Abstract
Interferon-tau (IFNT), which plays a major role in maternal recognition of pregnancy in cattle, is transcribed from multiple genes. Moreover, there are at least 12 cDNA variants, many presumably allelic. Although the IFNT locus is autosomal, Day 8 female blastocysts produced approximately twice as much antiviral activity as males. The questions addressed here are whether male and female blastocysts differed in the kind and number of IFNT they expressed, and whether this pattern changed over development. Day 8, in vitro-produced blastocysts were bisected, and one half of each was sexed by PCR. Demi-embryos (n = 64) were grouped according to whether they were male or female, to provide two pools of each sex. Individual cDNA were sequenced after RT-PCR amplification and shot-gun cloning to provide comparisons between male and female blastocysts, elongating conceptuses of various developmental ages (Days 14–19), and a female trophoblast cell line (CT-1). A total of 376 cDNA clones were sequenced. Six additional cDNA were identified, in addition to the forms described earlier. There were no differences between male and female blastocysts (P = 0.54), and between blastocysts and a trophoblast model system (CT-1 cells; P = 0.24) in the IFNT transcripts expressed, indicating that sexual dimorphism wass not correlated with particular IFNT variants. There were differences in variant frequencies (P < 0.001) among conceptuses of different age, although two, representing boIFN1a and boIFN3c, predominated throughout development. Notably, no alteration in overall IFNT variant diversity was detected in CT-1 cells over time (P = 0.124).
Keywords: Interferon-tau, IFN-tau, Pregnancy, Maternal recognition of pregnancy, Cattle
1. Introduction
Interferon-tau (IFNT) is a Type I IFN under unique transcriptional control that limits its expression to ruminant trophoblasts prior to implantation [1,2]. A major, but probably not the sole role of the cytokine, is to mute the pulsatile release of prostaglandin-F2α (PFG2α) from maternal uterine endometrium, thereby, blocking luteolysis [3]. Interestingly, at least 12 distinct IFNT cDNA have been identified from bovine conceptuses, despite the apparent singular responsibility of the cytokine in pregnancy maintenance [4,5]. It is unclear how many of these cDNA represent allelic forms of a limited number of genes, since IFNT number has never been fully clarified for any ruminant species, although several of the bovine genes are localized on chromosome 8, in close association with genes for IFN-omega (IFNW) and IFN-alpha (IFNA) [6]. Perhaps since IFNT diverged from the IFNW (~36 million years ago [7,8]), gene duplication has created the opportunity for new IFNT variants to be established and for evolutionary selection to act on these variants to create progressively more potent signaling molecules. Conversely, based on recent microarray data, IFNT regulates a large number of genes in the endometrium, including ones required for such vital functions as angiogenesis, growth factor production, and matrix deposition [9], raising the possibility that different variants have acquired somewhat specialized roles, with time of onset of expression reflecting this specialization. A third potential reason for why there are so many variants is that multiple genes are required for large-scale production of IFNT at a critical time during early pregnancy.
Furthermore, IFNT could play a role in sex selection of progeny, as Day 8 female bovine blastocysts (Day 0 = fertilization), produced approximately twice as much antiviral activity as males [10,11]. As this activity arises solely from the production of IFNT [12], females may possess a survival advantage relative to males, in terms of triggering maternal responses. This female advantage has been envisaged to be offset by the better development rate of male conceptuses, as glucose concentrations are raised [11,13,14]. Indeed, increased energy intake of the mother in the period around conception has been proposed as providing a uterine environment enriched in glucose that favors males over females for many mammalian species [15]. The basis for the excess IFNT production by female blastocysts remains an enigma [16]. Females may produce either greater quantities of IFNT or more potent forms of the cytokine, particularly as mixed populations of blastocysts are known to express numerous IFNT variants [5].
This paper addresses whether male and female bovine conceptuses differed in the number and kind of IFNT transcripts they expressed and whether transcript quality changed over development. For comparison with whole conceptuses, we also surveyed the IFNT transcript variants found in CT-1 cells, a continuously proliferating bovine cell line [17], which can serve as a useful model for examining IFNT expression [18], to verify whether this model system mimicked the sex and developmental age of the trophoblast cells from which it was derived. Although CT-1 cells are known to secrete IFNT and other pregnancy-associated proteins over multiple passages, no systematic analysis of the system has been previously made at the level of individual IFNT variants. Slight shifts in either the developmental age of the CT-1 cells, induced by time or minor changes in culture conditions, could be evident through changes in the IFNT variants expressed that would otherwise be missed.
2. Materials and methods
2.1. Experimental design
Three experimental analyses were performed: 1) Relative variant expression was compared among female Day 8 blastocysts (n = 32), male Day 8 blastocysts (n = 32), and CT-1 cells; 2) IFNT transcript variation was compared among in vivo derived conceptuses on Days 14, 16, 18, and 19 of development; and 3) IFNT transcript variation was compared in two sets of CT-1 cells, one of which had undergone approximately 80 more culture passages than the other.
2.2. RT-PCR amplification of IFNT transcripts
As PCR with even the best proof-reading polymerases is known to incorporate sequence errors into IFNT amplicons approximately once every 1091 bases for a 30 cycle reaction [5], it was necessary to perform multiple RT reactions and multiple PCR reactions for each RT product to avoid classifying PCR errors as IFNT variants. An overview of the cDNA amplification protocol used for each analysis is described below.
2.2.1. Relative variant expression between sexes
Embryos were derived from abbatoir-origin ovaries (see below) and cultured until the morning of Day 8 following their removal from insemination medium [12,19]. Blastocysts with an expanded cavity were selected and split into halves. One half was sexed (see below). The other halves were stored individually at −80 °C until RNA isolation. After sexing had been completed, the frozen demi-blastocysts were combined into two male and two female groups (n = 16/group). Two reverse transcription (RT) reactions were performed from the pooled RNA from each group, and two PCR reactions were performed for each RT reaction, providing four sets of amplicons for each group.
2.2.2. Relative variant expression over time
Two conceptuses were analyzed for each selected day of development. For this, RNA was isolated from each blastocyst; two RT reactions were performed per RNA sample and two PCR reactions were performed per RT reaction, to provide four sets of amplicons per conceptus.
2.2.3. Relative variant expression from CT-1 cells after passaging
The RNA was isolated from CT-1 cells cultured in 2001 and in 2005 (>80 passages from 2001 to 2005). Four RT reactions were performed for each CT-1 RNA sample and one PCR reaction was performed per RT reaction.
2.3. In vitro maturation and fertilization of oocytes and embryo culture
Oocyte collection, maturation, and fertilization were performed as previously described [12,19]. Zygotes were then cultured in 25 μL drops of D-glucose-free SOF medium supplemented with amino acids and 0.5% (w/v) bovine serum albumin (Sigma, St. Louis, MO, USA) overlaid with mineral oil (Sigma) at 39 °C in 5% CO2, 5% O2, and 90% N2 [11]. On Day 8, expanded blastocysts were split into two, approximately equal, halves. One half of the blastocyst was stored in RNA STAT-60 reagent (Tel-test, Friendswood, TX, USA) at −80 °C for RNA isolation and the other half of the blastocyst underwent PCR sexing.
2.4. CT-1 tissue culture
Continuously cultured bovine trophectoderm cells (CT-1) [17] were grown on a Matrigel (BD Biosciences, San Jose, CA, USA) substratum on a medium conditioned by STO cells (CRL-1503; American Type Tissue Culture, Rockville, MD, USA) [20]. They were sub-cultured at a 1:4 ratio every 2–3 wk. Immediately prior to RNA isolation or PCR sexing, CT-1 cells were dislodged from the substratum by repeated pipetting and collected by centrifugation at 500 x g.
2.5. Sexing of blastocysts and CT-1 cells by PCR
The sex of the CT-1 cells and blastocysts was determined through PCR amplification of a bovine Y-specific sequence [21] as described previously [11]. A bovine satellite sequence present in both sexes was simultaneously amplified to confirm the presence of bovine genomic DNA in each sample. The 301 nucleotide bovine Y-specific sequence and 216 nucleotide bovine satellite were PCR amplified for 30 cycles (96 °C 1 min, 62 °C 1 min, 72 °C 1 min). Bovine male genomic DNA (isolated from a liver biopsy), bovine female genomic DNA (isolated from an ovary), CT-1 medium, STO cells (that originated from murine spleen cells), and water were included as controls and treated in the same manner as blastocyst and CT-1 cell samples. Two individuals independently examined all gels, and each sample was labeled as “male”, “female”, or “unclear banding pattern.”
2.6. RNA isolation, RT-PCR for IFNT variants, and cloning cDNA
In vivo conceptuses from specific day points were provided by the MU Bovine Genome Project (http://genome.rnet.missouri.edu/Bovine) [22]. Total RNA from CT-1 cells, IVM/IVF blastocysts, and in vivo conceptuses was isolated with RNA STAT-60 reagent. Amino acid sequence, minus the signal peptide for all translated IFNT previously recorded in Genbank and all novel IFNT identified in this study, were aligned through ClustalW, with IFNT-1a as the reference sequence in the alignment.
RNase-free Turbo DNase (Ambion, Foster City, CA, USA) was added to all RNA preparations for 30 min at 37 °C to remove genomic DNA. The DNase was then inactivated by incubation with TURBO DNase inactivation reagent for 3 min at room temperature, according to the manufacturer’s instructions.
The RNA was reverse transcribed for 60 min at 50 °C by using SuperScript III (Invitrogen, Carlsbad, CA, USA), 250 μM of each dNTP, and an IFNT specific primer (5′-CTGAAATGAACACAGGTGAG-3′). The IFNT specific RT primer is complementary to a 19 nucleotide region within the 3′UTR that is 41 base pairs (bp) downstream of the translational stop codon. The IFNT cDNA were then PCR-amplified with primers specific for a 5′untranslated region and the transcriptional start site (5′-GGATCCCATCTTC CCCATGGCCTTC-3′) and a 3′ untranslated region (5′-GATTCCATCTTAGTCAGCGAGAGTC-3′), which is 4 bp downstream of the translational stop codon. The regions corresponding to the RT and PCR primers are conserved among all known IFNT. Thirty-two cycles of PCR (94 °C for 45 s, 62 °C for 30 s, and 68 °C for 2 min) were completed in the presence of 250 μM of each dNTP, the PCR primers described above, and KOD HotStart polymerase (Novagen, Gibbstown, NJ, USA), a high fidelity polymerase used to reduce incorporation of nucleotide errors. Reactions identical to the above, minus the addition of Superscript III during reverse transcription, were performed for each sample to verify that no amplification of genomic IFNT occurred.
The amplified PCR products were purified with the Wizard SV Gel and PCR Clean-up System (Promega, Madison, WI, USA). A single dATP overhang was incorporated into the PCR products through incubation with 5 U Taq (AB Peptides, St. Louis, MO, USA) and 200 μM dATP for 30 min at 70 °C. After a second PCR clean-up, the PCR products were ligated into pCRII TOPO vector (Invitrogen), and the resultant plasmids transformed into One Shot TOP10 chemically competent E. coli (Invitrogen), according to the manufacturer’s directions. Cells were plated on Luria broth plates containing ampicillin (50 μg/mL), X-gal (40 μg/mL), and isopropyl β-D-1-thiogalactopyranoside (IPTG) (1 mM) and incubated at 37 °C overnight. Colonies were picked through blue/white selection and were grown overnight in Luria broth containing ampicillin (50 μg/mL). Plasmid from the selected colonies was isolated by QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA, USA) and presence of insert verified by restriction digestion with Xho I and BamH I (Promega). Insert was sequenced at the University of Missouri-Columbia DNA Core Facility (http://biotech.rnet.missouri.edu/dnacore/), with an M13 forward primer.
2.7. Sequence analysis
All IFNT variants sequenced were imported into BioEdit version 7.09 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and aligned through ClustalW [23] with previously identified IFNT recorded in Genbank (Table 1). All sequences were translated and compared with the previously identified IFNT shown in Table 1. To account for PCR-induced sequence errors, sequences were only included in the analyses if they filled one or more of the following criteria: 1) the sequence had previously been identified and entered into Genbank; 2) the sequence had been identified in the annotated bovine genome database; and 3) the sequence occurred in at least two separate RT-PCR reactions. It was assumed that the incorporation of errors would be a relatively rare, e.g. one base in ~1000 and any errors not position biased, i.e. they occurred randomly [5]. Accordingly, the same error would be very unlikely to re-occur at the same base position.
Table 1.
IFNT variants previously annotated in Genbank.
| Gene | Accession no. | Gene | Accession no. |
|---|---|---|---|
| IFNT-1a* | M31557 | IFNT-3a* | AF196324 |
| IFNT-1b* | M60913 | IFNT-3b* | AF196325 |
| IFNT-1c* | AF196320 | IFNT-3c* | AF196326 |
| IFNT-1d* | M60908 | IFNT-3d* | AF196327 |
| IFNT-1f | EU828775 | IFNT-3e* | AF270471 |
| IFNT-1g | EU828776 | IFNT-3f | EU828780 |
| IFNT-2b* | AF196322 | IFNT-3g | EU828779 |
| IFNT-2c* | AF196323 | IFNT-3h | EU828777 |
Transcripts that matched the previously identified IFNT are shown (with asterisks) in the above table with their Genbank numbers. Transcripts that either matched IFNT identified in the bovine genome database or that were identified in two separate RT-PCR reactions are also included.
2.8. Statistical analysis
Sex ratios of IVM/IVF blastocysts were compared to an expected 50:50 ratio by a Chi square procedure in GraphPad Prism 4 [11]. Blastocysts with an “unclear banding pattern” were excluded from the statistical analysis of the sex ratio.
Expression frequency of the variants was calculated by counting the number of times each variant appeared within an RT-PCR reaction. The variant frequencies were small and distributed in a Poisson fashion, meaning the variance was proportional to the mean counts. In order to stabilize the variance, the expression frequency was normalized through the following equation: normalized expression (NE) = √(Count +1) [24]. Data are represented as NE ± Standard Error Measurement (SEM), where each observational group (n) represented one RT reaction. Comparisons of expression frequency for each analysis were performed in Statistical Analysis System (SAS) version 9.1 through ANOVA, with P < 0.05 considered significant.
2.9. Phylogenetic reconstruction
The coding sequences for all bovine IFNT variants listed in Genbank and novel IFNT identified in this study were aligned through ClustalW in BioEdit version 7.09. A consensus tree was constructed in MEGA version 4 (http://www.megasoftware.net/) through the Maximum Parsimony method [25], based on the 99 most parsimonious trees. Branches corresponding to partitions reproduced in <50% of the trees were collapsed. Branch lengths were calculated by using “the average pathway” method and based on the number of amino acid changes over the whole sequence. A total of 172 amino acid positions were analyzed, of which nine were parsimony informative.
3. Results
3.1. Sexing of blastocysts and CT-1 cells
Of 90 Day 8 blastocysts sexed, 32 were female, 44 male, and 14 indeterminate, i.e. had an “unclear banding pattern.” The sex ratio, i.e. fraction males was 0.58 (n = 76) and not different from 0.5, i.e. a 50:50 ratio (P = 0.329), thereby supporting previous data illustrating males and females are able to make the transition to the expanded blastocyst stage in glucose-free medium at approximately the same rate [11]. On the basis of these data, 64 half-blastocysts that had been stored frozen were sorted into equal groups of 16 single sex embryos, to determine IFNT transcript expression frequencies.
PCR sexing of CT-1 cells clearly proved that the cells were female in origin. As there was concern that contamination from the STO cells, which are spleen-derived murine fibroblasts and used to condition the culture medium, might confound PCR sexing, STO cells were included among the standard controls for the PCR sexing. These cells did not provide a PCR signal.
3.2. Relative variant expression between sexes
A total of 176 full length sequences were analyzed from the two male and two female groups of demi-blastocysts. Thirty of the 176 were discarded, either because they did not meet the selection criteria or because sequencing the full length clone failed. Consequently, we cannot rule out with certainty that some of these discards were not novel isoforms. For the statistical analysis, 80 full length sequences derived from males and 66 full length sequences from females were compared.
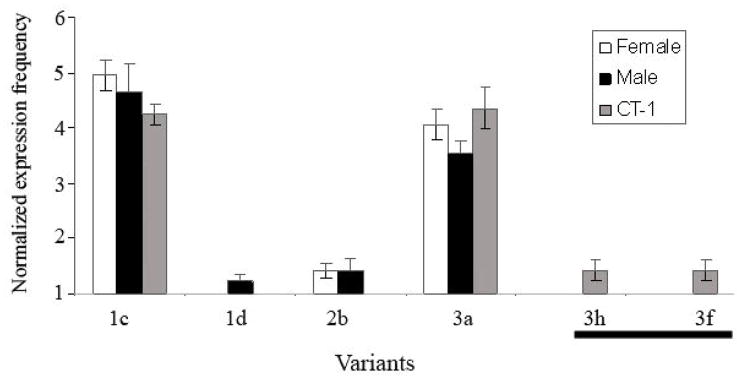
Four variants (IFNT-1c, 1d, 2b, and 3a) were identified in Day 8 blastocysts. The majority of the transcripts, whether male or female, were IFNT-1c and IFNT-3a (Fig. 1). The normalized expression for these two variants on a scale in which “1” equals no expression, was 4.80 and 4.75 for IFNT-1c, and 4.06 and 3.54 for IFNT-3a for females and males, respectively. In contrast, the mean expression of the next most highly expressed variant (IFNT-2b) was 1.41 in both sexes. The predominance of IFNT-1c and IFNT-3a in this analysis is generally consistent with previous studies on polymorphic bovine IFNT expression, although fewer variants were identified [5]. The relative degree of variant expression between Day 8 IVM/IVF male and female blastocysts was not different (P = 0.543). In other words, the same IFNT variants were expressed in males as in females and were present in similar proportions, thereby ruling out the possibility that females were selectively expressing forms with higher potential anti-viral activity.
Fig. 1.
IFNT variant expression among male and female Day 8 bovine blastocysts and CT-1 cells. The normalized expression frequency was plotted for each IFNT variant sequenced. There as no difference in IFNT variant expression frequency between male and females blastocysts (P = 0.543). CT-1 cells, which were derived from a Day 11 female blastocyst, did not have a different IFNT expression profile than IVM/IVF Day 8 blastocysts (P = 0.240). Two novel variants, 3f and 3h, were sequenced from CT-1 cells, and are indicated by a line below the X-axis on the right side of the graph.
The overall expression frequency of IFNT variants by CT-1 cells, which had been derived from a Day 11 blastocyst [20], was not different than that of Day 8 IVF/IVM blastocysts (P = 0.240). The CT-1 cells expressed the same molecular markers as the early stage blastocyst from which they were derived. However, two previously unidentified variants were also expressed in minor amounts by CT-1 cells. These two variants, termed IFNT-3h and IFNT-3f, only differed by one non-synonymous nucleotide change from previously identified IFNT variants. In that regard, IFNT-3h was identical to IFNT-1b, except for a single adenine (A) to guanine (G) change that would provide a glycine to aspartic acid conversion at codon 126. IFNT-3f differed from IFNT-3e by one nucleotide change (A to G), which converts a methionine to a valine codon at position 146 of the ORF. IFNT-3h reappeared in the Day 14 and Day 16 in vivo derived conceptuses described in the next section; it was also the only variant identified that matched an IFNT from the bovine genome database assembly 2.1, which was based on a single Hereford bull. Surprisingly, IFNT-3f never reoccurred among the 453 sequences analyzed in our study.
3.3. Temporal expression of IFNT variants from in vivo derived conceptuses
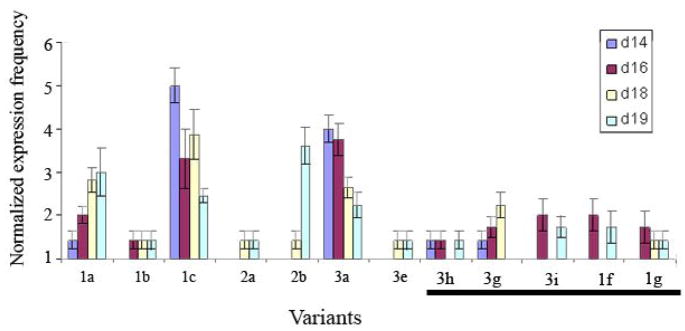
Of 185 sequences derived from eight older conceptuses distributed across 4 d of development, 31 were rejected, but again could possibly include rare variants. There was a difference (P < 0.001) in relative IFNT variant expression across days (Fig. 2). IFNT-1c and IFNT-3a were again the most frequently expressed variants overall, but their relative expression decreased over time (from 5 to 2.34 for IFNT-1c and from 4.14 to 2.24 for IFNT-3a from Day 14 to Day 19, respectively). The decrease in relative expression of IFNT-1c and 3a was compensated for by increases in the expression frequencies of IFNT-1a, members of the class 2 family, and four previously unidentified IFNT variants. Interestingly, IFNT-1a (normalized relative expression 3.0) and 2b (normalized relative expression 3.6) became the commonest transcripts, surpassing IFNT-1c and 3a by Day 19. The four novel transcripts each contained a single non-synonymous nucleotide change that differentiated them from previously known variants. We inferred that the pattern of IFNT expression established in the blastocyst was not sustained as the conceptus elongated.
Fig. 2.
Normalized expression frequency of IFNT variants from in vivo derived from bovine Day 14, 16, 18, and 19 conceptuses. The overall IFNT variant expression frequency profile was different (P < 0.001) across days. IFNT-1c and IFNT-3a decreased in expression frequency from Days 14 to 19. Conversely, IFNT-1a, members of the class 2 family, and several novel variants, increased in expression frequency from Days 14 to 19. The novel variants are indicated by a line below the X-axis.
3.4. Expression of IFNT variants from CT-1 cells analyzed over time
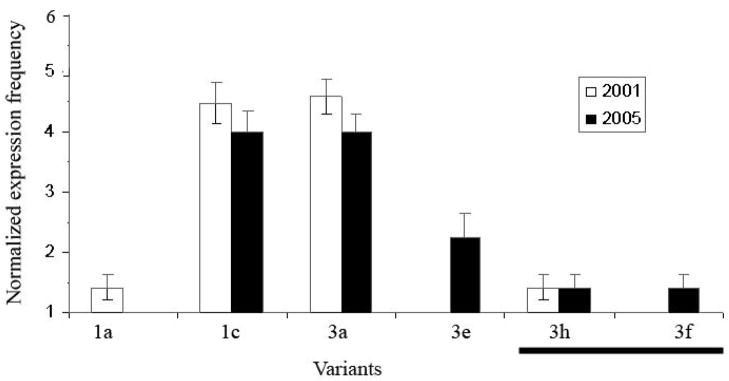
RNA was collected from CT-1 cells in 2001 and 2005, a period over which the culture had been passaged ~80 times, in order to verify that the trophoblast phenotype was relatively stable. For this analysis, we compared a total of 41 sequences from 2001 cells and 35 from 2005 cells (after discarding 16 sequences that did not meet our criteria). A total of six distinct transcripts (IFNT-1a, 1c, 3a, 3e, 3f, and 3h) were identified from CT-1 cells, of which IFNT-1c and 3a occurred most commonly. Variant 1c had normalized expressions of 4.49 in the 2001 isolate and 4.00 in 2005, whereas the corresponding values for 3a were 4.69 and 4.00. The next most frequently identified variant IFNT-3e, with a normalized expression of 2.24, was only identified in the 2005 isolate. Despite this one notable difference, no alteration in overall IFNT variant diversity was noted (P = 0.124) (Fig. 3). Thus, by this statistical criterion, the CT-1 cells had not shown a shift in phenotype over 4 y. Conversely, the appearance of a major variant, 3e, in the 2005 isolate was difficult to explain; perhaps the IFNT expression can alter as the cells are maintained over time.
Fig. 3.
A comparison of IFNT variant expression frequency in CT-1 cells over increased passage. Normalized expression frequency of IFNT variants from cells cultured in 2001 was compared to cells cultured in 2005. There was no difference (P = 0.124) in IFNT variant expression profile by CT-1 cells after 4 y of culturing (>80 passages). The novel variants are indicated by a line below the X-axis.
3.5. Novel IFNT variants
Several novel IFNT variants were identified during the course of this study. All of the variants, except IFNT-3f, were sequenced from separate conceptus groups and met our requirement only to accept novel IFNT variants derived from separate RT-PCR reactions. IFNT-3f, as previously discussed, was identified as one of three IFNT genes in bovine genome assembly 2.1. Interestingly, the same variant did not appear in the most current bovine genome assembly (3.1).
Each of the novel IFNT transcripts reported here bore a strikingly close nucleotide sequence identity to previously identified IFNT (Supplemental Fig. 1; on-line version only). Single non-synonymous nucleotide changes were responsible for each of the new “variants” identified. Indeed, only nine codons out of 190, the so-called variable sites, were not conserved in all of the 18 known IFNT variants so far reported. Moreover, the variation in amino acid in the translated sequence at each site was limited to two. The codon positions of the variable sites with their respective amino acids were as follows: 5 (D/N), 46 (N/S), 65 (L/F), 69 (Y/H), 70 (T/I), 102 (P/Q), 105 (G/E), 126 (G/D), and 146 (V/M).
Bovine IFNT variants were previously separated into three classes (1, 2, and 3) through a Maximum Parsimony phylogenetic reconstruction (Supplemental Fig. 2A; on-line version only) [5]. A new Maximum Parsimony phylogenetic tree was constructed here based on the amino acid sequence of all variants so far recorded. Unfortunately, the bootstrapping for most branches scored below 10, due to similarity of amino acid identity among variants. As bootstrapping scores below 20 have little statistical value in understanding phylogenetic relationships [26], we chose to use a slightly different approach, the Maximum Parsimony method, to create initial trees with a consensus tree derived from the 99 most parsimonious trees. This approach collapsed some branches present in the original tree (Supplemental Fig. 2B; on-line version only). It also divided sub-group 1 into two branches, one containing IFNT-1a and two of the novel variants, 1f and 1g, and the second comprised of IFNT-1c and 1d. IFNT-1b could not be placed with confidence in any grouping, and appeared to be intermediate between classes 1 and 3. Class 2 branched at the same place in all 99 of the parsimonious trees analyzed, supporting the continued placement of the three known IFNT2 variants into their own class. Class 3 also remained and contained several new members, IFNT-3f, 3g, 3h, and 3i, which had been identified in the present study.
4. Discussion
Our main goals were first to conduct an extensive sequence coverage of IFNT transcripts from bovine embryos of various developmental ages to define as completely as possible the full range of sequence variation of IFNT in the Bos taurus genome. It was also hoped that this exercise would provide insight into how many transcriptionally active IFNT are present in the genome, as this issue remains controversial, with the most recent assembly (3.1) suggesting that there are only three. A second goal was to determine whether male and female blastocysts could be distinguished according to the complement of transcripts they expressed. In particular, we sought an explanation for why female blastocysts have almost twice as high antiviral activity in their culture medium as their male counterparts at the blastocyst stage of development [10,11,16]. The well-documented increase in antiviral activity from female blastocysts could either be explained by the expression of more potent IFNT variants or more total IFNT. In this work, we focused on the first possibility and addressed the specific IFNT variants expressed in each sex. A third goal was to determine whether the pattern of IFNT expression changed over development age. For example, are there “early” and “late” genes that might reflect the need for certain variants at particular stages of trophoblast development? All data were based on mRNA expression, because no antibodies currently exist that can differentiate bovine IFNT isoforms. Our approach also allowed us to identify previously uncharacterized IFNT variants that would not have been detected through other methods.
Previous cDNA sequencing efforts had identified at least twelve IFNT variants expressed in bovine conceptuses over a range of developmental ages, raising the possibility that there were multiple genes and probably also considerable allelic variation. Of these variants, IFNT-1c and 3a predominated [5], an outcome that suggested that all genes and their allelic variants were not transcribed at similar rates. A similar heterogeneous pattern of IFNT variant expression was observed in the current study, in which fourteen IFNT variants were confirmed, six of them new. Again the predominately expressed variants were IFNT-1c and 3a. Four variants [IFNT-2c (AF196323), 3b (AF196325), 3c (AF196326), and 3d (AF196327)] previously reported by Ealy et al. [5] in an experiment that also controlled for the possibility of PCR error were not found in the present study, suggesting 18 protein variants actually exist in cattle. As pointed out above, it seemed likely that some, possibly the majority, of these variants were allelic and possibly breed-specific. Based on genomic analysis obtained through in situ hybridization and restriction fragment length hybridization, others have inferred six IFNT to be present in a mixed breed cow [6,27]. However, the recent annotation of Type I IFN locus in the bovine genome suggested that only three genes are present in the genome of the Hereford bull analyzed. One possibility is that the genes are so similar in sequence that some have been “lost” during the assembly process (particularly as genes have come and gone between assemblies). A careful examination of the original traces obtained in the 7.1-fold coverage of the bovine genome, revealed that 45 IFNT sequences were read, raising the possibility that the true number of IFNT is higher than three (A. Walker & R.M. Roberts, unpublished observations). Regardless, at least six individual IFNT variants have been identified here from a single Angus conceptus and six from CT-1 cells, a cell line derived from a single female blastocyst of unknown breed (Fig. 3), which was consistent with there being at least three polymorphic IFNT in the bovine genome. Unfortunately, the phylogenetic analysis (Supplementary Fig. 2B) provided little further insight into the number of genes present. We had hoped, despite the close similarity of the variant forms, that the main branches of the tree might represent genes and that the variants within each branch might represent alleles. On such a basis, up to six genes might be present, although only three groups (1, 2, and 3) can be defined with confidence. Oddly, one of them, group 2, was not represented at all in the current genome assembly and was not among the sequences represented in the 45 original traces that contained IFNT sequences. Indeed group 2 variants, although a well defined class based on their sequences, were expressed by some conceptuses, but not at all by others [5]. What was more surprising has been the failure to identify group 2 variants among any of the 176 sequences analyzed from Day 8 blastocysts. The most likely explanation is that group 2 genes are actually absent in some animals but present in others, and that when present they are “late” genes. Group 2 IFNT are clearly not a specific marker for the Angus breed as class 2 variants were identified in some conceptuses but not others. Such variability in gene complement has been reported for other multi-gene families, including the large family of human odorant genes [28].
The distribution of IFNT variant transcripts among male and female blastocysts did not explain the sexual dimorphism in IFNT production. Females neither expressed more kinds of transcripts than males nor a different group of transcripts. Males and females predominantly expressed IFNT-1c and 3a, whereas the other variants, although not ubiquitous, showed no bias towards one sex or the other. Although these data were obtained from in vitro derived blastocysts, similar IFNT expression profiles were observed in the earliest stage in vivo derived conceptuses examined (Day 14), which suggests the in vitro IFNT expression mirrors the in vivo expression prior to Day 14. As proposed by Kimura et al. [14,16], increased antiviral activity in the female culture medium must result from increased overall IFNT expression and not from the presence of more potent IFNT variants being expressed by females.
The most notable feature of IFNT variant expression in relation to conceptus age was that the relative quantities of IFNT-1c and 3a declined over time, whereas the total number of less common variants increased their representation. It would be inappropriate to evaluate the expression of each of the “rare” IFNT isoforms individually based on day because the number of sequences evaluated in each pool was only sufficient to make comparisons between the most abundant cDNAs present in the sample. However, the overall profile of IFNT variants expressed significantly altered as the conceptuses developed. It is unclear how to interpret these observations; although it is conceivable that the different isoforms possess somewhat different downstream targeting specificities and that the more unusual variants have been selected for specialized roles in modulating endometrial gene expression, which cannot be matched by the two commoner isoforms. Among human IFN-alpha (IFNA), it is well established that the various variants can display different activities, e.g. as antiviral and anti-proliferative agents, despite operating through the same Type I IFN receptor [29–31]. Similarly, ovine IFNT variants are known to vary in antiviral activities [32,33], although it is difficult to rule out the possibility that some of the disparity is due to refolding efficiency of the bacterial recombinant products. Perhaps more relevant to the present work, is the observation that individual bovine IFNT variants differentially regulate prostaglandin synthesis in endometrial cells [34]. Whereas bovine IFN-2b inhibited both prostaglandin PGF2α and PGE2 production in primary uterine epithelial cells, bovine IFNT-3b had an opposite effect; it induced more prostaglandin formation. Interestingly, bovine IFNT-1a inhibited prostaglandin synthesis at low concentrations (<1 μg/mL), but promoted synthesis at high concentrations (>1 μg/mL). The downstream actions of IFNT can clearly be complex, possibly through the distinctive abilities of variant forms to trigger the assorted signaling pathways that ultimately guide endometrial responses.
Changes in relative variant expression based on the age of the conceptus have important implications for any model system used in the study of IFNT. The most commonly employed model system for bovine trophectoderm in this and other laboratories is a bovine line of continuously cμμtured trophectoderm (CT-1) cells derived from an outgrowth of a single Day 11 blastocyst [17,18,35]. We, therefore, asked whether CT-1 cells shifted the relative expression of IFNT variants over increased passage to provide a pattern analogous to that observed for elongating conceptuses. The results of this experiment were equivocal. The overall expression profile of CT-1 cells resembled that of the Day 8 blastocysts. However, one new variant, which was relatively strongly expressed, did arise in the late passage cells. Whether this observation was the result of chance or a point mutation is not clear. It should also be emphasized that the CT-1 cells, although derived from a single blastocyst, were not clonal. Shifts in the population structure with passage could provide a substantial proportion of cells expressing the IFNT-3e variant, whose gene was silent in the population group that dominated in 2001.
The mechanism that leads to differential expression of individual IFNT variants is difficult to explain. As far as can be discerned by conventional analyses, the main control elements for the IFNT lie in the proximal 5′UTR [36], and are concentrated within a complex enhancer element centered around the binding sites for three transcription factors (DLX3, ETS2, and AP1) [2,37,38]. The promoters of four bovine IFNT variants, IFNT-1a (M60903), 1c (AF339094), 2b (AF339095), and 3b (AF339096), have been fully sequenced for approximately 430 bp upstream of their transcriptional start site. All four retain the complete DLX3/ETS2/AP1 enhancer and are >99% conserved over the entire 430 bp sequence [5]. IFNT promoters annotated from the bovine genome database assembly 3.1 have also been compared and are 98.3 to 99.9% identical to one another for at least 900 bp upstream of the transcriptional start site (A. Walker & R.M. Roberts, unpublished observation). These data no doubt attest, at least in part, to the recent origins of the duplications that produced the variant genes. The 5′UTR of sequenced ovine IFNT are also highly conserved over >450 bp of the translation start site [38]. Such conservation is in itself remarkable, particularly as it retained across diverse ruminant species [39,40]. How then can genes with identical sequence within their transcriptional control regions and with such overall conserved structure exhibit such profound differences in expression? Perhaps a cis-oriented LINE/SINE tandem sequence present 660 bp upstream of the transcriptional start site supplements the directive action of the proximal promoter region, as described for some other genes [41,42]. However, these two repetitive elements are present at identical positions in all three sequenced genes of the bovine genome assembly and are almost identical in sequence, making a role in transcriptional control less feasible. Another possibility is that an additional level of transcriptional control, particularly temporal control, operates from a site upstream of the entire IFNT locus, much as happens for the cis-acting, locus control regions (LCR) of the β-globin gene locus during human and mouse embryonic development [43]. As most LCRs have been discovered through experiments with transgenic mice, when normal, “physiological” expression could only be achieved by including far-upstream elements in the transgene construct, it will be extremely difficult to conduct a similar analysis in cattle. Consequently, the controls operating on IFNT are likely to remain mysterious for some time to come.
Supplementary Material
Acknowledgments
We thank Dr. Mark Ellersieck for his assistance on the statistical analysis and the MU Bovine Genome Project for providing conceptus samples from individual day points. This work was supported by NIH grant HD21896 and USDA NRI grant 2003-35205-12812.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ezashi T, Ghosh D, Roberts RM. Repression of Ets-2-induced transactivation of the tau interferon promoter by Oct-4. Mol Cell Biol. 2001;21:7883–91. doi: 10.1128/MCB.21.23.7883-7891.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ezashi T, Roberts RM. Regulation of interferon-tau (IFN-tau) gene promoters by growth factors that target the Ets-2 composite enhancer: a possible model for maternal control of IFN-tau production by the conceptus during early pregnancy. Endocrinology. 2004;145:4452–60. doi: 10.1210/en.2004-0606. [DOI] [PubMed] [Google Scholar]
- 3.Hansen TR, Austin KJ, Perry DJ, Pru JK, Teixeira MG, Johnson GA. Mechanism of action of interferon-tau in the uterus during early pregnancy. J Reprod Fertil Suppl. 1999;54:329–9. [PubMed] [Google Scholar]
- 4.Rasmussen TA, Ealy AD, Kubisch HM. Identification of bovine and novel interferon-tau alleles in the American plains bison (Bison bison) by analysis of hybrid cattle x bison blastocysts. Mol Reprod Dev. 2005;70:228–34. doi: 10.1002/mrd.20198. [DOI] [PubMed] [Google Scholar]
- 5.Ealy AD, Larson SF, Liu L, Alexenko AP, Winkelman GL, Kubisch HM, Bixby JA, Roberts RM. Polymorphic forms of expressed bovine interferon-tau genes: relative transcript abundance during early placental development, promoter sequences of genes and biological activity of protein products. Endocrinology. 2001;142:2906–15. doi: 10.1210/endo.142.7.8249. [DOI] [PubMed] [Google Scholar]
- 6.Ryan AM, Gallagher DS, Womack JE. Somatic cell mapping of omega and trophoblast interferon genes to bovine syntenic group U18 and in situ localization to chromosome 8. Cytogenet Cell Genet. 1993;63:6–10. doi: 10.1159/000133491. [DOI] [PubMed] [Google Scholar]
- 7.Roberts RM, Liu L, Alexenko A. New and atypical families of type I interferons in mammals: comparative functions, structures, and evolutionary relationships. Prog Nucleic Acid Res Mol Biol. 1997;56:287–325. doi: 10.1016/s0079-6603(08)61008-9. [DOI] [PubMed] [Google Scholar]
- 8.Roberts RM, Liu L, Guo Q, Leaman D, Bixby J. The evolution of the type I interferons. J Interferon Cytokine Res. 1998;18:805–16. doi: 10.1089/jir.1998.18.805. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Green JA, Antoniou E, Ealy AD, Mathialagan N, Walker AM, Avalle MP, Rosenfeld CS, Hearne LB, Roberts RM. Effect of interferon-tau administration on endometrium of nonpregnant ewes: a comparison with pregnant ewes. Endocrinology. 2006;147:2127–37. doi: 10.1210/en.2005-1310. [DOI] [PubMed] [Google Scholar]
- 10.Kimura K, Spate LD, Green MP, Murphy CN, Seidel GE, Jr, Roberts RM. Sexual dimorphism in interferon-tau production by in vivo-derived bovine embryos. Mol Reprod Dev. 2004;67:193–9. doi: 10.1002/mrd.10389. [DOI] [PubMed] [Google Scholar]
- 11.Larson MA, Kimura K, Kubisch HM, Roberts RM. Sexual dimorphism among bovine embryos in their ability to make the transition to expanded blastocyst and in the expression of the signaling molecule IFN-tau. Proc Natl Acad Sci U S A. 2001;98:9677–82. doi: 10.1073/pnas.171305398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Ledezma JJ, Sikes JD, Murphy CN, Watson AJ, Schultz GA, Roberts RM. Expression of bovine trophoblast interferon in conceptuses derived by in vitro techniques. Biol Reprod. 1992;47:374–80. doi: 10.1095/biolreprod47.3.374. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Adan A, Granados J, Pintado B, De La Fuente J. Influence of glucose on the sex ratio of bovine IVM/IVF embryos cultured in vitro. Reprod Fertil Dev. 2001;13:361–5. doi: 10.1071/rd00039. [DOI] [PubMed] [Google Scholar]
- 14.Kimura K, Spate LD, Green MP, Roberts RM. Effects of D-glucose concentration, D-fructose, and inhibitors of enzymes of the pentose phosphate pathway on the development and sex ratio of bovine blastocysts. Mol Reprod Dev. 2005;72:201–207. doi: 10.1002/mrd.20342. [DOI] [PubMed] [Google Scholar]
- 15.Cameron EZ, Lemons PR, Bateman PW, Bennett NC. Experimental alteration of litter sex ratios in a mammal. Proc Biol Sci. 2008;275:323–7. doi: 10.1098/rspb.2007.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura K, Spate LD, Green MP, Roberts RM. Effects of oxidative stress and inhibitors of the pentose phosphate pathway on sexually dimorphic production of IFN-tau by bovine blastocysts. Mol Reprod Dev. 2004;68:88–95. doi: 10.1002/mrd.20053. [DOI] [PubMed] [Google Scholar]
- 17.Talbot NC, Caperna TJ, Edwards JL, Garrett W, Wells KD, Ealy AD. Bovine blastocyst-derived trophectoderm and endoderm cell cultures: interferon tau and transferrin expression as respective in vitro markers. Biol Reprod. 2000;62:235–47. doi: 10.1095/biolreprod62.2.235. [DOI] [PubMed] [Google Scholar]
- 18.Ezashi T, Das P, Gupta R, Walker A, Roberts RM. Biol Reprod. 2008. The role of Homeobox Protein Distal-Less 3 and its interaction with ETS2 in regulating Bovine Interferon-Tau Gene Expression-Synergistic Transcriptional Activation with ETS2. [DOI] [PubMed] [Google Scholar]
- 19.Kubisch HM, Larson MA, Roberts RM. Relationship between age of blastocyst formation and interferon-tau secretion by in vitro-derived bovine embryos. Mol Reprod Dev. 1998;49:254–60. doi: 10.1002/(SICI)1098-2795(199803)49:3<254::AID-MRD5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Talbot NC, Caperna TJ, Edwards JL, Garrett W, Wells KD, Ealy AD. Bovine blastocyst-derived trophectoderm and endoderm cell cultures: interferon tau and transferrin expression as respective in vitro markers. Biol Reprod. 2000;62:235–47. doi: 10.1095/biolreprod62.2.235. [DOI] [PubMed] [Google Scholar]
- 21.Peura T, Hyttinen JM, Turunen M, Janne J. A reliable sex determination assay for bovine preimplantation embryos using the polymerase chain reaction. Theriogenology. 1991;35:547–55. doi: 10.1016/0093-691x(91)90451-i. [DOI] [PubMed] [Google Scholar]
- 22.McHughes C, Springer GK, Spate L, Li R, Woods R, Green M, Korte SW, Murphy CN, Green JA, Prather RS. Identification and quantification of differentially represented transcripts in preimplantation bovine embryos. Mol Reprod Dev. 2008 doi: 10.1002/mrd.20929. (in press) [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snedecor G, Cochran, William . Statistical Methods. 8. Ames, Iowa: Iowa State University Press; 1989. [Google Scholar]
- 25.Erck R, Dayhoff, Margaret . Atlas of Protein Sequence and Structure. Silver Springs, MD: National Biomedical Research Foundation; 1966. [Google Scholar]
- 26.Sanderson MJ, Wojciechowski MF. Improved bootstrap confidence limits in large-scale phylogenies, with an example from Neo-Astragalus (Leguminosae) Syst Biol. 2000;49:671–85. doi: 10.1080/106351500750049761. [DOI] [PubMed] [Google Scholar]
- 27.Iannuzzi L, Gallagher DS, Ryan AM, Di Meo GP, Womack JE. Chromosomal localization of omega and trophoblast interferon genes in cattle and river buffalo by sequential R-banding and fluorescent in situ hybridization. Cytogenet Cell Genet. 1993;62:224–7. doi: 10.1159/000133482. [DOI] [PubMed] [Google Scholar]
- 28.Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci U S A. 2007;104:20421–6. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaoka T, Kojima S, Ichi S, Kashiwazaki Y, Koide T, Sokawa Y. Biologic and binding activities of IFN-alpha subtypes in ACHN human renal cell carcinoma cells and Daudi Burkitt’s lymphoma cells. J Interferon Cytokine Res. 1999;19:1343–9. doi: 10.1089/107999099312803. [DOI] [PubMed] [Google Scholar]
- 30.Foster GR, Finter NB. Are all type I human interferons equivalent? J Viral Hepat. 1998;5:143–52. doi: 10.1046/j.1365-2893.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 31.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 32.Winkelman GL, Roberts RM, James Peterson A, Alexenko AP, Ealy AD. Identification of the expressed forms of ovine interferon-tau in the periimplantation conceptus: sequence relationships and comparative biological activities. Biol Reprod. 1999;61:1592–1600. doi: 10.1095/biolreprod61.6.1592. [DOI] [PubMed] [Google Scholar]
- 33.Niswender KD, Li J, Powell MR, Loos KR, Roberts RM, Keisler DH, Smith MF. Effect of variants of interferon-tau with mutations near the carboxyl terminus on luteal life span in sheep. Biol Reprod. 1997;56:214–20. doi: 10.1095/biolreprod56.1.214. [DOI] [PubMed] [Google Scholar]
- 34.Parent J, Villeneuve C, Alexenko AP, Ealy AD, Fortier MA. Influence of different isoforms of recombinant trophoblastic interferons on prostaglandin production in cultured bovine endometrial cells. Biol Reprod. 2003;68:1035–43. doi: 10.1095/biolreprod.102.008250. [DOI] [PubMed] [Google Scholar]
- 35.Michael DD, Wagner SK, Ocon OM, Talbot NC, Rooke JA, Ealy AD. Granulocyte-macrophage colony-stimulating-factor increases interferon-tau protein secretion in bovine trophectoderm cells. Am J Reprod Immunol. 2006;56:63–7. doi: 10.1111/j.1600-0897.2006.00390.x. [DOI] [PubMed] [Google Scholar]
- 36.Leaman DW, Cross JC, Roberts RM. Multiple regulatory elements are required to direct trophoblast interferon gene expression in choriocarcinoma cells and trophectoderm. Mol Endocrinol. 1994;8:456–68. doi: 10.1210/mend.8.4.8052267. [DOI] [PubMed] [Google Scholar]
- 37.Das P, Ezashi T, Gupta R, Roberts RM. Combinatorial roles of protein kinase A, Ets2, and 3′,5′-cyclic-adenosine monophosphate response element-binding protein-binding protein/p300 in the transcriptional control of interferon-tau expression in a trophoblast cell line. Mol Endocrinol. 2008;22:331–43. doi: 10.1210/me.2007-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ezashi T, Ealy AD, Ostrowski MC, Roberts RM. Control of interferon-tau gene expression by Ets-2. Proc Natl Acad Sci U S A. 1998;95:7882–7. doi: 10.1073/pnas.95.14.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leaman DW, Roberts RM. Genes for the trophoblast interferons in sheep, goat, and musk ox and distribution of related genes among mammals. J Interferon Res. 1992;12:1–11. doi: 10.1089/jir.1992.12.1. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Leaman DW, Roberts RM. The interferon-tau genes of the giraffe, a nonbovid species. J Interferon Cytokine Res. 1996;16:949–51. doi: 10.1089/jir.1996.16.949. [DOI] [PubMed] [Google Scholar]
- 41.Britten RJ. Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol Phylogenet Evol. 1996;5:13–7. doi: 10.1006/mpev.1996.0003. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro JA. A 21st century view of evolution: genome system architecture, repetitive DNA, and natural genetic engineering. Gene. 2005;345:91–100. doi: 10.1016/j.gene.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–86. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.