Abstract
In vivo gene silencing using RNAi plays an important role in target validation and is advancing towards the development of RNAi-based therapeutics. RNAs were thought to have just two broad functions in cells as messenger RNAs (mRNAs) and ribosomal RNAs, but recently the relevance of microRNAs is becoming more clearly understood. Messenger RNA molecules transmit information between DNA and protein and, as such, are vital intermediaries for gene expression. Ribosomal and transfer RNAs have structural, catalytic and information-decoding roles in the process of protein synthesis, whereas microRNAs are regulators of gene expression. This review presents the early and intriguing successes of using siRNAs for in vivo gene silencing and its use as a possible cancer therapeutics.
Introduction
Observed in the majority of eukaryotes, RNA interference (RNAi) is an evolutionarily conserved process that inhibits gene expression primarily by targeting mRNAs. RNAi targets, including RNA from viruses and transposons, play roles in regulating development and genome maintenance. Small interfering RNA (siRNA) strands are the key to RNAi process as siRNA have nucleotide sequences complementary to the targeted RNA strand. Specific RNAi pathway proteins are guided by the siRNA to the targeted messenger RNA (mRNA) where they “cleave” the target and break it down into smaller portions that can no longer be translated into protein (Fig. 1). The discovery of RNA interference (RNAi) is revolutionary and is transforming biological research in ways not seen before. Short double-stranded RNA (dsRNA) molecules are easily and rapidly available today. These dsRNA molecules can downregulate the expression of a target mRNA molecule in a sequence-specific manner (Dorsett and Tuschl, 2004;Novina and Sharp, 2004).
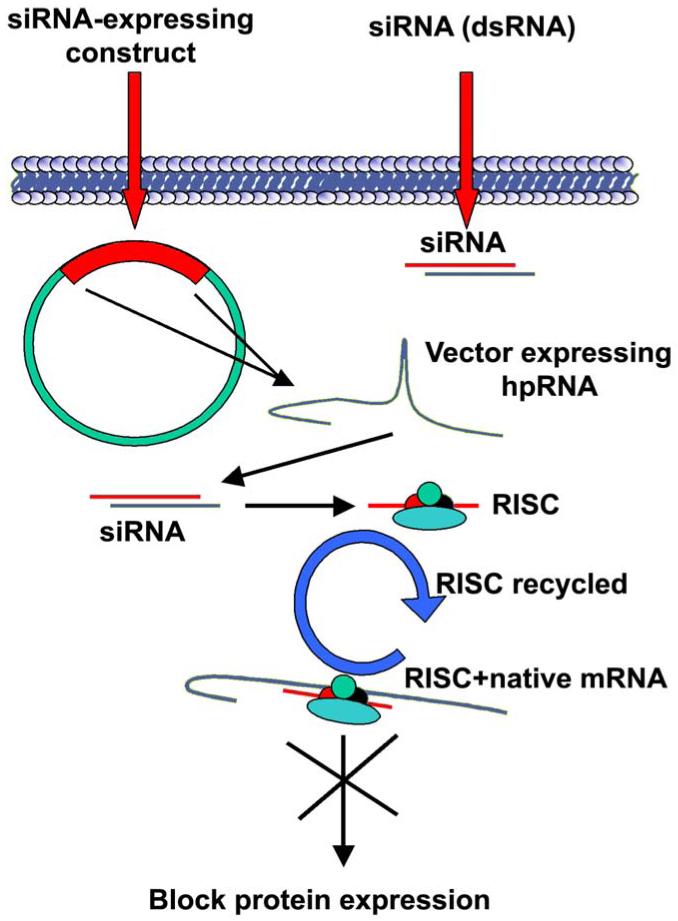
Figure 1. Mechanism of RNAi using siRNA molecules or siRNA expressing molecules.
RNAi involves the use of specific siRNA molecules that form a complex with proteins to form the RNA-induced silencing complex (RISC). In RNAi, one siRNA molecule is capable of targeting multiple mRNA molecules.
RNAs were thought to have just two broad functions in cells as messenger (Apirion, 1975) RNAs and ribosomal and transfer RNAs. Messenger RNAs (mRNAs) are vital intermediaries in gene expression, transmitting information between DNA and protein. Ribosomal and transfer RNAs have structural, catalytic and information-decoding roles in the process of protein synthesis. This concept changed in 1998 when Fire et al. (Fire et al, 1998) published their discovery of “potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.” RNAi is now widely accepted as ‘the’ method of choice for rapidly assessing biological mechanisms and pathways through loss-of-function. RNAi libraries, which allow functional phenotypic screening in mammalian cells, have been established in recent years (Berns et al, 2004). Such libraries take advantage of the ability to express short hairpin structures from a variety of vector systems, which are processed to generate the functional unit, a small interfering RNA (siRNA). Both short hairpin RNAs (shRNAs) and siRNAs can be used to downregulate mRNA expression in cells. SiRNAs are RNA duplexes commonly composed of two 21-mer oligonucleotides with a 19 nt complementary and a 2 nt single-stranded overhang at each 3′ end. Alternative structures of siRNAs also show in vitro function (Czauderna et al, 2003). Small RNA oligos are easily synthesized and annealed to form functional siRNA molecules, whereas small hairpin RNA can also be chemically synthesized as inverted repeats and allowed to self anneal. The increased length (more than 50 bases) makes the chemical synthesis less efficient. ShRNAs can be efficiently produced in large scale by plasmids expressing inverted repeats. Both siRNAs and shRNAs work similarly and cause efficient knockdown of target mRNAs. The silencing by plasmid based shRNAs is long lasting when compared to siRNAs because of the continuous synthesis of shRNA molecules in situ by the plasmids (Huang et al, 2008). For in vitro use, siRNA can be readily introduced into cell lines by a variety of techniques, including lipid complexes, liposomes, electroporation, etc (Li and Szoka, Jr., 2007). One of the major hurdles for using RNAi in vivo is efficient intracellular delivery of a highly charged and structurally rigid siRNA molecule. Progress is being been made in delivering siRNA in vivo, first through hydrodynamic methods and more recently through local or targeted administration (Herweijer and Wolff, 2007;Li et al, 2006). Delivery can be enhanced either by improving siRNA attributes directly through chemical modifications of the duplex or by using different formulations, such as liposomes and ionic polymers (Li and Szoka, Jr., 2007;Yang et al, 2006). Significant success has already been seen in using siRNA in many localized settings such as the eye, lungs, and the central nervous system. In addition, encouraging data have been generated in a variety of other in vivo settings, including xenograft models and viral infections. All of these studies demonstrate the potential for the systemic delivery of siRNA-based therapeutics.
In vivo delivery strategies
Delivery of siRNA in vivo can basically be divided into two classes, systemic and local. Systemic delivery involves the delivery of siRNA into the whole organism and usually requires large quantity of siRNA to achieve the desired downregulation. In contrast, localized delivery of siRNA to a specific site (e.g., brain, eye or a tumor) is more achievable since large quantities of siRNA are not required and subsequently do not affect the whole organism (Pardridge, 2007;Pushparaj et al, 2008;Shen, 2008). Numerous in vivo delivery approaches have been published, ranging from simple direct injection of siRNA to complex procedures involving structurally modified molecules (Alshamsan et al, 2009;Watts et al, 2008;Zhang et al, 2009). Animal studies using siRNA have either employed saline-based formulations or used conjugates, including liposome/lipoplexes and complexes with peptides, polymers, or antibodies (Aigner, 2008;Law et al, 2008;Wang et al, 2008;Whitehead et al, 2009;Xia et al, 2008). Local delivery has particular advantage in that, as with any pharmacologic approach, doses of siRNA required for efficacy are substantially lower when siRNAs are injected into or administered at or near the target tissue (Aigner, 2006;Li and Huang, 2008). Direct delivery also allows for more focused delivery of siRNA, which might circumvent any theoretical, undesired side effects resulting from systemic delivery.
Systemic delivery of siRNA, especially with cholesterol conjugates, liposomes, and polymer-based nanoparticle approaches, has also been widely explored with moderate success. Other approaches using antibodies, peptides, and aptamers have also been reported (Neidhardt et al, 2005;Turner et al, 2007). The various delivery methods developed to date are represented in Table 1 (Takeshita and Ochiya, 2006). Delivery of siRNA can vary from direct to indirect. The direct method can be classified as hydrodynamic delivery, which involves naked siRNA molecules and employs brute force to deliver siRNA to an organism. This method is only partially effective when large volumes are used (Herweijer and Wolff, 2003). As delivery methods are developed, the use of carriers of genetic material (adenoviruses, lentiviral vectors, adeno associated viruses) has gained popularity (Whitehead et al, 2009). The use of viruses as a delivery method was questioned when undesirable side effects related to antiviral response, hyper immune reactions and viral genome integration into host genome were reported (Braun, 2006). These drawbacks paved the way for the development of lipids, lipid complexes and chemically modified siRNA molecules as delivery agents (Fig. 2).
Table 1.
Small interfering RNA for cancer therapy in vivo*
| Carriers | Routes | Type of cancer (cell line) | Implanted site (target organ) | Target gene | Reference |
|---|---|---|---|---|---|
| Naked siRNA | i.p., i.v., s.c., i.t. | Fibrosarcoma (JT8) | s.c. | VEGF | (Filleur et al, 2003) |
| Naked siRNA + gemcitabine | i.v. | Pancreatic adenocarcinoma (PANC1, MIAPaCa2, BxPC3) | Orthotopic pancreas | FAK | (Duxbury et al, 2003) |
| Naked siRNA | i.v. | Pancreatic adenocarcinoma (BxPC3) | s.c., Orthotopic pancreas | CEACAM6 | (Duxbury et al, 2004c) |
| Naked siRNA | i.v. | Pancreatic adenocarcinoma (PANC1, MIAPaCa2, BxPC3, Capan2) | s.c., Orthotopic pancreas, liver metastasis | EphA2 | (Duxbury et al, 2004a) |
| Naked siRNA + gemcitabine | i.v. | Pancreatic adenocarcinoma (PANC1, MIAPaCa2, BxPC3, Capan2) | s.c., Liver metastasis | RRM2 | (Duxbury et al, 2004b) |
| Naked siRNA | i.v. | Breast cancer(MDA-MB-231) | Lung metastasis | CXCR4 | (Liang et al, 2005) |
| Liposome | i.p. | Colon cancer(HTC116) | s.c., i.p. | β -Catenin | (Verma et al, 2003) |
| Liposome | i.v. | Liver metastatic spleen cancer (A549) | Liver | bcl-2 | (Yano et al, 2004) |
| Liposome | i.t. | Bladder cancer (UM-UC-3-LUC) | Bladder | PLK-1 | (Nogawa et al, 2005) |
| Liposome | i.t. | Pancreatic carcinoma (Capan-1) | s.c. | Somatostatin | (Carrere et al, 2005). |
| CCLA (NeoPhectin-AT) | i.v. | Prostate cancer (PC-3) | s.c. | Raf-1 | (Pal et al, 2005) |
| CCLA | i.v. | Breast cancer (MDA-MB-231) | s.c. | c-raf | (Chien et al, 2005) |
| shRNA plasmid + pegylated immunoliposome | i.v. | Glioma (U87) | Brain | EGFR | (Zhang et al, 2004b) |
| PEI | i.p. | Ovarian carcinoma cells (SKOV-3) | s.c. | HER-2 | (Urban-Klein et al, 2005) |
| shRNA plasmid + PEI | i.t. | Ewing’s sarcoma (TC71) | s.c. | VEGF | (Guan et al, 2005) |
| Adenovirus vector | i.t. | Cervical adenocarcinoma, colon cancer (HeLa, HTC116) | s.c. | HIF-1a | (Zhang et al, 2004a) |
| Adenovirus vector | i.t. | Lung cancer (ACC-LC-172) | s.c. | Skp-2 | (Sumimoto et al, 2005) |
| shRNA plasmid | it | Meningioma IOMM-LEE | Brain osmotic pumps | uPAR+MMP-9 | (Tummalapalli et al, 2007) |
| shRNA plasmid | it | Breast (MDA MB 231) | Orthotopic breast | uPAR+MMP9 | (Kunigal et al, 2007) |
| shRNA plasmid | ip | Glioblastoma (SNB19) | Brain | uPAR+uPA | (Gondi et al, 2007) |
| shRNA plasmid | ip | Glioblastoma (SNB19) | Brain | uPAR+MMP-9 | (Lakka et al, 2005c) |
| shRNA plasmid | ip | Glioblastoma (SNB19) | Brain | uPAR+uPA+MMP-9 | (Gondi et al, 2004a) |
| shRNA plasmid | i.t. | Glioblastoma (SNB19) | Brain | MMP-9 + cathepsin B | (Lakka et al, 2004) |
| shRNA plasmid | i.t. | Glioma (SNB19) | Brain | Cathepsin B, uPAR | (Gondi et al, 2004b) |
| shRNA plasmid + ATA | i.v. | Cervical adenocarcinoma, lung cancer (HeLa S3, A549) | s.c. | PLK1 | (Spankuch et al, 2004) |
| PEI-PEG-RGD | i.v. | Neuroblastoma (N2A) | s.c. | VEGF-R2 | (Schiffelers et al, 2004) |
| CDP-AD-PEG-transferrin | i.v. | Ewing’s sarcoma (TC71) | Multiple organ metastasis | EWS-FLI1 | (Hu-Lieskovan et al, 2005) |
| HVJ envelope vector + cisplatin | i.t. | Cervical adenocarcinoma (HeLa) | Intradermally | Rad51 | (Ito et al, 2005) |
| ErbB2-protamine fusion protein | i.t., i.v. | Melanoma (B16) | s.c. | c-myc MDM2 VEGF (mix) | (Song et al, 2005) |
| Atelocollagen | i.t. | Prostate cancer (PC-3) | s.c. | VEGF | (Takei et al, 2004) |
| Atelocollagen | i.t. | Germ-cell tumor (NEC8) | Testis | FGF-4 | (Minakuchi et al, 2004) |
| Atelocollagen | i.v. | Prostate cancer (PC-3M-Luc) | Bone metastasis | EZH2, p110a | (Takeshita et al, 2005) |
AD-PEG, adamantane-PEG5000; ATA, aurintricarboxylic acid, nuclease inhibitor; CCLA, cationic cardiolipin analog-based liposome; CDP, cyclodextrin-containing polycations; i.p., intraperitoneal; i.t., intratumoral; i.v., intravenous; PEG, polyethylene glycol; PEI, polyethylenimine; RGD, Arg-Gly-Asp; s.c., subcutaneous; HVJ, Hemagglutinating virus of Japan.
Modified from (Takeshita and Ochiya, 2006).
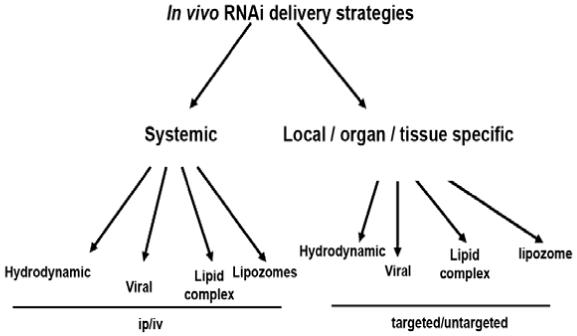
Figure 2. In vivo siRNA delivery strategies.
Delivery of siRNA in vivo can be accomplished through direct injections systemically or locally; however, direct delivery of unmodified siRNA cannot be targeted. Other methods of delivery that incorporate the use of viruses, lipids or lipid complexes have potential for use as targeted strategies.
Direct delivery
Hydrodynamic delivery involves rapid large-volume intravenous injection of siRNA equivalent to the total blood volume. The first successful in vivo delivery of siRNA into cells used a hydrodynamic methodology that was initially developed to deliver gene therapy vectors into hepatocytes (Song et al, 2003). In preliminary applications of siRNA in vivo, combinations of enhanced green fluorescent protein (EGFP) (Lewis et al, 2002) or luciferase (McCaffrey et al, 2002) encoding plasmids together with the siRNA yielded significant downregulation of target gene expression in the liver, suggesting that once delivered to hepatocytes, siRNAs were functional in vivo. Using this approach, several therapeutically relevant hepatic targets were downregulated, including Fas (Song et al, 2003) and caspase 8 (Zender et al, 2003). In both of these cases, the target genes were downregulated by 60-90%. Downregulation of Fas was sustained for 10 days and resulted in protection from hepatitis and liver failure. Similarly, hydrodynamically delivered siRNA directed toward hepatitis B inhibited viral replication in mice when co-injected with HBV-expressing plasmids (Giladi et al, 2003). More recently, 2′-F modified siRNAs with enhanced serum stability showed efficacy in silencing luciferase expressed from a co-injected plasmid, thereby demonstrating that chemically modified siRNAs are active in vivo (Layzer et al, 2004). Also, plasmids using H1 or U6 promoters have been used with promising results. Hao et al. have demonstrated that p53 downregulation efficiencies of H1- and U6-driven shRNAs were almost identical (Hao et al, 2005). Recently, Takahashi et al. have studied the gene silencing kinetics of short hairpin RNA (shRNA)-expressing plasmid DNA (pDNA) driven by U6, H1 or tRNA promoter (pU6-shLuc, pH1-shLuc, and ptRNA-shLuc) in melanoma cells expressing firefly luciferase (Takahashi et al, 2008). These studies have demonstrated that U6 promoter-driven shRNA expressing pDNA is the most effective in inducing gene silencing effect as far as the intensity and duration of RNAi effect is concerned, they. The main drawback of using Pol III promoters is their inability to process multi-cistronic constructs efficiently unlike CMV, which is a Pol II promoter. Recently, researchers have also described tRNA(Lys3)-shRNA chimeric expression cassettes, which produce siRNAs with comparable efficacy and strand selectivity to U6-expressed shRNAs and show that their activity is consistent with processing by endogenous 3′ tRNAse (Scherer et al, 2007).
Although hydrodynamic injection provides an important proof-of-concept demonstration that RNAi can function in vivo, it does not represent a therapeutically viable method due to the large volumes required. Interestingly, when plasmid vectors expressing siRNAs are used, large quantities of injection are not required due to plasmid transcription activity. We used plasmid-mediated RNAi targeting uPAR and uPA, which did not induce 2′,5′-oligoadenylate synthetase 1 (OAS1) expression, indicating no non-specific activation of the antiviral interferon response pathway in mice with pre-established intracranial tumors. Furthermore, the small interfering RNA plasmid targeting uPAR and uPA caused regression of pre-established intracranial tumors when compared with controls (Gondi et al, 2007). We have also demonstrated that intraperitoneal injections of RNAi expressing plasmids targeting uPAR, MMP-9, cathepsin B and uPA are capable of regressing intracranial tumors. In these studies, we used novel bi- or multi-cistronic plasmid construction method where a single plasmid was developed to generate multiple RNAi molecules mediated via hpRNAs driven by a single CMV promoter and which were processed to siRNAs via internal RNAi machinery (Gondi et al, 2004a;Gondi et al, 2004b;Gondi et al, 2007;Lakka et al, 2004). Given that the RNAi effect is limited only by the half life of the plasmids in vivo, the advantage of using multi-cistronic constructs over single constructs is that a single plasmid molecule can produce multiple RNAi molecules specific to more than one target, thereby enhancing the RNAi effect and resulting in higher efficiency. In contrast, methods utilizing the direct injection of siRNA molecules are limited by the volume that can be injected.
Viral delivery
Currently, researchers are exploring the use of both viral and nonviral vectors for siRNA delivery. The viability of inserting genetic material into a vector encoding shRNA, which efficiently blocks production of a specific protein, has been demonstrated (Zentilin and Giacca, 2004). Retroviral vectors have been designed to produce siRNA driven by either U6 or H1-RNA promoters for efficient, uniform delivery and immediate selection of stable knockdown in cells. Adenoviral vectors have been demonstrated to mediate gene silencing in in vitro lung and breast models (Shen et al, 2003) and to induce RNAi in a range of animal tissues. Our studies have shown effective downregulation of MMP-2 using RNAi mediated by adenovirus in lung and meduloblastoma models (Lakka et al, 2005b;Lakka et al, 2005a). Recently, Choy et al. have demonstrated a new set of plasmid vectors driven by promoters of the Epstein-Barr virus (EBV)-encoded small RNAs (EBERs). The EBERs are the most abundant transcript in infected nasopharengial carcinoma cells and are transcribed by Pol III. The authors also showed that the EBER promoters were able to drive the expression of shRNA fusion transcripts (Choy et al, 2008). Even though no in vivo studies using these constructs were conducted, the potential for in vivo use is very promising. Researchers using a lentiviral transfection system have shown that the U6 promoter is more efficient than H1 in GFP silencing in vitro, leading to 80% GFP knockdown at an average of one integrated vector genome per target cell genome. In addition, the U6 promoter was shown to be superior to H1 in vivo and led to stable GFP knockdown in mouse brain for at least 9 months, thereby indicating that LV-mediated RNAi is a powerful gene-silencing method for the long-term inhibition of gene expression in vitro and in vivo (Makinen et al, 2006). Other viral methods are also being developed using retroviral vectors but their in vivo use has not been fully explored (Hao et al, 2005;Wadhwa et al, 2004). The potential to use viral vectors as targeting agents carrying siRNA-expressing constructs is very promising if issues relating to immunogenicity and toxicity are addressed. However, no conclusive reports employing targeted viruses carrying siRNA-expressing constructs have been published to date.
Lipid-based complex delivery
The use of lipids as carriers of genetic material is not new (Ledley, 1994). With the advent of RNAi, research in the use of liposomes to deliver drugs into cells has surged. Liposomes are vesicles that consist of an aqueous compartment enclosed in a phospholipid bilayer with the drug typically entrapped in the center aqueous layer. The bilayer is composed of multicomponents, typically comprising a lipid component, cholesterol, and polyethylene glycol (PEG)-lipid. The liposomes that are formed yield particles with stable physicochemical characteristics, thus making them suitable drug delivery vehicles. In contrast, lipoplexes are spontaneously formed with the interaction of cationic lipids and negatively charged nucleic acids and are typical of most commercial transfection agents, such as Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and TransIT-TKO. Lipoplexes tend to be structurally more heterogeneous and unstable, aggregating over time in solution, and as a result, lipoplexes must be prepared immediately before use. These represent major disadvantages from the standpoint of reproducibility, manufacturing, and drug administration.
Significant and repeated success with liposome-mediated delivery has been reported in vivo. One of the most important advances in the development of RNAi as a therapeutic came with the demonstration that systemically delivered siRNA in a stable nucleic-acid-lipid form targeting ApoB was able to silence ApoB in mice and non-human primates (Zimmermann et al, 2006). While most lipid-based nucleic acid delivery systems make use of a cationic lipid to associate with the negatively charged siRNA backbone, neutral liposomal delivery systems have also proven effective (Spagnou et al, 2004;Welz et al, 2000). Using the same dioleoyl phosphatidylcholine (DOPC)-based delivery system, studies using siRNAs directed against EphA2 (Landen, Jr. et al, 2005) and focal adhesion kinase (Halder et al, 2006) were able to demonstrate specific target protein knockdown and inhibition of tumor growth in an orthotropic mouse model of ovarian cancer. In both studies, animals were treated with formulated siRNA at a dose of 150 mg/kg twice a week for 3 weeks. More recently, the authors have reported the ability of neuropilin-2 siRNA formulated in DOPC liposomes to inhibit the growth of colorectal carcinoma cells implanted in the murine liver (Gray et al, 2008). As neutral lipid-based formulations are generally well tolerated, these results are encouraging. No conclusive studies using siRNA plasmids or expression cassettes employing the use of lipids or lipid complexes have been reported yet in an in vivo setting. These types of studies could aid in the faster development of therapy models for effective in vivo siRNA delivery.
Other delivery methods
In addition to the in vivo delivery methods already described, other methods are also being developed and show significant potential as efficient strategies for the delivery of therapeutic siRNAs. Rozema et al. (Rozema et al, 2007) have demonstrated an ingenious targeted delivery method using dynamic polyconjugates and also shown effective delivery of siRNA in vivo in mice by silencing endogenous ApoB and peroxisome proliferator-activated receptor [PPAR]-α mRNA in hepatocytes. Although this study did not deal with cancer, it is a very significant demonstration of a novel method that could be used to deliver siRNAs of interest to cancer therapeutics. Another polymer approach involves using transferrin-targeted, cyclodextrin-containing polycation nanoparticles. These nanoparticles have demonstrated targeted silencing of the EWS-FLI1 gene product in transferrin receptor-expressing Ewing’s sarcoma tumor cells (Hu-Lieskovan et al, 2005). The siRNA formulated in these nanoparticles was well tolerated in non-human primates (Heidel et al, 2007). Both of these delivery strategies are appealing in that they incorporate rational approaches using targeted delivery and endosomal escape mechanisms.
In addition to these methods, there are several other methods that are being developed for the delivery of siRNA in vivo, such as the direct siRNA conjugation method that utilizes the conjugation of the siRNA molecules to larger molecules like cholesterol (Wolfrum et al, 2007). However, the utility of this method for cancer therapy has not been explored fully. Conjugation of siRNA molecules with proteins or peptides is being explored, especially conjugation with signal peptides targeting specific cells (Kumar et al, 2007). The concept of conjugating antibodies to siRNA molecules has also been proposed (Peer et al, 2007) and holds great promise as a targeted therapeutic approach for various cancers.
One of the earlier methods showing promise as a delivery strategy involves the use of atelocollagen. Atelocollagen was the first biomaterial with the potential for use as a carrier for gene delivery (Ochiya et al, 1999). Atelocollagen has been obtained from type I collagen of calf dermis by pepsin treatment (Ochiya et al, 2001;Sano et al, 2003). Atelocollagen obtained by pepsin treatment is low in immunogenicity because it is free from telopeptides, and is used clinically for a wide range of purposes. The surface of atelocollagen molecules is positively charged, and the molecules can bond electrostatically with negatively charged nucleic acid molecules. The size of the complex particles can be changed by altering the ratio of nucleic acid to atelocollagen. When the concentration of atelocollagen is high, the complex persists locally for a long time, which functions as a sustained release carrier. One problem for systemic treatment in vivo is that, although most endogenous RNases are inactive against dsRNA, some serum RNases can degrade siRNA. However, siRNA complexed with atelocollagen is resistant to nucleases and is transduced efficiently into cells, thereby allowing long-term gene silencing at the site of delivery (Minakuchi et al, 2004).
Clinical Trials
RNAi has rapidly advanced from research discovery to clinical trials with several new RNAi-based drugs expected to enter trials in 2008. Many researchers have developed strategies to target specific organs or tissues (Pirollo and Chang, 2008). Initial RNAi involving trials have focused on direct local delivery of siRNA and on well-validated therapeutic targets, such as the VEGF pathway for the treatment of the wet form of age-related macular degeneration (AMD) (Grunweller and Hartmann, 2005) and the respiratory syncytial virus (RSV) genome for the treatment of RSV (Whitehead et al, 2009) infection. The RNAi therapeutics being developed for ophthalmic indications all employ direct injection into the vitreal cavity to efficiently target the siRNA to the back of the eye whereas the RSV RNAi therapeutic uses direct delivery to the lung.
The development of RNAi-based therapies targeted against cancer is quickly progressing. Calando Pharmaceuticals has initiated the first anti-cancer, RNAi-based clinical trial (Clinical Trial #NCT00689065, 2008) using CALAA-01, which is a targeted molecule designed to inhibit tumor growth and/or reduce tumor size. The active ingredient in CALAA-01 is a small interfering RNA (siRNA) and inhibits tumor growth by targeting the expression of the M2 subunit of ribonucleotide reductase (RRM2). It was originally developed for pancreatic adenocarcinoma resistant to gemcitabine and to assess the efficacy of siRNA directed against RRM2 in suppressing resistance to gemcitabine (Duxbury et al, 2004b). The CALAA-01 siRNA is protected from nuclease degradation within a stabilized nanoparticle targeted to tumor cells. The outcome of this study will initiate a new era in cancer therapies.
Future of RNAi in cancer therapy
The growth of RNAi technology and the potential of this phenomenon to control gene expression very precisely have opened avenues for the management of diseases and metabolic conditions previously thought unmanageable. Cancer is the alteration of gene expression causing uncontrolled gene expressions. The identification of these altered expression or mutations is the key for the precise targeting of these molecules using RNAi. Chemotherapy drugs are broad based and usually not targeted to one particular molecule. Even if target-specific these drugs are plagued with side effects causing a deterioration in the quality of life in many cases is worse than the disease itself (Jain, 2006). So far RNAi had shown no such unmanageable side effects and the development of efficient delivery methods will allow for the development of individualized anti-cancer RNAi molecules suited to a particular patient. This growth and development of RNAi delivery systems will assist in controlling cancer to a more manageable disease (Li and Huang, 2008;Moreira et al, 2008;Racz and Hamar, 2008).
Conclusion
The effective targeted delivery of specific siRNA molecules to cancer cells in vivo is the key to controlling and, to a certain extent, curing most malignancies. From the direct delivery method to the development of complex targeted liposomes harboring multi-cistronic siRNA expressing plasmids, cancer researchers have focused on RNAi. The discovery of new cancer-specific molecules and pathways would provide a greater impetus for the further development of targeted therapies. The use of multi-cistronic constructs in plasmids expressing multiple siRNA molecules that can be specifically targeted to cancer cells would allow for the treatment cancer as a manageable disease if not a complete cure.
Acknowledgments
This research was supported by National Cancer Institute Grant CA75557, CA92393, CA95058, CA116708, N.I.N.D.S. NS47699, NS57529, and NS61835 and Caterpillar, Inc., OSF St. Francis, Inc. Peoria, IL (to J.S.R.). Contents are sole the responsibility of the authors and do not necessarily represent the official views of NIH.
Literature Cited
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Aigner A. Cellular delivery in vivo of siRNA-based therapeutics. Curr Pharm Des. 2008;14:3603–3619. doi: 10.2174/138161208786898815. [DOI] [PubMed] [Google Scholar]
- Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Formulation and Delivery of siRNA by Oleic Acid and Stearic Acid Modified Polyethylenimine. Mol Pharm. 2009;6:121–133. doi: 10.1021/mp8000815. [DOI] [PubMed] [Google Scholar]
- Apirion D. The fate of mRNA and rRNA in Escherichia coli. Brookhaven Symp Biol. 1975:286–306. [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Braun A. Biosafety in handling gene transfer vectors. Curr Protoc Hum Genet. 2006 doi: 10.1002/0471142905.hg1201s50. Chapter 12:Unit. [DOI] [PubMed] [Google Scholar]
- Carrere N, Vernejoul F, Souque A, Asnacios A, Vaysse N, Pradayrol L, Susini C, Buscail L, Cordelier P. Characterization of the bystander effect of somatostatin receptor sst2 after in vivo gene transfer into human pancreatic cancer cells. Hum Gene Ther. 2005;16:1175–1193. doi: 10.1089/hum.2005.16.1175. [DOI] [PubMed] [Google Scholar]
- Chien PY, Wang J, Carbonaro D, Lei S, Miller B, Sheikh S, Ali SM, Ahmad MU, Ahmad I. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005;12:321–328. doi: 10.1038/sj.cgt.7700793. [DOI] [PubMed] [Google Scholar]
- Choy EY, Kok KH, Tsao SW, Jin DY. Utility of Epstein-Barr virus-encoded small RNA promoters for driving the expression of fusion transcripts harboring short hairpin RNAs. Gene Ther. 2008;15:191–202. doi: 10.1038/sj.gt.3303055. [DOI] [PubMed] [Google Scholar]
- Safety study of CALAA-01 to treat solid tumor cancers. 2008. Clinical Trial # NCT00689065. Calando PharmaceuticalsLast updated September 19, 2008. [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Benoit E, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting focal adhesion kinase enhances pancreatic adenocarcinoma gemcitabine chemosensitivity. Biochem Biophys Res Commun. 2003;311:786–792. doi: 10.1016/j.bbrc.2003.10.060. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004a;23:1448–1456. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004b;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Ann Surg. 2004c;240:667–674. doi: 10.1097/01.sla.0000140755.97224.9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S, Courtin A, it-Si-Ali S, Guglielmi J, Merle C, Harel-Bellan A, Clezardin P, Cabon F. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919–3922. [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Dinh D, Olivero W, Gujrati M, Rao JS. Downregulation of uPA, uPAR and MMP-9 using small, interfering, hairpin RNA (siRNA) inhibits glioma cell invasion, angiogenesis and tumor growth. Neuron Glia Biology. 2004a;1:165–176. doi: 10.1017/s1740925x04000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. RNAi-mediated inhibition of cathepsin B and uPAR leads to decreased cell invasion, angiogenesis and tumor growth in gliomas. Oncogene. 2004b;23:8486–8496. doi: 10.1038/sj.onc.1207879. [DOI] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Intraperitoneal injection of an hpRNA-expressing plasmid targeting uPAR and uPA retards angiogenesis and inhibits intracranial tumor growth in nude mice. Clin Cancer Res. 2007;13:4051–4060. doi: 10.1158/1078-0432.CCR-06-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Van BG, Dallas NA, Xia L, Wang X, Yang AD, Somcio RJ, Lin YG, Lim S, Fan F, Mangala LS, Arumugam T, Logsdon CD, Lopez-Berestein G, Sood AK, Ellis LM. Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver. J Natl Cancer Inst. 2008;100:109–120. doi: 10.1093/jnci/djm279. [DOI] [PubMed] [Google Scholar]
- Grunweller A, Hartmann RK. RNA interference as a gene-specific approach for molecular medicine. Curr Med Chem. 2005;12:3143–3161. doi: 10.2174/092986705774933489. [DOI] [PubMed] [Google Scholar]
- Guan H, Zhou Z, Wang H, Jia SF, Liu W, Kleinerman ES. A small interfering RNA targeting vascular endothelial growth factor inhibits Ewing’s sarcoma growth in a xenograft mouse model. Clin Cancer Res. 2005;11:2662–2669. doi: 10.1158/1078-0432.CCR-04-1206. [DOI] [PubMed] [Google Scholar]
- Halder J, Kamat AA, Landen CN, Jr., Han LY, Lutgendorf SK, Lin YG, Merritt WM, Jennings NB, Chavez-Reyes A, Coleman RL, Gershenson DM, Schmandt R, Cole SW, Lopez-Berestein G, Sood AK. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin Cancer Res. 2006;12:4916–4924. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao DL, Liu CM, Dong WJ, Gong H, Wu XS, Liu DP, Liang CC. Knockdown of human p53 gene expression in 293-T cells by retroviral vector-mediated short hairpin RNA. Acta Biochim Biophys Sin (Shanghai) 2005;37:779–783. doi: 10.1111/j.1745-7270.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herweijer H, Wolff JA. Progress and prospects: naked DNA gene transfer and therapy. Gene Ther. 2003;10:453–458. doi: 10.1038/sj.gt.3301983. [DOI] [PubMed] [Google Scholar]
- Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Huang C, Li M, Chen C, Yao Q. Small interfering RNA therapy in cancer: mechanism, potential targets, and clinical applications. Expert Opin Ther Targets. 2008;12:637–645. doi: 10.1517/14728222.12.5.637. [DOI] [PubMed] [Google Scholar]
- Ito M, Yamamoto S, Nimura K, Hiraoka K, Tamai K, Kaneda Y. Rad51 siRNA delivered by HVJ envelope vector enhances the anti-cancer effect of cisplatin. J Gene Med. 2005;7:1044–1052. doi: 10.1002/jgm.753. [DOI] [PubMed] [Google Scholar]
- Jain KK. Commercial potential of RNAi. Mol Biosyst. 2006;2:523–526. doi: 10.1039/b611485g. [DOI] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Kunigal S, Lakka SS, Gondi CS, Estes N, Rao JS. RNAi-mediated downregulation of urokinase plasminogen activator receptor and matrix metalloprotease-9 in human breast cancer cells results in decreased tumor invasion, angiogenesis and growth. Int J Cancer. 2007;121:2307–2316. doi: 10.1002/ijc.22962. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lakka SS, Chittivelu S, Joseph P, Rao JS. Adenovirus-mediated siRNA targeting gelatinase B inhibits lung tumor growth and reduces systemic metastasis of human lung cancer cells in nude mice. 2005a. p. 79. [Google Scholar]
- Lakka SS, Dinh D, Olivero W, Gujrati M, Rao JS. Adenovirus-mediated MMP-2 siRNA delivery downregulates tumor growth and invasion in medulloblastoma. 2005b. p. 698. [Google Scholar]
- Lakka SS, Gondi CS, Dinh DH, Olivero WC, Gujrati M, Rao VH, Sioka C, Rao JS. Specific interference of uPAR and MMP-9 gene expression induced by double-stranded RNA results in decreased invasion, tumor growth and angiogenesis in gliomas. J Biol Chem. 2005c;280:21882–21892. doi: 10.1074/jbc.M408520200. [DOI] [PubMed] [Google Scholar]
- Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr., Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Law M, Jafari M, Chen P. Physicochemical characterization of siRNA-peptide complexes. Biotechnol Prog. 2008;24:957–963. doi: 10.1002/btpr.13. [DOI] [PubMed] [Google Scholar]
- Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledley FD. Non-viral gene therapy. Curr Opin Biotechnol. 1994;5:626–636. doi: 10.1016/0958-1669(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle. 2006;5:2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- Li SD, Huang L. Targeted delivery of siRNA by nonviral vectors: lessons learned from recent advances. Curr Opin Investig Drugs. 2008;9:1317–1323. [PubMed] [Google Scholar]
- Li W, Szoka FC., Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- Makinen PI, Koponen JK, Karkkainen AM, Malm TM, Pulkkinen KH, Koistinaho J, Turunen MP, Yla-Herttuala S. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J Gene Med. 2006;8:433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Minakuchi Y, Takeshita F, Kosaka N, Sasaki H, Yamamoto Y, Kouno M, Honma K, Nagahara S, Hanai K, Sano A, Kato T, Terada M, Ochiya T. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32:e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JN, Santos A, Moura V, Pedroso de Lima MC, Simoes S. Non-viral lipid-based nanoparticles for targeted cancer systemic gene silencing. J Nanosci Nanotechnol. 2008;8:2187–2204. doi: 10.1166/jnn.2008.319. [DOI] [PubMed] [Google Scholar]
- Neidhardt J, Wycisk K, Klockener-Gruissem B. Viral and nonviral gene therapy for treatment of retinal diseases. Ophthalmologe. 2005;102:764–771. doi: 10.1007/s00347-005-1245-z. [DOI] [PubMed] [Google Scholar]
- Nogawa M, Yuasa T, Kimura S, Tanaka M, Kuroda J, Sato K, Yokota A, Segawa H, Toda Y, Kageyama S, Yoshiki T, Okada Y, Maekawa T. Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J Clin Invest. 2005;115:978–985. doi: 10.1172/JCI23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Ochiya T, Nagahara S, Sano A, Itoh H, Terada M. Biomaterials for gene delivery: atelocollagen-mediated controlled release of molecular medicines. Curr Gene Ther. 2001;1:31–52. doi: 10.2174/1566523013348887. [DOI] [PubMed] [Google Scholar]
- Ochiya T, Takahama Y, Nagahara S, Sumita Y, Hisada A, Itoh H, Nagai Y, Terada M. New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet. Nat Med. 1999;5:707–710. doi: 10.1038/9560. [DOI] [PubMed] [Google Scholar]
- Pal A, Ahmad A, Khan S, Sakabe I, Zhang C, Kasid UN, Ahmad I. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int J Oncol. 2005;26:1087–1091. [PubMed] [Google Scholar]
- Pardridge WM. shRNA and siRNA delivery to the brain. Adv Drug Deliv Rev. 2007;59:141–152. doi: 10.1016/j.addr.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci U S A. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. doi: 10.1158/0008-5472.CAN-07-5810. [DOI] [PubMed] [Google Scholar]
- Pushparaj PN, Aarthi JJ, Manikandan J, Kumar SD. siRNA, miRNA, and shRNA: in vivo applications. J Dent Res. 2008;87:992–1003. doi: 10.1177/154405910808701109. [DOI] [PubMed] [Google Scholar]
- Racz Z, Hamar P. SiRNA technology, the gene therapy of the future? Orv Hetil. 2008;149:153–159. doi: 10.1556/OH.2008.28289. [DOI] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci U S A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano A, Maeda M, Nagahara S, Ochiya T, Honma K, Itoh H, Miyata T, Fujioka K. Atelocollagen for protein and gene delivery. Adv Drug Deliv Rev. 2003;55:1651–1677. doi: 10.1016/j.addr.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Buck AK, Liu X, Winkler M, Reske SN. Gene silencing by adenovirus-delivered siRNA. FEBS Lett. 2003;539:111–114. doi: 10.1016/s0014-5793(03)00209-6. [DOI] [PubMed] [Google Scholar]
- Shen Y. Advances in the development of siRNA-based therapeutics for cancer. IDrugs. 2008;11:572–578. [PubMed] [Google Scholar]
- Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- Spankuch B, Matthess Y, Knecht R, Zimmer B, Kaufmann M, Strebhardt K. Cancer inhibition in nude mice after systemic application of U6 promoter-driven short hairpin RNAs against PLK1. J Natl Cancer Inst. 2004;96:862–872. doi: 10.1093/jnci/djh146. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Yamagata S, Shimizu A, Miyoshi H, Mizuguchi H, Hayakawa T, Miyagishi M, Taira K, Kawakami Y. Gene therapy for human small-cell lung carcinoma by inactivation of Skp-2 with virally mediated RNA interference. Gene Ther. 2005;12:95–100. doi: 10.1038/sj.gt.3302391. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Yamaoka K, Nishikawa M, Takakura Y. Quantitative and temporal analysis of gene silencing in tumor cells induced by small interfering RNA or short hairpin RNA expressed from plasmid vectors. J Pharm Sci. 2008;98:74–80. doi: 10.1002/jps.21398. [DOI] [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y, Hanai K, Kato T, Sano A, Ochiya T. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci U S A. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummalapalli P, Gondi CS, Dinh DH, Gujrati M, Rao JS. RNA interference-mediated targeting of urokinase plasminogen activator receptor and matrix metalloproteinase-9 gene expression in the IOMM-Lee malignant meningioma cell line inhibits tumor growth, tumor cell invasion and angiogenesis. Int J Oncol. 2007;31:5–17. [PMC free article] [PubMed] [Google Scholar]
- Turner JJ, Jones S, Fabani MM, Ivanova G, Arzumanov AA, Gait MJ. RNA targeting with peptide conjugates of oligonucleotides, siRNA and PNA. Blood Cells Mol Dis. 2007;38:1–7. doi: 10.1016/j.bcmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9:1291–1300. [PubMed] [Google Scholar]
- Wadhwa R, Kaul SC, Miyagishi M, Taira K. Vectors for RNA interference. Curr Opin Mol Ther. 2004;6:367–372. [PubMed] [Google Scholar]
- Wang XL, Xu R, Lu ZR. A peptide-targeted delivery system with pH-sensitive amphiphilic cell membrane disruption for efficient receptor-mediated siRNA delivery. J Control Release. 2008 doi: 10.1016/j.jconrel.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Welz C, Neuhuber W, Schreier H, Repp R, Rascher W, Fahr A. Nuclear gene targeting using negatively charged liposomes. Int J Pharm. 2000;196:251–252. doi: 10.1016/s0378-5173(99)00433-0. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- Xia CF, Boado RJ, Pardridge WM. Antibody-Mediated Targeting of siRNA via the Human Insulin Receptor Using Avidin-Biotin Technology. Mol Pharm. 2008 doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Wang Y, Powell R, Chan P. Polymeric core-shell nanoparticles for therapeutics. Clin Exp Pharmacol Physiol. 2006;33:557–562. doi: 10.1111/j.1440-1681.2006.04408.x. [DOI] [PubMed] [Google Scholar]
- Yano J, Hirabayashi K, Nakagawa S, Yamaguchi T, Nogawa M, Kashimori I, Naito H, Kitagawa H, Ishiyama K, Ohgi T, Irimura T. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clin Cancer Res. 2004;10:7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]
- Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, Waltemathe M, Gosling T, Flemming P, Malek NP, Trautwein C, Manns MP, Kuhnel F, Kubicka S. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentilin L, Giacca M. In vivo transfer and expression of genes coding for short interfering RNAs. Curr Pharm Biotechnol. 2004;5:341–347. doi: 10.2174/1389201043376742. [DOI] [PubMed] [Google Scholar]
- Zhang C, Newsome JT, Mewani R, Pei J, Gokhale PC, Kasid UN. Systemic delivery and pre-clinical evaluation of nanoparticles containing antisense oligonucleotides and siRNAs. Methods Mol Biol. 2009;480:65–83. doi: 10.1007/978-1-59745-429-2_5. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kon T, Wang H, Li F, Huang Q, Rabbani ZN, Kirkpatrick JP, Vujaskovic Z, Dewhirst MW, Li CY. Enhancement of hypoxia-induced tumor cell death in vitro and radiation therapy in vivo by use of small interfering RNA targeted to hypoxia-inducible factor-1alpha. Cancer Res. 2004a;64:8139–8142. doi: 10.1158/0008-5472.CAN-03-2301. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang YF, Bryant J, Charles A, Boado RJ, Pardridge WM. Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin Cancer Res. 2004b;10:3667–3677. doi: 10.1158/1078-0432.CCR-03-0740. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]