Abstract
As an alternative to conventional, target-oriented drug discovery, we report a strategy that identifies compounds on the basis of the state that they induce in a signaling network. Immortalized human cells are grown in microtiter plates and treated with compounds from a small-molecule library. The target network is then activated and lysates derived from each sample are arrayed onto glass-supported nitrocellulose pads. By probing these microarrays with antibodies that report on the abundance or phosphorylation state of selected proteins, a global picture of the target network is obtained. As proof of concept, we screened 84 kinase and phosphatase inhibitors for their ability to induce different states in the ErbB signaling network. We observed functional connections between proteins that match our understanding of ErbB signaling, indicating that state-based screens can be used to define the topology of signaling networks. Additionally, compounds sort according to the multidimensional phenotypes they induce, suggesting that state-based screens may inform efforts to identify the targets of biologically active small molecules.
The signal transduction networks that control human cellular physiology typically comprise tens to hundreds of proteins that interrelate in a complex, nonlinear fashion. Defects in these systems underlie many human pathologies, including cancer1, autoimmunity2 and developmental abnormalities3. One of the principal challenges of systems biology is to understand how information flows through these networks and how we can best intervene to halt or to redirect the flow of aberrant signaling. Small molecules that modulate the activity of signaling proteins are useful both as tools to dissect protein function and as potential therapeutics. Currently, most efforts to discover such compounds are target-based: active compounds are identified by their ability to modulate the function of a specific protein of interest. Altering the flow of information through a network, however, may require nonintuitive solutions; it may even require molecules that target more than one protein.
To address this need, we developed a method that identifies compounds by their ability to induce different states in a network. Here we define the ‘state’ of a network as the quantitative levels of its components and assume that an informative picture can be obtained by measuring a subset of these components in cell lysates. We refer to this strategy as ‘state-based discovery’. Having the ability to push networks into different states will provide us with tools to dissect how information is directed, and redirected, through these systems in real time.
To facilitate state-based discovery, we need technologies that can measure the abundance and activation of multiple proteins and are compatible with high-throughput methods. Automated fluorescence microscopy4 and multiplexed flow cytometry5 offer potential solutions: both techniques provide the ability to track several proteins and have been adapted to a microtiter-plate format. Both methods, however, achieve their multiplexing with different colored fluorophores. To track more than a dozen proteins, additional samples must be generated. By contrast, microarray technology enables a single sample to be replicated thousands of times on separate arrays and is therefore easily scaled6,7. It has been shown that microarrays of tissue lysates can be used to study many proteins in microdissected biopsies8. That study and subsequent efforts9,10 used low-throughput strategies to generate lysates and fabricated single arrays on 2.5 × 7.5 cm slides at low spatial densities (≤400 spots per cm2). We reasoned that if this technology could be further miniaturized and adapted to a high-throughput format, it could provide a general solution for state-based discovery that is extensible to extremely high levels of multiplexing.
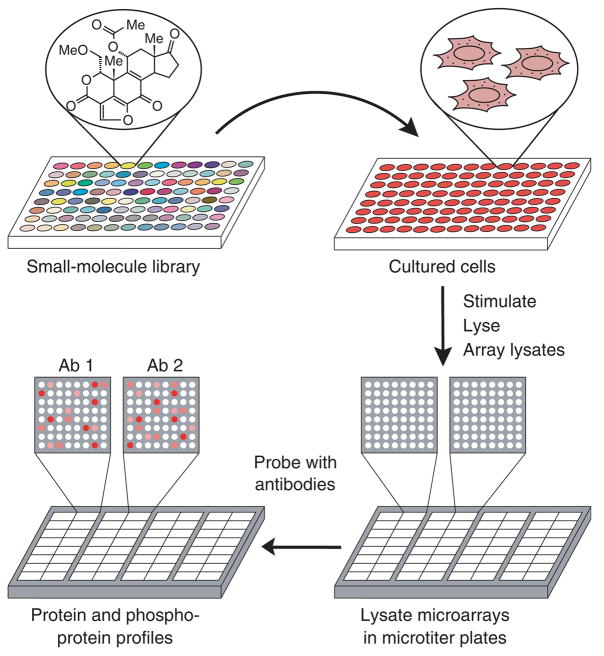
Our strategy is illustrated in Figure 1. Immortalized cells are grown in microtiter plates, compounds from a library are applied to each well, and the target network is stimulated with an appropriate ligand. The cells are then lysed in a small volume (~30 μl) and the resulting samples are arrayed at high spatial density (1,600 spots per cm2) onto glass-supported nitrocellulose pads arranged in a microtiter-plate format. Each lysate microarray is probed with a different antibody and the cognate antigens are quantified by a fluorescence-based method. Because each spot consumes ~250 pl of lysate, the number of proteins that can be studied is limited only by the availability of appropriate antibodies.
Figure 1.
State-based screening using lysate microarrays. Cells are grown in 96-well microtiter plates, treated with small molecules (one compound per well), stimulated with a growth factor, and lysed. The resulting lysates are arrayed onto glass-supported nitrocellulose pads and the arrays are assembled into a microtiter-plate format. Each array is probed with a different pan- or phosphorylation-specific antibody, yielding a quantitative snapshot of the state of the signaling network induced by each small molecule.
RESULTS
Antibody screening and validation
To develop our strategy for state-based discovery, we focused on ErbB signaling. When epidermal growth factor (EGF) and its relatives bind to ErbB receptors, they trigger diverse intracellular pathways that collectively elicit the appropriate physiological response11. Because these pathways are intimately interconnected, it is difficult to predict how genetic or pharmacological perturbations will affect the overall system and hence the cellular response. The ErbB network is thus an ideal target for state-based discovery.
To implement our strategy, we began by screening antibodies directed at sites of phosphorylation on proteins known to be activated by EGF. We also included pan-specific antibodies for the EGF receptor (EGFR, also known as ErbB1) and ErbB2 to highlight molecules that interfere with receptor trafficking and degradation12. To assess the performance of each antibody, we created a standardized set of cell lysates: pan-specific antibodies were evaluated with lysates derived from six cancer cell lines, and phosphorylation-specific antibodies were evaluated with lysates derived from A431 epidermoid carcinoma cells stimulated for different times with EGF.
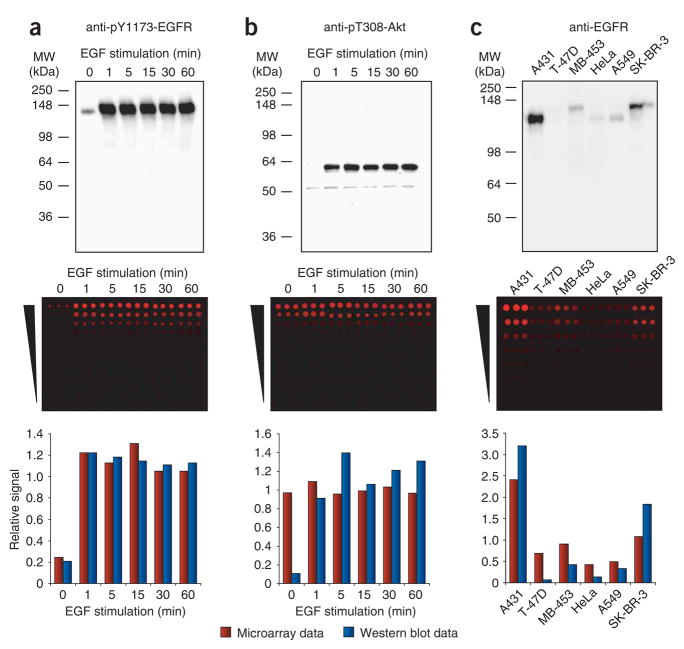
Whereas immunoblots are somewhat forgiving of antibody cross-reactivity because off-target proteins are separated from the protein of interest before detection, lysate microarrays are not. Because size separation is not practical in a high-throughput screen, only highly selective antibodies can be used. Of the 61 antibodies that we tested, 34 yielded a single dominant band on immunoblots of our control lysates. We then assessed these antibodies by using microarrays fabricated with the same lysates (Fig. 2 and Supplementary Fig. 1 online). A quantitative comparison of the immunoblots and the microarrays identified three classes of antibodies. The first class comprised four antibodies that behaved well: the relative signals that they produced on the microarrays matched those observed on the immunoblots (Fig. 2a). By contrast, the second class comprised 22 antibodies that showed no discernible change in signal on the microarrays, even though immunoblotting revealed substantial differences (Fig. 2b). It seems that, despite our initial screen, the off-target signal of these antibodies on the microarrays overwhelmed their cognate signal. Previous studies8–10 using lysate microarrays have either ignored antibody cross-reactivity or have used immunoblotting alone to select appropriate antibodies. Our results show that many antibodies do not produce meaningful data on lysate microarrays and it is therefore essential to evaluate antibodies under microarray conditions.
Figure 2.
Evaluation of antibodies for use in lysate microarrays. (a) Antibody to pY1173-EGFR shows no significant cross-reactivity on lysate microarrays. (b) Antibody to pT308-Akt antibody shows only nonspecific signals on lysate microarrays. (c) Antibody to EGFR shows some cross-reactivity on lysate microarrays, resulting in compressed signal ratios relative to the immunoblots. Cellular lysates were analyzed at one concentration on immunoblots (top) and at seven different concentrations on lysate microarrays (twofold dilution series; middle). Lysates in a and b were derived from A431 cells stimulated for different durations with 200 ng/ml of EGF; lysates in c were derived from six cancer cell lines. Quantitative data from the immunoblots and the lysate microarrays were normalized relative to their respective mean values and compared (bottom).
The third class comprised eight antibodies that produced microarray data with the same pattern as observed on immunoblots, but with compressed signal ratios (Fig. 2c). The cross-reactivity of these antibodies was sufficiently low to allow a reasonable assessment of how their antigens are affected by EGF stimulation. Overall, we identified 12 antibodies that produce reliable data in a microarray format (Supplementary Fig. 1 and Supplementary Table 1 online). These antibodies enabled us to assess the abundance of EGFR and ErbB2, and the following ten phosphorylation states: EGFR phosphorylated on Tyr845 (pY845-EGFR), pY1068-EGFR, pY1173-EGFR, pY1139-ErbB2, pS473-Akt, pT202/pY204-Erk, pS217/pS221-MEK, pS380-p90RSK, pY896–IRS-1 and pY1179–IRS-1.
State-based screen of a small-molecule library
With these reagents in hand, we conducted a pilot screen in a microtiter-plate format using a library of 84 kinase and phosphatase inhibitors (Supplementary Table 2 online). As controls, we included six wells that received no stimulation to record the basal phosphorylation level of each protein, and six wells that received EGF but no compound to obtain a measure of the unperturbed network.
We grew A431 cells to 50% confluence in 96-well plates and serum-starved them for 24 h. Then we preincubated them with the library compounds for 30 min and stimulated them for 5 min with EGF. All liquid handling was done robotically. The cells were lysed in situ, and the lysates were then arrayed in quadruplicate onto nitrocellulose-coated 16-pad slides and assembled into a microtiter-plate format using plastic wells and silicone gaskets. We used two arrays for each of 12 antibodies, yielding a quantitative, 12-dimensional readout of the network state for each of the 84 compounds. We also probed two arrays with an antibody to β-actin to determine the relative concentration of each lysate and normalized all signals accordingly.
To visualize the proteins, we used a multistep, catalyzed reporter deposition protocol based on the tyramide signal amplification system13. This procedure offers excellent sensitivity (all antigens were detected in lysates diluted to 0.01 mg/ml), but produces a sigmoidal, rather than linear, relationship between analyte concentration and fluorescence9. To correct for this nonlinearity, we generated calibration curves on each microarray using twofold serial dilutions of a control lysate (Fig. 3). To define the reliability of this technology, we did the screen in triplicate. A detailed description of the data processing steps and a treatment of how these steps affect uncertainty in the data are available in the Supplementary Note online. Overall, we found that the assay was reproducible: the average coefficient of variation (c.v.) of replicate spots in an array was 3.5%; the average c.v. for the same sample in replicate arrays was 5.0%; and the average c.v. across replicate experiments was 12.4%.
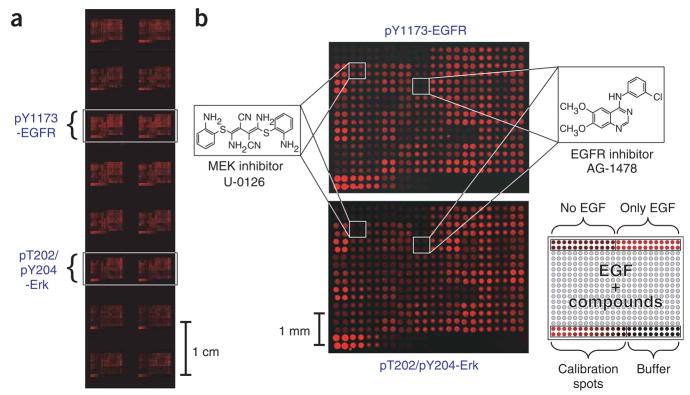
Figure 3.
Lysate microarrays resulting from a state-based screen of 84 kinase and phosphatase inhibitors. (a) A 2.5 × 7.5 cm glass slide with 16 identical lysate microarrays probed with eight different antibodies. Arrays probed with antibodies to pY1173-EGFR and to pT202/pY204-Erk are highlighted. (b) Enlarged views of microarrays shown in a. Top, array probed with antibody to pY1173-EGFR; bottom, array probed with antibody to pT202/pY204-Erk. Locations of lysates derived from cells treated with U-0126 and AG-1478 are highlighted. The EGFR inhibitor AG-1478 causes a decrease in the EGF-induced phosphorylation of EGFR and Erk, whereas the MEK inhibitor U-0126 causes a decrease in phosphorylation of Erk but does not affect that of EGFR. The layout of each microarray is shown in bottom right.
Visual inspection of the microarrays (Fig. 3) revealed specific patterns of small-molecule inhibition and/or activation for all ten phosphorylated antigens. For example, the EGFR inhibitor AG-1478 (ref. 14) caused a strong decrease in the phosphorylation of all the signaling proteins, whereas the MEK inhibitor U-0126 (ref. 15) caused a strong reduction in the phosphorylation of downstream proteins (Erk and p90RSK), but left upstream proteins such as EGFR and ErbB2, and proteins in other pathways (Akt and IRS-1) unchanged. To substantiate the data collected in our high-throughput screen, we selected five active compounds and repeated the multidimensional assay by immunoblot analysis. In all five cases, the results obtained by immunoblotting matched the high-throughput data (Supplementary Fig. 2 online).
Network connectivity and network states
To identify discrete ‘states’ induced by the small molecules and to investigate network connectivity, we calculated relative inhibition values for each small-molecule–antigen pair and subjected the resulting data to unsupervised hierarchical clustering in both dimensions (compounds and antibodies; Fig. 4a). Because none of the compounds had a statistically significant effect on the abundance of EGFR or ErbB2, data from the pan-specific antibodies were not included. We averaged first the results from quadruplicate spots, then those from duplicate arrays, and finally those from the three independent experiments. Because the greatest variation came from replicate experiments, we used this variation as a measure of data quality. On closer inspection, we noticed that the average c.v. was different for each antibody and correlated with the extent of antibody cross-reactivity. Moreover, for any given antibody, the extent of variation differed among the compounds. To ensure that clustering was driven by relevant differences, we designed an algorithm that weights the data according to their reliability. The algorithm uses weighted euclidean distance as the similarity metric, and the reciprocal of the standard deviation of three experiments as the weighting factor (840 weighting factors, one for each compound-antigen pair). Omitting these factors resulted in a less interpretable outcome.
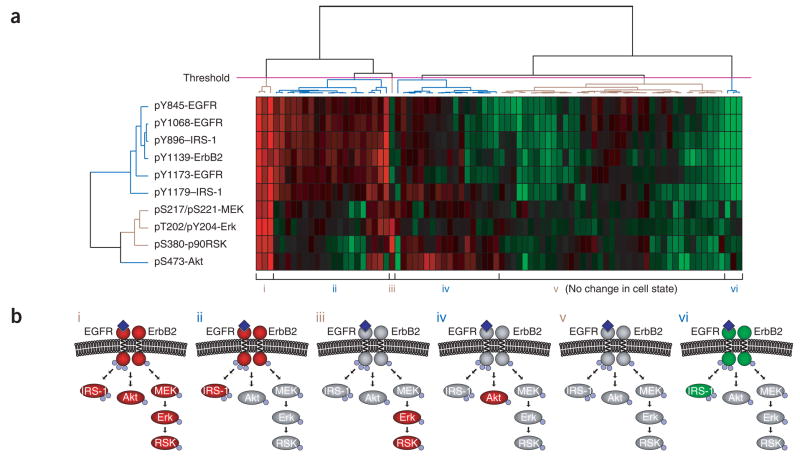
Figure 4.
Clustering of data from a state-based screen. (a) Heat map obtained by hierarchical clustering of relative inhibition values for each small-molecule–antigen pair. Positive inhibition values, indicating a decrease in phosphorylation, range from 0 (black; no inhibition) to 1 (red; 100% inhibition). Negative inhibition values, indicating an increase in phosphorylation, range from 0 (black; no inhibition) to −1 (green; twofold increase in phosphorylation). (b) Simplified view of the six network states defined by the threshold (magenta line) shown in a. Red ovals represent proteins with decreased phosphorylation; green ovals represent proteins with increased phosphorylation; gray ovals represent proteins whose phosphorylation was unaffected. Small blue spheres represent sites of phosphorylation. Dark blue diamonds represent EGF bound to the extracellular domain of EGFR.
The dendrogram produced by clustering the signaling proteins (antibodies) provides insight into the structure of the network. Our analysis segregated sites of phosphorylation into two main groups (Fig. 4a). One group comprises all phosphorylation sites on the ErbB receptors and both sites on the insulin receptor substrate-1 (IRS-1). Although it is not surprising that phosphorylated EGFR and phosphorylated ErbB2 are closely linked (the two receptors physically associate and phosphorylate each other), we found that pY896 of IRS-1 was as similar to the EGFR phosphorylation sites as any of those sites were to each other. Provided that we are measuring phosphorylation under linear, nonsaturating conditions, proteins that are substrates of the same kinase should be indistinguishable in our screen. The fact that inhibition profiles for pY1139 of ErbB2, pY896 of IRS-1 and the three sites on EGFR behave identically suggests that these sites are phosphorylated by the same kinase. This finding is consistent with both a study implicating pY896 of IRS-1 as a direct substrate of EGFR16 and our finding that the phosphotyrosine-binding domain of IRS-1 recognizes sites of phosphorylation on EGFR and ErbB2 (ref. 17).
The second cluster of signaling proteins comprises phosphorylation sites on downstream proteins, none of which is phosphorylated by the ErbB receptors. In this group, the mitogen-activated protein kinase cascade proteins MEK, Erk and p90RSK cluster closely, whereas Akt is further away. This observation shows that information about pathway connectivity can be obtained from state-based discovery efforts, even without prior knowledge of the signaling network or compound selectivity. We anticipate that data arising from broader screens will inform efforts to define functional connections in less well-understood networks.
From the perspective of the compounds, our clustering analysis identifies discrete network states (Fig. 4a,b). The number of states is determined by an arbitrary threshold placed across the dendrogram. For example, the threshold in Figure 4a highlights six biologically interpretable states: the unperturbed network (state v) and five others. More stringent thresholds lead to further subdivision of these states, but many of these subdivisions arise as much from noise as from meaningful biological differences.
To simplify interpretation, we reduced the quantitative data to a ternary representation in which each protein had one of three fates: increased phosphorylation, decreased phosphorylation, or unchanged phosphorylation relative to the unperturbed network (Fig. 4b). The five perturbed states are understandable from our knowledge of the small molecules that induce them. For example, staurosporine, a broad-specificity kinase inhibitor18, induces a state in which phosphorylation of all the proteins is decreased (state i). This state is also induced by tyrphostin AG 1478, a selective inhibitor of the EGFR kinase14. The phosphatidylinositol 3-kinase (PI3K) inhibitors LY294002 (ref. 19) and wortmannin20, by contrast, induce a state in which only Akt phosphorylation is reduced (state iv), whereas the MEK inhibitor U-0126 induces a state in which only Erk and p90RSK phosphorylation are reduced (state iii). One state (state vi) was characterized by a robust increase in phosphorylation of EGFR, ErbB2 and IRS-1. The molecules that induce this state include the broadly selective phosphatase inhibitor RK-682 (ref. 21). Other molecules also fall into these clusters, and it seems reasonable to propose that they achieve their effects through similar mechanisms. We anticipate that an expanded version of our assay that covers a diverse range of signaling molecules will, like other multidimensional approaches4,22,23, inform investigations into the mechanism of action of bioactive compounds whose targets are unknown.
From a systems perspective, state ii is particularly interesting in that the ErbB receptors and IRS-1 show decreased phosphorylation, whereas Akt and the MAPK cascade proteins do not. This observation shows that in A431 cells, which overexpress EGFR, partial inhibition of the receptor has little effect on signaling through these canonical pathways. Consistent with this observation, quantitative immunoblotting revealed that phosphorylation of proteins in these pathways is saturated even by relatively small amounts of activated receptor (data not shown). Thus, inhibiting EGFR might not be an efficient way to block signaling in these cells. By contrast, state ii shows that phosphorylation of IRS-1 is affected by partial inhibition of the receptors, consistent with the observation that IRS-1 phosphorylation is saturated only by very large amounts of activated EGFR (data not shown).
Dose-response behavior of the network
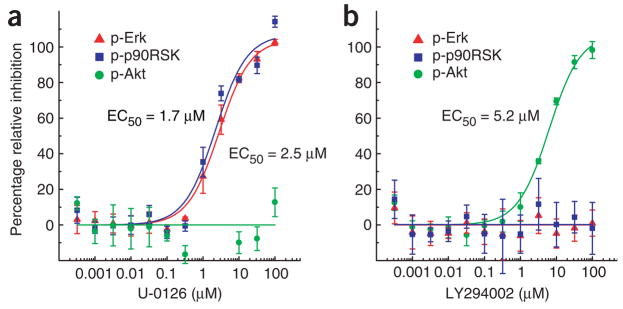
In addition to providing primary screening data, our method can be used to determine the dose-response behavior of active compounds on multiple proteins simultaneously. To test this application, we treated A431 cells, in a microtiter-plate format, with different concentrations of U-0126 or LY294002 and stimulated the cells for 5 min with EGF. We then determined the phosphorylation of Erk, p90RSK, and Akt at each concentration of inhibitor by lysate microarrays (Fig. 5). Consistent with the results of our primary screen, the compounds showed orthogonal response profiles: U-0126 inhibited Erk and p90RSK phosphorylation, and LY294002 inhibited phosphorylation of Akt.
Figure 5.
Dose-response curves determined with lysate microarrays. (a) MEK inhibitor U-0126; (b) PI3K inhibitor LY294002. For each antibody, normalized intensity values and fit curves were scaled relative to the maximum and minimum asymptotic values of the fit. Error bars represent the s.d. from three replicate wells.
In addition, the effector concentration for half-maximal response (EC50) values obtained for U-0126 on Erk and p90RSK phosphorylation (2.5 μM and 1.7 μM, respectively) were almost indistinguishable, as were the Hill coefficients, indicating that sensitivity to MEK inhibition does not change from Erk to p90RSK. We anticipate that determining the dose-response behavior for different small-molecule inhibitors on a network-wide basis will be a useful means to dissect pathway connectivity and to study signal amplification or attenuation.
DISCUSSION
The current emphasis on system-wide investigations of protein function is beginning to yield a more integrated understanding of signaling networks and how they regulate cell fate24,25. Although it is in its infancy, systems biology promises to reorient our thinking to focus on cellular states, rather than on individual proteins. Here we have provided a link between systems biology and small-molecule discovery that similarly redirects our focus away from one-dimensional phenotypes. Expansion of our efforts to monitor proteins in other signaling networks should provide us with a general tool to identify compounds that affect a wide range of biological processes.
By far the largest obstacle to overcome is the identification of appropriate antibodies. So far, our efforts have relied on screening commercially available antibodies, but we submit that a more effective strategy would be to screen antibody clones at an earlier stage in their production for their ability to function in this assay. Presently, most monoclonal antibodies are obtained by screening the supernatants of hybridoma clones using an enzyme-linked immunosorbant assay. By replacing this one-dimensional screen with one that uses microarrays of control lysates, it is likely that more antibodies will be identified that perform well in this assay.
Ultimately, we anticipate that state-based discovery not only will identify interesting compounds that would otherwise be over-looked by target-oriented screens, but will uncover functional connections between proteins in signaling networks and help to inform efforts to explain the mechanism of action of compounds whose targets are unknown.
METHODS
Small-molecule library experiments
We split A431 cells into 96-well tissue-culture plates (BD Biosciences), grew them to ~50% confluence and serum-starved them for 24 h. We prepared two 10-μMdilutions of the kinase and phosphatase inhibitor library (BIOMOL) in DMEM, one with and one without 400 ng/ml of EGF. We aspirated medium from the cells and replaced it with the diluted library compounds in DMEM, and incubated the cells at 37 °C for 30 min. An equal amount of library in DMEM supplemented with 400 ng/ml of EGF was added by a Biomek FX 96-channel pipetting robot (Beckman Coulter). As controls, wells A1–A6 contained neither EGF nor small molecules, and wells A7–A12 contained EGF but no small molecules. After a 5-min incubation at 37 °C, we washed the cells with ice-cold phosphate-buffered saline (PBS) using an Elx405 microtiter-plate washer (Bio-Tek). After aspiration of the buffer, we added 30 μl of cold 2% SDS lysis buffer11 to each well and lysed the cells for 30 min on ice. We clarified the lysates by centrifugation through a 96-well 0.2-μm filter plate (Pall Corporation) and stored them at −80 °C.
Microarray experiments
Nitrocellulose-coated 16-pad slides, 16-well ProPlate slide modules and silicone gaskets were a gift of Grace Bio-Labs. For antibody validation experiments, we prepared twofold serial dilutions of lysates in 2% SDS buffer and printed each sample in triplicate on nitrocellulose slides with a Biochip Arrayer (Perkin Elmer). For small-molecule library experiments, we spotted the lysates in quadruplicate in the same layout used for the small-molecule library plate. To correct for the nonlinearity of tyramide signal amplification, twofold serial dilutions of a control lysate (A431 cells stimulated for 5 min with 200 ng/ml of EGF) were printed at the bottom of each microarray and used as an in-well calibration standard. Lysates were arrayed with a pitch of 250 μm and an average spotted volume of 250 pl. For antigen detection, ProPlate wells with silicone gaskets were fixed to the slides, which were then incubated with PBS plus 0.2% Tween-20 (vol/vol). After this and all subsequent probing steps, we washed slides three times with PBST (PBS plus 0.1% Tween-20 (vol/vol)). We incubated slides with primary antibodies dissolved in PBST at a working dilution of 1:500, then with biotinylated secondary antibodies at 1:1,000 dilution in PBST and finally with streptavidin-HRP at 1:200 dilution in PBST. Tyramide signal amplification was carried out as recommended by the manufacturer using a 1:50 dilution of biotinyl-tyramide. We then incubated slides with a 1:200 dilution of streptavidin-Alexafluor647 in PBST and washed them thoroughly after removal of the ProPlate wells. We dried the slides by centrifugation and imaged themat 5-μm resolution with a GenePix 4000B microarray scanner (Molecular Devices).
Data analysis
For library experiments, we determined calibration curves for each array by fitting signal intensities from the control dilution series to equation (1):
| (1) |
This represents a Boltzmann sigmoidal curve, where x is the relative concentration of lysates in the serial dilution (1, 0.5, 0.25, …). By using the parameters obtained, all signal intensities in the array were corrected for nonlinearity by applying the inverse function (2).
| (2) |
After normalizing each array to its mean signal intensity, we averaged all replicate spots from duplicate arrays. We normalized signal intensities to the respective β-actin signals. We calculated relative inhibition for each compound and antigen according to equation (3).
| (3) |
Because each signal intensity is the sum of specific and cross-reactive signals, this strategy eliminates signals of nonspecific origin.
During our investigations, we noticed a positional dependence of signal intensities in each 96-well plate: edge wells had, on average, lower signal values. These wells fell into three distinct groups: wells A7–A12 (wells A1–A6 did not receive EGF and were considered unaffected), wells H1–H12, and columns 1 and 12 (excluding the corner wells). To correct for this effect, we adjusted signal values for each of these groups of wells to the average signal intensity of the interior wells.
For antibody validation experiments, we assigned the lysate showing the highest signal intensity in each array a value of 1. Then we determined a calibration curve from a dilution series of this lysate, and the signal intensities for the other lysates were adjusted as described above. We normalized signals to the respective β-actin signals and compared them to immunoblot data. To facilitate this comparison, we scaled both array and immunoblot data relative to their mean values.
Clustering analysis
We collected data from three independent library experiments and calculated average relative inhibition values and standard deviations for each compound-antigen pair. We calculated weighting factors for each compound-antigen pair as the reciprocal of the standard deviation across replicate experiments. To cluster the molecules, we calculated the weighted euclidean distance between any two molecules, k and l, by equation (4):
| (4) |
where d is the weighted euclidean distance, n is the number of measurements for each molecule (number of antigens), wxy is the weighting factor for compound x and antigen y, and Ixy is the average relative inhibition for compound x and antigen y. Hierarchical clustering was done by the maximum linkage method. We used the same approach to cluster the antigens. In both cases, we determined the final ordering of columns and rows by a ‘nearest-neighbor’ approach: starting with the least-distant pair of rows and columns and working outward, we placed clusters next to each other in a way that juxtaposed the most similar rows and columns.
Additional methods
A detailed description of reagents, cell culture, immunoblotting and dose-response experiments is available in the Supplementary Methods online.
Supplementary Material
Acknowledgments
We thank the Bauer Center for Genomics Research at Harvard University for support with instrumentation and automation. This work was supported by awards from the Arnold and Mabel Beckman Foundation and the W.M. Keck Foundation. M.S. is the recipient of an Alfred and Isabel Bader fellowship and a Jacques-Émile Dubois fellowship.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Published online at http://www.nature.com/naturemethods/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi PS. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat Rev Immunol. 2002;2:427–438. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 3.Casalini P, Iorio MV, Galmozzi E, Menard S. Role of HER receptors family in development and differentiation. J Cell Physiol. 2004;200:343–350. doi: 10.1002/jcp.20007. [DOI] [PubMed] [Google Scholar]
- 4.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 5.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20:155–162. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 6.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 7.Shalon D, Smith SJ, Brown POA. DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 8.Paweletz CP, et al. Reverse phase protein microarrays which capture disease progression show activation of prosurvival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 9.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Protein microarrays for multiplex analysis of signal transduction pathways. Nat Med. 2004;10:1390–1396. doi: 10.1038/nm1139. [DOI] [PubMed] [Google Scholar]
- 10.Nishizuka S, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 12.Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284:78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 13.Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J Immunol Methods. 1989;125:279–285. doi: 10.1016/0022-1759(89)90104-x. [DOI] [PubMed] [Google Scholar]
- 14.Levitzki A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 15.Favata MF, et al. Identification of a novel inhibitor of mitogen-activated protein kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 16.Gaudet S, et al. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 18.Tamaoki T, et al. Staurosporine, a potent inhibitor of phospholipid/Ca2+-dependent protein kinase. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 19.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H–1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 20.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamaguchi T, Sudo T, Osada H. RK-682, a potent inhibitor of tyrosine phosphatase, arrested the mammalian cell cycle progression at G1 phase. FEBS Lett. 1995;372:54–58. doi: 10.1016/0014-5793(95)00953-7. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel EJ, et al. An integrative biology approach for analysis of drug action in models of human vascular inflammation. FASEB J. 2004;18:1279–1281. doi: 10.1096/fj.04-1538fje. [DOI] [PubMed] [Google Scholar]
- 23.Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 24.Janes KA, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 25.Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.