Abstract
Exposure to intense noise induces apoptosis in hair cells in the cochlea. To identify the molecular changes associated with noise-induced apoptosis, we used quantitative real-time PCR to evaluate the changes in 84 apoptosis related genes in cochlear samples from the sensory epithelium and lateral wall. Sprague Dawley rats exposed to a continuous noise at 115 dB SPL for 2 h. The exposure caused a 40–60 dB threshold shift 4 h post-exposure that decreased to 20–30 dB 7 days post-exposure. These functional changes were associated with apoptotic markers including nuclear condensation and fragmentation and TUNEL staining. Immediately after the noise exposure, 12 genes were downregulated, whereas only one gene (Traf4) was upregulated. At 4 h post-exposure, 8 genes were upregulated; 3 (Tnrsf1a, Tnfrsf1b, Tnfrst5) belonged to the Tnfrsf family, 3 (Bir3, Mcl1 and Prok2) have anti-apoptotic properties and 1 (Gadd45a) is a target of p53. At 7 d post-exposure, all the upregulated genes returned to pre-noise levels. Interestingly, the normal control cochlea had high constitutive levels of several apoptosis-related genes. These constitutively expressed genes, together with the inducible genes, may participate in the induction of cochlear apoptotic activity.
Keywords: mRNA Expression, Rt-PCR, Noise Trauma, Hair Cells
Introduction
Exposure to intense noise traumatizes the cochlea and can lead to cell death primarily through apoptosis and necrosis (Hu et al., 2000, Nicotera et al., 2001, Wang et al., 2002, Ylikoski et al., 2002, Niu et al., 2003, Shibuya et al., 2003, Han et al., 2006, Bohne et al., 2007 ) with apoptosis being the primary cell death pathway (Hu et al., 2002b, Yang et al., 2004). Apoptosis begins immediately after a noise exposure (Hu et al., 2006) and continues to emerge for several days after the noise exposure (Yang et al., 2004). Several apoptotic events have been identified including activation of caspases-3, -8 and -9 (Nicotera et al., 2003), release of cytochrome c from the mitochondria to the cytosol (Nicotera et al., 2003), and translocation of EndoG and AIF from the mitochondria to nuclei (Yamashita et al., 2004, Han et al., 2006). In addition, the involvement of several apoptotic molecules has been reported including c-Jun-N-terminal kinase (Pirvola et al., 2000), transcriptional factor activator protein-1 (Matsunobu et al., 2004), BAD (Vicente-Torres and Schacht, 2006), Bcl-xL and Bak (Yamashita et al., 2008) and TNF-α (Fujioka et al., 2006).
Several studies have screened the expression of a large number of genes in noise-traumatized cochleae using gene array techniques. Taggart (Taggart et al., 2001) exposed chinchillas to a moderate level of noise (95 dB SPL, 3 or 6 h) and found expression changes in genes associated with metabolism, cytoskeletal proteins, calcium balance, and heat shock protein. However, no apoptosis-related genes were specifically reported possibly due to insufficient level of noise exposure needed to induce apoptosis. Another gene array study (Cho et al., 2004) reported that exposure to an intense noise induced the expression of the early genes that encode transcription factors (c-FOS, EGR1, NUR77/TR3) and cytokines (PC3/BTG2, LIF and IP10). Some of these genes have been linked to apoptosis. Differential gene expression in rat cochleae has also been examined after impulse noise exposure (Kirkegaard et al., 2006). This study revealed expression of several gene families including genes associated with regulation of transcription, cell cycle/differentiation, metal ion homeostasis, inflammatory response, and response to oxidative stress.
It is now recognized that noise-induced cochlear apoptosis involves complex signaling pathways including the extrinsic and intrinsic signaling cell death pathways. To date, a handful of apoptotic proteins and genes have been implicated in noise-induced apoptosis. To more fully quantify and characterize the role of apoptotic genes in noise-induced hearing loss, we screened a panel of 84 apoptosis-related genes using quantitative real-time PCR array, a method that features a high degree of sensitivity, selectivity and accuracy. Our results identified 22 genes that significantly increased or decreased expression following noise exposure. Many of the genes have heretofore not been linked to noise-induced apoptosis. In addition, a strong constitutive expression of apoptosis-related genes was observed in the cochlea.
Experimental Procedures
Animals
Young Sprague Dawley rats (210–300g, male, Charles River Laboratories, Wilmington, MA) were used. The procedures involving use and care of animals were reviewed and approved by the State University of New York at Buffalo Institutional Animal Care and Use Committee.
Noise exposure
Rats were exposed for 2 h to a continuous noise (1–7 kHz) at 115 dB SPL (re 20 μPa). This noise level was selected because it induced mainly temporary hearing loss and sub-lethal cell damage, but was not strong enough to immediately destroy a large number of sensory or supporting cells. The goal was to avoid immediate and massive cell loss that would significantly alter the subpopulation of cells used to obtain the mRNA for analysis. The noise was generated digitally using a real-time signal processor (RP2.1, TDT). The signal was routed through an attenuator (PA5 TDT), and a power amplifier (Crown XLS 202) to a loud speaker (Eminence LT250). The loudspeaker was suspended approximately 20 cm directly above the animal holding cage. The noise level in the sound field was calibrated using a sound level meter (Larson Davis 800B), a pre-amplifier (Larson and Davis model 825), and a condenser microphone (Larson and Davis, LDL 2559). The sound field was calibrated by placing the microphone within the cage at the level of the animal’s head.
Auditory Brainstem Responses (ABR)
ABR measurements were conducted individually for the right and left ears to determine the hearing sensitivity of the animals before and 4 h and 7 days post-exposure. Stainless steel needle electrodes were placed subdermally over the vertex (noninverting input) and behind the ears (inverting input and ground) of the animal. During testing, the animal was lightly anesthetized by inhalation of isoflurane mixed with oxygen (2–3%, flow rate 1.5 L/min for induction and 0.8 L/min for maintenance) or by intramuscular injection of a mixture of ketamine (87 mg/kg) and xylazine (3 mg/kg). The ABR was elicited with tone bursts generated digitally (TDT, SigGen) using a D/A converter (TDT, RP2.1, 100 kHz sampling rate) and fed to a programmable attenuator (TDT PA5), an amplifier (TDT, SA1) and a calibrated closed field loudspeaker (TDT, CF1). Tone bursts were presented at 5, 10, 20, 30 and 40 kHz (0.5 ms rise/fall Blackman ramp, 1 ms duration, alternating phase) at the rate of 21 per second. The output of the electrodes were led to the head stage of a differential amplifier (TDT, RA4LI), followed by an amplifier (TDT, RA16PA), and then routed by a fiber optic cable to a real-time processor (TDT, RP2.1). The auditory evoked response averaging system was controlled by TDT software (BioSig). Input signals were amplified (~50,000×), filtered (100–3000 Hz), processed with the artifact reject software set at 80% of full scale, averaged (250 sweeps), and then stored and displayed on a computer. Stimulus level was decreased in 5 dB steps. The ABR threshold was defined as the lowest intensity that reliably elicited a detectable response.
Histology
Hair cell pathology in the organ of Corti was examined at 10 min., 4 h and 7 days post-exposure. For the 10-min, and 4-h time points, nuclear morphology was assessed by propidium iodide staining to identify cells with condensed and/or fragmented nuclei, morphological characteristics of cells undergoing apoptosis. Apoptosis was confirmed by the TUNEL assay. For the 7-day time point, the integrity of the cuticular plates of hair cells, together with the nuclear morphology, was examined to quantify both missing cells and apoptotic cells.
Propidium iodide staining
Animals were sacrificed and the cochleae quickly removed, opened and perfused with the propidium iodide solution (Invitrogen, Inc., 5 μg/ml in 10 mM PBS) through the round window. The solution was allowed to remain in the cochleae for 10 min. at room temperature and then fixed with 10% buffered formalin.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
A TUNEL assay was used to detect nuclear DNA fragmentation as previously described (Nicotera et al., 2003). Briefly, cochleae were fixed with 10% buffered formalin and the organs of Corti were carefully dissected, transferred to ice-cold 70% (v/v) ethanol and stored overnight at −20°C. Tissues were washed and then incubated with DNA-labeling solution (Molecular Probes, Inc.) containing 10 μl of reaction buffer, 0.75 μl of TdT enzyme, 8.0 μL of BrdUTP, and 31.25 μL of dH2O for 16 h at room temperature. The tissues were stained with Alexa Fluor 488 dye–labeled anti-BrdU antibody (5 μL of antibody plus 95 μL of washing buffer) at room temperature for 1 h. After labeling, the tissues were stained with propidium iodide.
F-actin staining
F-actin staining was used to quantify the number of missing hair cells as previously described (Hu et al., 2002a). Briefly, after completion of the cochlear dissection, the organ of Corti was transferred to freshly-prepared staining solution containing FITC-phalloidin (1:250, Sigma), 0.25% Triton-X-100, and 1% bovine serum albumin in PBS. The tissues were incubated at room temperature for 30 min.
All specimens were thoroughly examined with a fluorescence microscope to identify hair cell lesions. The lesions were further examined using confocal microscopy (Zeiss LSM 510 META). The numbers of apoptotic and missing hair cells were quantified. The criteria for identification of apoptotic, and missing hair cells have been reported previously (Nicotera et al., 2003, Yang et al., 2004). Briefly, a cell showing a condensed nucleus together with positive TUNEL labeling was counted as an apoptotic cell. A lack of F-actin staining in the cuticular plate region was counted as a missing cell. Hair cell loss was assembled into a cochleogram showing the frequency-place correlation for the rat (Muller, 1991).
mRNA expression levels of apoptosis-related genes
The expression levels of 84 known apoptotic genes (see Table 1) were examined using RT2 Profiler™ PCR Array (SuperArray Bioscience Corporation). The genes on the array participate in various apoptotic pathways.
Table 1.
Apoptosis-related genes
| Symbol | Name | Symbol | Name |
|---|---|---|---|
| Apaf1 | Apoptotic peptidase activating factor 1 | Dffb | DNA fragmentation factor, beta sununit |
| Api5_predicted | Apoptosis inhibitor 5 (predicted) | ||
| Aven_predicted | Apoptosis, caspase activation inhibitor (predicted) | Fadd | Fas (TNFRSF6)-associated via death domain |
| Bad | Bcl2-associated death promoter | Faim | Fas apoptotic inhibitory molecule |
| Bag1_predicted | Bcl2-associated athanogene 1 (predicted) | Gadd45a | Growth arrest and DNA-damage-inducible 45 alpha |
| Bak1 | BCL2-antagonist/killer 1 | ll10 | Interleukin 10 |
| Bax | Bcl2-associated X protein | Lhx4_predicted | LIM homeobox protein 4 (predicted) |
| Bcl10 | B-cell CLL/lymphoma 10 | Lta | Lymphotoxin A |
| Bcl2 | B-cell leukemia/lymphoma 2 | Ltbr | Lymphotoxin B receptor |
| Bcl2a1 | B-cell leukemia/lymphoma 2 related protein A1 | Mapk8ip | Mitogen activated protein kinasa 8 interacting protein |
| Bcl2l1 | Bcl2-like 1 | Mcl1 | Myeloid cell leukemia sequence 1 |
| Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | Mfkb 1 | Nuclear factor of kappa light chain gene enhancer in B-cells 1, p 105 |
| Bcl2l2 | Bcl2-like 2 | ||
| Bclaf 1 | BCL2-associated transcription factor 1 | Nol3 | Nucleolar protein 3 (apoptosis repressor with CARD domain) |
| Bid | BH3 interacting domain death agonist | ||
| Bid3 | BH3 interacting (with BCL2 family) domain, apoptosis agonist | Prolb Prdx2 |
Polymerase (DNA directed), beta Peroxiredoxin 2 |
| Bik | Bcl2-interacting killer | Prlr | Prolactin receptor |
| Birc1b | Baculoviral IAP repeat-containing 1b | Prok2 | Prokineticin 2 |
| Birc3 | Baculoviral IAP repeat containing 3 | Pycard | PYD and CARD domain containing |
| Birc4 | Baculoviral IAP repeat-containing 4 | Ripk2 | Receptor (TNFRSF)-interacting serine- threonine kinase 2 |
| Birc5 | Baculoviral IAP repeat-containing 5 | ||
| Bnip1 | BCL2/adenovirus E1B 19kDa-interacting protein 1 | Sphk2 Tnf |
Sphingosine kinase 2 Tumor necrosis factor (TNF superfamily, member 2) |
| Bnip2_predicted | BCL2/adenovirus E1B 19kDa-interacting protein 1, NIP2 (predicted) | Tnfrsf 10b_predicted | Tum or necrosis factor receptor superfamily, member 10b (predicted) |
| Bnip3 | BCL2/adenovirus E1B 19 kDa-interacting protein 3 | Tnfrsf11b | Tumor necrosis factor receptor superfamily, member 11b (osteoproteqerin) |
| Bok | Bcl-2-related ovarian killer protein | ||
| Card 10_predicted | Caspase recruitment domain family, member 10 (predicted) | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a |
| Card6_predicted | Caspase recruitment domain family, member 6 (predicted) | Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b |
| Casp 1 | Caspasa 1 | ||
| Casp 4 | Caspase 4, apoptosis-related cysteine peptidase | Tnfrsf5 | Tumor necrosis factor receptor superfamily, member 5 |
| Casp12 | Caspasa 12 | Tnfrsf6 | Tumor necrosis factor receptor superfamily, member 6 |
| Casp 14_predicted | Caspase 14 (predicted) | ||
| Casp2 | Caspase 2 | Tnfsf10 | Tumor necrosis factor (ligand) superfamily, member 10 |
| Casp3 | Caspase 3, apoptosis related cysteine protease | Tnfsf12 | Tumor necrosis factor ligand superfamily member 12 |
| Casp6 | Caspase 6 | ||
| Casp7 | Caspase 7 | Cd40lg | CD40 ligand |
| Casp8 | Caspase 8 | Faslg | Fas ligand (TNF superfamily, member 6) |
| Casp8ap2_predicted | Caspase 8 associated protein 2 (predicted) | Tp53 | Tumor protein p53 |
| Casp9 | Caspase 9 | Tradd | TNFRSF1A-associated via death domain |
| Cflar | CASP8 and FADD-like apoptosis regulator | Traf1 | Transcribed locus, strongly similar to NP_033447.2 Tnf receptor-associated factor 1 [Mus musculus] |
| Cidea_predicted | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A (predicted) | Traf2_predicted | Tnf receptor-associated factor 2 (predicted) |
| Cideb_Predicted | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector B (predicted) | Traf3_predicted | Tnf receptor-associated factor 3 (predicted) |
| Cradd | CASP2 and RIPK1 domain containing adaptor with death domain (predicted) | Traf4_predicted | Tnf receptor associated factor 4 (predicted) |
| Dad1 | Defender against cell death 1 | Trp53bp2_predicted | Transformation related protein 53 binding protein 2 (predicted) |
| Dapk1_predicted | Death associated protein kinase 1 (predicted) | Trp63 | Transformation related protein 63 |
| Dffa | DNA fragmentation factor, alpha subunit | Trp73_predicted | Transformation related protein 73 (predicted) |
Total RNA
Animals were anesthetized with CO2, decapitated and the cochleae quickly removed, opened and perfused through the round window with RNAlater (Qiagen). Then, the cochleae were carefully dissected and the sensory epithelia and the lateral walls were collected. The cochlear tissues from both cochleae of one animal were pooled to generate one sample. Each sample was run separately for the qRT-PCR analysis.
The hippocampal tissues were collected from 3 normal rats and used to compare the relative abundance of apoptosis gene in the brain versus the cochlea. The animals were sacrificed and the hippocampi from both the right and left sides of the brain were dissected out on a plate pretreated with the RNaseZap (Ambion, Inc, CA), an RNase inhibitor. The tissue from one animal was used to generate one sample for the qRT-PCR analysis; three hippocampal samples were run separately for the analysis.
Total RNA was extracted using an RNA extraction kit (RNeasy Mini Kit, Qiagen) as per manufacturer’s protocols. The extracted RNA solution was treated with RNase-Free DNase (Qiagen, Catalog# 79254) to remove DNA contamination. After the initial extraction, the RNA solution was cleaned up using RT2 qPCR-Grade RNA Isolation Kit (SuperArray, Catalog # PA-001). The quantity and integrity of total RNA were evaluated with a spectrophotometer (Beckman Coulter DU 640) or an Agilent Bioanalyzer 2001 (Agilent Technologies). The average amount of total RNA extracted from both cochleae from each rat was 0.44±0.8 μg. The total RNA extracted from the hippocampi of one animal was 1 μg.
qRT-PCR array analysis
The RT2 Profiler™ PCR Array (SuperArray Bioscience Corporation) was used to measure the expression levels of apoptosis-related genes. Upon completion of total RNA extraction and quality assessment, first strand cDNA was synthesized using oligo-dT primed reverse transcription supplied with the RT2 first strand kit (SuperArray). This kit contains genomic DNA elimination buffer and a built-in external RNA control. First strand cDNA synthesis was performed according to the manufacturer’s instructions. qRT-PCR was performed using the Bio-Rad MyiQ Single Color Real Time PCR System. The cDNA solution was mixed with SuperArray RT qPCR Master Mix and then loaded on to a 96-well array. The PCR reaction was run with a two-step cycling program. Upon completion of the PCR run, the Ct values were calculated.
Experimental procedures
The animals were sacrificed at 10 min, 4 h or 7 day post-exposure for assessment of cochlear pathologies and mRNA expression levels. The first two time points (10 min and 4 h post-exposure) represent the acute phase of cochlear pathogenesis, and the last time point (7 d post-exposure) represents the recovery phase of cochlear pathogenesis. Selection of these time points allowed us to assess the temporal patterns of gene expression changes at different phases of cochlear pathogenesis.
After completing the baseline hearing tests, the animals were randomly divided into one of three group with increasing post-exposure survival times (G-1:10-min; G-2: 4-h; or G-3: 7-day) or a control group (G-4). G-1, G-2, and G-3 were exposed to the 115-dB noise for 2 h. ABR measurements were obtained from animals in G-2 and G-3 groups just before the time of sacrifice at 4 h and 7 days post-exposure. Because of time constraints, animals in G-1 were sacrificed at 10 min post-exposure without collecting ABR data. The cochleae were processed for either histological evaluation or mRNA measurement as described above. The cochleae from G-4 controls were processed for assessment of hair cell morphology or assessment of mRNA levels using procedures identical to those used for the noise-exposed groups. Table 2 shows the numbers of animals used for each experimental group.
Table 2.
The numbers of animals used for each condition
| Group | Conditions | Pathological examination | Rt-PCR array |
|---|---|---|---|
| G-1 | 10-min post exposure | 5 | 6 |
| G-2 | 4-h post-exposure | 5 | 4 |
| G-3 | 7-day post-exposure | 5 | 5 |
| G-4 | Control | 6 | 7 |
Note: (1) For the mRNA analysis, three animals (one from the 10-min group, one from 7-d group and one from the control group) were excluded from the final analysis due to sample contamination. (2) Three additional rats were used for the mRNA analysis of apoptosis-related genes with the hippocampal tissues.
Data analyses
Average ABR thresholds at the three time points (pre-exposure, 4-h and 7-days post-exposure) and five test frequencies (5, 10, 20, 30 and 40 kHz) were compared using a two-way ANOVA. The average numbers of apoptotic cells quantified at the three time points (10 min, 4 h and 7 days post-exposure) were compared using a one-way ANOVA.
mRNA expression analyses were conducted for assessment of the expression patterns of apoptosis-related genes in the normal and the noise-traumatized cochleae. For the samples from the normal cochleae, the fold differences in the expression levels between the apoptotic genes and the housekeep genes were calculated to evaluate the relative abundance of of apoptosis-related genes under normal conditions. First, the expression levels of the three housekeeping genes (Hprt, Rpl13a and Actb) of a given sample were averaged. For each sample, the expression levels of the apoptosis-related genes were individually compared with the average expression level of the three housekeeping genes to determine the fold differences each apoptosis gene and the three housekeeping genes. Finally, the fold differences between each apoptotic gene and three housekeeping genes derived from the 6 samples were averaged. The fold differences reflect the relative expression levels of the apoptosis-related genes normalized to the housekeeping genes in the normal cochlea. When an apoptotic gene was expressed at a level greater than the expression level of the housekeeping genes, the value was defined as positive. When an apoptotic gene was expressed at a lower level, the value was expressed as negative.
To determine whether the pattern of apoptotic gene expression in normal cochlear tissues was similar to or different from that of normal brain tissue, the relative expression levels of the apoptotic genes were calculated for the hippocampal tissues using the same methods described above for cochlear tissues. A linear regression of the relative expression levels of the apoptotic genes for the cochlear tissues versus hippocampal tissues was plotted and computed using Prism 5. The genes that were outside ± 95% confidence interval of the linear regression line were considered to have substantial differences in expression levels between cochlear tissue and hippocampal tissue.
To evaluate the variation in mRNA expression levels across individual animals, the coefficient of variation (CV) of the expression levels for each gene was calculated using data of the 6 biological replications of the 6 control animals using a method that has previously been described (Livak and Schmittgen, 2001). Specifically, the expression level of each apoptotic gene was first normalized to the average expression level of three housekeeping genes. Then, the mean and standard deviation of the expression level of each gene relative to the housekeeping genes from the 6 samples were calculated. The CV is reported as a percentage and calculated from the mean and standard deviation of the relative expression level where: CV = (standard deviation/mean) × 100.
For analyses of noise-induced expression changes, a relative quantification method (Livak and Schmittgen, 2001, Stankovic and Corfas, 2003) was used to evaluate change in expression levels of mRNA following the exposure. The expression level of a given gene was first normalized to the average level of three housekeeping genes, Hprt, Actb and Rpl13a to generate the ΔCt of each apoptosis genes, where Ct represents the cycle threshold. Then, the ΔΔCt was calculated with the formula: ΔΔCt = ΔCt (noise group) − ΔCt (control group), where the control group was G-4 and the noise group was G-1, G-2 or G-3. The statistical analysis of the PCR data was accomplished with a web-based software package provided by SuperArray Bioscience Corporation with the p-value set at 0.05. Only fold-changes equal to or greater than two-fold were considered biologically significant.
Results
ABR threshold shifts
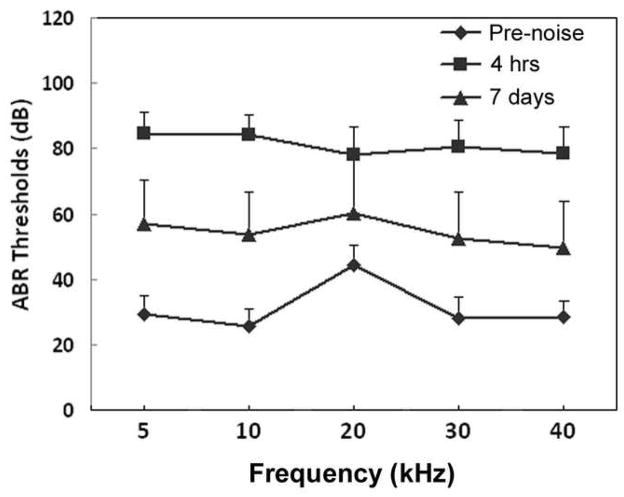
The average ABR thresholds measured pre-exposure and 4 h and 7-days post-exposure are presented in Fig. 1. The pre-exposure ABR thresholds varied from 30 to 45 dB between 5 and 40 kHz consistent with a previous study (Chen and Fechter, 2003). ABR thresholds at 4 h post-exposure were elevated significantly relative to pre-exposure thresholds (p<0.01). The average threshold shift across the frequency range was 50 ± 9.3 dB (mean ± SD). At 7-days post-exposure, thresholds had partially recovered leaving an average threshold shift across frequency of 24 ± 5.2 dB, which was also statistically different from baseline (p<0.01). These results indicate that the 115-dB noise induced a severe hearing loss across a broad range of frequencies at 4 h post-exposure which only partially recovered by 7 days post-exposure.
Figure 1.
Average ABR thresholds measured pre-exposure and 4 h and 7 days post-exposure. ABR thresholds were elevated 4 h post-exposure and partially recovered 7 days post-exposure. Values are means + standard deviation.
Apoptosis and hair cell loss
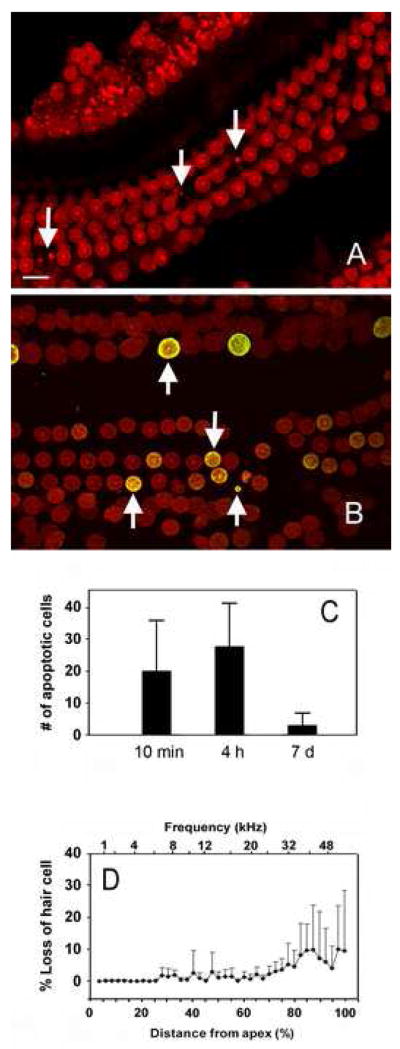
At 10-min and 4-h post-exposure, apoptotic cells, involving both inner hair cells and outer hair cells, were clear visible 20% to 60% of the distance from the apex of the cochlea. Fig. 2A is a typical example of hair cells with apoptotic features 10 min post-exposure. Arrows point to the outer hair cells with very condensed nuclei. Fig. 2B shows TUNEL staining in a cochlea with a focal hair cell lesion 4 h post-exposure. Note that hair cells having condensed nuclei also exhibit TUNEL fluorescence, confirming that these cells were dying by apoptosis. The numbers of apoptotic hair cells were quantified in cochleae examined 10 min and 4 h post-exposure. There were more apoptotic hair cells 4 h post-exposure than 10 min post-exposure; however, the difference was not statistically significant (Fig. 2C). At 7 days post-exposure, the number of apoptotic cells was markedly reduced presumably because hair cells that were previously in the process of dying were now missing (Fig. 2C). Indeed, the cochleogram measured 7 days post-exposure (Fig. 2D) shows that many outer hair cells were missing in the basal third of the cochlea.
Figure 2.
Analyses of cochlear pathology. 2A: A typical example of hair cells with apoptotic features 10 min post-exposure. The specimen was stained with propidium iodide. Arrows point to the outer hair cells with very condensed nuclei. 2B: TUNEL staining in a cochlea with a focal hair cell lesion 4 h post-exposure. Note that hair cells having condensed nuclei also exhibit TUNEL fluorescence, confirming that these cells were dying by apoptosis. 2C: Comparison of the numbers of apoptotic hair cells quantified at the three post-exposure times. Note that the number of apoptotic cells is significantly reduced at 7 days post-exposure. Values plotted are the mean (n=5 for each time point) + SD. 2D: The cochleogram showing the percentage of missing cells measured 7 days post-exposure. Missing hair cells are more evident in the middle and basal portions of the organ of Corti. Values plotted are the mean (n=5) + SD.
Housekeeping genes
The expression levels of five housekeeping genes, Rplp1, Hprt, Rpl13a, Ldha, and Actb, in the PCR array were evaluated to determine if they remained stable following the noise exposure. Table 3 shows the mean fold change in expression of each housekeeping gene at the three post-exposure times compared to expression levels in the control samples. Among the five genes, Hprt, Rpl13a and Actb were very stable, with average fold changes equal or less than +/− 0.37 fold at all three time points. The remaining two genes showed fold changes of 2.87 (Ldha) and 4.86 (Rplp1) at one time point. Therefore, we used the average of Hprt, Rpl13a and Actb to normalize the expression levels of apoptotic genes.
Table 3.
Fold changes in five housekeeping genes following exposure to the noise
| Symbol | 10 min | 4 hrs | 7 days | |||
|---|---|---|---|---|---|---|
| Fold change | p Value | Fold change | p Value | Fold change | p Value | |
| Rplp1 | 4.86 | (0.08) | −1.08 | (0.85) | 1.24 | (0.70) |
| Rpl13a | 1.25 | (0.09) | 1.08 | (0.40) | 1.07 | (0.64) |
| Ldha | −1.40 | (0.09) | −2.87 | (0.01) | 1.02 | (0.99) |
| Actb | −1.29 | (0.09) | −1.94 | (0.40) | −1.03 | (0.64) |
| Hprt | −1.04 | (0.87) | 1.02 | (0.90) | −1.04 | (0.80) |
Apoptosis genes in normal cochleae
The constitutive expression levels of apoptosis-related genes were evaluated in the normal, unexposed cochleae (6 biological replicates). Using the average expression level of the three stable housekeeping genes (Hprt, Rpl13a and Actb) as the reference, the relative expression levels of apoptosis-related genes were calculated. Table 4 presents the fold differences between the apoptotic genes and the housekeeping genes. The three most highly expressed genes of the 84 genes tested, Tnfrsf11b, Prdx2 and Mapk8ip, had expression levels similar to the mean of the three housekeeping genes (−2 < fold difference < +2). Interestingly, other highly-expressed genes (10 of the11 most highly genes, fold difference > −5.9 to +1.0) have the anti-apoptotic property (marked by asterisks). In contrast, many genes that had very low, or virtually no expression in the normal cochleae are pro-apoptotic such as Dffb, Bcl2l11, Prlr, Trp63, Lta, Casp14-predicted, and Trp73-predicted. Table 4 also shows the CV values of the apoptosis-related genes. Note that the CV values vary among the genes. Since each measurement was based on data from a single animal, the CV values may be dominated by between-subject differences in expression levels of these genes.
Table 4.
Fold differences between apoptosis-related genes and housekeeping genes in normal cochleae
| Fold difference | % CV values | Fold difference | % CV values | ||
|---|---|---|---|---|---|
| Bcl-2 Family: | Traf Family: | ||||
| Bad (Bbc2) | −24.8 | 95.52 | Traf1 | −310.1 | 26.20 |
| *Bag1 | −4.4 | 48.09 | Traf2 | −22.7 | 21.47 |
| Bak1 | −45.4 | 71.98 | Traf3 | −31.2 | 49.65 |
| Bax | −9.9 | 32.12 | Traf4 | −62.4 | 43.53 |
| Bcl2 | −14.7 | 26.06 | Card Family | ||
| Bcl2a1 | −647.3 | 47.39 | Apaf1 | −198.1 | 78.72 |
| *Bcl2l1 | −2.3 | 35.79 | Bcl10 | −26.9 | 36.69 |
| Bcl2l2 | −19.4 | 71.70 | Card6 | −11.7 | 20.65 |
| Bcl2l11 | −2421.5 | 92.41 | Card10 | −35.3 | 42.48 |
| Bclaf1 | −7.3 | 26.89 | Cradd | −66.5 | 27.21 |
| Bld | −31 | 48.97 | Nol3 | −73.3 | 59.14 |
| Bid3 (Hrk) | −89.7 | 75.42 | Pycard (Asc) | −26.9 | 34.32 |
| Blk | −30.8 | 47.35 | Ripk2 | −13.9 | 38.52 |
| Bnip1 | −23.1 | 25.52 | Death Domain Family: | ||
| Bnlp2 | −18.3 | 25.01 | Dapk1 | −35.2 | 39.08 |
| *Bnlp3 | −3 | 22.05 | Fadd | −20.3 | 16.76 |
| Bok | −19.6 | 51.55 | Tradd | −30.4 | 33.62 |
| *Mcl1 | −4.6 | 34.10 | CIDE Domain Family: | ||
| TNF Ligand and Receptor Family: | Cidea | −41.5 | 46.15 | ||
| Lta | −4973.3 | 136.81 | Cideb | −353.8 | 95.87 |
| Tnf | −495.7 | 59.90 | Dffa | −58.6 | 30.70 |
| Tnfsf10 | −67.5 | 29.48 | Dffb | −2288.2 | 55.08 |
| Tnfs12 | −26.3 | 50.94 | p53 and DNA Damage-Induced Apeptosis: | ||
| Cd40lg (Tnfsf5) | −8974.8 | 125.23 | Gadd45a | −.30.3 | 82.30 |
| Faslg (Tnfsf6) | −855.1 | 54.81 | Tp53 (p53) | −29 | 43.75 |
| Ltbr | −13.4 | 45.01 | Trp53bp2 | −35.4 | 58.62 |
| Tnfrsf10b | −564.2 | 57.95 | Trp63 | −2906.4 | 76.92 |
| Tnfrsf11b | 1.6 | 17.92 | TAP Family: | ||
| Tnfrsf1a | −19.1 | 33.24 | Birc1b | −1093.7 | 50.56 |
| Tnfrsf1b | −354.2 | 28.20 | Birc3 | −502 | 74.13 |
| Tnfrsf5 | −489.4 | 27.17 | *Birc4 | −5.9 | 45.48 |
| Tnfrsf6 | −826 | 35.40 | Birc5 | −248.7 | 26.45 |
| Caspase Family: | Others with anti-apoptotic function: | ||||
| Casp1 | −197.9 | 68.78 | Trp73 | −12245.8 | 68.13 |
| Casp2 | −34.6 | 32.67 | *Apl5 | −4.2 | 21.55 |
| Casp3 | −14 | 27.27 | Aven | −29.3 | 42.03 |
| Casp6 | −16.1 | 13.36 | *Dad1 | −4.2 | 32.11 |
| Casp7 | −238.3 | 50.61 | Falm | −45.8 | 33.88 |
| Casp8 | −129.6 | 39.86 | ll10 | −1619.9 | 79.89 |
| Casp8ap2 | −296.1 | 102.86 | Lhx4 | −1335.7 | 100.69 |
| Casp9 | −24.4 | 17.93 | *Mapk8lp | −1.9 | 38.97 |
| Casp4 (Casp11) | −8.5 | 39.47 | Nfkb1 | −10.5 | 38.41 |
| Casp12 | −10.5 | 38.26 | Polb | −74.5 | 50.60 |
| Casp14 | −12245.8 | 43.65 | *Prdx2 | 1 | 30.36 |
| Death Effecter Domain Family: | Prlr | −2804.1 | 50.63 | ||
| *Cflar | −5 | 16.80 | Prok2 | −2873 | 60.98 |
| Sphk2 | −10.5 | 33.71 | |||
Note: Asterisks point to 1he 10 antl-apoptolic genes
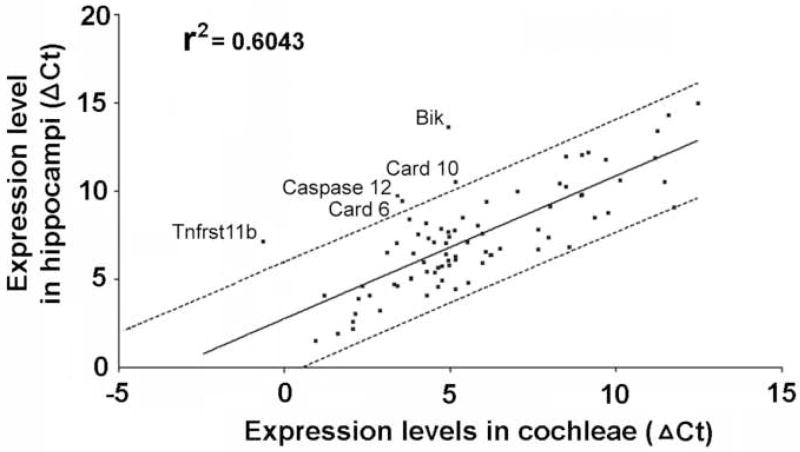
To determine whether the pattern of mRNA expression in the normal cochlea is organ-specific, we compared the expression levels of apoptosis-related genes in the hippocampus with those in the cochlea using the same procedures. Fig. 3 plots the relative expression level of each gene in the hippocampus versus that in the cochlea. The data were fit with a linear regression line; the dashed line shows the 95% confidence interval. Many of the genes have a similar level of expression in both the cochlea and hippocampus and therefore lie within the 95% confidence interval. However, five genes are outside the 95% confidence interval and therefore show a significant difference in expression level between the cochlea and hippocampus. The five genes with significantly lower Ct values (higher expression levels) in the cochlea versus the hippocampus are Bik, Caspase 12, Card 10, Card 6 and Tnfrst11b.
Figure 3.
Scatterplot shows the relative expression of each gene in the hippocampi versus the cochlea. Data fit with linear regression (solid line); dashed line shows 95% confidence interval. The expression levels of apoptotic genes were normalized to the housekeeping genes. Five genes above the 95% confidence interval were: Bik, Caspase 12, Card 10, Card 6 and Tnfrst11b; these five genes had high expression levels in the cochlea (lower Ct values) than in the hippocampus.
Noise-induced gene expression changes
Differential expression of the apoptotic genes was examined at 10 min, 4 h and 7 days post-exposure. The first two time points represent the acute phase of cochlear pathogenesis and the last time point represents the late recovery phase of cochlear pathogenesis.
10 min post-exposure
At 10 min post-exposure, 12 genes were significantly (p<0.05) downregulated with fold decreases ranging from 52.9 (Api5) to 2.1 (Tnfrsf6) (see Table 5). Api5 not only had the largest fold decrease, but also was highly expressed in the normal cochlea. Although Tnfrsf6 was expressed at very low levels in the normal cochlea it exhibited a highly significant, 2.1 fold decrease at the 10 minute time point. Among the downregulated genes, eight are classified as pro-apoptotic and four genes are classified as anti-apoptotic. At this time point, only one pro-apoptotic gene, Traf4, was upregulated.
Table 5.
Differential ex press in of apoplosis-relaled genes
| 10 min (fold change, p value) | 4 hr (fold change, p value) | 7 day (fold change, p value) | ||
|---|---|---|---|---|
| Unregulated | *Traf4 (2.6. 0.03) | Birc3 (17.54, 0.0001) | ||
| *Gadd45a (5.56. 0.0266) | ||||
| Mcl1 (3.15. 0.0014) | ||||
| Prok2 (5.66. 0.0019) | ||||
| Tnfrsf1b (2.72. 0.0008) | ||||
| Tnfrsf1a (5.90. 0.0001) | ||||
| Tnfrsf5 (2.28. 0.0024) | ||||
| *Traf4 (2.21. 0.0159) | ||||
|
| ||||
| Downregulated | Api5 (−52.9. 0.040) | Casp9 (−6.25. 0.04) | ||
| *Bok (−13.0. 0.003) | Dffb (−4.60. 0.04) | *Card 10 (−2.01. 0.0339) | *Bct2l2 (−30.29. 0.02) | |
| *Bcl2l2 (−11.0. 0.020) | Bik (−3.66. 0.025) | Casp2 (−2.04. 0.0104) | *Bok (−60.34. 0.0066) | |
| Bclaf1 (−7.54. 0.049) | Prlr (−3.60. 0.03) | Casp9 (−2.23. 0.0006) | *Card10 (−15.50. 0.03) | |
| Casp1 (−7.2. 0.040) | Cidea (−2.90. 0.04) | Tnfsf 10(−2.91. 0.0090) | Gadd45a (−24.20. 0.05) | |
| *Card10(−6.3. 0.042) | Tnfrsf6(−2.10. 0.01) | Pycard (−4.10. 0.04) | ||
Note: asterisks indicate the genes that are altered at multiple time points.
4 h post-exposure
At 4 h post-exposure, eight genes were significantly (p<0.05) upregulated (Table 5). Among these upregulated genes, three (Tnfrsf1a, Tnfrsf1b, Tnfrst5) belong to the tumor necrosis factor receptor superfamily and are pro-apoptotic. Another, pro-apoptotic gene, Traf4, that was upregulated at 10min post exposure (2.6 fold) remained at an elevated level at the 4 h time point. Three genes, Bir3, Mcl1 and Prok2, have anti-apoptotic properties. Gadd45a, the remaining gene that was upregulated at 4 h, is a p53 target gene which possesses both pro- and anti-apoptotic properties. Also, at this time point, four genes, Card 10, Casp2, Casp9 and Tnfsf 10, were significantly downregulated. All the downregulated genes are classified as pro-apoptotic.
7 days post–exposure
At 7 day post-exposure, all of the apoptotic genes that were upregulated at the 10-min or 4-h time points had returned to their pre-exposure level and none of the 84 genes tested were expressed above control levels. Five genes were significantly downregulated (Table 5). Four were pro-apoptotic (Bok, Card10, Gadd45 and Pycard), and one was anti-apoptotic (Bcl2l2).
Genes altered at multiple time points
Five genes showed significant changes in expression at two or more time points. Upregulation of Traf4 was observed at 10 min and 4 h post-exposure. Bcl2l2 and Bok were downregulated at 10 min and 7 days post-exposure. Card 10 was downregulated at all three time points. Gadd45 exhibited a biphasic change consisting of an initial rise and a subsequent fall at 4 h and 7 days post-exposure.
Discussion
We exposed rats for 2 h to broadband noise at 115 dB SPL. The exposure caused a 40–60 dB hearing loss over a wide frequency range 4 h post-exposure. Morphological assessment of the cochlea at this time revealed a small portion (approximately 1% of the total hair cell population) of hair cells with apoptotic features, specifically condensed nuclei and TUNEL positive staining. At 7 days post-exposure, the hearing loss had decreased to 20–30 dB. Few hair cells with apoptotic features were seen at this time suggesting that hair cells with apoptotic features had either recovered or completely degenerated. The mean cytocochleograms measured 7 day post-exposure showed a mean hair cell loss of 10% or less in the base of the cochlea (80–100% distance from apex). Although the broadband noise caused a broad hearing loss, hair cell loss was largely confined to the base of the cochlea. The basal turn loss was not unexpected, possibly due to less antioxidant capacity of cells in this region (Sha et al., 2001). It is important to note that relatively few hair cells were missing at 7 day post-exposure; this means that the samples of mRNA harvested 7 days post-exposure were not biased by massive loss of cells of a particular type or region of the cochlea.
We examined the changes in expression of 84 apoptosis-related genes in the organ of Corti and lateral wall of the rat cochlea using qRTPCR at 10 min. 4 h and 7 days post-exposure. A total of 22 genes among the 84 examined increased and/or decreased significantly following the noise exposure. These genes belong to several apoptotic gene families and participate in multiple signaling pathways each related to cell survival or cell death. Many have not been implicated in noise-induced cochlear pathogenesis before. In addition to the induced changes, a strong constitutive expression of apoptosis-related genes was present in normal cochleae.
Noise-induced change in apoptotic genes
Time-dependent changes in mRNA expression levels were observed in the noise-traumatized cochleae. The upregulation of apoptotic genes occurred primarily at 4 h post-exposure. This finding is associated with severe, widespread hearing loss at this time. The upregulated genes can be grouped into several apoptotic families, specifically the tumor necrosis factor receptor (TNFR) family, the B-cell leukemia/lymphoma 2 (Bcl2) family, the tumor necrosis factor receptor associated factor (TRAF) family and the inhibitor of apoptosis protein (IAP) family. The TNFR family members that were upregulated included Tnfrsf1a, Tnfrst1b and Tnfrst5. These TNFR members participate in a variety of biological activities including apoptotic stress response (Locksley et al., 2001, Liu, 2005). In certain cell types, Tnfrst1a mediates cell death, whereas Tnfrst1b acts to enhance TNFR1-mediated cell death (Chan et al., 2000). A common ligand for these two TNF receptors is TNF-α. TNF-α has been implicated in noise-induced cochlear damage. Fujioka et al. (Fujioka et al., 2006) reported a transient upregulation of TNF-α within 6 h post-exposure, consistent with the time frame showing upregulation of its receptor mRNA level observed in the current study. The involvement of both TNF-α and its receptors suggest that TNFs are important players in the initiation of acute cochlear apoptosis.
One of the signaling pathways downstream of TNFRs are TRAFs, a class of intracellular adaptor protein (Inoue et al., 2000). TRAF4, which was upregulated 10 min and 4 h post-exposure, is involved in the mitogen-activated protein kinase (MAPK) pathway which activates JNK (Xu et al., 2002, Abell and Johnson, 2005). The JNK pathway has also been implicated in noise-induced apoptosis in the cochlea (Pirvola et al., 2000). The pharmacological inhibition of the JNK pathway protects against noise induced-hearing loss (Wang et al., 2003, Coleman et al., 2007). Another upregulated gene that interacts with TNFR members is Birc3, a member of the IAP family. Birc3 encodes a protein that inhibits apoptosis in various cell types. The mechanism behind Birc3 inhibition of apoptosis is associated with Tnfrsf 1a and Tnfrsf 1b which interfere with activation of IL-1beta- converting enzyme (ICE) -like proteases (Rothe et al., 1995).
The upregulated Bcl2 family member, Mcl1, is localized predominantly in the mitochondrial membranes where it regulates the permeability of cytochrome c and hence the downstream activation of the apoptosome complex that initiates a caspase cascade leading to apoptotic phenotypes (Zhou et al., 1997, Nijhawan et al., 2003). Two mRNA splice variants of Mcl1 have been described in human tissues: a full-length form (Mcl1-L), which has homology in the C-terminal transmembrane region to Bcl-2 and confers an anti-apoptotic function, and a short form (Mcl1-S) with a pro-apoptotic function (Bae et al., 2000, Bingle et al., 2000). Although it is not clear which variant is involved in noise-induced apoptotic activity, the strong upregulation of Mcl1 (17.5 folds) suggests an important role in cochlear pathogenesis.
In addition to genes that are related to TNF and Bcl2 families, Gadd45a and Prok2 were significantly upregulated. Gadd45a belongs to a family of three Gadd 45 genes which possess apoptotic properties (Takekawa and Saito, 1998, Sheikh et al., 2000). However, several studies show that this gene also participates in cell survival (Gupta et al., 2005). Gadd45a is a p53 target gene. We have shown that p53 is upregulated in hair cells and supporting cells following acoustic trauma and cisplatin ototoxicity (Zhang et al., 2003, A.R. Fetoni, et.al., unpublished observation). Gadd45a also interacts with the JNK pathway (Harkin et al., 1999). Although there is a clear correlation between Gadd45a expression and apoptosis, it is unclear whether Gadd45a expression is a cause or an effect of this complex signaling program. Prok2, which shows little expression in the normal cochlea, was upregulated 5.6 fold post exposure. Although Prok2 has anti-apoptotic properties (Ngan et al., 2007), the action of this gene in the cochlea is unknown.
In noise-induced hair cell apoptosis, activation of both caspases-8 and -9 has been observed after noise exposure (Nicotera et al., 2003) suggesting the involvement of both the membrane and mitochondrial pathways in noise-induced cell death. The involvement of both membrane and mitochondrial cell death pathways is consistent with the current observation showing upregulation of genes involved in both the membrane death pathway involving TNF genes and the mitochondrial pathway related to the Bcl2 gene family. Our results shows that a greater number of genes involved in noise-induced hearing loss were related to the membrane pathway than the mitochondrial pathway suggesting a differential contribution of these two pathways to noise induce cell death. However, this interpretation needs to be tempered by the fact that only a subset of gene involved in cell death are represented on the arrays used in this study.
In contrast to the marked upregulation of apoptotic genes at the 4-h time point, the 10 min and 7 days time points were characterized by downregulation of apoptosis-related genes. At 10 min post-exposure, two-thirds of the 12 downregulated genes were classified as pro-apoptotic. This suggests that the initial global response of the cochlea to noise may be to promote cell survival by suppressing the apoptotic response. However, as traumatic events unfold or accelerate the global response of the cochlea shifts predominantly to apoptotic at 4-h post-exposure. However, as the apoptotic cells die off, pro-apoptotic signaling would be expected to decline. This is consistent with previous noise studies showing that hair cell loss peaks a few days post-exposure and drops off rapidly thereafter (Hu et al., 2002b, Yang et al., 2004). Thus, the 7-day time point represents the recovery phase of cochlear pathogenesis. No significant upregulation of apoptosis-related genes was found and several apoptosis related genes were downregulated. This result is consistent with our TUNEL observations showing a lack of apoptotic activity at this time. A previous study has shown that the hearing sensitivity in Sprague Dawley rats became stable by 7 days following exposure to an octave band noise at 124 dB SPL for 2 h (Fujioka et al., 2006). Taken together, these observations indicate that the apoptotic response is most active in the early phase of cochlear pathogenesis.
It is important to note that we are not suggesting that regulation of apoptosis genes is confined to the period of temporary threshold changes sampled in this study. It will be especially interesting to analyzed changes in gene regulation that occur as the cochlea shifts from a state of temporary to permanent threshold shift. It is possible to speculate that there may in fact be a shift toward signals contributing more and more toward extrinsic apoptotic pathways as the lesion on the organ of Corti grows during this period.
The methodology used in the current study for the mRNA analyses is unable to define the site of changes in mRNA expression within specific groups of cells or regions on the cochlea. We are cognizant of the fact that it is important to identify changes in gene expression in particular cell types within the cochlea or indeed within a single hair cell, neuron or supporting cell. Therefore, future investigation on the spatial pattern of apoptotic gene expression in the cochlea is warranted.
Apoptotic gene expression in normal cochleae
The current study revealed strong constitutive expression of certain apoptosis-related genes in normal cochleae. Many of these highly-expressed genes possess anti-apoptotic properties (10 genes, marked by asterisks in Table 4). Because sound is always present in the environment, the hair cells, supporting cells and neurons are continually being activated resulting in a high level of succinate dehydrogenase, an enzyme involved in aerobic metabolism, in hair cells. In order to suppress cell death from oxidative stress, it is possible that these anti-apoptotic genes are normally expressed at high levels to maintain cochlear homeostasis. Surprisingly, the normal cochlea also exhibits strong expression of Tnfrsf11b, a pro-apoptotic gene. In addition, several pro-apoptotic genes (Bik, Caspase 12, Card 10, Card 6 and Tnfrst11b) show higher expression levels (lower Ct values) in the cochlea than in the hippocampus. Although the biological roles of these pro-apoptotic genes in preservation of the cochlear homeostasis are not clear, we suspect that the high expression level may allow for rapid induction of apoptosis. Our previous study has shown that exposure to intense impulse noise activates cochlear apoptosis a few minutes after the beginning of the noise exposure (Hu et al., 2006). This rapid onset of cochlear apoptosis may be due to the involvement of the constitutively expressed apoptotic molecules. It is important to note that the confirmation of the constitutive expression of apoptotic genes in the normal cochlea requires the analyses of the protein expression levels and functions of these genes. Addressing this question warrants future quantitative analyses of protein expression levels.
Another interesting finding of the current study is the variation in expression levels of apoptosis-related genes across individual animals. Some genes are expressed consistent levels across subjects (small CV values), whereas others are quite variable. It is possible that the variation in gene expression simply reflects random variation in the measurement technique. To assess the technical repeatability of the array method, we ran several repetitions with a single sample in a previous observation using the same type of the apoptosis PCR array from the same company (unpublished observation). The results showed a relatively-consistent expression level across individual runs, indicating that the PCR arrays results are reliable. Another intriguing possibility for the large CV values is that the variability reflects real differences in expression of these apoptosis genes and that these differences make some animals more or less susceptible to noise induced cochlear damage. Moreover, some genes may show significant day to day variation whereas others are maintained at a relatively stable level. A better understanding how the level of these constitutively expressed apoptotic gene contributes to noise-induced hearing loss and apoptosis warrants further study.
Acknowledgments
The research was supported by R01DC00909101, R01DC006630, R03DC006181-03 and New Faculty Startup funds from College of Arts and Sciences, University at Buffalo.
List of abbreviations
- ABR
Auditory Brainstem Responses
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- CV
The coefficient of variation
- ANOVA
Analysis of variance
- TNFR
The tumor necrosis factor receptor
- Bcl2
The B-cell leukemia/lymphoma 2
- TRAF
The tumor necrosis factor receptor associated factor
- IAP
The inhibitor of apoptosis protein
- MAPK
The mitogen-activated protein kinase
- ICE
IL-1beta- converting enzyme
Footnotes
Presented in part at the 38th annual meeting of the Society for Neuroscience, held Nov. 15 to 19, 2008 in Washington, DC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abell AN, Johnson GL. MEKK4 is an effector of the embryonic TRAF4 for JNK activation. J Biol Chem. 2005;280:35793–35796. doi: 10.1074/jbc.C500260200. [DOI] [PubMed] [Google Scholar]
- Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- Bohne BA, Harding GW, Lee SC. Death pathways in noise-damaged outer hair cells. Hear Res. 2007;223:61–70. doi: 10.1016/j.heares.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Chan KF, Siegel MR, Lenardo JM. Signaling by the TNF receptor superfamily and T cell homeostasis. Immunity. 2000;13:419–422. doi: 10.1016/s1074-7613(00)00041-8. [DOI] [PubMed] [Google Scholar]
- Chen GD, Fechter LD. The relationship between noise-induced hearing loss and hair cell loss in rats. Hear Res. 2003;177:81–90. doi: 10.1016/s0378-5955(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gong TW, Kanicki A, Altschuler RA, Lomax MI. Noise overstimulation induces immediate early genes in the rat cochlea. Brain Res Mol Brain Res. 2004;130:134–148. doi: 10.1016/j.molbrainres.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Coleman JK, Littlesunday C, Jackson R, Meyer T. AM-111 protects against permanent hearing loss from impulse noise trauma. Hear Res. 2007;226:70–78. doi: 10.1016/j.heares.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Kanzaki S, Okano HJ, Masuda M, Ogawa K, Okano H. Proinflammatory cytokines expression in noise-induced damaged cochlea. J Neurosci Res. 2006;83:575–583. doi: 10.1002/jnr.20764. [DOI] [PubMed] [Google Scholar]
- Gupta M, Gupta SK, Balliet AG, Hollander MC, Fornace AJ, Hoffman B, Liebermann DA. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene. 2005;24:7170–7179. doi: 10.1038/sj.onc.1208847. [DOI] [PubMed] [Google Scholar]
- Han W, Shi X, Nuttall AL. AIF and endoG translocation in noise exposure induced hair cell death. Hear Res. 2006;211:85–95. doi: 10.1016/j.heares.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, Haber DA. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- Hu BH, Guo W, Wang PY, Henderson D, Jiang SC. Intense noise-induced apoptosis in hair cells of guinea pig cochleae. Acta Otolaryngol. 2000;120:19–24. [PubMed] [Google Scholar]
- Hu BH, Henderson D, Nicotera TM. F-actin cleavage in apoptotic outer hair cells in chinchilla cochleas exposed to intense noise. Hear Res. 2002a;172:1–9. doi: 10.1016/s0378-5955(01)00361-6. [DOI] [PubMed] [Google Scholar]
- Hu BH, Henderson D, Nicotera TM. Involvement of apoptosis in progression of cochlear lesion following exposure to intense noise. Hear Res. 2002b;166:62–71. doi: 10.1016/s0378-5955(02)00286-1. [DOI] [PubMed] [Google Scholar]
- Hu BH, Henderson D, Nicotera TM. Extremely rapid induction of outer hair cell apoptosis in the chinchilla cochlea following exposure to impulse noise. Hear Res. 2006;211:16–25. doi: 10.1016/j.heares.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Inoue J, Ishida T, Tsukamoto N, Kobayashi N, Naito A, Azuma S, Yamamoto T. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp Cell Res. 2000;254:14–24. doi: 10.1006/excr.1999.4733. [DOI] [PubMed] [Google Scholar]
- Kirkegaard M, Murai N, Risling M, Suneson A, Jarlebark L, Ulfendahl M. Differential gene expression in the rat cochlea after exposure to impulse noise. Neuroscience. 2006;142:425–435. doi: 10.1016/j.neuroscience.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Liu ZG. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27. doi: 10.1038/sj.cr.7290259. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Matsunobu T, Ogita K, Schacht J. Modulation of activator protein 1/DNA binding activity by acoustic overstimulation in the guinea-pig cochlea. Neuroscience. 2004;123:1037–1043. doi: 10.1016/j.neuroscience.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Muller M. Frequency representation in the rat cochlea. Hear Res. 1991;51:247–254. doi: 10.1016/0378-5955(91)90041-7. [DOI] [PubMed] [Google Scholar]
- Ngan ES, Sit FY, Lee K, Miao X, Yuan Z, Wang W, Nicholls JM, Wong KK, Garcia-Barcelo M, Lui VC, Tam PK. Implications of endocrine gland-derived vascular endothelial growth factor/prokineticin-1 signaling in human neuroblastoma progression. Clin Cancer Res. 2007;13:868–875. doi: 10.1158/1078-0432.CCR-06-2176. [DOI] [PubMed] [Google Scholar]
- Nicotera T, Henderson D, Hu BH, Zheng XY. Noise exposure and mechanisms of hair cell death. In: Henderson D, et al., editors. Noise Induced Hearing Loss: Basic Mechanisms, Prevention and Control. London: Noise Research Network Publications; 2001. pp. 99–117. [Google Scholar]
- Nicotera TM, Hu BH, Henderson D. The caspase pathway in noise-induced apoptosis of the chinchilla cochlea. J Assoc Res Otolaryngol. 2003;4:466–477. doi: 10.1007/s10162-002-3038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Shao R, Canlon B. Suppression of apoptosis occurs in the cochlea by sound conditioning. Neuroreport. 2003;14:1025–1029. doi: 10.1097/01.wnr.0000070830.57864.32. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Xing-Qun L, Virkkala J, Saarma M, Murakata C, Camoratto AM, Walton KM, Ylikoski J. Rescue of hearing, auditory hair cells, and neurons by CEP-1347/KT7515, an inhibitor of c-Jun N-terminal kinase activation. J Neurosci. 2000;20:43–50. doi: 10.1523/JNEUROSCI.20-01-00043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Sha SH, Taylor R, Forge A, Schacht J. Differential vulnerability of basal and apical hair cells is based on intrinsic susceptibility to free radicals. Hear Res. 2001;155:1–8. doi: 10.1016/s0378-5955(01)00224-6. [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Hollander MC, Fornance AJ., Jr Role of Gadd45 in apoptosis. Biochem Pharmacol. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- Shibuya H, Kato Y, Saito M, Isobe T, Tsuboi R, Koga M, Toyota H, Mizuguchi J. Induction of apoptosis and/or necrosis following exposure to antitumour agents in a melanoma cell line, probably through modulation of Bcl-2 family proteins. Melanoma Res. 2003;13:457–464. doi: 10.1097/00008390-200310000-00004. [DOI] [PubMed] [Google Scholar]
- Stankovic KM, Corfas G. Real-time quantitative RT-PCR for low-abundance transcripts in the inner ear: analysis of neurotrophic factor expression. Hear Res. 2003;185:97–108. doi: 10.1016/s0378-5955(03)00298-3. [DOI] [PubMed] [Google Scholar]
- Taggart RT, McFadden SL, Ding DL, Henderson D, Jin X, Sun W, Salvi R. Gene Expression Changes in Chinchilla Cochlea from Noise-Induced Temporary Threshold Shift. Noise Health. 2001;3:1–18. [PubMed] [Google Scholar]
- Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- Vicente-Torres MA, Schacht J. A BAD link to mitochondrial cell death in the cochlea of mice with noise-induced hearing loss. J Neurosci Res. 2006;83:1564–1572. doi: 10.1002/jnr.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dib M, Lenoir M, Vago P, Eybalin M, Hameg A, Pujol R, Puel J. Riluzole rescues cochlear sensory cells from acoustic trauma in the guinea-pig. Neuroscience. 2002;111:635–648. doi: 10.1016/s0306-4522(02)00004-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Van De Water TR, Bonny C, de Ribaupierre F, Puel JL, Zine A. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J Neurosci. 2003;23:8596–8607. doi: 10.1523/JNEUROSCI.23-24-08596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YC, Wu RF, Gu Y, Yang YS, Yang MC, Nwariaku FE, Terada LS. Involvement of TRAF4 in oxidative activation of c-Jun N-terminal kinase. J Biol Chem. 2002;277:28051–28057. doi: 10.1074/jbc.M202665200. [DOI] [PubMed] [Google Scholar]
- Yamashita D, Miller JM, Jiang HY, Minami SB, Schacht J. AIF and EndoG in noise-induced hearing loss. Neuroreport. 2004;15:2719–2722. [PubMed] [Google Scholar]
- Yamashita D, Minami SB, Kanzaki S, Ogawa K, Miller JM. Bcl-2 genes regulate noise-induced hearing loss. J Neurosci Res. 2008;86:920–928. doi: 10.1002/jnr.21533. [DOI] [PubMed] [Google Scholar]
- Yang WP, Henderson D, Hu BH, Nicotera TM. Quantitative analysis of apoptotic and necrotic outer hair cells after exposure to different levels of continuous noise. Hear Res. 2004;196:69–76. doi: 10.1016/j.heares.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Ylikoski J, Xing-Qun L, Virkkala J, Pirvola U. Blockade of c-Jun N-terminal kinase pathway attenuates gentamicin-induced cochlear and vestibular hair cell death. Hear Res. 2002;166:33–43. doi: 10.1016/s0378-5955(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liu W, Ding D, Salvi R. Pifithrin-alpha suppresses p53 and protects cochlear and vestibular hair cells from cisplatin-induced apoptosis. Neuroscience. 2003;120:191–205. doi: 10.1016/s0306-4522(03)00286-0. [DOI] [PubMed] [Google Scholar]
- Zhou P, Qian L, Kozopas KM, Craig RW. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood. 1997;89:630–643. [PubMed] [Google Scholar]