Abstract
Human APOBEC3G and other APOBEC3 cytidine deaminases inhibit a variety of retroviruses, including Vif-deficient HIV-1. These host proteins are packaged into viral particles and inhibit the replication of virus in new target cells. A3G and A3F are known to be efficiently packaged into HIV-1 virions by binding to 7SL RNA through the Gag NC domain; however, the packaging mechanisms of other APOBEC3 proteins are poorly defined. We have now demonstrated that APOBEC3C (A3C) can be efficiently packaged into HIV-1 virions that are deficient for viral genomic RNA. Inhibition of the encapsidation of 7SL RNA into HIV-1 virions blocked the packaging of A3G, but not A3C. While the NC domain is required for efficient packaging of A3G, deletion of this domain had little effect on A3C packaging into HIV-1 Gag particles. A3C interacted with HIV-1 Gag which was MA domain-dependent and RNA-dependent. Deletion of the MA domain of HIV-1 Gag inhibited A3C but not A3G packaging into HIV-1 Gag particles. Thus, A3G and A3C have evolved to use distinct mechanisms for targeting retroviruses.
Keywords: APOBEC3G, Vif, HIV-1, Cytidine deaminase, APOBEC3C
Introduction
Human cytidine deaminase apolipoprotein B mRNA-editing catalytic polypeptide-like 3G (APOBEC3G, A3G) and other APOBEC3 proteins (Jarmuz et al., 2002) are related to a family of proteins that also includes apolipoprotein B-editing catalytic subunit 1 (APOBEC1), APOBEC2, and activation-induced cytidine deaminase (AID). These proteins have cytidine deaminase activities that modify RNA or DNA. A3G was the first AID/APOBEC proteins to be identified as a potent inhibitor of HIV-1 in the absence of Vif (Sheehy et al., 2002). Subsequently, various APOBEC3 proteins were shown to have broad antiviral activity against a wide range of retroviruses (Bieniasz, 2004; Chiu and Greene, 2006; Cullen, 2006; Goff, 2004; Harris and Liddament, 2004; Malim, 2006; Navarro and Landau, 2004; Rose et al., 2004; Turelli and Trono, 2005; Yu, 2006), endogenous retroviruses (Esnault et al., 2005), LINE-1 and Alu retrotransposons (Bogerd et al., 2006; Chen et al., 2006; Chiu et al., 2006; Hulme et al., 2007; Muckenfuss et al., 2006; Niewiadomska et al., 2007; Stenglein and Harris, 2006; Turelli et al., 2004b), hepatitis B virus (Bonvin et al., 2006; Rosler et al., 2005; Turelli et al., 2004a,b; Xu et al., 2007; Zhang et al., 2008), adeno-associated virus (Chen et al., 2006), and DNA virus promoter activities (Zhang et al., 2008).
APOBEC3 proteins can be packaged into diverse retroviruses that mediate potent antiviral functions in newly infected target cells. Virion-packaged A3G molecules induce cytidine deamination of viral DNA during reverse transcription in newly infected target cells (Harris et al., 2003; Lecossier et al., 2003; Mangeat et al., 2003; Mariani et al., 2003; Suspene et al., 2004; Xu et al., 2007; Yu et al., 2004b; Zhang et al., 2003). A3G and another APOBEC3 protein, A3F, also reduce the accumulation of viral reverse transcription products (Bishop et al., 2006; Guo et al., 2006; Iwatani et al., 2007; Kaiser and Emerman, 2006; Li et al., 2007; Luo et al., 2007; Mbisa et al., 2007; Yang et al., 2007) and inhibit proviral DNA formation (Luo et al., 2007; Mbisa et al., 2007).
To suppress the restriction imposed by APOBEC3 factors, HIV-1 encodes a regulatory protein, Vif, which triggers the degradation of APOBEC3 proteins through polyubiquitination and proteasomal degradation (Conticello et al., 2003; Liu et al., 2005, 2004; Marin et al., 2003; Mehle et al., 2004; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003). Vif recruits Cul5 and ElonginB/C E3 ubiquitin ligase (Kobayashi et al., 2005; Liu et al., 2005; Luo et al., 2005; Mehle et al., 2004; Yu et al., 2003, 2004c) through an HCCH motif (Luo et al., 2005) and virus-specific BC-box (Mehle et al., 2004; Yu et al., 2004c), respectively. The amino-terminal region of HIV-1 Vif interacts with the A3G or A3F through distinct interfaces (Marin et al., 2003; Russell and Pathak, 2007; Schrofelbauer et al., 2006; Simon et al., 2005; Tian et al., 2006), and three functional domains in the carboxy-terminal region of this Vif protein interact with Cul5 and ElonginB/C. The HCCH motif (Luo et al., 2005), which binds zinc (Mehle et al., 2006; Xiao et al., 2007a, 2006) and contains a consensus sequence Hx2YFx-CFx4Φx2AΦx7-8Cx5H (Xiao et al., 2007b), is required for Cul5 binding. The SLQXLA motif, a virus-specific BC-box (Mehle et al., 2004; Yu et al., 2004c) that mediates the interaction with ElonginsB/C, is conserved between HIV-1 and other lentiviral Vif molecules. Another Cul5-binding motif, LPx4L, is located downstream of this BC-box. In addition to mediating APOBEC3 degradation, Vif is also thought to reduce the translation of A3G and to suppress APOBEC3 through a ubiquitin/proteasome-independent mechanism (Opi et al., 2007; Stopak et al., 2003).
Encapsidation of APOBEC3G (A3G) into HIV-1 particles is mediated by retroviral Gag molecules (Alce and Popik, 2004; Cen et al., 2004; Douaisi et al., 2004; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Zennou et al., 2004). In particular, the nucleocapsid domain (NC) of HIV-1 Gag is known to be required for efficient A3G packaging. The interaction that occurs between HIV-1 Gag and A3G in virus-producing cells is believed to be the driving force behind A3G packaging; this also requires cellular RNA(s) (Douaisi et al., 2004; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Svarovskaia et al., 2004; Zennou et al., 2004). Since HIV-1 Gag particles lack viral genomic RNA, it appears that viral genomic RNA is not required for A3G packaging. However, this issue is still controversial (Khan et al., 2005; Svarovskaia et al., 2004).
Our recent data indicate that 7SL RNA is selectively packaged into HIV-1 through an interaction with the NC domain of HIV-1 Gag (Tian et al., 2007) and the virion packaging of A3G or A3F involves a selective interaction between 7SL RNA and A3G or A3F (Wang et al., 2007; Wang et al., 2008). However, the packaging mechanisms of other APOBEC3 proteins have not been well characterized to date. APOBEC3C (A3C), the single domain APOBEC3 protein, has been detected in HIV-1 virions (Langlois et al., 2005; Yu et al., 2004a). It has weak anti-HIV-1 activity (Dang et al., 2006; Langlois et al., 2005; Yu et al., 2004a), induces less cytidine deamination in HIV-1 DNA than A3G (Yu et al., 2004a), and has reduced ability to inhibit HIV-1 reverse transcription and integration compared to A3G (Luo et al., 2007).
In the present study, we have explored the viral determinants of A3C packaging. Our data indicate that the packaging of A3C differs from that of A3G in that the NC domain of HIV-1 Gag is apparently not important for A3C packaging. Reducing 7SL RNA encapsidation into HIV-1 virions also did not affect A3C packaging. However, we found that the MA domain of HIV-1 Gag is important for the interaction between Gag and A3C and for the efficient packaging of A3C; in the case of A3G, it is not essential for either of these processes. Thus, these two, and perhaps other APOBEC3 cytidine deaminases have evolved to use distinct mechanisms to gain access into retroviral particles.
Results
Viral genomic RNA is not required for A3C virion packaging
The viral determinant of A3C virion packaging has not yet been identified. To address the role of viral genomic RNA in the incorporation of A3C into virions, we first compared the levels of A3C in ΔPolΔEnv particles and in particles containing only Gag molecules (GagINS). The pΔPolΔEnv construct contains all the 5′RNA sequences upsteam of the gag coding sequence, including TAR, PolyA, PBS, DIS, SD, and the packaging signal (Fig. 1A); it does not express Pol, Env, Vif, Vpr, Vpu, or Nef. The Gag amino acid sequence of pGagINS is identical to that of pΔPolΔEnv and could be expressed in the absence of Rev (Qiu et al., 1999). However, pGagINS lacks all the 5′ viral RNA sequences, including the viral RNA packaging sequences upstream of the Gag coding region (Fig. 1A). Both constructs produce VLPs consisting only of Gag protein of viral origin.
Fig. 1.
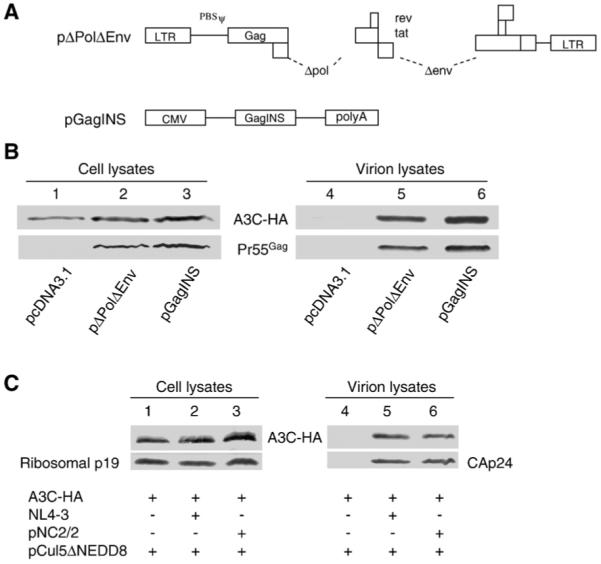
HIV-1 RNA is not required for A3C packaging. (A) Diagrams of the HIV-1 constructs ΔPolΔEnv and pGAGINS. (B) Immunoblot analysis of the virion incorporation of A3C-HA into ΔPolΔEnv and GAGINS. (C) Immunoblot analysis of the packaging of A3C into wild-type and NC mutant HIV-1 virions. pNL4-3 and pNL2/2 were co-transfected with the A3C-HA expression vector plus pCul5ΔNEDD8 into 293T cells as indicated. 48 h after transfection, cell lysates and virion pellets were prepared and analyzed by immunoblotting with an MAb against CAp24 to detect viral Gag proteins and an MAb against the HA tag to detect A3C-HA.
We found that the expression of A3C-HA in all types of transfected cells was comparable (Fig. 1B, lanes 1-3). Gag VLPs were produced from both pΔPolΔEnv- and pGagINS-transfected 293T cells (Fig. 1B, lanes 5 and 6). VLP-associated A3C-HA was comparable in Gag VLP from pΔPolΔEnv-transfected cells (Fig. 1B, lane 5) and from pGagINS-transfected cells (Fig. 1B, lane 6). As expected, no A3C-HA was detected in the supernatant of control vector-transfected cells, which did not generate VLPs (Fig. 1B, lane 4). Similar results for A3G has been previously reported (Luo et al., 2004).
To further address whether viral genomic RNA plays a role in virion incorporation of A3C, we compared the levels of virion-associated A3C in wild-type pNL4-3 virus and the viral genomic RNA packaging mutant pNC2/2. pNC2/2 contains five amino acid substitutions in the N-terminal zinc finger of the HIV-1 NC region and packages only approximately 15% of the viral genomic RNA. Again, the expression of A3C-HA was comparable in both types of transfected cells (Fig. 1C, lanes 1-3). A3C degradation was inhibited by co-expression of Cul5ΔNEDD8 in this experiment (Fig. 1C). A3C was detected in the pNL4-3 virions (Fig. 1C, lane 5) and to a similar degree in the pNC2/2 mutant virions (Fig. 1C, lane 6). As expected, no A3C-HA was detected in the supernatant of control vector-transfected cells, which did not generate HIV-1 virions (Fig. 1C, lane 4). Similar results for A3G has been previously reported (Luo et al., 2004). Thus, these results indicated that A3C packaging is independent of viral genomic RNA. Moreover, like A3G, VLP containing only Gag molecules could still package human A3C.
The NC domain of Gag is important for the packaging of A3G, but not A3C, into HIV-1 particles
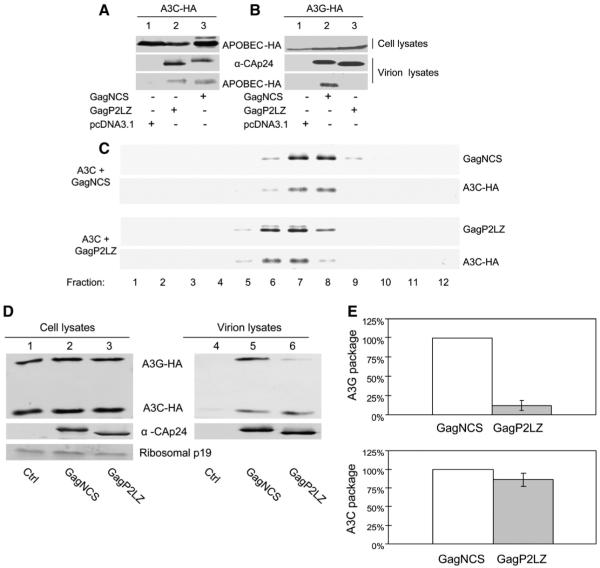
Previous studies have demonstrated that A3G binds to the nucleocapsid (NC) domain of the Gag polyprotein during virion assembly and that the packaging of A3G into HIV-1 particles requires the Gag NC domain (Alce and Popik, 2004; Cen et al., 2004; Douaisi et al., 2004; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Zennou et al., 2004). To examine the role of the NC domain in A3C packaging, we compared the encapsidation of A3C (Fig. 2A) and A3G (Fig. 2B) into HIV-1 Gag VLPs containing NC (GagNCS) and into Gag derivatives that lacked NC (GagP2LZ) but could still produce VLPs (Luo et al., 2004). 293T cells were co-transfected with a plasmid encoding A3C-HA or A3G-HA plus either pGagNCS or pGagP2LZ. The cell lysates of transfected cells were then analyzed by immunoblotting with an anti-HA antibody to detect A3C-HA (Fig. 2A) or A3G-HA (Fig. 2B), and the viral products were analyzed by immunoblotting with an anti-CAp24 antibody. This analysis indicated that similar levels of A3C-HA were packaged into pGagNCS and pGagP2LZ particles (Fig. 2A). In contrast, packaging of A3G-HA into pGagP2LZ particles was significantly reduced when compared to that into pGagNCS particles (Fig. 2B).
Fig. 2.
Virion incorporation of A3C is NC-independent. (A) Immunoblot analysis of the packaging of A3C-HA into HIV-1 Gag NCS or P2LZ particles. (B) Immunoblot analysis of the packaging of A3G-HA into HIV-1 Gag NCS or P2LZ particles. (C) Co-sedimentation of A3C-HA with HIV-1 Gag NCS or P2LZ particles in sucrose density gradients. GagNCS or P2LZ constructs were co-transfected with an A3C-HA expression vector into 293T cells. At 48 h after transfection, culture supernatants were separated from cell debris by filtration and ultracentrifugation through a 20% sucrose cushion. Gag particle pellets were resuspended in 0.5 ml PBS and layered onto a five-step sucrose density gradient (20-60%), centrifuged in an SW41 rotor (Beckman) at 22,000 rpm for 16 h at 4 °C. The fractions were collected and analyzed by immunoblotting with an MAb against the HA tag to detect A3C-HA and an MAb against CAp24 to detect NCS and P2LZ Gag molecules. (D) Expression vectors for A3C-HA and A3G-HA were co-transfected with NCS or P2LZ into 293T cells. At 48 h after transfection, cell and viral lysates were prepared and analyzed by immunoblotting using an MAb against the HA tag to detect A3C-HA or A3G-HA and an MAb against CAp24 to detect NCS and P2LZ Gag molecules. (E) Quantification of A3C-HA or A3G-HA packaging into Gag NCS or P2LZ particles. Packaging of A3C-HA or A3G-HA into Gag NCS particles was set to 100%.
To further determine whether A3C could be packaged into NC-deficient HIV-1Gag VLPs, 293T cells were co-transfected with A3C-HA and either pGagNCS or pGagP2LZ. VLPs in the supernatant were collected and resolved on sucrose density gradients, as described previously (Dettenhofer and Yu, 1999). Gradient fractions were analyzed by immunoblotting with anti-HA or anti-CAp24 antibody. As shown in Fig. 2C, the peak distribution of A3C-HA coincided with that of GagNCS. VLPs formed by GagP2LZ molecules (Fig. 2C, bottom panel) were slightly lighter than GagNCS particles (Fig. 2C, top panel). However, A3C-HA still co-fractionated with the GagP2LZ molecules (Fig. 2C, bottom panel). These data indicate that A3C molecules were indeed packaged into NC-deleted Gag particles and shared the same density as the corresponding Gag particles.
To further investigate this apparent requirement of NC for A3G but not A3C packaging, we co-transfected 293T cells with A3C-HA and A3G-HA expression vectors with pGagNCS or pGagP2LZ. The cell lysates of transfected cells and Gag VLP were analyzed by Immunoblotting (Fig. 2D), with anti-HA (top panel) and anti-CAp24 (bottom panel). Clearly, packaging of A3G-HA was significantly reduced in the pGagP2LZ particles (Fig. 2D, lane 6) when compared to the GagNCS particles (lane 5). In contrast, little difference was detected between GagNCS and GagP2LZ particles in terms of A3C-HA packaging (Fig. 2D). In repeated experiments, A3G packaging into GagP2LZ particles was reduced by approximately 85% when compared to GagNCS particles (Fig. 2E), but A3C packaging into GagP2LZ particles was not significantly reduced (Fig. 2E). Apparently, the viral determinants for the packaging of A3C and A3G are different.
Inhibition of 7SL RNA packaging reduces A3G, but not A3C, packaging into virions
7SL (SRP) RNA is the RNA component of the signal recognition particle (SRP) RNP complex, which also contains one copy each of the SRP72, SRP68, SRP54, SRP19, SRP14, and SRP9 proteins. 7SL RNA, but not the SRP proteins, is packaged into HIV-1 virions (Onafuwa-Nuga et al., 2006), suggesting that HIV-1 packages free 7SL RNA but not 7SL RNA that is part of an SRP RNP complex. Overexpression of the 7SL RNA binding protein SRP19 inhibits 7SL RNA packaging into HIV-1 virions (Wang et al., 2007). Consistent with our previous observations, overexpression of SRP19 in the present study inhibited A3G packaging into HIV-1 virions (Fig. 3A). However, its overexpression had little effect on the packaging of A3C (Fig. 3B). SRP19 overexpression also did not affect the intracellular expression of A3C (Fig. 3B, lanes 1 to 3). Examination of the virions revealed that the packaging of A3C into HIV-1 virions in the absence of SRP19 (Fig. 3B, lane 2) was not significantly different from that into HIV-1 virions produced in the presence of SRP19 (Fig. 3B, lane 3). Thus, inhibiting 7SL RNA packaging into HIV-1 virions did not affect A3C packaging. Deletion of the NC domain of HIV-1 Gag also did not affect A3C packaging. Since both A3G and 7SL RNA packaging require the NC domain of HIV-1 Gag, these data indicate that A3C packaging does not rely on 7SL RNA.
Fig. 3.
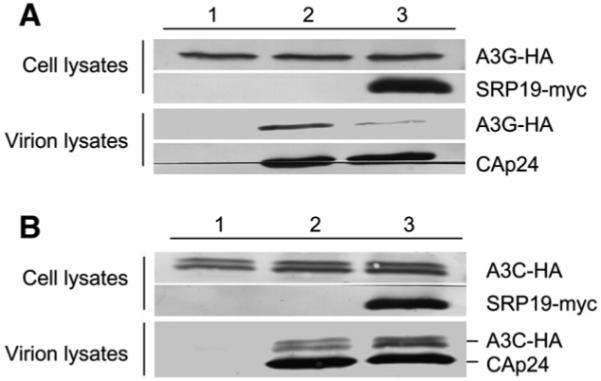
Overexpression of SRP19 proteins impairs the packaging of A3G, but not A3C. (A) Overexpression of SRP19 proteins inhibits A3G encapsidation into HIV-1 virions. HIV-1ΔVif virions plus A3G-HA were produced from transfected 293T cells in the absence (lane 2) or presence of exogenous SRP19-myc proteins (lane 3). Packaging of A3G-HA into HIV-1ΔVif virions was analyzed by immunoblotting with an anti-HA Mab to detect A3G-HA and an anti-CAp24 Mab to detect viral Gag proteins. SRP19-myc proteins were detected with an anti-myc Mab. Secretion of A3G-HA in the absence of HIV-1ΔVif was used as a negative control (lane 1). (B) Overexpression of SRP19 proteins does not inhibit A3C encapsidation into HIV-1 virions. HIV-1ΔVif virions plus A3C-HA were produced from transfected 293T cells in the absence (lane 2) or the presence of exogenous SRP19-myc proteins (lane 3). Packaging of A3C-HA into HIV-1ΔVif virions was analyzed by immunoblotting with the antibodies described in (A). Secretion of A3C-HA in the absence of HIV-1ΔVif was used as a negative control (lane 1).
The MA domain of HIV-1 Gag is required for efficient A3C packaging
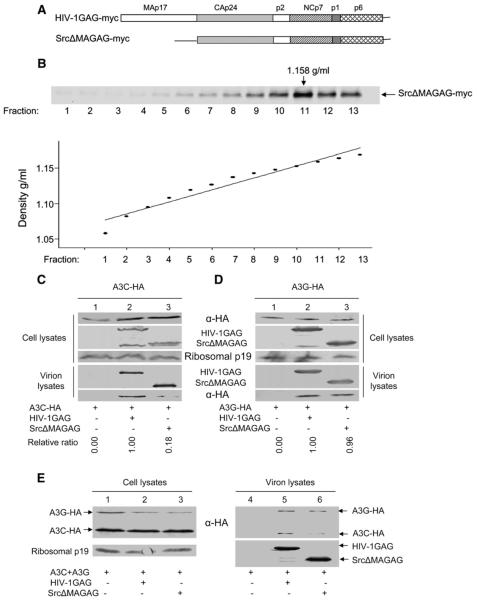
Since GagP2LZ lacks NC, P1, and P6 domains of HIV-1 Gag and still packages A3C, we then asked whether the matrix (MA) domain within HIV-1 Gag plays a role in its packaging. For this purpose, we analyzed the packaging of A3C in the presence of full-length HIV-1 Gag (GAG-myc) and an MA deletion construct Src ΔMA GAG-myc. SrcΔMA GAG-myc contains an in-frame deletion of the MA domain, and the membrane targeting function of MA is replaced by the v-src myristylation signal (Dong et al., 2005). SrcΔMA GAG-myc produced Gag VPL particles (Fig. 4B, upper panel) with densities of ∼1.15-1.16 g/ml (Fig. 4B, lower panel). Both the full-length Gag and MA-deficient Gag produced Gag particles efficiently (Fig. 4C), and the expression of A3C-HA was comparable in both cell types (Fig. 4C, top lanes 1-3). As expected, no A3C protein was detected in the supernatant of control vector-transfected cells, which did not generate Gag particles (Fig. 4C, lane 1). A3C-HA packaging into SrcΔMAGAG-myc particles was clearly reduced when compared to that into GAG-myc particles (Fig. 4C, compare lanes 2 and 3). In contrast, no difference in A3G packaging was detected between the GAG-myc and SrcΔMAGAG-myc particles (Fig. 4D, compare lanes 2 and 3). No A3G protein was detected in the supernatant of control vector-transfected cells, which did not generate Gag particles (Fig. 4D, lane 1).
Fig. 4.
Influence of HIV-1 MA on A3C and A3G packaging. (A) Diagrams of HIV-1 full-length Gag and MA deletion SrcΔMAGAG-myc constructs. (B) The SrcΔMAGAG-myc forms VLP. The culture supernatants of SrcΔMAGAG-myc transfected 293T cells were harvested by filtration and ultracentrifugation through 20% sucrose cushion, and then centrifugation at 100,000 ×g on a 20-60% linear sucrose gradient for 15 h. Analysis was via Immunoblot analysis by using mAb against CAp24 (upper panel). Density analysis is show below (lower panel). (C) Immunoblot analysis of the packaging of A3C-HA into HIV-1 Gag-myc or SrcΔMAGAG-myc. (D) Immunoblot analysis of the packaging of A3G-HA into HIV-1 Gag-myc or SrcΔMAGAG-myc. Quantitative analysis, calculated as the relative ratio of A3C-HA and A3G-HA in SrcΔMAGAG VLP as compared to A3C-HA and A3G-HA in HIV-1 GAG VLP, is shown below. The VLP encapsidation of APOBEC-HA protein was analyzed after normalizing the myc antigen content. Values for the HIV-1 GAG VLP incorporation of APOBEC-HA protein were set to 1. (D) Comparison of the packaging of A3C-HA and A3G-HA into MA-deleted Gag particles from co-expressing cells. Expression vectors for A3C-HA and A3G-HA were co-transfected with Gag-myc or SrcΔMAGAG-myc into 293T cells. At 48 h after transfection, cell and viral lysates were prepared and analyzed by immunoblotting using MAb against the HA tag to detect A3C-HA or A3G-HA and an MAb against myc to detect HIV-1 full-length Gag-myc and truncated SrcΔMAGAG-myc molecules.
To further compare the packaging of A3C and A3G into MA-deficient Gag particles, we co-transfected A3C-HA and A3G-HA with GAG-myc or SrcΔMAGAG-myc into 293T cells, then analyzed the cell lysates of the transfected cells and Gag particles by immunoblotting. Although the intracellular expression of A3C-HA in SrcΔMAGAG-myc-expressing cells (Fig. 4E, lane 3) was similar to that in GAG-myc-expressing cells (Fig. 4E, lane 2), a significantly lower level of A3C-HA was detected in SrcΔMAGAG-myc particles (Fig. 4E, lane 6) than in GAG-myc particles (Fig. 4E, lane 5). At the same time, the packaging of A3G-HA into these two types of particles was not significantly affected (Fig. 4E, compare lane 5 and 6). Since A3C-HA and A3G-HA were co-expressed with either GAG-myc or SrcΔMAGAG-myc, it is clear that the packaging of A3C and A3G was differentially affected by the MA-domain deletion.
The interaction of A3C with HIV-1 Gag is MA domain-dependent and RNA-dependent
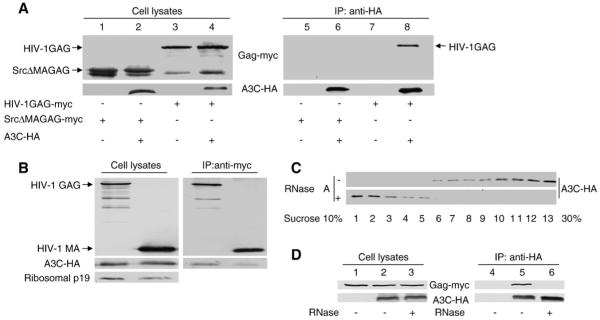
Our data indicate that A3G binds HIV-1 Gag but not to NC domain-deleted Gag molecules, and this inefficient interaction with NC-deleted Gag is also correlated with poor packaging of A3G into these Gag mutant particles (Luo et al., 2004). To determine whether A3C also interacts with HIV-1 Gag, we used co-immunoprecipitation experiments to characterize the intracellular interaction between Gag and A3C. HIV-1 GAG-myc or SrcΔMAGAG-myc was co-transfected with A3C-HA into 293T cells. At 48 h after transfection, A3C-HA was immunoprecipitated from the cell lysates using an anti-HA antibody. The results indicated that the full-length HIV-1 GAG-myc was co-precipitated with A3C-HA (Fig. 5A, lane 8). As a control, GAG-myc was not detected in the absence of A3C-HA (Fig. 5A, lane 7), indicating a specific interaction between HIV-1 Gag and A3C. In contrast to the full-length Gag, SrcΔMAGAG-myc did not interact efficiently with A3C-HA (Fig. 5A, lane 6), despite the fact that SrcΔMAGAG-myc was expressed at a level comparable to that of Gag-myc (Fig. 5A, compare lanes 2 and 4). Thus, A3C packaging into HIV-1 Gag particles appears to be mediated through an interaction between A3C and HIV-1 Gag, and the MA domain of Gag is required for efficient interaction. However, MA domain alone interacted less efficiently with A3C than the full length Gag molecule (Fig. 5B).
Fig. 5.
(A) A3C-HA binds full-length HIV-1 Gag but not MA-deleted Gag molecules. A3C-HA or control plasmids were cotransfected with HIV-1 Gag-myc or SrcΔMAGAG-myc. At 48 h after transfection, cell lysates were prepared and immunoprecipitated with an anti-HA antibody conjugated to beads. Co-precipitated samples were analyzed by immunoblotting using an MAb against the HA tag to detect A3C-HA and an MAb against myc to detect HIV-1 full-length Gag-myc and truncated SrcΔMAGAG-myc molecules. (B) A3C-HA binds HIV-1 MA protein but weaker than binds full-length HIV-1 Gag. A3C-HA was cotransfected with HIV-1 Gag-myc or HIV-1 MA-myc. 48 h after transfection, cell lysates were prepared and immunoprecipitated with an anti-myc antibody. Co-precipitated samples were analyzed by immunoblotting using an MAb against the HA tag to detect A3C-HA and an MAb against myc to detect HIV-1 full-length Gag-myc and MA-myc molecules. (C) Sedimentation of human A3C and A3G in sucrose gradients. 293T cells which transfected A3C-HA expression vector. 48 h after transfection, cell lysates were prepared were lysed, and cell debris was removed by centrifugation at 10,000 ×g for 15 min. Supernatants were treated with RNAse A or RNase inhibitor for 30 min at 37 °C and centrifuged at 260,800 ×g for 4 h. Different fractions were analyzed by immunoblotting with a MAb against the HA tag for the detection of APOBEC-HA. (D) A3C-HA binds HIV-1 Gag-myc and is disrupted by RNase treatment. Co-immunoprecipitation was performed as described above. One aliquot of the sample was first treated with RNase (1.5 μg/ml) at 37 °C for 30 min before co-immunoprecipitation.
Cellular RNA may play a role in A3C packaging into HIV-1 Gag particles since we observed that A3C was associated with high-molecular-weight complexes in 293T cell lysates when analyzed through a sucrose velocity gradient (Fig. 5C, top panel, fractions 10 to 13). After treatment with RNase A, A3C-HA was largely detected in fraction 1 to 3 of the sucrose velocity gradient (Fig. 5C, bottom panel), suggesting that A3C is largely associated with cellular complexes that contain RNA. The interaction between HIV-1 Gag and A3C was also sensitive to RNase A treatment (Fig. 5D). Interaction between A3C and HIV-1 Gag (Fig. 5D, lane 5) was largely disrupted when the sample was first treated with RNase A (Fig. 5D, lane 6). The MA domain of HIV-1 Gag has been reported to have RNA binding activity (Burniston et al., 1999; Lochrie et al., 1997; Ott et al., 2005; Purohit et al., 2001). Thus, the interaction between HIV-1 Gag and A3C is MA-dependent and requires the presence of RNA.
Discussion
The packaging of APOBEC3 proteins into HIV-1 virions is believed to be essential to their anti-viral activity in newly infected target cells; however, the viral determinants of APOBEC3 packaging have not been well characterized. Although it has previously been reported that A3G packaging is dependent upon an interaction with the nucleocapsid (NC) domain of HIV-1 Gag and that this interaction requires an RNA bridge (Douaisi et al., 2004; Khan et al., 2005; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Svarovskaia et al., 2004; Wang et al., 2007; Zennou et al., 2004). The data reported here indicate that the viral determinants for the packaging of A3C are distinct from those for A3G. HIV-1 genomic RNA appeared to be dispensable for the packaging of A3C (Fig. 1) as well as A3G (Alce and Popik, 2004; Burnett and Spearman, 2007; Cen et al., 2004; Douaisi et al., 2004; Khan et al., 2005; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Wang et al., 2008; Zennou et al., 2004), however, although the Gag molecule could mediate both A3C and A3G packaging, we found that the MA domain was required for the efficient packaging of A3C but not A3G into Gag particles (Fig. 4). On the other hand, A3G packaging requires the NC domain of Gag, which was dispensable for A3C packaging (Figs. 2).
We and others have previously shown that the virion encapsidation of A3G depends on the NC domain of HIV-1 Gag (Alce and Popik, 2004; Burnett and Spearman, 2007; Cen et al., 2004; Douaisi et al., 2004; Luo et al., 2004; Navarro et al., 2005; Schafer et al., 2004; Zennou et al., 2004). Recently, we have observed that A3G specifically interacts with 7SL RNA (Wang et al., 2007), which is likely a key mediator of its packaging; this 7SL RNA is bound by A3G in both virus-producing cells and mature virions (Tian et al., 2007; Wang et al., 2007). 7SL RNA is more selectively packaged into HIV-1 virions than other abundant cellular RNAs (Tian et al., 2007). In particular, the NCp7 domain of HIV-1 Gag, and specifically the N-terminal basic region and the basic linker region, were found to be important for efficient 7SL RNA packaging (Tian et al., 2007). We and others have further showed that these regions of NC are important for the virion packaging of cytidine deaminase A3G (Burnett and Spearman, 2007; Luo et al., 2004). Inhibiting 7SL RNA encapsidation also inhibited A3G and A3F packaging into HIV-1 virions (Wang et al., 2007, 2008). Thus it is likely that 7SL RNA serves as a bridging factor between Gag and A3G and mediate A3G packaging into the virions.
In contrast to the results obtained for A3G, deletion of the NC domain had little effect on A3C packaging in the present study (Fig. 2). The particles produced by NC-deleted Gag molecules had a lighter density than the Gag particles containing NC (Fig. 2). In both cases, A3C co-peaked with the respective Gag particles (Fig. 2). This lack of requirement for NC in A3C packaging was more clearly demonstrated by co-expression of A3C and A3G in the same virus-producing cells. In this case, a clear requirement for NC was seen in the case of A3G but not A3C (Fig. 2D). Thus, it is likely that the NC domain and 7SL RNA do not mediate A3C packaging. This conclusion is further supported by the observation that limiting 7SL encapsidation inhibited the packaging of A3G but not A3C (Fig. 3).
In the present study, we have also made the observation that deletion of the MA domain of HIV-1 Gag inhibited the packaging of A3C but not A3G. There are several possible reasons why the MA domain might be critical for A3C packaging. One is that the MA domain targets Gag molecules to certain intracellular sites during virus assembly that have a high concentration of A3C molecules, resulting in a passive packaging of A3C into virus; in the same way, MA-deleted Gag molecules could be targeted to different sites that have a low concentration of A3C, resulting in reduced A3C packaging. Alternatively, HIV-1 Gag may interact with A3C either directly or indirectly to recruit A3C into virions. Consistent with this interpretation, we have observed an interaction between HIV-1 Gag and A3C in virus-producing cells. In this scenario, the MA domain would be important for mediating the Gag-A3C interaction, a prediction supported by our finding that MA-deleted Gag showed a reduced level of interaction with A3C. Along the observations that MA domain of Gag is important for its interaction with A3C, we have also found that the interaction is RNA-dependent. Treatment of RNase A disrupted the interaction between Gag and A3C. (Fig. 5D, lane 6) Although NC domain is the most prominent RNA binding domain in Gag and is largely responsible for selective packaging of genomic RNA (Berkowitz et al., 1996), MA domain was shown to be able to bind genomic viral and non-viral mRNA and tRNAs in the absence of NC (Bukrinskaya et al., 1992; Ott et al., 2005). It is likely that MA domain plays a redundant role in Gag RNA binding activity (Ott et al., 2005). It is reasonable to speculate that cellular RNA(s) may serve as a bridging factor between Gag and A3C and mediate A3C packaging. We have shown in previous studies that 7SL RNA mediates A3G and A3F packaging through NC domain of Gag (Wang et al., 2007, 2008), and it would be interesting to further identify which cellular RNA is involved in A3C packaging.
It is possible that in addition to the MA domain of HIV-1 Gag, other viral determinant(s) also mediate A3C virion packaging. We have observed that the packaging of A3C into HIV-1 Gag particles was less efficient than that of A3G (Figs. 2D and 4E). However, other people have reported that both A3C and A3G are packaged efficiently into HIV-1 virions (Yu et al., 2004a). Further study will be required to determine whether other viral determinant(s) in addition to Gag also mediate A3C virion packaging.
Taken together, our results indicate that human APOBEC3 proteins have apparently evolved to use distinct mechanisms to target retroviruses. Understanding the precise mechanisms involved in virion packaging of APOBEC3 proteins may lead to novel developments that can enhance the packaging of APOBEC3 proteins and the anti-viral activities of these molecules.
Materials and methods
Plasmid construction
pNL4-3 was obtained from the AIDS Research Reagents Program, Division of AIDS, NIAID, NIH (Cat.# 114). The HIV-1 construct ΔPolΔEnv (Lee and Yu, 1998) and Gag expression vectors pGAGINS (Qiu et al., 1999), pGagNCS, and pGagP2LZ (Luo et al., 2004), Gag-myc and SrcΔMAGag-myc (Dong et al., 2005) have been previously described. A3G-HA and SRP19-myc expression vectors have been previously described (Wang et al., 2007). The HIV-1 MA was amplified by RT-PCR using HIV-1 Gag MA forward primer: 5′ GTACGCTAGCGCCATGGGAGCCCGCGCC AGC-3′; reverse primer: 5′-GTAC AAGCTTTCAAAGATCTTCTTCTGATATGAGTTTTTGTTCGTAGTTCTGGCT CACCTG-3′ containing NheI and Hind III sites, respectively, and a C-terminal c-myc tag. The PCR product was cloned into pcDNA3.1 to generate HIV-1 MA-myc. A3C-HA was generously provided by Dr. Michael Malim, and pNC2/2 was generously provided by Dr. Robert Gorelick.
Antibodies and cell lines
The following antibodies were used for this study: Anti-CAp24 monoclonal antibody (mAb), obtained from the AIDS Research Reagents Program, Division of AIDS, NIAID, NIH; anti-myc mAb (Sigma, Cat. #M5546), and anti-HA mAb (Covance, Cat. # MMS-101R-10000). 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 10% fetal bovine serum and penicillin/streptomycin (D-10 medium) and passaged upon confluence.
Transfection and virus purification
DNA transfections were carried out using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. At 48 h after transfection, virion-associated viral proteins were prepared from cell culture supernatants and separated from cellular debris by centrifugation at 3000 rpm for 30 min in a Sorvall RT 6000B centrifuge and filtration through a 0.22-μm pore-size membrane. Virus particles were concentrated by centrifugation through a 20% sucrose cushion at 100,000 ×g for 1.5 h in a Sorvall Ultra80 ultracentrifuge.
Immunoblot analysis
Cells were collected 48 h after transfection. Cell and viral lysates were prepared as previously described (Tian et al., 2007). Cells (1×105) were lysed in 1× loading buffer (0.08 M Tris, pH 6.8, with 2.0% SDS, 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue). The samples were boiled for 10 min, and proteins were separated by SDS-PAGE. For virion lysates, cell culture supernatants were collected 72 h after transfection by removal of cellular debris through centrifugation at 3000 rpm for 10 min in a Sorvall RT 6000B and filtration through a 0.2-μm pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 ×g for 2 h in a Sorvall Ultra80 ultracentrifuge. Proteins were transferred onto nitrocellulose membranes, and the membranes were probed with various primary antibodies against proteins of interest. Secondary antibodies were alkaline phosphatase-conjugated anti-human and anti-mouse (Jackson Immunoresearch, Inc) antibodies, and staining was carried out with 5-bromo-4-chloro-3indolyl phosphate (BCIP) and nitro-blue tetrazolium (NBT) solutions prepared from chemicals obtained from Sigma. Blots were imaged using the FujiFilm LAS-1000 Image Station, and protein band densities were analyzed using the spot density analysis software Image Gauge V3.41.
Immunoprecipitation
For APOBEC-HA immunoprecipitations, transfected 293T cells were harvested and washed twice with cold PBS, then lysed with PBS containing 0.5% Triton X-100 and protease inhibitor cocktail (Roche, Basel, Switzerland) at 4 °C for 1 h. Cell lysates were clarified by centrifugation at 10,000 ×g for 30 min at 4 °C. Anti-HA agarose (Roche) was mixed with the pre-cleared cell lysates and incubated at 4 °C for 3 h. The reaction mixture was then washed three times with cold PBS and eluted with 0.1 M glycine-HCl buffer, pH 2.0. The eluted materials were subsequently analyzed by immunoblotting.
Sucrose gradient analysis
To identify the incorporation of APOBEC3C into Gag virus-like particles (VLPs), sucrose gradients were prepared as follows: Gag VLPs in the culture supernatants of APOBEC transfected 293T cells were separated from cell debris by filtration and ultracentrifugation through a 20% sucrose cushion, and the VLP pellet was resuspended, then layered onto a sucrose density gradient (20-60%). Samples were then centrifuged at 100,000 ×g at 4 °C. The fractions of 1 ml each were collected from the top for immunoblotting analysis.
Acknowledgments
We thank Dr. Michael Malim for A3C-HA vector, Anjie Zhen and Lindi Tan for technical assistance and thoughtful discussions, and Dr. Deborah McClellan for editorial assistance. This work was supported by grants from the NIH (AI062644 and AI071769), a grant from the Johns Hopkins Center for AIDS Research (CFAR), and funding from the National Science Foundation of China (NSFC-30425012) and Cheung Kong Scholars Program Foundation of the Chinese Ministry of Education to X-F.Yu.
References
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279(33):34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5(11):1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80(17):8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea K,S, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 2006;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43(6):1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- Bukrinskaya AG, Vorkunova GK, Tentsov Y. HIV-1 matrix protein p17 resides in cell nuclei in association with genomic RNA. AIDS Res. Hum. Retrovir. 1992;8(10):1795–1801. doi: 10.1089/aid.1992.8.1795. [DOI] [PubMed] [Google Scholar]
- Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81(10):5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 1999;73(10):8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279(32):33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. APOBEC3 cytidine deaminases: distinct antiviral actions along the retroviral life cycle. J. Biol. Chem. 2006;281(13):8309–8312. doi: 10.1074/jbc.R500021200. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13(22):2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 2006;80(3):1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006;80(21):10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 1999;73(2):1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, Spearman P. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Douaisi M, Dussart S, Courcoul M, Bessou G, Vigne R, Decroly E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 2004;321(3):566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433(7024):430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Goff SP. Retrovirus restriction factors. Mol. Cell. 2004;16(6):849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006;80(23):11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev., Immunol. 2004;4(11):868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(12):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35(21):7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79(3):285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 2006;80(2):875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79(9):5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 2005;280(19):18573–18578. doi: 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33(6):1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300(5622):1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lee YM, Yu XF. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243(1):78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 2007;282(44):32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Liu B, Yu X, Luo K, Yu Y, Yu XF. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 2004;78(4):2072–2081. doi: 10.1128/JVI.78.4.2072-2081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sarkis PT, Luo K, Yu Y, Yu XF. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 2005;79(15):9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Waugh S, Pratt DG, Jr., Clever J, Parslow TG, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25(14):2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78(21):11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 2007;81(13):7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 2005;102(32):11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH. Natural resistance to HIV infection: The Vif-APOBEC interaction. C. R. Biol. 2006;329(11):871–875. doi: 10.1016/j.crvi.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114(1):21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9(11):1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 2007;81(13):7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18(23):2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 2006;281(25):17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Navarro F, Landau NR. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 2004;16(4):477–482. doi: 10.1016/j.coi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 2007;81(17):9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. Rna. 2006;12(4):542–546. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81(15):8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Gagliardi TD. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 2005;79(22):13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit P, Dupont S, Stevenson M, Green MR. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. Rna. 2001;7(4):576–584. doi: 10.1017/s1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JT, Song R, Dettenhofer M, Tian C, August T, Felber BK, Pavlakis GN, Yu XF. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 1999;73(11):9145–9152. doi: 10.1128/jvi.73.11.9145-9152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose KM, Marin M, Kozak SL, Kabat D. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 2004;10(6):291–297. doi: 10.1016/j.molmed.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Rosler C, Kock J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsacker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42(2):301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 2007;81(15):8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328(2):163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 2006;80(12):5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9(11):1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1(1):e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12(3):591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32(8):2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279(34):35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 2006;80(6):3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang T, Zhang W, Yu XF. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007;35(21):7288–7302. doi: 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Trono D. Editing at the crossroad of innate and adaptive immunity. Science. 2005;307(5712):1061–1065. doi: 10.1126/science.1105964. [DOI] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004a;303(5665):1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 2004b;279(42):43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007;81(23):13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 2008;375(4):1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349(2):290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 2007a;21(1):217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. Characterization of a novel cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 2007b;373(3):541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B × antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46(6):1810–1820. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- Yang B, Chen K, Zhang C, Huang S, Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007;282(16):11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- Yu X. Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr. Opin. HIV AIDS. 2006;1(3):187–193. doi: 10.1097/01.COH.0000221590.03670.32. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004a;279(51):53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minusstrand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004b;11(5):435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004c;18(23):2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78(21):12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424(6944):94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Tian C, Wang T, Sarkis PT, Fang Y, Zheng S, Yu XF, Xu R. Cytidine deaminase APOBEC3B interacts with heterogeneous nuclear ribonucleoprotein K and suppresses hepatitis B virus expression. Cell. Microbiol. 2008;10(1):112–121. doi: 10.1111/j.1462-5822.2007.01020.x. [DOI] [PubMed] [Google Scholar]