Abstract
Endogenous ligands for cannabinoid receptors (“endocannabinoids”) include the lipid transmitters anandamide and 2-arachidonoylglycerol (2-AG). Endocannabinoids modulate a diverse set of physiological processes and are tightly regulated by enzymatic biosynthesis and degradation. Termination of anandamide signaling by fatty acid amide hydrolase (FAAH) is well-characterized, but less is known about the inactivation of 2-AG, which can be hydrolyzed by multiple enzymes in vitro, including FAAH and monoacylglycerol lipase (MAGL). Here, we have taken a functional proteomic approach to comprehensively map 2-AG hydrolases in the mouse brain. Our data reveal that ~85% of brain 2-AG hydrolase activity can be ascribed to MAGL, with the remaining 15% being mostly catalyzed by two uncharacterized enzymes ABHD6 and ABHD12. Interestingly, MAGL, ABHD6, and ABHD12 display distinct subcellular distributions, suggesting that they may control distinct pools of 2-AG in the nervous system.
The lipid transmitters anandamide (AEA) and 2-arachidonoylglycerol (2-AG) are referred to as ‘endocannabinoids’ because they serve as natural ligands for the cannabinoid (CB) receptors, which are also targets of the psychoactive component of marijuana, Δ9-tehtrahydrocannibinol [1, 2]. Endocannabinoids are biosynthesized from membrane phospholipid precursors in an activity-dependent manner, stimulate CB receptors in the vicinity of their production, and are inactivated by rapid enzymatic hydrolysis. Endocannabinoid signaling has been found to modulate a number of physiological processes, including pain sensation [3], appetite [4], and cognitive and emotional state [5, 6].
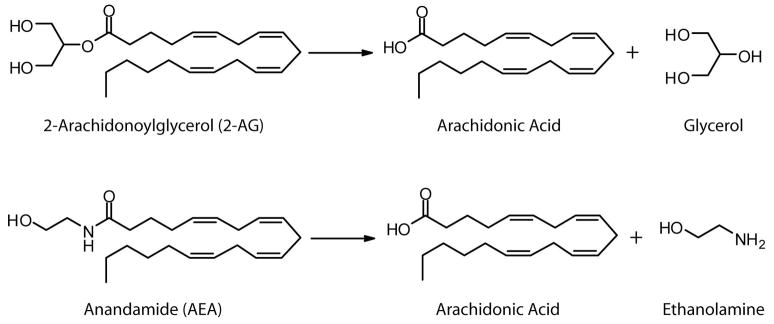
Endocannabinoid levels are tightly controlled by enzymatic biosynthesis and degradation in vivo, and, as such, the enzymes responsible for these processes are considered central components of endocannabinoid signaling networks [7–10]. The hydrolysis of AEA to arachidonic acid and ethanolamine is principally mediated by a single enzyme in the nervous system - the integral membrane enzyme fatty acid amide hydrolase (FAAH) (Figure 1) [11, 12]. Brain tissue from FAAH-knockout mice [13] or rodents treated with FAAH inhibitors [14] are essentially void of AEA-hydrolyzing activity. Moreover, endogenous brain levels of AEA and other bioactive fatty acid amides are dramatically elevated in FAAH-disrupted animals, leading to a variety of CB receptor-dependent behavioral phenotypes [3, 13–16]. In contrast, much less is understood about the enzymes that terminate 2-AG signaling in vivo. The hydrolysis of 2-AG to arachidonic acid and glycerol (Figure 1), can be performed by multiple enzymes in vitro including FAAH [17], neuropathy target esterase (NTE) [18], and the cytosolic enzymes monoacylglycerol lipase (MAGL) [19] and hormone sensitive lipase (HSL) [20]. Whether these enzymes constitute the principle or, for that matter, only 2-AG hydrolases in mammalian tissues, however, remains unknown.
Figure 1.
Structures of 2-arachidonoylglycerol (2-AG), anandamide (AEA) and their hydrolysis products.
It has generally been assumed that MAGL is the main contributor to 2-AG hydrolysis in the brain [9]. MAGL is a serine hydrolase originally purified and cloned from adipose tissue [21], where it is thought to catalyze the final step of triglyceride metabolism. MAGL is also abundant in brain tissue [19], where it localizes to presynaptic terminals [22]. Multiple studies have provided evidence for the involvement of MAGL in 2-AG hydrolysis in the nervous system. Overexpression of MAGL in rat cortical neurons was found to reduce the activity-dependent accumulation of 2-AG [19]. Immunodepletion of MAGL in cytosolic rat brain fractions decreased 2-AG hydrolysis by ~50% [23]. Similarly, treatment of rat cerebellar membranes with maleimide reagents that irreversibly inhibit MAGL decreased 2-AG hydrolysis by ~85% [24]. Most recently, first-generation MAGL inhibitors have been reported to raise brain 2-AG levels and produce anti-hyperalgesic effects in rodents [3, 25]. However, the efficacy and/or selectivity of these inhibitors have since been called into question [26, 27]. Taken together, these results suggest that MAGL likely plays an important role in 2-AG hydrolysis in the brain. That MAGL inhibition or immunodepletion did not completely eliminate 2-AG hydrolysis in brain preparations, however, indicates the existence of additional pathways for 2-AG degradation in the nervous system. Consistent with this idea, the murine microglial BV2 cell line was recently shown to possess 2-AG hydrolase activity despite lacking MAGL expression [28].
Here, we have adopted a functional proteomic strategy to assemble a complete and quantitative profile of mouse brain enzymes that display 2-AG hydrolase activity. Our studies confirm that MAGL is a primary 2-AG hydrolase in brain tissue and also identify two previously uncharacterized enzymes, ABHD6 and ABHD12, that possess this activity. MAGL, ABHD6, and ABHD12 collectively account for at least 98% of the total 2-AG hydrolase activity in brain. We further show that each enzyme exhibits a distinct subcellular distribution, suggesting that they may regulate distinct pools of 2-AG in the nervous system.
Results
A Comprehensive Profile of Candidate 2-AG Hydrolases in Mouse Brain
Previous studies demonstrated that 2-AG hydrolase activity in rodent brain is sensitive to the general serine hydrolase inhibitors methyl arachidonyl fluorophosphonate (MAFP) and phenylmethylsulphonyl fluoride (PMSF) [29]. Based on these results, we hypothesized that all 2-AG hydrolases in mouse brain could be collectively enriched and identified using the activity-based proteomic probe fluorophosphonate-biotin (FP-biotin), which broadly targets the serine hydrolase superfamily [30].
We first determined that FP-biotin (5 μM, 1 hr) blocked greater than 98% of the 2-AG hydrolase activity in mouse brain membrane or soluble proteomes (Figure 2). We then enriched brain targets of FP-biotin by avidin chromatography and identified these proteins using an advanced liquid chromatography-mass spectrometry (LC-MS) platform termed ABPP-MudPIT (activity-based protein profiling-multidimensional protein identification technology) [31]. Briefly, following binding to avidin beads, FP-biotin-labeled proteins were digested on-bead with trypsin, analyzed by multidimensional LC-MS, and identified using the SEQUEST search algorithm [32]. ABPP-MudPIT analysis identified a total of 32 metabolic serine hydrolases in mouse brain membrane (Table 1) and soluble (Table S1) proteomes. Among these hydrolases were several well-characterized enzymes (e.g., FAAH, MAGL, HSL, acetylcholinesterase), as well as many enzymes of unknown function. Serine proteases, components of the proteasome and fatty acid synthase were also identified as targets of FP-biotin, but these proteins were not regarded as likely contributors to brain 2-AG hydrolase activity and therefore excluded from subsequent analyses.
Figure 2.

Mouse brain 2-AG hydrolase activity is completely inhibited by the activity-based proteomic probe FP-biotin. The 2-AG hydrolase activity of membrane and soluble mouse brain fractions was reduced by >98% following treatment with FP-biotin (5 μM, 1 hr). 2-AG hydrolase activity was measured under the following conditions: 50 mM Tris-HCl, pH 7.5, 0.05 μg protein/mL, 100 μM 2-AG, 200 μL reaction volume, 10 min, room temperature. Results represent the average values ± standard errors of the mean (SEM) for 3 independent experiments.
Table 1.
List of the metabolic serine hydrolases identified in the mouse brain membrane proteome by ABPP-MudPIT. Peptide spectral counts of serine hydrolases in the membrane fraction are reported as the average values of 3 individual ABPP-MudPIT experiments ± SEM. Spectral count values for serine hydrolases identified in the soluble fraction are listed in Table S1.
| Ensemble Identifier | Common Name | Abbreviation | Average Spectral Counts | SEM |
|---|---|---|---|---|
| ENSMUSG00000027698 | KIAA1363 | KIAA1363 | 163 | 26 |
| ENSMUSG00000025277 | α/β-hydrolase 6 | ABHD6 | 149 | 7 |
| ENSMUSG00000033174 | Monoacylglycerol lipase | MAGL | 123 | 19 |
| ENSMUSG00000032046 | α/β-hydrolase 12 | ABHD12 | 110 | 20 |
| ENSMUSG00000034171 | Fatty acid amide hydrolase | FAAH | 99 | 36 |
| ENSMUSG00000007036 | HLA-B associated transcript 5 | BAT5 | 51 | 5 |
| ENSMUSG00000021996 | Esterase 10/Esterase D | ES10 | 37 | 5 |
| ENSMUSG00000070889 | GPI deacylase | GPID | 33 | 8 |
| ENSMUSG00000036257 | Similar to calcium independent phospholipase A2 | iPLA2-2 | 32 | 2 |
| ENSMUSG00000047368 | Cgi67 | CGI67 | 31 | 6 |
| ENSMUSG00000002475 | α/β-hydrolase 3 | ABHD3 | 30 | 6 |
| ENSMUSG00000033157 | α/β-hydrolase 10 | ABHD10 | 30 | 2 |
| ENSMUSG00000023913 | Phospholipase A2 group VII | PLA2g7 | 25 | 2 |
| ENSMUSG00000072949 | Acyl-coenzyme A thioesterase 1 | ACOT1 | 24 | 5 |
| ENSMUSG00000040997 | α/β-hydrolase 4 | ABHD4 | 24 | 1 |
| ENSMUSG00000003346 | BC005632 | BC0 | 20 | 2 |
| ENSMUSG00000004565 | Neuropathy target esterase | NTE | 17 | 7 |
| ENSMUSG00000038459 | RIKEN cDNA 2210412D01 gene | R2D01 | 15 | 2 |
| ENSMUSG00000023328 | Acetylcholinesterase | AChE | 14 | 1 |
| ENSMUSG00000028670 | Acyl-protein thioesterase 2 | APT2 | 14 | 1 |
| ENSMUSG00000031903 | Lysophospholipase 3 | LYPLA3 | 14 | 1 |
| ENSMUSG00000040532 | α/β-hydrolase 11 | ABHD11 | 14 | 2 |
| ENSMUSG00000030718 | Protein phosphatase methylesterase 1 | PME1 | 13 | 1 |
| ENSMUSG00000039246 | Lysophospholipase-like protein 1 | LPL1 | 13 | 4 |
| ENSMUSG00000025903 | Acyl-protein thioesterase 1 | APT1 | 12 | 1 |
| ENSMUSG00000001229 | Dipeptidyl peptidase 9 | DPP9 | 9 | 1 |
| ENSMUSG00000027428 | Retinoblastoma-binding protein 9 | RBBP9 | 8 | 2 |
| ENSMUSG00000032590 | Acyl peptide hydrolase | APEH | 7 | 1 |
| ENSMUSG00000005447 | Platelet-activating factor acetylhydrolase IB subunit gamma | PAFAH Ibg | 6 | 1 |
| ENSMUSG00000003123 | Hormone sensitive lipase | HSL | 5 | 1 |
Evaluation of the 2-AG Hydrolase Activity of Recombinantly Expressed Brain Serine Hydrolases
To determine which of the 32 brain serine hydrolases displayed 2-AG hydrolase activity, we recombinantly expressed each enzyme in COS-7 cells by transient transfection. Successful expression of 30 of the 32 proteins as active enzymes was confirmed by ABPP analysis using a rhodamine-tagged FP probe (FP-rhodamine; Figure 3 and Figure S1). The only exceptions were Ca2+-insensitive phospholipase A2 gamma (iPLA2γ) and glycerophosphoinositol deacylase (GPID); however, since both of these enzymes have been ascribed metabolic functions distinct from 2-AG hydrolysis [33], [34], we suspected that their contributions to this activity would be negligible. Serine hydrolase-transfected cell homogenates were assayed for 2-AG hydrolase activity relative to a mock-transfected homogenate (transfected with empty vector). The data were further normalized to account for differences in the expression levels of active enzymes, which were measured by in-gel fluorescence scanning of FP-rhodamine labeling intensities. This analysis identified seven enzymes that displayed significant 2-AG hydrolase activity: MAGL, FAAH, HSL, NTE, platelet-activating factor subunit 1B gamma (PAFAH IBγ), α/β-hydrolase-6 (ABHD6) and α/β-hydrolase-12 (ABHD12) (Figure S2).
Figure 3.
Recombinant expression of mouse brain serine hydrolases. COS-7 cells transiently transfected with mouse brain serine hydrolase cDNAs were labeled with FP-rhodamine (2 μM, 1 hr), separated by SDS-PAGE, and analyzed by in-gel fluorescence scanning to confirm expression of active enzymes. Expression efficiency of active enzyme was calculated from the integrated fluorescence intensities of the asterisked bands. Fluorescent gel show in grayscale.
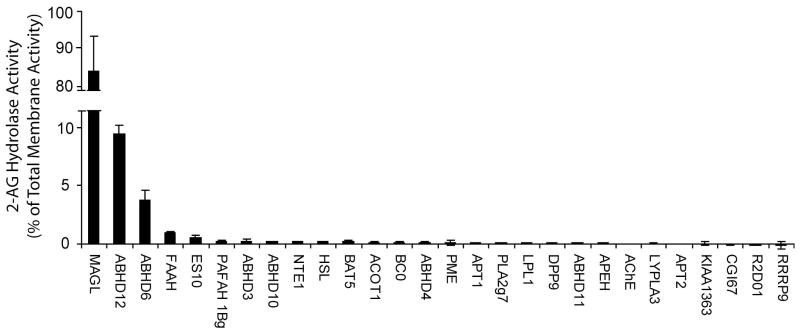
In order to estimate the relative contribution that each of these seven enzymes made to total brain 2-AG hydrolase activity, we needed to account for their relative expression levels in brain tissue. Since brain 2-AG hydrolase activity was primarily localized to membranes (~90% of total activity; Figure 2), we focused our attention on this proteomic fraction. Expression levels of serine hydrolases in the brain membrane proteome were estimated by first quantifying their average spectral counts in ABPP-MudPIT data sets of FP-biotin-enriched samples (Table 1). These values were then normalized to account for differences in the number of theoretical tryptic peptides per protein (largely a reflection of differences in molecular mass). This “isotope-free” method for protein quantification has been shown to provide accurate estimates of the absolute expression levels of proteins in proteomic samples [35]. The resulting profile of normalized brain 2-AG hydrolase activities is shown in Figure 4. Consistent with previous studies [24], MAGL was found to be the principal 2-AG hydrolase in mouse brain, accounting for approximately 85% of the total membrane activity. Interestingly, however, none of other known 2-AG hydrolases (FAAH, HSL, NTE) made substantial contributions to the remaining brain activity, which instead was mostly attributable to the two uncharacterized enzymes ABHD12 (~9%) and ABHD6 (~4%). FAAH was identified as the next largest contributor to 2-AG hydrolysis, accounting for ~1% of total membrane activity.
Figure 4.
The relative 2-AG hydrolase activities for mouse brain membrane serine hydrolases. SH-transfected cell homogenates were assayed for 2-AG hydrolase activity (50 mM Tris-HCl, pH 7.5, 0.05 μg protein/mL, 100 μM 2-AG, 10 min, room temperature), and these values were normalized to account for differences in enzyme expression in transfected cells. To determine the relative contribution of each enzyme to total brain membrane 2-AG hydrolase activity, the results were further normalized based on mouse brain expression levels for each serine hydrolase, as estimated by their average spectral count values from the ABPP-MudPIT analysis (Table 1) corrected for the number of theoretical tryptic peptides per enzyme. Results represent the average values ± SEM of 2 independent experiments for 2 separate transfections per enzyme (n = 4).
Pharmacological Characterization of 2-AG Hydrolases in Mouse Brain
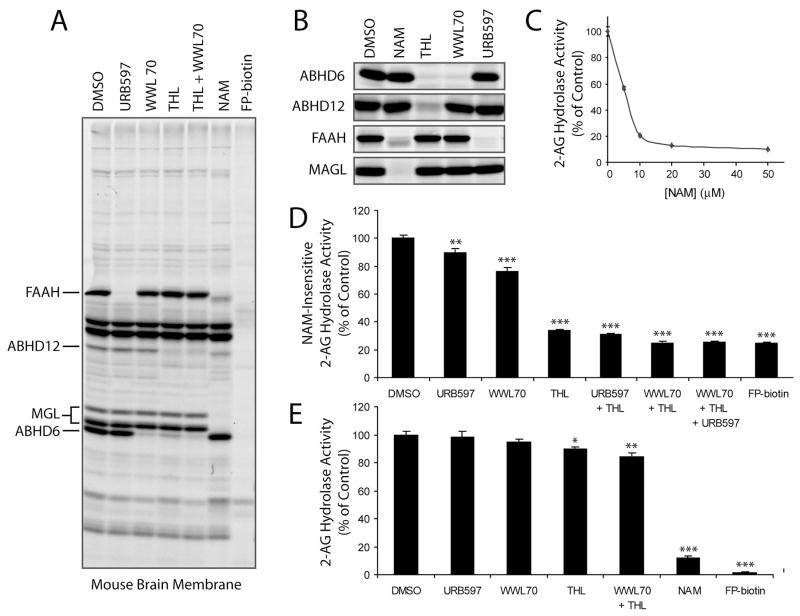
Our functional proteomic analysis designated MAGL, ABHD12, and ABHD6 as the major 2-AG hydrolases in mouse brain membranes, with a minor contribution also being made by FAAH. To confirm these results, we sought to identify inhibitors that could discriminate among these enzymes. The FAAH inhibitor URB597 [14] has previously been shown to specifically target this enzyme in brain membrane proteomes [36, 37]. We recently reported a selective inhibitor of ABHD6, WWL70, that was discovered in a competitive ABPP screen of a carbamate library [38]. Consistent with these previous studies, we observed selective blockade of FP-rhodamine labeling of FAAH and ABHD6 in brain membrane proteomes treated with URB597 (10 μM) and WWL70 (10 μM), respectively (Figure 5A). Although specific inhibitors of MAGL and ABHD12 are not yet available, we determined by competitive ABPP [39] that N-arachidonoyl maleimide (NAM) [24] and the lipase inhibitor tetrahydrolipstatin (THL) [40] inactivated these enzymes, respectively, without targeting a large number of additional serine hydrolases in brain membrane proteomes (Figure 5A). In addition to blocking FP-rhodamine labeling of MAGL, NAM appeared to at least partially affect FAAH and ABHD12, especially at higher inhibitor concentrations (10–50 μM), where both proteins showed altered migration by SDS-PAGE (presumably due to covalent modification of free cysteine residues by the NAM inhibitor) (Figure 5A and Figure S3). THL (20 μM), on the other hand, completely blocked FP-rhodamine labeling of ABHD12 and partially blocked ABHD6, but did not inhibit MAGL or FAAH (Figure 5A and Figure S3). We confirmed these general inhibitor sensitivity profiles using recombinantly expressed enzymes (Figure 5B).
Figure 5.
Effects of inhibitors of MAGL, ABHD12 and ABHD6 on brain membrane 2-AG hydrolase activity. (A) The selectivity profiles of NAM (50 μM), WWL70 (10 μM), THL (20 μM), URB597 (10 μM), and FP-biotin (5 μM) as judged by competitive ABPP analysis [1 hr pre-incubation with inhibitors, followed by 1 hr labeling with FP-rhodamine (2 μM)] in the mouse brain membrane proteome (A) and transfected COS-7 proteomes (B). Note that MAGL migrates as two distinct protein bands in the brain proteome, consistent with previous findings [23, 48]. (C) NAM treatment (0 – 50 μM, 1 hr) inhibited up to 85% of the 2-AG hydrolase activity of the mouse brain membrane proteome. (D) Effects of inhibitors of ABHD12, ABHD6, and FAAH [with THL (20 μM), WWL70 (10 μM), and URB597 (10 μM), respectively] on the “NAM-resistant” 2-AG hydrolase activity of the mouse brain membrane proteome. Assays were conducted in proteomic samples pre-treated with NAM (50 μM, 10 min) to block MAGL activity. (E) Effects of inhibitors on total brain membrane 2-AG hydrolase activity. For C-E, results represent the average ± SEM of 3–5 individual experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 for inhibited versus control (DMSO) treated samples.
We next tested the effects of inhibitors on the 2-AG hydrolase activity of brain membrane preparations. Consistent with previous studies [24], NAM (20–50 μM) blocked ~85% of the total brain membrane 2-AG hydrolase activity (Figure 5C). THL (20 μM), WWL70 (10 μM), and URB597 (10 μM) individually blocked 66, 24, and 10% of the remaining “NAM-resistant” activity (Figure 5D). Combined treatment of NAM, THL and WWL70, which completely blocked MAGL, ABHD6, and ABHD12 (as judged by ABPP; Figure S3) decreased the total membrane 2-AG hydrolase activity by ~98%, lowering it to a level equivalent to inhibition by FP-biotin (Figure 5D). We also applied THL, WWL70, and URB597 to untreated (i.e., NAM-free) brain membrane proteome. THL (20 μM) significantly decreased 2-AG hydrolysis by 10.5 +/1.9% (Figure 5E). URB597 and WWL70 caused more modest reductions in 2-AG hydrolysis activity (1.9 +/− 4.3 and 5.1 +/− 2.3%, respectively) (Figure 5E), but these values did not reach statistical significance compared to control reactions performed in the absence of inhibitor. Combined treatment of the brain membrane proteome with THL and WWL70 decreased 2-AG hydrolysis by 15.6 +/− 2.7% (Figure 5E), matching remarkably well the residual level of activity observed in MAGL-inhibited (i.e., NAM-treated) samples. Collectively, these data provide further evidence that the hydrolysis of 2-AG in brain membranes is dictated by the combined action of three enzymes – MAGL, ABHD6, and ABHD12.
MAGL, ABHD6 and ABHD12 Display Distinct Subcellular Distributions
Why might the brain possess multiple 2-AG hydrolases? One possibility is that these enzymes exhibit different cellular and/or subcellular distributions, which could impart upon them the ability to regulate distinct pools of 2-AG in the brain. Consistent with this premise, hydropathy plots predicted that both ABHD6 and ABHD12 are integral membrane enzymes (Figure S4). In contrast, MAGL is a soluble enzyme that associates with membranes in a peripheral manner [12]. We confirmed these general distributions by ABPP analysis of cellular fractions of transfected COS-7 proteomes (Figure 6A), as well as by analysis of the original ABPP-MudPIT data sets of brain membrane and soluble proteomes (Table S1). Interestingly, treatment of transfected proteomes with the general N-linked glycosidase PNGaseF resulted in a significant shift in gel migration for ABHD12, but not ABHD6 or MAGL (Figure 6A). These data indicate that ABHD12 is an integral membrane enzyme with its active site oriented toward the luminal/extracellular compartments of the cell, while ABHD6 is an integral membrane enzyme that faces the cytoplasm (Figure 6B). These data thus invoke a model where each of the major 2-AG hydrolases in brain membranes resides in a distinct subcellular compartment – integral membrane, lumenal/extracellular orientation (ABHD12); integral membrane, cytoplasmic orientation (ABHD6); and soluble/peripheral membrane, cytoplasmic orientation (MAGL).
Figure 6.
MAGL, ABHD12 and ABHD6 exhibit distinct subcellular distributions. (A) The distribution of enzymes in the membrane and soluble fractions of transfected COS-7 cells, as judged by ABPP analysis. PNGaseF treatment revealed that ABHD12, but not MAGL or ABHD6, is a glycoprotein, indicating a luminal/extracellular orientation for this enzyme. (B) Cartoon model representing the predicted orientations of the principal 2-AG hydrolases in mouse brain.
Discussion
The hydrolysis of monoacylglycerides, and 2-AG in particular, has been ascribed as an activity to many enzymes in vitro, including several that are expressed in the nervous system (e.g., MAGL, FAAH, HSL, and NTE). These findings starkly contrast with the degradation of AEA, which is principally dictated by a single brain enzyme, FAAH, and raise the pertinent question – which enzymes in the nervous system make the most significant contribution to 2-AG hydrolysis in vivo? To answer this question, a more complete and quantitative understanding of the brain enzymes that exhibit 2-AG hydrolase activity is required. Recognizing that virtually all 2-AG hydrolase activity in brain tissue is sensitive to inhibition by general serine hydrolase inhibitors, such as MAFP or PMSF [29], we adopted a functional proteomic approach to fully inventory the 2-AG hydrolases expressed in mouse brain. This approach took advantage of the activity-based probe FP-biotin [30] coupled with advanced LC-MS methods [31] to assemble a list of 32 serine hydrolases expressed in brain. Recombinant expression of these enzymes identified seven proteins that hydrolyzed 2-AG. Using ‘isotope-free’, spectral counting methods to quantify the relative expression levels of these enzymes in brain tissue, we were able to estimate their respective contributions to total 2-AG hydrolase activity. These estimates were confirmed by pharmacological studies with inhibitors that showed distinct selectivity profiles for the brain 2-AG hydrolases.
MAGL was found to mediate ~85% of total brain membrane 2-AG hydrolase activity. This value matches remarkably well previous reports that used the MAGL inhibitor NAM to block 80–85% of cerebellar membrane 2-AG hydrolase activity [24]. These data, in conjunction with our competitive ABPP studies (e.g., see Figure 5A), further argue that NAM, despite containing a highly reactive maleimide group, exhibits rather high selectivity for MAGL relative to other brain serine hydrolases. MAGL has been modeled to contain a non-catalytic cysteine residue in its active site [24], which could account for its unusual sensitivity to maleimide reagents. We initially anticipated that the remaining 15% of brain 2-AG hydrolase activity might be due to other enzymes known to hydrolyze 2-AG, such as FAAH, HSL, and NTE. However, our data indicate that none of these enzymes makes substantial contributions to total brain 2-AG hydrolase activity. Instead, the ‘MAGL-independent’ 2-AG hydrolase activity is largely mediated by two enzymes of previously uncharacterized function – ABHD12 (~9%) and ABHD6 (~4%), with the remaining ~2% activity presumably being performed by FAAH and/or other enzymes. These percent contribution values were determined at near physiologic pH (pH 7.5), and it is important to note that they may be pH-dependent.
When contemplating why the brain might contain multiple enzymes with 2-AG hydrolase activity, we consider the following points. First, these enzymes could exhibit distinct cellular or subcellular distributions, or undergo different forms of regulated expression. Indeed, the metabolism of other classes of bioactive molecules, including acetylcholine [41], monoamines [42], and prostaglandins [43], has been shown to be regulated by multiple enzymes or multiple isoforms of the same enzyme. This “redundancy” presumably offers cells greater versatility to tailor the magnitude and duration of small-molecule signaling events to meet specific physiological objectives. For example, neurons that express membrane-bound versus secreted acetylcholinesterase isoforms display differences in synaptic signal strength due to distinct rates of acetylcholine degradation at the synapse [41]. In this context, it is noteworthy that MAGL, ABHD6, and ABHD12 each displayed a distinct subcellular distribution. We speculate that these enzymes may have preferred access to distinct pools of 2-AG in vivo, which could in turn shape the signaling activity of this endocannabinoid at different synapses throughout the nervous system. Recent work from the Parsons group also argues for the existence of distinct pools of 2-AG in the brain. These authors determined by in vivo microdialysis that extracellular levels of 2-AG are approximately 200-fold lower than total brain levels of this lipid [44], indicating that only a small fraction of total 2-AG may be “signaling-competent”. Of course, elucidating the respective roles of MAGL, ABHD12, and ABHD6 as regulators of 2-AG signaling in vivo will require selective genetic and/or pharmacological tools to perturb their individual functions. In this regard, it is noteworthy that both ABHD6 and ABHD12 were inactivated by the lipase inhibitor THL. This finding indicates that ABHD6 and ABHD12 likely share active site structural similarity, despite showing very low sequence homology (< 20%). Active site relatedness among enzymes from the serine hydrolase family that lack sequence identity has been noted previously [39].
It is also possible that ABHD6 and/or ABHD12 may play a more dominant role in 2-AG hydrolysis in cells that lack MAGL. It will be interesting, for example, to determine whether these enzymes contribute to 2-AG hydrolysis in microglial cells, which have recently been shown to possess this activity despite lacking MAGL [28]. Finally, it is also possible that ABHD6 and ABHD12 metabolize endogenous substrates that are distinct from 2-AG. Such has proven to be the case with FAAH, which despite hydrolyzing 2-AG in vitro [17], is primarily responsible for degrading fatty acid amide substrates in vivo [45], [46]. On this subject, however, we do believe it is instructive to place the 2-AG hydrolase activities of ABHD6 and ABHD12 in perspective by noting that they exceed the rate of FAAH-catalyzed hydrolysis of AEA in brain tissue by ~10–20-fold (2.6 and 5.9 nmol/min/mg vrs 0.3 nmol/min-mg [13]). Thus, although ABHD12 and ABHD6 only contribute to ~15% of the total 2-AG hydrolysis in brain, their activities are still quite high compared to other pathways for lipid transmitter degradation.
In summary, we have performed herein the first comprehensive characterization of brain enzymes that hydrolyze 2-AG. These studies both confirm the role of established 2-AG hydrolases, such as MAGL, and designate the previously uncharacterized enzymes ABHD12 and ABHD6 as potential regulators of endocannabinoid signaling pathways. Assuming that one or more of these enzymes is confirmed to regulate 2-AG degradation in vivo, they might constitute useful therapeutic targets for a range of nervous system disorders. More generally, we suggest that the functional proteomic strategy put forth in this manuscript could be employed to comprehensively inventory enzymes that possess other hydrolytic activities of relevance to mammalian signaling and physiology, including, for example, diacyl- and triacylglyceride metabolism and the production and/or degradation of bioactive lipids such as lysophosphatidic acid.
Significance
Endocannabinoids are lipid transmitters that modulate a wide range of physiological processes. The two principal endocannabinoids in the nervous system, anandamide (AEA) and 2-arachidonoyl glycerol (2-AG), are regulated by distinct biosynthetic and degradative pathways. Identifying the enzymes that control AEA and 2-AG metabolism is imperative to achieve a mechanistic understanding of endocannabinoid signaling networks and to control these pathways for therapeutic gain. While the degradation of AEA has been shown to be principally mediated by the integral membrane enzyme fatty acid amide hydrolase (FAAH), multiple enzymes, including FAAH and monoacylglycerol lipase (MAGL), are known to hydrolyze 2-AG in vitro. Here, we have adopted a functional proteomic strategy to comprehensively inventory the 2-AG hydrolases expressed in mouse brain. We find that virtually all brain 2-AG hydrolase activity (> 98%) can be accounted for by three enzymes – MAGL (~85% of total), ABHD12 (~9% of total), and ABHD6 (~4% of total). These results thus confirm that MAGL is the principal, but not sole 2-AG hydrolase in mammalian brain. The discovery of ABHD12 and ABHD6 as novel 2-AG hydrolases with subcellular distributions distinct from MAGL designates these enzymes as potentially new components of the endocannabinoid system. We speculate that MAGL, ABHD12, and ABHD6 may regulate different cellular or subcellular pools of 2-AG, thereby making unique contributions to endocannabinoid signaling in vivo. More generally, we believe that the functional proteomic methods detailed herein can be applied to globally inventory enzymes that possess any hydrolytic activity of relevance to mammalian physiology.
Experimental Procedures
Materials
2-arachidonoylglycerol (2-AG), pentadecanoic acid, N-arachidonoyl maleimide (NAM) and URB597 were purchased from Cayman Chemical (Ann Arbor, MI). Tetrahydrolipostatin (THL) was purchased from Sigma-Aldrich. FP-biotin and FP-rhodamine were prepared as previously described [30, 47].
Preparation of Mouse Brain Proteomes
Brains were harvested from wild-type C57Bl/6J mice and immediately frozen on dry ice. The brains were then Dounce-homogenized in Tris buffer (50 mM Tris-HCl pH 7.5) with 150 mM NaCl, sonicated, and centrifuged at slow speed (1000 g for 10 min at 4°C) to remove debris. The supernatant was centrifuged at high speed (145,000 g for 45 min at 4°C) and this supernatant was saved as the cytosolic proteome. The pellet was resuspended in Tris buffer, sonicated, incubated at 4°C with rotation and centrifuged at high speed (145,000 g for 45 min at 4°C). This wash procedure was performed twice and the final pellet was resuspended in Tris buffer and saved as the membrane proteome. The total protein concentration of each proteome was determined using the Bio-Rad Dc Protein Assay kit. Aliquots of the proteomes were stored at −80°C until use.
ABPP-MudPIT Analysis of Mouse Brain Proteomes
Mouse brain proteomes (1 mg in 1 ml Tris buffer) were treated with 5 μM FP-biotin for 1 hr at room temperature. Preparation of the labeled samples for ABPP-MudPIT analysis was performed as previously described [31] except that the Lys-C digestion step was omitted. MudPIT analysis was performed as previously described [31] on an LTQ ion trap mass spectrometer (ThermoFisher) coupled to an Agilent 1100 series HPLC. The tandem MS data were searched against the mouse IPI database using the SEQUEST search algorithm, and results were filtered and grouped with DTASELECT. Peptides with cross-correlation scores greater than 1.8 (+1), 2.5 (+2), 3.5 (+3) and delta CN scores greater than 0.08 were included in the spectral counting analysis. Only proteins for which an average of 10 or more spectral counts were identified in either soluble or membrane samples were included in the subsequent analysis. Spectral counts are reported as the average of three samples with the standard error (SE) of the mean.
Recombinant Expression of Brain Serine Hydrolases in COS-7 Cells
Full-length cDNAs encoding mouse serine hydrolases identified by MudPIT analysis were purchased from OpenBioSystems (Huntsville, AL), with the exception of APH1 and RBBP9, for which rat and human cDNAs were purchased, respectively. cDNAs were either transfected directly (if available in a eukaryotic expression vector) or subcloned into pcDNA3 (Invitrogen). Transient transfections were performed as follows. COS-7 cells were grown to ~70% confluence in 100-mm dishes in complete medium (DMEM with L-glutamine, nonessential amino acids, sodium pyruvate, and FBS) at 37°C and 5% CO2. The cells were transiently transfected using the appropriate cDNA or empty vector control (“mock”) and the FUGENE 6 (Roche Applied Science) or Lipofectamine (Invitrogen) transfection reagents following the manufacturers’ protocols. After 48 h, the cells were washed twice with phosphate-buffered saline (PBS), collected by scraping, resuspended in 250 μl Tris buffer, and lysed by sonication. The lysates were either used in assays as whole cell homogenates or centrifuged at 145,000 g for 45 min at 4°C to isolate the cytosolic and membrane fractions. Protein concentrations were determined using the Bio-rad Dc protein assay and aliquots of the homogenates were stored at −80°C until use. Successful overexpression was confirmed by treatment of the cell homogenates (50 μg in 50 μl Tris buffer) with 2 μM FP-rhodamine for 1 hr at room temperature. Reactions were quenched with 4x SDS/PAGE loading buffer (reducing), separated by SDS/PAGE (10% acrylamide) and visualized in-gel with a Hitachi FMBio IIe flatbed fluorescence scanner (MiraiBio). Relative expression efficiency of active enzyme was determined by calculating integrated band intensities of the labeled proteins. For deglycosylation studies of ABHD6, ABHD12, FAAH and MAGL, a portion of the FP-labeled cytosolic or membrane lysates was treated with PNGase F (New England Biolabs) for 45 min before SDS/PAGE analysis.
Enzyme Activity Assays
Enzyme assays of whole cell lysates were performed in Tris buffer in a total volume of 200 μl using 10 μg total protein (except for MAGL, where 0.1 μg of MAGL-transfected cell lysate was diluted into 9.9 μg mock-transfected lysate). The reactions were incubated for 5 min at room temperature with 100 μM of synthetic 2-AG (2 μl of 10 mM stock in DMSO). The reactions were quenched by the addition of 500 μl of chloroform and 200 μl of MeOH, vortexed to mix and centrifuged for 5 min at 1,400 g to separate phases. The organic phase was extracted and 50 μl was injected onto an Agilent 1100 series LC-MS. Briefly, chromatography was performed on a 50 × 4.60 mm 5 micron Gemini C18 column (Phenomenex), products and standards were eluted with a 5 min gradient of 0 to 100% Buffer B in Buffer A (Buffer A: 95% H2O, 5% MeOH, 0.1% ammonium hydroxide; Buffer B: 60% iPrOH, 35% MeOH, 5% H2O, 0.1% ammonium hydroxide) and mass analyzed in negative mode Arachidonic acid release was measured by comparison with a pentadecanoic acid standard. The relative activity of each enzyme was calculated by subtracting mock activity and normalizing for expression efficiency in COS-7 using the integrated band intensity from the SDS/PAGE gel of the FP-rhodamine labeled cell lysates. In order to account for the relative expression of serine hydrolases in the mouse brain, their 2-AG hydrolase activities were normalized using the average spectral count data from ABPP-MudPIT analysis corrected for the number of theoretical tryptic peptides per protein. The theoretical number of tryptic peptides for each serine hydrolase was calculated based on amino acid sequence, and tryptic peptides either less than 6 or greater than 39 amino acids in length were excluded from the numerical estimate. This analysis of the data also assumes that each individual 2-AG hydrolase was labeled to completion by FP-biotin, a premise that was validated by the complete blockade of brain 2-AG hydrolase activity under the standard labeling conditions used for ABPP-MudPIT (see Figure 2). Assays were conducted in duplicate for each of two separate transfections, and the error bars represent the standard error of the mean (SEM).
For activity assays of mouse brain, membrane fractions were diluted to 1 mg/ml in Tris buffer and treated with DMSO, 5–50 μM NAM, 20 μM THL, 10 μM WWL70, 10 μM URB597, 5 μM FP-biotin or combinations thereof for 1 hr at room temperature. A portion of this reaction was labeled with FP-rhodamine as described above. 2-AG hydrolysis activity assays were performed essentially as described above. Inhibitor-treated mouse brain proteome (10 μg) diluted in Tris buffer was incubated with 100 μM of synthetic 2-AG for either 10 or 60 min (reaction time was adjusted to give ~15% hydrolysis for the most active sample). The reactions were quenched and analyzed by LC-MS as described above. Assays were conducted with n ≥ 3 and the error bars represent the SEM.
Supplementary Material
Acknowledgments
We thank B.Q. Wei for assistance in generating programs for normalization of spectral counting data and the Cravatt group for helpful discussions and careful reading of the manuscript. This work was supported by the National Institutes of Health grants DA015197 and DA017259 (B.F.C), the Daniel Koshland Fellowship in Enzyme Biochemistry (G.M.S.), a Fletcher Jones Foundation Scholarship (J.L.B.), the Skaggs Institute for Chemical Biology, and the Helen L. Dorris Institute for the Study of Neurological and Psychiatric Disorders of Children and Adolescents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–884. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- 2.Di Marzo V, Bisogno T, De Petrocellis L. Endocannabinoids and Related Compounds: Walking Back and Forth between Plant Natural Products and Animal Physiology. Chemistry & Biology. 2007;14:741–756. doi: 10.1016/j.chembiol.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, Krey JF, Walker JM, Holmes PV, Crystal JD, Duranti A, Tontini A, Mor M, Tarzia G, Piomelli D. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- 4.Di Marzo V, Goparaju SK, Wang L, Liu J, Batkai S, Jarai Z, Fezza F, Miura GI, Palmiter RD, Sugiura T, Kunos G. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature. 2001;410:822–825. doi: 10.1038/35071088. [DOI] [PubMed] [Google Scholar]
- 5.Marsicano G, Wotjak CT, Azad SC, Bisogno T, Rammes G, Cascio MG, Hermann H, Tang J, Hofmann C, Zieglgansberger W, Di Marzo V, Lutz B. The endogenous cannabinoid system controls extinction of aversive memories. Nature. 2002;418:530–534. doi: 10.1038/nature00839. [DOI] [PubMed] [Google Scholar]
- 6.Varvel SA, Lichtman AH. Evaluation of CB1 receptor knockout mice in the Morris water maze. J Pharmacol Exp Ther. 2002;301:915–924. doi: 10.1124/jpet.301.3.915. [DOI] [PubMed] [Google Scholar]
- 7.Patricelli MP, Cravatt BF. Proteins regulating the biosynthesis and inactivation of neuromodulatory fatty acid amides. Vitam Horm. 2001;62:95–131. doi: 10.1016/s0083-6729(01)62002-8. [DOI] [PubMed] [Google Scholar]
- 8.Ueda N. Endocannabinoid hydrolases. Prostaglandins Leuko Essent Fatty Acids . 2002;68–69:521–534. doi: 10.1016/s0090-6980(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 9.Lambert DM, Fowler CJ. The endocannabinoid system: drug targets, lead compounds, and potential therapeutic applications. J Med Chem. 2005;48:5059–5087. doi: 10.1021/jm058183t. [DOI] [PubMed] [Google Scholar]
- 10.Kogan NM, Mechoulam R. The chemistry of endocannabinoids. J Endocrinol Invest. 2006;29:3–14. [PubMed] [Google Scholar]
- 11.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 12.McKinney MK, Cravatt BF. Structure and function of Fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 13.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 15.Lichtman AH, Shelton CC, Advani T, Cravatt BF. Mice lacking fatty acid amide hydrolase exhibit a cannabinoid receptor-mediated phenotypic hypoalgesia. Pain. 2004;109:319–327. doi: 10.1016/j.pain.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Varvel SA, Cravatt BF, Engram AE, Lichtman AH. Fatty acid amide hydrolase (−/−) mice exhibit an increased sensitivity to the disruptive effects of anandamide or oleamide in a working memory water maze task. J Pharmacol Exp Ther. 2006;317:251–257. doi: 10.1124/jpet.105.095059. [DOI] [PubMed] [Google Scholar]
- 17.Goparaju SK, Ueda N, Yamaguchi H, Yamamoto S. Anandamide amidohydrolase reacting with 2-arachidonoylglycerol, another cannabinoid receptor ligand. FEBS Lett. 1998;422:69–73. doi: 10.1016/s0014-5793(97)01603-7. [DOI] [PubMed] [Google Scholar]
- 18.van Tienhoven M, Atkins J, Li Y, Glynn P. Human Neuropathy Target Esterase Catalyzes Hydrolysis of Membrane Lipids. J Biol Chem. 2002;277:20942–20948. doi: 10.1074/jbc.M200330200. [DOI] [PubMed] [Google Scholar]
- 19.Dinh TP, Freund TF, Piomelli D. A role for monoacylglyceride lipase in 2-arachidonoylglycerol inactivation. Chem Phys Lipids. 2002;2002:149–158. doi: 10.1016/s0009-3084(02)00150-0. [DOI] [PubMed] [Google Scholar]
- 20.Belfrage P, Jergil B, Strålfors P, Tornqvist H. Hormone-sensitive lipase of rat adipose tissue: identification and some properties of the enzyme protein. FEBS Lett. 1977;75:259–264. doi: 10.1016/0014-5793(77)80099-9. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. cDNA Cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J Biol Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 22.Gulyas AI, Cravatt BF, Bracey MH, Dinh TP, Piomelli D, Boscia F, Freund TF. Segregation of two endocannabinoid-hydrolyzing enzymes into pre- and postsynaptic compartments in the rat hippocampus, cerebellum and amygdala. Eur J Neurosci. 2004;20:441–458. doi: 10.1111/j.1460-9568.2004.03428.x. [DOI] [PubMed] [Google Scholar]
- 23.Dinh TP, Kathuria S, Piomelli D. RNA Interference Suggests a Primary Role for Monoacylglycerol Lipase in the Degradation of the Endocannabinoid 2-Arachidonoylglycerol. Mol Pharmacol. 2004 doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- 24.Saario SM, Salo OM, Nevalainen T, Poso A, Laitinen JT, Jarvinen T, Niemi R. Characterization of the sulfhydryl-sensitive site in the enzyme responsible for hydrolysis of 2-arachidonoyl-glycerol in rat cerebellar membranes. Chem Biol. 2005;12:649–656. doi: 10.1016/j.chembiol.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Makara JK, Mor M, Fegley D, Szabo SI, Kathuria S, Astarita G, Duranti A, Tontini A, Tarzia G, Rivara S, Freund TF, Piomelli D. Selective inhibition of 2-AG hydrolysis enhances endocannabinoid signaling in hippocampus. Nat Neurosci. 2005;8:1139–1141. doi: 10.1038/nn1521. [DOI] [PubMed] [Google Scholar]
- 26.Saario SM, Palomaki V, Lehtonen M, Nevalainen T, Jarvinen T, Laitinen JT. URB754 has no effect on the hydrolysis or signaling capacity of 2-AG in the rat brain. Chem Biol. 2006;13:811–814. doi: 10.1016/j.chembiol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Vandevoorde S, Jonsson KO, Labar G, Persson E, Lambert DM, Fowler CJ. Lack of selectivity of URB602 for 2-oleoylglycerol compared to anandamide hydrolysis in vitro. Br J Pharmacol. 2006;150:186–191. doi: 10.1038/sj.bjp.0706971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muccioli GG, Xu C, Odah E, Cudaback E, Cisneros JA, Lambert DM, Lopez Rodriguez ML, Bajjalieh S, Stella N. Identification of a Novel Endocannabinoid-Hydrolyzing Enzyme Expressed by Microglial Cells. J Neurosci. 2007;27:2883–2889. doi: 10.1523/JNEUROSCI.4830-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saario SM, Savinainen JR, Laitinen JT, Jarvinen T, Niemi R. Monoglyceride lipase-like enzymatic activity is responsible for hydrolysis of 2-arachidonoylglycerol in rat cerebellar membranes. Biochemical Pharmacology. 2004;67:1381–1387. doi: 10.1016/j.bcp.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: The serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 32.Eng J, McCormack AL, Yates JR. An approach to correlate MS/MS data to amino acid sequences in a protein database. J Amer Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 33.Mancuso DJ, Jenkins CM, Gross RW. The Genomic Organization, Complete mRNA Sequence, Cloning, and Expression of a Novel Human Intracellular Membrane-associated Calcium-independent Phospholipase A2. J Biol Chem. 2000;275:9937–9945. doi: 10.1074/jbc.275.14.9937. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka S, Maeda Y, Tashima Y, Kinoshita T. Inositol Deacylation of Glycosylphosphatidylinositol-anchored Proteins Is Mediated by Mammalian PGAP1 and Yeast Bst1p. J Biol Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- 35.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proceedings of the National Academy of Sciences. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander JP, Cravatt BF. The putative endocannabinoid transport blocker LY2183240 is a potent inhibitor of FAAH and several other brain serine hydrolases. J Am Chem Soc. 2006;128:9699–9704. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]
- 38.Li W, Blankman JL, Cravatt BF. A Functional Proteomic Strategy to Discover Inhibitors for Uncharacterized Hydrolases. J Am Chem Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 39.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 40.Borgstrom B. Mode of action of tetrahydrolipstatin: a derivative of the naturally occuring lipase inhibitor lipstatin. Biochim Biophys Acta. 1988;962:308–316. doi: 10.1016/0005-2760(88)90260-3. [DOI] [PubMed] [Google Scholar]
- 41.Legay C. Why so many forms of Acetylcholinesterase? Microsc Res Tech. 2000;49:56–72. doi: 10.1002/(SICI)1097-0029(20000401)49:1<56::AID-JEMT7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Shih JC, Chen K, Ridd MJ. Monoamine oxidase: from genes to behavior. Annu Rev Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzpatrick FA. Cycooxygenase enzymes: regulation and function. Curr Pharm Des. 2004;10:577–588. doi: 10.2174/1381612043453144. [DOI] [PubMed] [Google Scholar]
- 44.Caille S, Alvarez-Jaimes L, Polis I, Stouffer DG, Parsons LH. Specific Alterations of Extracellular Endocannabinoid Levels in the Nucleus Accumbens by Ethanol, Heroin, and Cocaine Self-Administration. J Neurosci. 2007;27:3695–3702. doi: 10.1523/JNEUROSCI.4403-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lichtman AH, Hawkins EG, Griffin G, Cravatt BF. Pharmacological activity of fatty acid amides is regulated, but not mediated, by fatty acid amide hydrolase in vivo. J Pharmacol Exp Ther. 2002;302:73–79. doi: 10.1124/jpet.302.1.73. [DOI] [PubMed] [Google Scholar]
- 46.Osei-Hyiaman D, Depetrillo M, Harvey-White J, Bannon AW, Cravatt BF, Kuhar MJ, Mackie K, Palkovits M, Kunos G. Cocaine- and amphetamine-related transcript is involved in the orexigenic effect of endogenous anandamide. Neuroendocrinology. 2005;81:273–282. doi: 10.1159/000087925. [DOI] [PubMed] [Google Scholar]
- 47.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteome using fluorescent active site-directed probes. Proteomics. 2001;1:1067–1071. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson M, Reue K, Xia YR, Lusis AJ, Langin D, Tornqvist H, Holm C. Exon-intron organization and chromosomal localization of the mouse monoglyceride lipase gene. Gene. 2001;272:11–18. doi: 10.1016/s0378-1119(01)00559-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.