled the identification of proteins showing true changes in carbonylation in aging skeletal muscle. Because analysis of quantitative changes in enriched carbonylated proteins on its own does not rule out the possibility that the observed changes are actually due to a change in overall abundance, we complemented our proteomic method for studying carbonylation changes with quantitative proteomic analysis of changes in overall abundance of mitochondrial proteins from young and old animals (Figure 2). Comparative analysis of these data sets enabled the identification of proteins showing true changes in their carbonylation state with age. This two-pronged proteomic strategy identified 15 proteins showing significant changes in carbonylation with age, while showing no similar change in the overall abundance of the protein.
Bioinformatic Analysis Reveals Connections Between Age-Dependent Protein Carbonylation and Impaired Biochemical Functions in Aging Muscle
Bioinformatic analysis of our novel data sets using IPA provided numerous examples of possible connections between protein carbonylation and cellular functions associated with age-dependent skeletal muscle decline. One example is the detection of carbonylated voltage-dependent anion channel (VDAC) selective protein from fast-twitch muscle, which maps to “Cellular function and maintenance” within the IPA environment. This protein showed a large increase in carbonylation in aged muscle (Table 3). Along with its binding partner, adenosine diphosphate (ADP)/ATP translocase protein, which also showed a marked increase in carbonylation with age (Table 3), VDAC enables transport of ions, such as calcium ions (Ca+2), across the inner mitochondrial membrane, critical to mitochondrial function (39). Impaired mitochondrial cycling of Ca+2 has been associated with aging skeletal muscle (40). One may hypothesize that increased carbonyl modification of these proteins critical to mitochondrial inner membrane transport may contribute to this impaired cellular function in aged muscle.
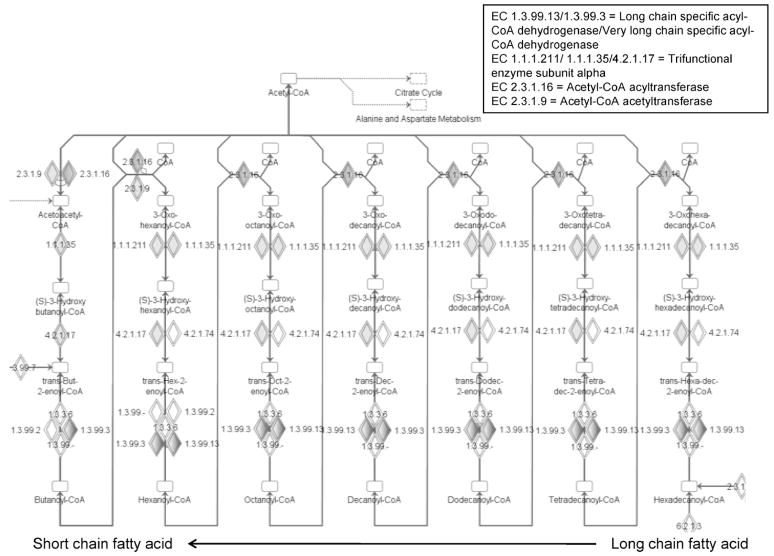
IPA also enabled identification of biochemical pathways represented by proteins showing changes in carbonylation with age, which are known to be impaired in aging muscle. The use of an advanced tool such as IPA is highly valuable, as it enables the identification of these pathways from our data set, which may not be apparent via manual interpretation of the data. One example is the “fatty acid metabolism” pathway (Table 2). The network of enzymatic reactions within the fatty acid metabolism pathway is shown in Figure 5. Beginning with long-chain fatty acids activated with coenzyme A (CoA; Figure 5, bottom right), these are oxidized in successive steps, with each step decreasing the fatty acid by two carbons and producing acetyl-CoA. In the final step, the shortened, four-carbon fatty acid chain (Figure 5, bottom left) is oxidized, producing a final molecule of acetyl-CoA. The acetyl-CoA produced from fatty acid metabolism enters the citrate cycle, ultimately driving ATP production. In our data set, proteins with enzymatic activity mapping to five of the steps in fatty acid metabolism show increased age-dependent carbonylation (lightly shaded diamonds in middle rows, larger shaded diamonds in bottom row), and one (acetyl-CoA acyltransferase [EC 2.3.1.16]) showed decreased abundance of its carbonylated form (grey-shaded diamonds in top row).
Figure 5.
Fatty acid metabolism pathways represented by carbonylated proteins. Proteins with enzymatic activities within this pathway and identified in our studies as having significant age-related changes in carbonylation are highlighted by the grey-shaded diamonds. Figure depicts successive steps of oxidation of long-chain fatty acids (starting at the bottom right and moving right to left), with each step reducing the fatty acid chain by two carbons, and producing acetyl-coenzyme A (acetyl-CoA). Enzyme commission (EC) numbers and the proteins identified with corresponding activities are shown.
Our identification of proteins showing susceptibility to carbonylation within the fatty acid metabolism pathway is interesting based on two known features of aging skeletal muscle. First, lipid content is known to increase in aging skeletal muscle (41). Given that breakdown of these lipids via ROS leads to formation of compounds such as HNE (42), which then may react with amino acid side chains and introduce reactive carbonyls into proteins, enzymes involved in processing of fatty acids are likely targets of carbonylation. Consistent with this reasoning, most of the proteins identified in our study and mapped to this pathway show increased carbonylation with aging.
Second, carbonylation of fatty acid proteins suggests a possible role of these modifications in decreasing the function of this pathway with age. A known feature of aging skeletal muscle is the decreased ability of this tissue to oxidize fatty acids for energy generation (43,44). The increased carbonylation of numerous proteins involved in fatty acid metabolism therefore points to a possible link between these modifications, causing a decrease in their function and leading to decreased efficiency in removing lipids. Supporting this hypothesis, previous work by members of our research group has shown that carbonylation with HNE can affect the binding of fatty acid substrates to proteins (45). Decreased fatty acid metabolism would increase the presence of toxic lipids within skeletal muscle tissue, leading to more protein carbonylation, setting up a feedback scenario by which carbonylation impairs function and leads to further lipid peroxidation and modification and dysfunction of these proteins.
Another pathway known to show decreased activity in aging skeletal muscle, the citrate cycle (46), was also significantly represented by the set of proteins with increased carbonylation with aging (Table 4). Among the proteins involved in the citrate cycle which we identified as showing age-dependent carbonylation increase is nicotinamide adenine dinucleotide phosphate (NADP)-isocitrate dehydrogenase, which is the only enzyme supplying reduced nicotinamide adenine dinucleotide phosphate (NADPH) in mitochondria. If carbonylation affects its function, it could impair not only energy generation via the citrate cycle, but also the regeneration of glutathione (47) and the NADPH-dependent thioredoxin system, thereby compromising the antioxidant system in mitochondria (48,49). Because this enzyme is regulated by binding HNE (48), one future investigation suggested by our results may determine (a) whether this protein is carbonylated via modification with HNE within skeletal muscle mitochondria, and (b) its possible functional effect.
Another interesting subset of proteins showing age-related changes in carbonylation are proteins involved in “cell signaling function” (Table 5). The heat shock proteins, HSP 60 kd and Stress-70 kd, which play an important role in helping cells cope with a number of stresses such as heat shock and oxidative stress, are included in this IPA function. There is a controversy as to whether aging is associated with an increased or a decreased level of HSPs (50,51). In our study, we did not find any significant change in Stress-70 overall protein abundance in mitochondria (Old:Young = 0.9, Table 3), but its carbonylation state was increased 6.4-fold in old rats relative to young rats. If carbonylation is affecting the function of these proteins, this may lead to loss of cell signaling regulation such as during apoptosis/necrosis (50), and potentially decreased protection conferred by these proteins against known age-related functional impairment (52,53). Investigating the possible disruption of functional complexes formed by these proteins (54,55) due to carbonylation would provide insights into the role of increased protein carbonylation in affecting the functions of these proteins in aging muscle.
Protein–Protein Relationships Revealed by IPA Generate Hypotheses on Mechanisms of Protein Carbonylation
As a final note, IPA not only helps in generating hypotheses relating protein carbonylation to biological functions that are affected with the aging process, but also provides information on known protein–protein relationships between members of large protein data sets through graphical representation of pathways. This information may be useful in identifying the mechanisms of protein carbonylation within the cellular milieu. As an example from our data, ubiquinol–cytochrome c reductase iron–sulfur protein and ubiquinol–cytochrome c reductase core protein 1 were both identified as being carbonylated (Table 1, entries 11 and 12, respectively). As made apparent by IPA, these proteins contain protein–protein interaction domains in the mitochondrial complex III. Because one of these proteins has iron–sulfur clusters, which are involved in superoxide generation (56,57), it is plausible that the other protein in the proximity of ROS production is therefore susceptible to carbonylation, consistent with our findings. Such information, combined with experimental evidence, can be useful in better understanding mechanisms leading to protein carbonylation.
Conclusion
We have presented results from a novel proteomic study investigating muscle-type dependence and age-related changes of carbonyl modifications in rat skeletal muscle, identifying about twice as many carbonylated proteins in the fast- versus the slow-twitch muscles. The relatively small number of proteins changing in their carbonylation in fast-twitch muscle is a powerful predictor of potential functions and pathways affected by protein carbonylation, demonstrated by the 17 biological functions and pathways mapped to these proteins using IPA. Our findings have generated new hypotheses on ROS-induced mechanisms of protein carbonylation, as well as possible connections between protein carbonyl modifications and cellular functions already known to be impaired in aging muscle. These numerous hypotheses provide targets for future testing, putting us a step closer to understanding the role of protein carbonylation in aging muscle decline.
Acknowledgments
This work was supported by the National Institutes of Health (AG025371, AG17768, and AG21626). We thank Kate O’Connor for providing rat skeletal muscle tissue used in this study. We also thank the University of Minnesota Mass Spectrometry and Proteomics facility and the Minnesota Supercomputing Institute for instrumentation and computational support, respectively. T. J. G. also thanks Eli Lilly and Company for support.
References
- 1.Ashok BT, Ali R. The aging paradox: free radical theory of aging. Exp Gerontol. 1999;34:293–303. doi: 10.1016/s0531-5565(99)00005-4. [DOI] [PubMed] [Google Scholar]
- 2.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 4.Bautista J, Mateos-Nevado MD. Immunological detection and quantification of oxidized proteins by labelling with digoxigenin. Biosci Biotechnol Biochem. 1998;62:419–423. doi: 10.1271/bbb.62.419. [DOI] [PubMed] [Google Scholar]
- 5.Fano G, Mecocci P, Vecchiet J, et al. Age and sex influence on oxidative damage and functional status in human skeletal muscle. J Muscle Res Cell Motil. 2001;22:345–351. doi: 10.1023/a:1013122805060. [DOI] [PubMed] [Google Scholar]
- 6.Mecocci P, Fano G, Fulle S, et al. Age-dependent increases in oxidative damage to DNA, lipids, and proteins in human skeletal muscle. Free Radic Biol Med. 1999;26:303–308. doi: 10.1016/s0891-5849(98)00208-1. [DOI] [PubMed] [Google Scholar]
- 7.Starke PE, Oliver CN, Stadtman ER. Modification of hepatic proteins in rats exposed to high oxygen concentration. FASEB J. 1987;1:36–39. doi: 10.1096/fasebj.1.1.2886388. [DOI] [PubMed] [Google Scholar]
- 8.Sohal RS, Agarwal S, Sohal BH. Oxidative stress and aging in the Mongolian gerbil (Meriones unguiculatus) Mech Ageing Dev. 1995;81:15–25. doi: 10.1016/0047-6374(94)01578-a. [DOI] [PubMed] [Google Scholar]
- 9.Sohal RS, Agarwal S, Dubey A, Orr WC. Protein oxidative damage is associated with life expectancy of houseflies. Proc Natl Acad Sci U S A. 1993;90:7255–7259. doi: 10.1073/pnas.90.15.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver CN, Ahn BW, Moerman EJ, Goldstein S, Stadtman ER. Age-related changes in oxidized proteins. J Biol Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 11.Goto S, Nakamura A, Radak Z, et al. Carbonylated proteins in aging and exercise: immunoblot approaches. Mech Ageing Dev. 1999;107:245–253. doi: 10.1016/s0047-6374(98)00133-x. [DOI] [PubMed] [Google Scholar]
- 12.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 14.Davies KJ, Delsignore ME, Lin SW. Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J Biol Chem. 1987;262:9902–9907. [PubMed] [Google Scholar]
- 15.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 16.Bruckdorfer KR. Lipid oxidation products and vascular function. Free Radic Res. 1998;28:573–581. doi: 10.3109/10715769809065813. [DOI] [PubMed] [Google Scholar]
- 17.Robinson CE, Keshavarzian A, Pasco DS, Frommel TO, Winship DH, Holmes EW. Determination of protein carbonyl groups by immunoblotting. Anal Biochem. 1999;266:48–57. doi: 10.1006/abio.1998.2932. [DOI] [PubMed] [Google Scholar]
- 18.Barreiro E, Gea J, Di Falco M, Kriazhev L, James S, Hussain SN. Protein carbonyl formation in the diaphragm. Am J Respir Cell Mol Biol. 2005;32:9–17. doi: 10.1165/rcmb.2004-0021OC. [DOI] [PubMed] [Google Scholar]
- 19.Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional gel electrophoresis using avidin-fluorescein affinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 20.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaei H, Regnier F. Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem. 2005;77:2386–2392. doi: 10.1021/ac0484373. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Liu W, Bemis A, et al. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum. 2007;56:3675–3684. doi: 10.1002/art.22876. [DOI] [PubMed] [Google Scholar]
- 23.Meneses-Lorente G, Watt A, Salim K, et al. Identification of early proteomic markers for hepatic steatosis. Chem Res Toxicol. 2006;19:986–998. doi: 10.1021/tx060007f. [DOI] [PubMed] [Google Scholar]
- 24.Mayburd AL, Martlinez A, Sackett D, et al. Ingenuity network-assisted transcription profiling: identification of a new pharmacologic mechanism for mk886. Clin Cancer Res. 2006;12:1820–1827. doi: 10.1158/1078-0432.CCR-05-2149. [DOI] [PubMed] [Google Scholar]
- 25.Griffin TJ, Xie H, Bandhakavi S, et al. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J Proteome Res. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (lc/lc-ms/ms) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by ms/ms and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie H, Griffin TJ. Trade-off between high sensitivity and increased potential for false positive peptide sequence matches using a two-dimensional linear ion trap for tandem mass spectrometry-based proteomics. J Proteome Res. 2006;5:1003–1009. doi: 10.1021/pr050472i. [DOI] [PubMed] [Google Scholar]
- 31.Requena JR, Chao CC, Levine RL, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci U S A. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson LV. Skeletal muscle adaptations with age, inactivity, and therapeutic exercise. J Orthop Sports Phys Ther. 2002;32:44–57. doi: 10.2519/jospt.2002.32.2.44. [DOI] [PubMed] [Google Scholar]
- 33.Bejma J, Ji LL. Aging and acute exercise enhance free radical generation in rat skeletal muscle. J Appl Physiol. 1999;87:465–470. doi: 10.1152/jappl.1999.87.1.465. [DOI] [PubMed] [Google Scholar]
- 34.Ahmadzadeh H, Johnson RD, Thompson LV, Arriaga EA. Direct sampling from muscle cross sections for electrophoretic analysis of individual mitochondria. Anal Chem. 2004;76:315–321. doi: 10.1021/ac034809g. [DOI] [PubMed] [Google Scholar]
- 35.Lowe DA, Warren GL, Snow LM, Thompson LV, Thomas DD. Muscle activity and aging affect myosin structural distribution and force generation in rat fibers. J Appl Physiol. 2004;96:498–506. doi: 10.1152/japplphysiol.00842.2003. [DOI] [PubMed] [Google Scholar]
- 36.Anderson EJ, Neufer PD. Type ii skeletal myofibers possess unique properties that potentiate mitochondrial h(2)o(2) generation. Am J Physiol Cell Physiol. 2006;290:C844–C851. doi: 10.1152/ajpcell.00402.2005. [DOI] [PubMed] [Google Scholar]
- 37.Reid MB, Haack KE, Franchek KM, Valberg PA, Kobzik L, West MS. Reactive oxygen in skeletal muscle. I. Intracellular oxidant kinetics and fatigue in vitro. J Appl Physiol. 1992;73:1797–1804. doi: 10.1152/jappl.1992.73.5.1797. [DOI] [PubMed] [Google Scholar]
- 38.Meydani M, Evans WJ, Handelman G, et al. Protective effect of vitamin e on exercise-induced oxidative damage in young and older adults. Am J Physiol. 1993;264:R992–R998. doi: 10.1152/ajpregu.1993.264.5.R992. [DOI] [PubMed] [Google Scholar]
- 39.Pi Y, Goldenthal MJ, Marin-Garcia J. Mitochondrial channelopathies in aging. J Mol Med. 2007;85:937–951. doi: 10.1007/s00109-007-0190-5. [DOI] [PubMed] [Google Scholar]
- 40.Weisleder N, Brotto M, Komazaki S, et al. Muscle aging is associated with compromised Ca2+ spark signaling and segregated intracellular Ca2+ release. J Cell Biol. 2006;174:639–645. doi: 10.1083/jcb.200604166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker MZ, Turcotte LP. Aging is associated with elevated muscle triglyceride content and increased insulin-stimulated fatty acid uptake. Am J Physiol Endocrinol Metab. 2003;285:E827–E835. doi: 10.1152/ajpendo.00222.2002. [DOI] [PubMed] [Google Scholar]
- 42.Leeuwenburgh C, Hansen P, Shaish A, Holloszy JO, Heinecke JW. Markers of protein oxidation by hydroxyl radical and reactive nitrogen species in tissues of aging rats. Am J Physiol. 1998;274:R453–R461. doi: 10.1152/ajpregu.1998.274.2.R453. [DOI] [PubMed] [Google Scholar]
- 43.Nagy TR, Goran MI, Weinsier RL, Toth MJ, Schutz Y, Poehlman ET. Determinants of basal fat oxidation in healthy Caucasians. J Appl Physiol. 1996;80:1743–1748. doi: 10.1152/jappl.1996.80.5.1743. [DOI] [PubMed] [Google Scholar]
- 44.Toth MJ, Tchernof A. Lipid metabolism in the elderly. Eur J Clin Nutr. 2000;54(Suppl 3):S121–S125. doi: 10.1038/sj.ejcn.1601033. [DOI] [PubMed] [Google Scholar]
- 45.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Pastoris O, Boschi F, Verri M, et al. The effects of aging on enzyme activities and metabolite concentrations in skeletal muscle from sedentary male and female subjects. Exp Gerontol. 2000;35:95–104. doi: 10.1016/s0531-5565(99)00077-7. [DOI] [PubMed] [Google Scholar]
- 47.Murakami K, Yoshino M. Aluminum decreases the glutathione regeneration by the inhibition of NADP-isocitrate dehydrogenase in mitochondria. J Cell Biochem. 2004;93:1267–1271. doi: 10.1002/jcb.20261. [DOI] [PubMed] [Google Scholar]
- 48.Benderdour M, Charron G, Comte B, et al. Decreased cardiac mitochondrial NADP+-isocitrate dehydrogenase activity and expression: a marker of oxidative stress in hypertrophy development. Am J Physiol. 2004;287:H2122–H2131. doi: 10.1152/ajpheart.00378.2004. [DOI] [PubMed] [Google Scholar]
- 49.Yang JH, Yang ES, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by lipid peroxidation products. Free Radic Res. 2004;38:241–249. doi: 10.1080/10715760310001657712. [DOI] [PubMed] [Google Scholar]
- 50.Chung L, Ng YC. Age-related alterations in expression of apoptosis regulatory proteins and heat shock proteins in rat skeletal muscle. Biochim Biophys Acta. 2006;1762:103. doi: 10.1016/j.bbadis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Donoghue P, Doran P, Dowling P, Ohlendieck K. Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. Biochim Biophys Acta. 2005;1752:166–176. doi: 10.1016/j.bbapap.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Deocaris CC, Kaul SC, Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones. 2006;11:116–128. doi: 10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broome CS, Kayani AC, Palomero J, et al. Effect of lifelong overexpression of hsp70 in skeletal muscle on age-related oxidative stress and adaptation after nondamaging contractile activity. FASEB J. 2006;20:1549–1551. doi: 10.1096/fj.05-4935fje. [DOI] [PubMed] [Google Scholar]
- 54.Deocaris CC, Widodo N, Ishii T, Kaul SC, Wadhwa R. Functional significance of minor structural and expression changes in stress chaperone mortalin. Ann N Y Acad Sci. 2007;1119:165–175. doi: 10.1196/annals.1404.007. [DOI] [PubMed] [Google Scholar]
- 55.Yaguchi T, Aida S, Kaul SC, Wadhwa R. Involvement of mortalin in cellular senescence from the perspective of its mitochondrial import, chaperone, and oxidative stress management functions. Ann N Y Acad Sci. 2007;1100:306–311. doi: 10.1196/annals.1395.032. [DOI] [PubMed] [Google Scholar]
- 56.Lenaz G, Bovina C, D’Aurelio M, et al. Role of mitochondria in oxidative stress and aging. Ann N Y Acad Sci. 2002;959:199–213. doi: 10.1111/j.1749-6632.2002.tb02094.x. [DOI] [PubMed] [Google Scholar]
- 57.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]