Abstract
We performed in situ hybridization to determine the cell type specific accumulation of the intron of the latency-associated transcript (LAT) in tissues in HSV-2 LAT transgenic mice in which LAT expression is driven by its native promoter. We identified LAT in multiple cell types in most tissues analyzed from HSV-2 LAT transgenic mice. While weak to moderate signals were seen in brain and spinal cord neurons, epithelial cells, and muscle cells, the strongest signals were detected in neurons from dorsal root and trigeminal ganglia. About 70–86% of neurons in these ganglia were LAT-positive with varying signal intensities, while cells surrounding the neurons were LAT-negative. The frequency of A5 or KH10-positive neurons was similar in LAT-positive and total neurons. These data indicate that HSV-2 LAT promoter activity is not restricted to neurons and that LAT accumulation in ganglionic neurons is likely regulated by cell-specific factors.
Keywords: herpes simplex virus 2, tissue specific gene expression, latency-associated transcript, transgenic mouse, in situ hybridization, neuron
Introduction
Human herpesvirus 2 or herpes simplex virus type 2 (HSV-2) is usually transmitted by sexual contact and causes genital lesions. After primary infection of the genital region, HSV-2 establishes life long latent infection in the dorsal root ganglia (DRG) that innervate the site of primary infection. The virus resides in sensory neurons and periodically reactivates to cause lytic infection in the peripheral tissues. During latent infection, the viral DNA persists in sensory neurons while its gene expression is extremely restricted. Only a single family of abundant viral RNAs, the HSV-2 latency-associated transcripts (LATs), accumulates to a level detectable by in situ hybridization (ISH) (Croen et al., 1991; Tenser et al., 1991).
The LAT locus is about 9 kb in length and is located in the long repeat segments flanking the unique long (UL) fragment of the HSV-2 genome. The LAT gene extends anti-sense to the ICP0 and ICP34.5 genes (McGeoch et al., 1991). HSV-2 LATs have not been consistently shown to encode protein products during virus infection. However, at least 2 species of LATs are detectable in latently infected ganglia. The primary transcript is about 8.3 kb long and can be detected by Northern blot (Krause et al., 1991). The most abundant HSV-2 LAT (the major LAT) is believed to be a 2.2 kb stable intron (LAT intron), like the major LAT of HSV-1, spliced from the primary transcript (McGeoch et al., 1991). The HSV-2 LAT is the only viral RNA that can be readily detected via ISH in latently infected neurons (Burke et al., 1991; Croen et al., 1991; Tenser et al., 1991) and has been recognized as the hallmark of HSV-2 latency. Deletion of a Not I fragment (nt −392 to +240 relative to the HSV-2 LAT transcription start site) diminishes the ability of the virus to reactivate from latency (Krause et al., 1995). The HSV-2 LAT encodes two microRNAs (miR-I, miR-II) that inhibit expression of the viral ICP34.5 gene, and a third microRNA (miR-III) that silences the expression of ICP0; these microRNAs may have important roles in viral latency (Tang et al., 2008; Tang et al., in press). The HSV-1 LAT gene has been studied more extensively. It has been reported that deletion of a 1.8 kb fragment beginning at the HSV-1 LAT promoter increased productive-cycle gene expression during acute and latent infection in mice (Chen et al., 1997; Garber et al., 1997) which may be due to the ability of the LAT gene to promote assembly of heterochromatin on viral lytic-gene promoters (Wang et al., 2005b), and/or to encode miRNAs that regulate viral genes (Umbach et al., 2008). HSV-1 LAT may also facilitate viral latency by blocking apoptosis of infected neurons (Perng et al., 2000; Thompson and Sawtell, 2001).
We have previously studied HSV-2 LAT gene expression both in vitro and in vivo. Transfection of cells with a reporter gene linked to a DNA fragment located between nucleotide −102 to +34, relative to the transcription initiation site of the LAT, results in constitutive transcription of the reporter gene in both neuronal and non-neuronal cell lines (Wang et al., 1995). Additionally, a region further upstream of the HSV-2 LAT promoter, nucleotides −392 to −102 relative to the transcription initiation site, strongly increases promoter activity in neuronal cell lines (Wang et al., 1995) and in latently infected sensory ganglia of guinea pigs (Wang et al., 1997).
By studying single neurons in human trigeminal ganglia that were latently infected with HSV-1, a virus very closely related to HSV-2, we found that HSV-1 LAT is not uniformly expressed in all latently infected neurons. Some neurons harbor many copies of HSV-1 genomic DNA, but accumulate no detectable LAT by ISH (Wang et al., 2005a). This finding suggests that regulation of LAT expression in neurons is complex and varies among different neurons in the same ganglion.
Previously we constructed mice transgenic for a 5.5 kb DNA fragment which contains the 5′ portion of the HSV-2 LAT gene beginning 1,167 bp upstream of the transcription initiation site, and ending 1,170 bp downstream of the splicing adapter site of the LAT intron (corresponding to nucleotide 118,332 to 123,608 of HSV-2 strain HG52) (Fig. 1). The accumulation of the LAT intron was detected in the nucleus of neurons in the trigeminal ganglia (TG) by ISH, and it was also detected in brain by Northern blot assay (Wang et al., 2001).
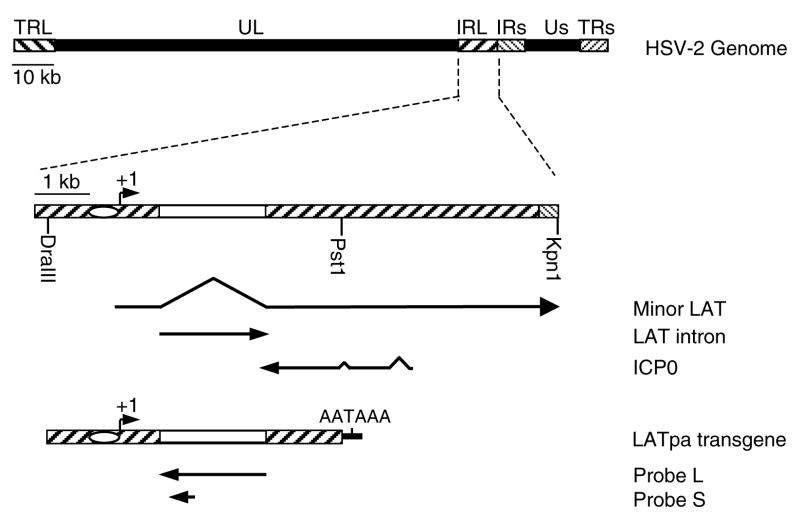
Fig. 1. Schematic representation of the HSV-2 genome, LAT gene region, and the LAT transgene cassette.
The top line represents the HSV-2 genome, with terminal (TR) and internal (IR) long (L) and short (s) repeats flanking the unique long and short segments (UL and Us, respectively). The second line shows the LAT gene region. The oval represents the LAT promoter and the open box represents the segment coding for the LAT intron. The minor and major LATs and the overlapping gene ICP0 are depicted below. The last line represents the LAT transgene cassette. The arrows below the transgene represent the RNA probes used for ISH. Probe L was used for ISH in figures 2, 3, 5, 6, 7 and probe S for ISH in figure 4.
While our transient expression results indicated that the HSV-2 LAT promoter is active in some non-neuronal cells in culture, it was unknown whether this promoter was active in different types of non-neuronal cells, including satellite cells, in vivo. To further characterize HSV-2 LAT promoter activity in various types of cells, we examined multiple tissues from HSV-2 LAT transgenic mice by ISH. Here we show that in HSV-2 LAT transgenic mice, the LAT intron is detectable by ISH not only in neurons, but also in several other tissues and cell types, and that HSV-2 LAT expression varies widely among different neurons. These findings suggest that LAT promoter activity and/or the stability of the LAT intron is regulated by different cell factors expressed in various cells including different sensory neurons.
Results
Accumulation of LAT intron in neurons is heterogenous
Trigeminal ganglia (TG), thoracic dorsal root ganglia (tDRG), sacral dorsal root ganglia (sDRG), brain, and spinal cord from LAT transgenic and wild-type mice were assayed by in situ hybridization (ISH) using a biotin labeled RNA probe antisense to the HSV-2 LAT intron (Fig. 1). LAT was detected in all of the nervous system tissues mentioned above; however, the intensity of the staining varied from tissue to tissue, and from cell to cell. About 70–86% of neurons in the sensory ganglia were positive for LAT, with blue staining distributed throughout the nuclei. The intensity of the signal ranged from strong (dark blue) to moderate (blue) to weak (light blue), and the strongest signals were seen in sDRG neurons (Fig. 2). Neurons with weak staining were scattered throughout the brain. More LAT positive neurons, about 60%, were seen with moderate to weak staining in hippocampus (Fig. 3B). Moderate signals were also detected in some of the neurons in the spinal cord (Fig. 3D). The ISH signal was present only in nuclei and interestingly was solely detected in neurons. Despite the fact that non-neuronal cells in the nervous system should have the same copy number of LAT transgenes as neurons, satellite cells and other non-neuronal cells had undetectable ISH signals. The differences in signal intensity noted among different cell types are due to differences in LAT expression and/or accumulation, and not to differences in copy number of the LAT transgene in different cell types in the transgenic line. While differences in transgene copy number in different cells have been reported in mosaic mice, this did not occur in the LAT transgenic lines since the copy numbers of the transgene in both line 5221 and line 5238 were maintained from founder mice to their offspring at levels consistent with Mendelian genetics.
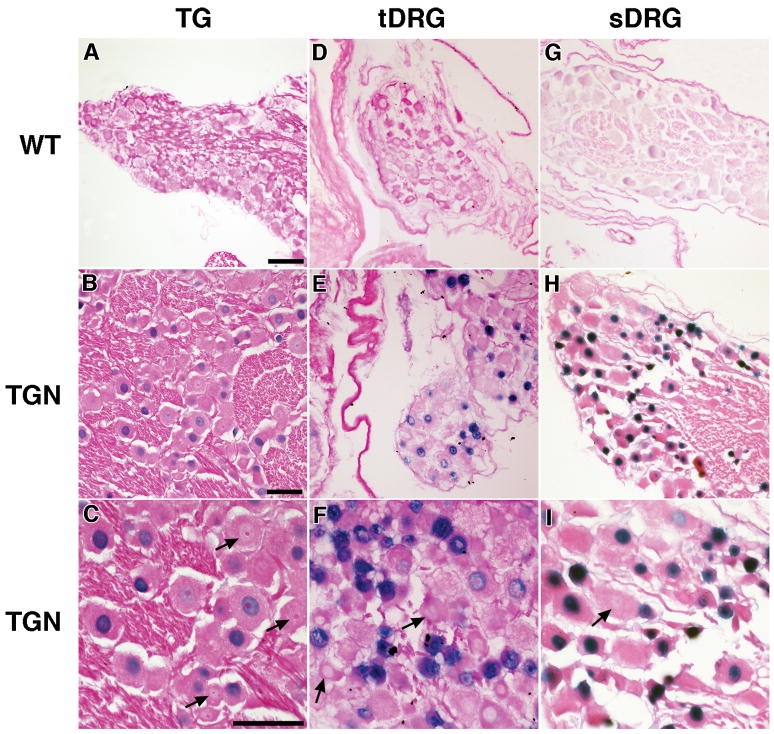
Fig. 2. LAT accumulation in sensory ganglion neurons is heterogeneous.
The LAT intron was detected in sensory ganglia by ISH. Panels A, D, and G show tissues from wild-type (WT) mouse trigeminal ganglia (TG), thoracic dorsal root ganglia (tDRG), and sacral dorsal root ganglia (sDRG), respectively, stained in parallel with sections from transgenic mice. ISH signal (blue staining) was seen in the nuclei of 70–86% of neurons in sections from HSV-2 LAT transgenic (TGN) mouse TG (B, C), tDRG (E, F), and sDRG (H, I). Images in each row are at the same magnification and the scale bar = 50 μm. Arrows point to neurons in which nuclei are clearly visible but with little or no detectable LAT accumulation.
Fig. 3. LAT is detectable in neurons of other nervous tissue.
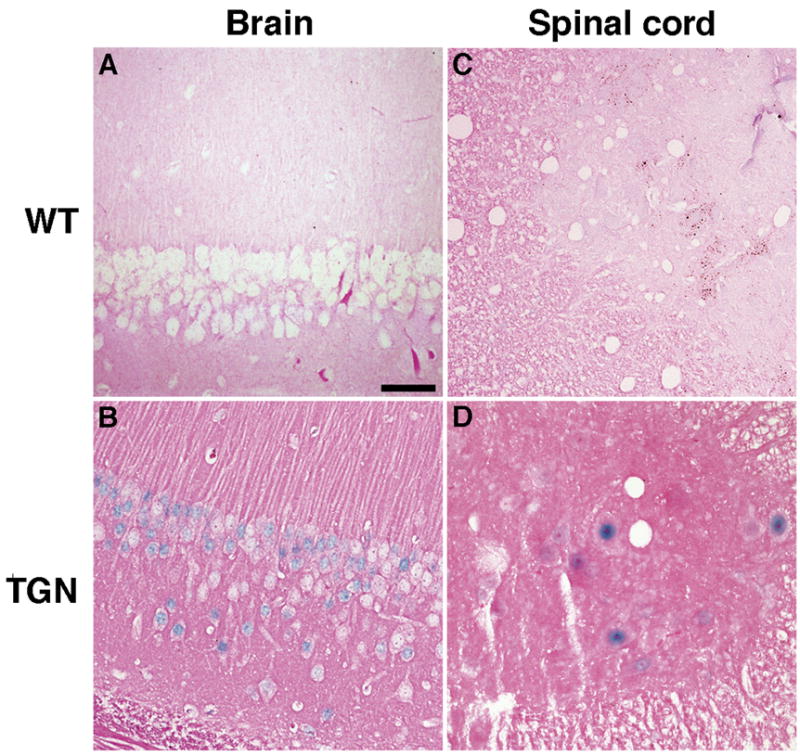
Sections from brain and spine were examined by ISH for HSV-2 LAT. Hippocampus from a wild-type mouse (A) or transgenic mouse (B), and spinal cord from a wild-type mouse (C) or transgenic mouse (D). Positive signals (blue staining) are seen in nuclei of neurons in sections from transgenic mouse. Scale bar = 50 μm.
To determine if accumulation of LAT in ganglia correlates with neuronal markers A5 or KH10, DRG sections were initially hybridized with a DIG-labeled LAT probe and visualized with Rhodamine-labeled anti-DIG-AP Fab fragments, followed by staining with a monoclonal antibody specific for neuronal markers A5 or KH10 and FITC-labeled secondary antibody (Fig. 4). Careful analysis of stained tissue sections revealed that 9.9% (105/1059) of neurons stained with monoclonal antibody A5 and 19.7% (161/816) of neurons stained with monoclonal antibody KH10. Amongst these sections we chose those areas with the best dual staining for both LAT and cell surface markers and found that 9.8% (21/215) of LAT-positive neurons also stained with monoclonal antibody A5 and 15% (35/233) of LAT-positive neurons stained with monoclonal antibody KH10.
Fig. 4. LAT accumulation in neurons does not correlate with neuronal markers A5 or KH10.
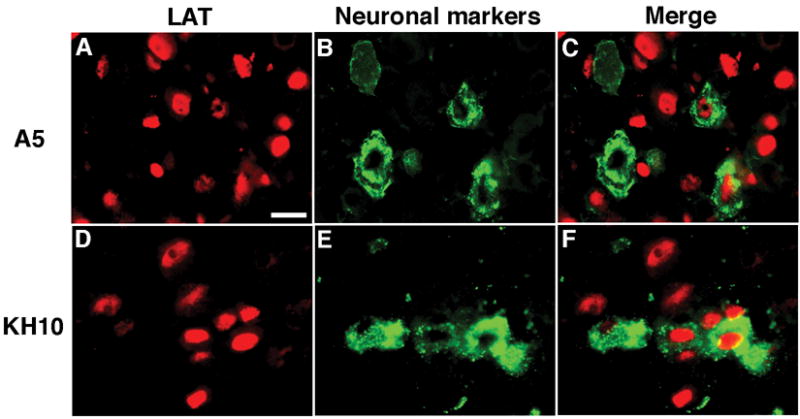
DRG sections were first hybridized with a DIG-labeled LAT probe and visualized with Rodamine-labeled anti-DIG-AP Fab fragments (A, D), then stained for neuronal markers A5 (B) or KH10 (E). Merged images are shown in C (LAT and A5) and in F (LAT and KH10). Scale bar = 20 μm.
The LAT intron is detectable in epithelial cells by ISH
In addition to the nervous system, LAT-positive cells were also detected by ISH in epithelial cells in many organs. Moderate ISH signals were seen in some epithelial cells lining the stomach of transgenic mice (Fig. 5B). Weak signals were found in some epithelial cells lining the small and large intestine (Fig. 5D, F). ISH signal was also detected in some epithelial cells lining the bladder, urethra of the penis, and vagina (Fig. 5H, J, L,). Very weak signal was noted in some epithelial cell in the uterus (Fig. 5N). The signal intensity increased gradually from the basement membrane to the mucosal surface, indicating that in these tissues mature cells accumulate more LAT than immature cells. Keratinized epithelial cells in the skin expressed considerably less LAT than other epithelial cells (Fig. 5P). LAT was also detected in the cells lining the ducts of the hair follicles (Fig. 5P). LAT was detected in about 30% of hepatocytes with signal intensities ranging from moderate to weak (Fig. 5R). Weak ISH signal was found in some cells in renal tubules (Fig. 5T). Moderate signals were seen in some epithelial cells lining the bronchioles of the lungs (Fig. 5V). In all of the epithelial cells, ISH signals were only detected in the nuclei of the cells.
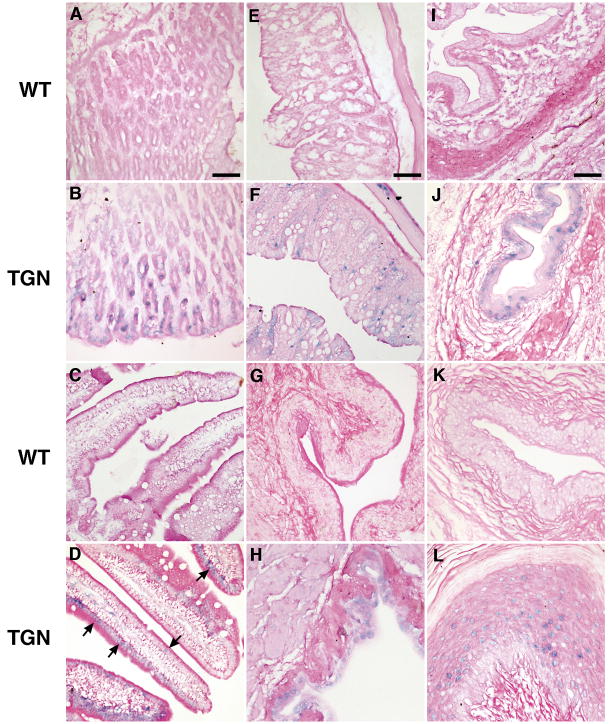
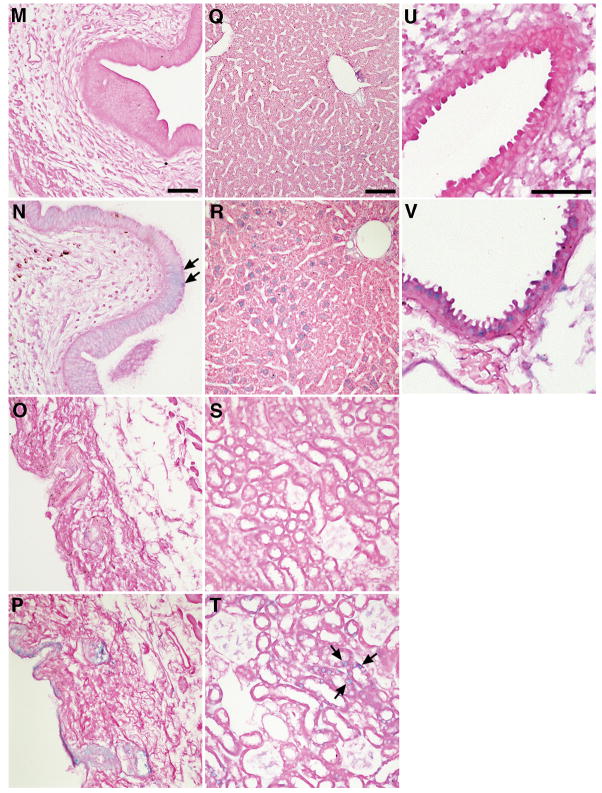
Fig. 5. LAT is detectable in epithelial cells by ISH.
Mouse organs were examined by ISH for LAT. LAT was detected (blue staining) in nuclei of epithelial cells of the stomach (B), small intestine (D), large intestine (F), bladder (H), urethra of the penis (J), vagina (L), and uterus (N) of transgenic (TGN) mice. LAT was also detectable in nuclei of epithelial cells of the skin (P), hepatocytes (R), renal tubules (T), and lungs (V) of transgenic mice, but was absent from corresponding tissues of wild-type (WT) mice (A, C, E, G, I, K, M, O, Q, S, U). Images in each column are at the same magnification and the scale bar = 50 μm.
The LAT intron is detected in other types of cells by ISH
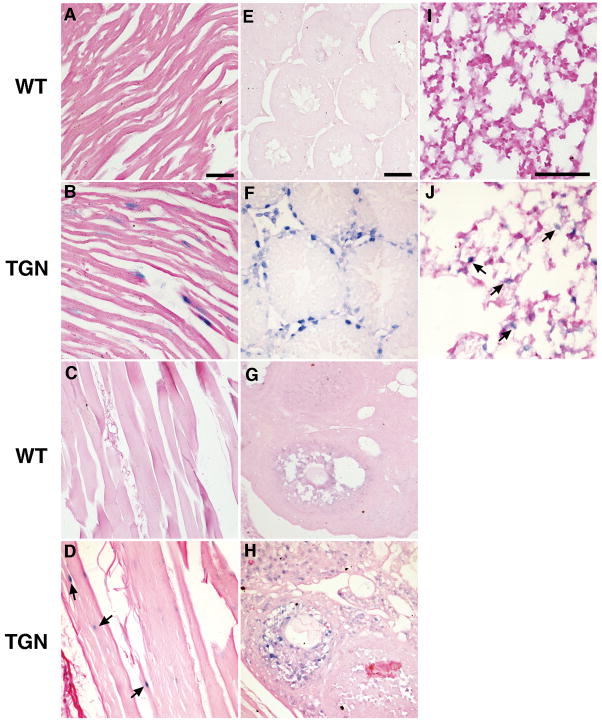
Moderate to weak signals for LAT were seen in cardiac and skeletal muscle cells (Fig. 6B, D). Moderate signals were also detected in basement membrane cells of the testes and follicular granulosa cells of the ovaries (Fig. 6F, H) and in the wall of lung alveoli (Fig. 6J). No ISH signal was detected in splenocytes and thymocytes (Fig. 7B, D).
Fig. 6. LAT is detected in other types of cells.
Sections from heart, skeleton muscle from the hind limb, testes, ovary, and lung were examined by ISH for HSV-2 LAT. Positive signals (blue staining) were visible in the nuclei of myocardial cells (B) and skeleton muscle (D) from transgenic (TGN) mice. LAT was also detected in basement membrane cells of the testes (F), in follicular granulosa cells of the ovaries (H), and in the wall of lung alveoli of transgenic mice, but was absent in the corresponding tissues (A, C, E, G, I) of wild-type (WT) mice. Images in each column are at the same magnification and the scale bar = 50 μm.
Fig. 7. LAT is not detected in splenocytes or in thymocytes.
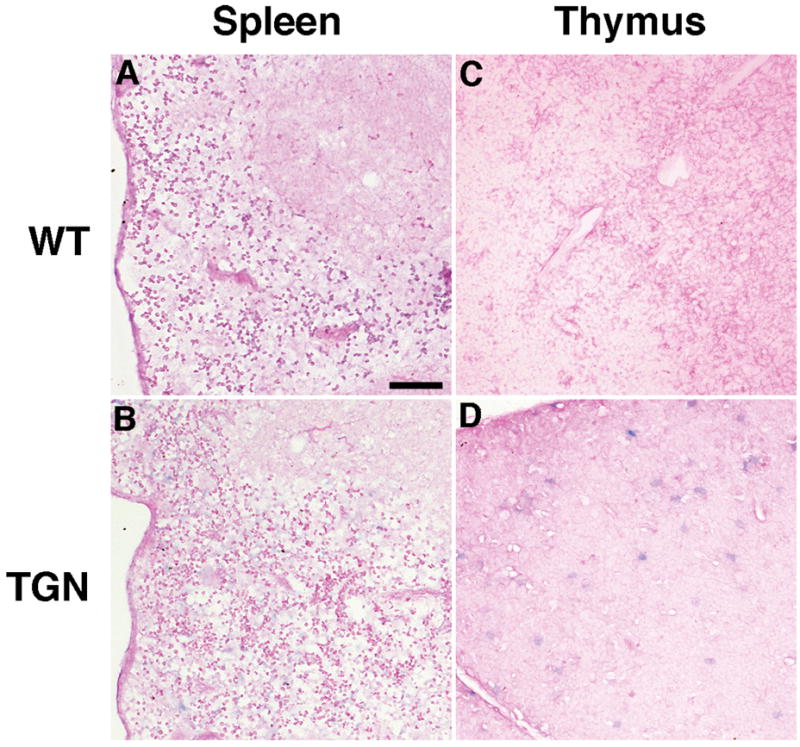
Sections from spleen and thymus were examined by ISH for HSV-2 LAT. No positive signals (blue staining) are visible in splenocytes (B) or thymocytes (D) of transgenic (TGN) and wild-type (WT) mice. Scale bar = 50 μm.
Discussion
To better understand how the LAT intron is expressed and accumulates in diverse tissues, we examined multiple tissues from HSV-2 LAT transgenic mice by ISH. In contrast to a prior report for HSV-1 LAT transgenic mice in which LAT was only detected in sensory ganglia by ISH (Gussow et al., 2006), HSV-2 LAT was detected by ISH at various signal intensities in nearly all tissues from HSV-2 LAT transgenic mice. We detected LAT in epithelial cells from multiple organs indicating that the LAT promoter can be recognized by transcription factor(s) common in epithelial cells. In addition, LAT expression in other cell types including muscle cells, follicular granulosa cells of the ovary, and basement membrane cells of the testes suggests that either the promoter of the HSV-2 LAT gene has basal activity in multiple cell types or that different cis-elements in the promoter can interact with different transcription factors in various cell types.
Despite the observation that the LAT intron was detected in multiple cell types outside the nervous system, LAT was not detected in non-neuronal cells in sensory ganglia of mice from both transgenic line 5221 and line 5238. The absence of LAT in non-neuronal cells in ganglia during latency is due to the lack of HSV genomes in these cells (Gressens and Martin, 1994; Mehta et al., 1995; Sawtell, 1997; Wang et al., 2005a). However, when the LAT transgene is present at 40 copies per cell in the transgenic mice, and LAT accumulates at high levels in the neurons, LAT is still not detected by in situ hybridization in the non-neuronal cells that also have the LAT transgene. Therefore, if the LAT promoter has strong basal activity (since LAT is detected in multiple cell types outside the nervous system), then non-neuronal cells in sensory ganglia may have factors that either inhibit expression from the LAT promoter or that degrade the LAT transcript. The lack of LAT expression and/or accumulation in satellite cells, which surround neurons, is particularly interesting. While satellite cells can be infected during acute HSV infection (Georgsson et al., 1987), latent HSV DNA has not been detected in these cells (Gressens and Martin, 1994; Mehta et al., 1995; Sawtell, 1997; Wang et al., 2005a). Based on findings in our study we speculate that lack of LAT expression and/or accumulation in satellite cells might be partly responsible for the failure of HSV to establish latency in these cells.
Our results with HSV-2 transgenic mice, in which LAT was detected by ISH in multiple tissue types, are different from what was reported for transgenic mice which contained the HSV-1 LAT promoter, first exon, and LAT intron where LAT was only detectable by ISH in neurons in trigeminal and dorsal root ganglia, and in small numbers of neurons in the thalamus and spinal cord (Gussow et al., 2006). While LAT RNA was detected in other tissues examined, including the heart, lung, kidney, thymus, liver, spleen, and skin in the HSV-1 transgenic mice using real time PCR; the spliced LAT intron was not detected by Northern blotting or ISH in non-neuronal tissues. Several reasons might explain the difference in the ISH results in these two studies. First, ISH is a complex procedure and the sensitivity of the assay might differ from one laboratory to another. Second, while HSV-1 and HSV-2 share a high level of nucleotide homology between their genomes, small differences in the promoter sequences such as the GC-rich cis-element in HSV-2 LAT promoter (Wang et al., 1995) which is not presented in the HSV-1 LAT promoter, may result in different promoter activities. In addition, the slightly longer upstream region in the HSV-2 LAT construct might also contribute to the different promoter activities. Lekstrom-Himes et al. (Lekstrom-Himes et al., 1998) showed that in transient transfection assays, the HSV-2 LAT promoter was 6- to 10-fold more active than the HSV-1 LAT promoter in driving reporter gene expression. Bertke and colleagues (Bertke et al., 2007) constructed chimeric HSV in which the HSV-2 LAT promoter or 2.5 kb of the HSV-2 LAT sequence was replaced with the corresponding region from HSV-1, and found that the LAT sequence, not the LAT promoter region, was required for type-specific reactivation of HSV-2 and virus neurotropism. Therefore, differences in the LAT promoter or LAT sequence between HSV-1 and HSV-2 might result in difference expression or accumulation in various tissues.
Other reasons might also explain the differences between our results with HSV-2 LAT transgenic mice and those of Gussow and colleagues (Gussow et al., 2006) with HSV-1 LAT transgenic mice. The insertion site(s) in the cellular genome and the copy number of transgenes per cell in the HSV-1 and HSV-2 transgenic mice might influence expression in various tissues. The transgenic mice we used (heterozygous mice of lines 5221 and 5238) have about 10 and 40 copies of the LAT transgene per cell (Wang et al., 2001), while the HSV-1 transgenic mice have about 1 copy of the transgene per cell (Gussow et al., 2006). Hoshino et al (Hoshino et al., 2008) showed that mice latently infected with HSV-2 have a median of 14 viral genomes per cell; since there are two copies of LAT per viral genome, there would be a median of 28 copies of LAT DNA per latently infected cell, which is similar to the number of LAT transgenes per cell in our mice. Similarly, mice latently infected with HSV-1 contain a median of 14–15 copies of HSV-1 genome per neuron (Sawtell et al., 1998); therefore, the low number of copies of HSV-1 LAT per neuron in the HSV-1 transgenic mice may have resulted in low LAT expression and failure to detect LAT in other tissues by ISH.
We found that LAT was expressed at different levels in individual neurons in sensory ganglia of the HSV-2 LAT transgenic mice. This is consistent with observations in HSV-1 latently infected mice and in human sensory ganglia that more neurons are positive for HSV DNA than for LAT (Gressens and Martin, 1994; Mehta et al., 1995; Sawtell, 1997; Wang et al., 2005a), and that expression of LAT does not correlate with the latent viral DNA load in the same neurons (Wang et al., 2005a). Our observation here further confirms that LAT accumulation in neurons is tightly regulated by factors in addition to the copy number of its genomic DNA. The population of neurons found within a single sensory ganglion is diverse in morphology, function, and gene expression. LAT may be expressed at different levels in certain type of neurons due to cellular features such as transcription factors, chromatin modifications, and possibly cellular miRNAs that target LAT. While a previous report noted that neuronal markers A5 and KH10 directly correlated with the accumulation of HSV-1 and HSV-2 LAT, respectively, in latently infected mouse ganglia (Margolis et al., 2007; Yang et al., 2000), we did not observe a similar correlation for KH10 in HSV-2 LAT transgenic mice. This finding supports the previous hypothesis that the correlation of LAT expression with A5 or KH10 expression in latently infected ganglia is due to which cell types are infected by HSV-2, rather than which cells are able to express HSV-2 LAT (Margolis et al., 2007).
Materials and Methods
HSV-2 LAT transgene and transgenic mice
HSV-2 LAT transgenic mice were previously described (Wang et al., 2001). The 5,511-bp LATpa transgene consists of a DraIII-PstI fragment from HSV-2 strain 333 genomic DNA (nucleotides 118,337 to 123,610 of HSV-2 genome based on the sequence of HSV-2 HG52) and begins 1,167 bp upstream of the LAT transcription initiation site, encompasses the LAT promoter and some of its upstream region, the first exon, the entire 2.2-kb LAT intron, and 1,170 bp downstream of the LAT intron splicing adapter site. The LAT transgene encompasses miR-III, a microRNA encoded by the HSV-2 LAT gene (Tang et al., in press). The polyadenylation signal of bovine growth hormone gene was added to the 3′ end of the viral fragment to complete the transcription unit, LATpa (Fig. 1). The LATpa cassette was introduced into C57BL/6 mice by microinjecting mouse pronuclei with the transgene DNA fragment. Two mouse colonies, lines 5221 and 5238, were established from two transgenic founders. Crossing homozygous mice of line 5221 or line 5238 with wild-type C57BL/6 mice yielded heterozygous mice which have 10 or 40 copies of the LAT transgene per cell, respectively. These heterozygous mice were used since it is less likely that a phenotype would be due to insertional inactivation of a mouse gene by the LAT transgene, because heterozygotes will have at least one copy of each wild-type mouse gene.
The amount of LAT accumulation in TG and liver was not gender or age dependent when RNA from 6-week to 2-year old mice were assayed by real-time PCR (unpublished data).
Preparation of tissues and slides
Six-week old mice (LAT transgenic or age and gender matched C57BL/6 mice) were perfused with10% formalin in PBS by cardiac puncture. Organs of interest were harvested and fixed in 10% formalin overnight. Tissues were paraffin embedded, sectioned to 5 μm in thickness, and mounted on positively charged; silanated glass slides with precautions to avoid RNase contamination. Each slide contained sections of the same organ from a wild-type mouse, and from transgenic lines 5221 and 5238. All organs were from female mice with exception of male reproductive organs.
Slides used for combined staining of fluorescent in situ hybridization (FISH) and immunofluorescence (IF) were prepared differently. Briefly, six-week old LAT transgenic mice were euthanized by carbon dioxide inhalation and thoracotomy. Cardiac perfusion with 0.1M phosphate-buffered saline (PBS, pH7.4) was performed immediately, followed by perfusion with 1% paraformaldehyde (PFA) in 0.1M phosphate-buffer (PB, pH7.4). Dissected dorsal root ganglia (DRGs) were immersion fixed in 1% PFA in 0.1M PB for 30 min at 4°C, followed by equilibration in 30% sucrose in 0.1M PBS at 4°C. Fixed tissue was then embedded in Tissue-Tek® O.C.T. compound (Sakura Finetechnical, Tokyo, Japan) and frozen in liquid nitrogen. Serial sections (7μm) were collected as four alternate sets onto Superfrost®/Plus microscope slides (Fisher Scientific, Pittsburgh, PA) and stored at −80°C. These studies were approved by the UCSF Committee on Animal Research.
Preparation of probes for in situ hybridization
Plasmid pLATin was constructed by inserting the entire HSV-2 LAT intron sequence into pcDNA3 (Invitrogen) between the Hind III and Xba I sites. This plasmid was used as template for in vitro transcription. Sense (identical to LAT intron) and anti-sense (reverse complement to LAT intron) cRNA were produced by T7 and SP6 polymerases (Roche), respectively, following the manufacturer’s instruction. The in vitro transcription products were subjected to partial alkaline hydrolysis to reduce the size of the RNA transcripts to approximately 200 nucleotides in length as previously described (Wang et al., 2005a). The RNAs were then biotinylated using a PhotoProbe Biotin labeling kit (Vector Labs) as described previously (Wang et al., 2005a).
The probe used for FISH was in vitro transcribed from plasmid pYz-2LAT, which contains a 451 bp fragment (nucleotides 120394–120844) of HSV-2 genomic DNA. To synthesize a DIG-labeled RNA probe, linearized plasmid was incubated with DIG RNA Labeling Mix (Roche) and T3 polymerase for 3 hours at 37°C. After treatment with DNase I, the probe was purified by passing through a G-50 Sephadex column (Roche) and dissolved into 50% formamide in nuclease-free water. The probe concentration was estimated using a DIG-labeled control RNA on a nylon membrane as described by the manufacturer (Roche).
In situ hybridization and evaluation of results
ISH was performed as previously described (Wang et al., 2005a) with slight modifications. Slides were de-waxed in two xylene baths for 5 min each, and rehydrated 1 min each in 100%, 95%, 70%, and 50% ethanol successively. Slides were then dipped in 0.2 N HCl for 20 min at room temperature (RT), digested with 25 μg/ml proteinase K in 10 mM Tris-Cl pH 7.4, 2 mM CaCl2 at 37°C for 15 min, incubated in 0.25% (v/v) acetic anhydride with 1.33% (v/v) triethanolamine at pH 8.0 for 10 min at RT, and rinsed with water between each step. Pre-hybridization was performed in solution containing 50% (v/v) formamide, 2xSSC, 50 mM Tris-Cl pH 7.4, 1 mM EDTA, 0.02% Ficoll, 0.02% polyvinylpyrrolidone, 500 μg/ml tRNA, 500 μg/ml BSA at 42°C for 2 hours. The biotinylated probe was denatured at 65°C for 15 min and added to the hybridization solution (20% (v/v) dextran sulfate, 50% (v/v) formamide, 300 mM NaCl, 50 mM Tris-Cl pH 7.4, 1 mM EDTA, 0.02% each of Ficoll and polyvinylpyrrolidone, 10 mM DTT, 500 μg/ml of tRNA and 500 μg/ml of BSA) to 100 ng/ml. After adding 30 μl of hybridization solution to cover the sections, coverslips were applied and its edges were sealed with rubber cement. Slides were then incubated at 55°C overnight. The next day, coverslips were removed, and slides were washed twice in 2xSSC at RT for 5 min, twice in solution with 2xSSC, 1 mM EDTA, and 0.1% (v/v) Triton X-100 at 55°C for 10 min each, and once in solution with 0.1xSSC, 1 mM EDTA, 0.1% (v/v) Triton X-100 at 55°C for 20 min. Slides were than treated with 4 μg/ml RNase A plus 1 U/ml RNase T1 in RNase buffer (10 mM Tris-Cl pH 7.4, 300 mM NaCl) at 37°C for 20 min and washed twice for 20 min each with 2xSSC at 55°C. To detect the hybridized probe, slides were incubated in blocking solution containing 1xcasein in TBST (100 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% (v/v) Tween 20) for 40 min. Horseradish peroxidase-streptavidin (Vector Laboratories) was added to the blocking solution to yield a final concentration of 4 mg/ml, and slides were incubated for another 30 min. After 3 washes in TBST, the slides were developed using a TMB Substrate Kit for Horseradish Peroxidase (Vector Laboratories) for 5 min, and then rinsed in water briefly. Slides were successively dehydrated in 70%, 95%, 100% ethanol, and again in 100% ethanol for 1 minute each, counterstained with Eosin Y-phloxine, rinsed in 100% ethanol, rinsed twice in xylene, air dried, and mounted with Permont and coverslips.
Each slide contained sections of the same organ from a wild-type mouse, and from transgenic lines 5221 and 5238. A TG section was included in each ISH experiment as a positive and signal intensity control. Signals in tissues from line 5238 were slightly stronger than those from the corresponding tissues of line 5221; therefore, line 5238 was used for all of the results reported here. Cells with blue staining were scored as LAT-positive, and semi-quantified as strong (dark blue), moderate (blue), or weak (light blue). Photomicrographs were taken at 200×, or 400× magnification. Combined FISH and IF staining was carried out as previously described (Margolis et al., 2007)
Acknowledgments
We thank Drs. Jerrold M. Ward, David Kleiner, and Stefania Pittaluga, for their help in determining cell types in which LAT signals were detected. This study was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bertke AS, Patel A, Krause PR. Herpes simplex virus latency-associated transcript sequence downstream of the promoter influences type-specific reactivation and viral neurotropism. J Virol. 2007;81 (12):6605–6613. doi: 10.1128/JVI.02701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RL, Hartog K, Croen KD, Ostrove JM. Detection and characterization of latent HSV RNA by in situ and northern blot hybridization in guinea pigs. Virology. 1991;181 (2):793–797. doi: 10.1016/0042-6822(91)90920-7. [DOI] [PubMed] [Google Scholar]
- Chen SH, Kramer MF, Schaffer PA, Coen DM. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71 (8):5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen KD, Ostrove JM, Dragovic L, Straus SE. Characterization of herpes simplex virus type 2 latency-associated transcription in human sacral ganglia and in cell culture. J Infect Dis. 1991;163 (1):23–28. doi: 10.1093/infdis/163.1.23. [DOI] [PubMed] [Google Scholar]
- Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71 (8):5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgsson G, Martin JR, Stoner GL, Webster HF. Virus spread and initial pathological changes in the nervous system in genital herpes simplex virus type 2 infection in mice. A correlative immunohistochemical, light and electron microscopic study. Acta Neuropathol (Berl) 1987;72 (4):377–388. doi: 10.1007/BF00687270. [DOI] [PubMed] [Google Scholar]
- Gressens P, Martin JR. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J Virol Methods. 1994;46 (1):61–83. doi: 10.1016/0166-0934(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Gussow AM, Giordani NV, Tran RK, Imai Y, Kwiatkowski DL, Rall GF, Margolis TP, Bloom DC. Tissue-specific splicing of the herpes simplex virus type 1 latency-associated transcript (LAT) intron in LAT transgenic mice. J Virol. 2006;80 (19):9414–9423. doi: 10.1128/JVI.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Qin J, Follmann D, Cohen JI, Straus SE. The number of herpes simplex virus-infected neurons and the number of viral genome copies per neuron correlate with the latent viral load in ganglia. Virology. 2008;372 (1):56–63. doi: 10.1016/j.virol.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PR, Ostrove JM, Straus SE. The nucleotide sequence, 5′ end, promoter domain, and kinetics of expression of the gene encoding the herpes simplex virus type 2 latency-associated transcript. J Virol. 1991;65 (10):5619–5623. doi: 10.1128/jvi.65.10.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PR, Stanberry LR, Bourne N, Connelly B, Kurawadwala JF, Patel A, Straus SE. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med. 1995;181 (1):297–306. doi: 10.1084/jem.181.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekstrom-Himes JA, Wang K, Pesnicak L, Krause PR, Straus SE. The comparative biology of latent herpes simplex virus type 1 and type 2 infections: latency-associated transcript promoter activity and expression in vitro and in infected mice. J Neurovirol. 1998;4 (1):27–37. doi: 10.3109/13550289809113479. [DOI] [PubMed] [Google Scholar]
- Margolis TP, Imai Y, Yang L, Vallas V, Krause PR. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J Virol. 2007;81 (4):1872–1878. doi: 10.1128/JVI.02110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Cunningham C, McIntyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72 (Pt 12):3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- Mehta A, Maggioncalda J, Bagasra O, Thikkavarapu S, Saikumari P, Valyi-Nagy T, Fraser NW, Block TM. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology. 1995;206 (1):633–640. doi: 10.1016/s0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287 (5457):1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- Sawtell NM. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J Virol. 1997;71 (7):5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell NM, Poon DK, Tansky CS, Thompson RL. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J Virol. 1998;72 (7):5343–5350. doi: 10.1128/jvi.72.7.5343-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105 (31):10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Patel A, Krause PR. Novel Less-Abundant Viral miRNAs Encoded by Herpes Simplex Virus 2 Latency-Associated Transcript and Their Roles in Regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009 doi: 10.1128/JVI.01723-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenser RB, Edris WA, Hay KA, de Galan BE. Expression of herpes simplex virus type 2 latency-associated transcript in neurons and nonneurons. J Virol. 1991;65 (5):2745–2750. doi: 10.1128/jvi.65.5.2745-2750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RL, Sawtell NM. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol. 2001;75 (14):6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454 (7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Krause PR, Straus SE. Analysis of the promoter and cis-acting elements regulating expression of herpes simplex virus type 2 latency-associated transcripts. J Virol. 1995;69 (5):2873–2880. doi: 10.1128/jvi.69.5.2873-2880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Lau TY, Morales M, Mont EK, Straus SE. Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal Ganglia at the single-cell level. J Virol. 2005a;79 (22):14079–14087. doi: 10.1128/JVI.79.22.14079-14087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Pesnicak L, Guancial E, Krause PR, Straus SE. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J Virol. 2001;75 (17):8166–8172. doi: 10.1128/JVI.75.17.8166-8172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Pesnicak L, Straus SE. Mutations in the 5′ end of the herpes simplex virus type 2 latency-associated transcript (LAT) promoter affect LAT expression in vivo but not the rate of spontaneous reactivation of genital herpes. J Virol. 1997;71 (10):7903–7910. doi: 10.1128/jvi.71.10.7903-7910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005b;102 (44):16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Voytek CC, Margolis TP. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J Virol. 2000;74 (1):209–217. doi: 10.1128/jvi.74.1.209-217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]