Abstract
Delta-like 1 (Dll1) is a mammalian ligand for Notch receptors. Interactions between Dll1 and Notch in trans activate the Notch pathway, whereas Dll1 binding to Notch in cis inhibits Notch signaling. Dll1 undergoes proteolytic processing in its extracellular domain by ADAM10. In this work, we demonstrate that Dll1 represents a substrate for several other members of the ADAM family. In co-transfected cells, Dll1 is constitutively cleaved by ADAM12, and the N-terminal fragment of Dll1 is released to medium. ADAM12-mediated cleavage of Dll1 is cell density-dependent, takes place in cis orientation, and does not require the presence of the cytoplasmic domain of ADAM12. Full length Dll1, but not its N- or C-terminal proteolytic fragment, co-immunoprecipitates with ADAM12. By using a Notch reporter construct, we show that Dll1 processing by ADAM12 increases Notch signaling in a cell-autonomous manner. Furthermore, ADAM9 and ADAM17 have the ability to process Dll1. In contrast, ADAM15 does not cleave Dll1, although the two proteins still co-immunoprecipitate with each other. Asn353 present in the catalytic motif of ADAM12 and other Dll1-processing ADAMs, but absent in ADAM15, is necessary for Dll1 cleavage. Dll1 cleavage is reduced in ADAM9/12/15−/− mouse embryonic fibroblasts (MEFs), suggesting that the endogenous ADAM9 and/or ADAM12 present in wild type MEFs contribute to Dll1 processing. Finally, the endogenous Dll1 present in primary mouse myoblasts undergoes cleavage in confluent, differentiating myoblast cultures and this cleavage is decreased by ADAM12 siRNAs. Our findings expand the role of ADAM proteins in the regulation of Notch signaling.
Notch signaling regulates cell fate decisions during development and in the adult (1-3). The signaling pathway is activated by direct interactions between Notch receptor, a transmembrane protein present at the surface of a signal-receiving cell, and a DSL (Delta/Serrate/Lag2) ligand, a transmembrane protein at the surface of a signal-sending cell. In mammals, there are four different Notch receptors (Notch 1 - 4), and five DSL ligands (Delta-like 1, 3, and 4, Jagged 1 and 2). Ligand-bound Notch undergoes proteolytic cleavage at the S2 site in the extracellular domain, which is mediated by ADAM10 or ADAM17 (4-6), members the ADAM family of metalloprotease-disintegrins (7, 8). This is followed by the cleavage at the S3 site in the transmembrane domain of Notch by a γ-secretase complex (9, 10). The intracellular domain of Notch translocates to the nucleus, where it interacts with CSL transcription factors and activates expression of target genes (11).
Similar to their receptors, Notch ligands also undergo ADAM-mediated cleavage in their extracellular domains, which is then followed by processing by γ-secretase (12-15). Two ADAMs were postulated to cleave Notch ligands in mammalian cells, ADAM10 and 17. ADAM10 was implicated in the processing of mouse (13) and rat Delta-like 1 (Dll1) (14), while ADAM17 was suggested to cleave rat Jagged 1 (14). Proteolytic cleavage of Dll1 in ADAM10−/− mouse embryonic fibroblasts (MEFs) still amounts to 50% of the processing observed in ADAM10+/+ MEFs (13), suggesting that ADAM10 is only partially responsible for Dll1 cleavage and other ADAMs may account for the remaining processing in ADAM10−/− cells. MEFs express several ADAM proteins with catalytically active metalloprotease domains, including ADAM9, 12, 15, and 17 (16). However, to date, none of these ADAM has been shown to be capable of cleaving Dll1.
Furthermore, while it is well established that proteolytic processing of Notch ligands down-regulates Notch signaling in neighboring cells (17, 18), the effect of ligand cleavage on the Notch pathway within the same cell is less clear. Notch receptors can associate with their ligands in a cell-autonomous manner (19-21). Associations between receptors and ligands in cis decrease Notch receptivity and attenuate the Notch pathway (19-22). In Drosophila, proteolytic processing of Delta by ADAM10-like (Kuzbanian-like; Kul) alleviates the inhibitory effect of Delta on Notch in the same cell (23). In contrast, processing of Jagged 1 in mammalian cells by ADAM17 was postulated to inhibit Notch signaling in cis due to competition of the C-terminal fragments of Jagged 1 and Notch for γ-secretase (14).
In this work, we have tested the ability of several ADAMs other than ADAM10 to cleave murine Dll1 and examined the effect of Dll1 cleavage on Notch signaling in the same cell. We show that ADAM12, which has not been previously implicated in Notch signaling, can efficiently process Dll1, but not Notch1. We demonstrate that Dll1 cleavage by ADAM12 activates Notch in a cell-autonomous manner. In addition, we show that two other ADAMs, ADAM9 and 17, are capable of Dll1 processing and we identify a residue within the ADAM catalytic motif that appears necessary for the cleavage of Dll1. The extent of processing of Dll1 transfected into ADAM9/12/15−/− MEFs is reduced when compared with the processing in wild type MEFs, suggesting that the endogenous ADAM9 and/or ADAM12 present in wild type MEFs contribute to Dll1 cleavage. Finally, we show that the endogenous Dll1 present in primary mouse myoblasts undergoes cleavage in confluent, differentiating myoblast cultures and this cleavage is decreased by ADAM12 siRNAs.
EXPERIMENTAL PROCEDURES
Expression constructs
Mouse Dll1 cDNA was amplified by PCR using a full length clone (ID 6402691; Invitrogen) as a template and cloned into pIRESpuro expression vector. c-Myc-Dll1 containing an internal c-Myc tag between amino acids 46 and 47, inserted into the Sac II site in Dll1 cDNA, was cloned into pcDNA3.1 vector. Mouse full length cDNAs of ADAM9, 12, 15, and 17 were cloned into pcDNA3.1 vector; these ADAMs contained c-Myc and 6xHis tags at their termini. An untagged ADAM12 was generated by introducing a stop codon after the C-terminal Lys residue. The E349Q and N353S ADAM12 mutants and the S354N ADAM15 mutant were generated by site-directed mutagenesis using QuickChange kit (Stratagene). Mouse Notch1 containing an intact extracellular domain and in which 348 C-terminal amino acids were replaced with six copies of c-Myc tag (pCS2+mN1FL6MT) was provided by R. Kopan (Washington University). CBF1 reporters containing four wild-type or mutated CBF1 binding sites (pJH23A and pJH25A, respectively) were provided by D. Hayward (Johns Hopkins School of Medicine); Notch reporter containing eight CBF1 binding sites (pJT123A) was provided by P. D. Ling (Baylor College of Medicine).
Cell culture and plasmid transfection
COS-7, NIH3T3, and CHO-K1 cells (American Tissue Culture Collection) were grown in Dulbecco's modified Eagle's medium (DMEM; COS-7 and NIH3T3) or F12K nutrient mixture (CHO-K1), supplemented with 10% FBS, at 37°C in the presence of 5% CO2 under a humidified atmosphere. Primary myoblasts were isolated from 2-3 day old C57BL/6 mice according to ref. 24 and were grown on collagen I-coated plates in DMEM supplemented with 20% FBS and 100 μg/ml each penicillin and streptomycin. Mouse embryonic fibroblasts (MEFs) isolated from ADAM9/12/15−/− (16) or from wild type mice and immortalized with simian virus 40 large T antigen were grown on gelatin-coated dishes in DMEM containing 10% FCS and 100 μg/ml each penicillin and streptomycin.
One day after plating, cells were transfected using Fugene6 (Roche). In most experiments, the amount of total DNA was 1 μg/well in a 6-well plate; for CBF1 reporter assays, 2.05 μg of DNA/well was used (see below). For stable expression of c-Myc-Dll1 in CHO-K1 cells, transfected cells were grown for two weeks in the presence of geneticin (800 μg/ml); a clone positive for c-Myc-Dll1 expression was isolated. Cells with the highest expression of cell-surface c-Myc were further selected by cell sorting using FACSCalibur (Becton Dickinson).
RNA interference
Three different Silencer siRNA duplexes specific for mouse ADAM12 and a negative control siRNA (control #1) were purchased from Ambion. The sequences of sense RNA strands were: CCAGAGAGGAGCUUACGAAtt (siRNA1); GCCAAUGAAAAACACCACUtt (siRNA2); GCAAAUUACCACAUUGUCUtt (siRNA3). Mouse primary myoblasts were trypsinized and transfected with individual siRNA duplexes (30 nM) using siPORT NeoFX transfection reagent (Ambion) as cells attached to plates. One day after transfection, confluent cells were transferred to differentiation medium (DMEM with 2% horse serum). The level of expression of ADAM12 and the amount of Dll1 cleavage were analyzed 40 h later.
Cell treatments
In some experiments, prior to harvesting, cells were incubated for 3 h with 5 μM lactacystin (a proteasomal inhibitor), for 6 h with 1 μM L685,458 (a γ-secretase inhibitor), for 1 h with 1 μM ionomycin, or for 1 h with 25 ng/ml phorbol myristate acetate (PMA). Cells were serum-starved for 30 min prior to and during the PMA treatment.
Western blotting
Cellular proteins were extracted with extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM 4-(2-aminoethyl)-benzene-sulfonylfluoride hydrochloride (AEBSF), 5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, 10 mM 1,10-phenanthroline; 0.5 ml extraction buffer/well in a 6-well plate). Cell extracts were centrifuged at 21,000g for 15 min, supernatants were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked with 3% (w/v) dry milk and 0.3% (v/v) Tween-20 in DPBS, then incubated with primary antibodies in blocking buffer, followed by incubation with horseradish peroxidase-labeled secondary antibodies and detection using the WestPico chemiluminescence kit (Pierce). To detect the endogenous ADAM12 in primary myoblasts, cell extracts were enriched for glycoproteins using concanavalin A agarose beads (20 μl bed volume/ml cell extract) prior to SDS-PAGE and Western blotting (25). The following primary antibodies were used: rabbit anti-ADAM12 cytoplasmic peptide antibody (1:3000), rabbit anti-ADAM12 disintegrin antibody (1:3000), rabbit anti-Dll1 (Santa Cruz Biotechnology, H-265; 0.2 μg/ml), mouse anti-c-Myc (clone 9E10, Upstate Biotechnology; 1 μg/ml). Secondary antibodies were horseradish peroxidase (HRP)-conjugated anti-rabbit IgG or anti-mouse IgG (heavy and light chain-specific) or HRP-conjugated anti-rabbit IgG (Fc fragment-specific, Jackson ImmunoResearch; 0.8 μg/ml; used in the experiment shown in Fig. 4D). Intensities of the bands in Western blots were determined by densitometry and quantified using ScionImage software. Each experiment involving quantitative determination of Dll1 cleavage was repeated at least three times.
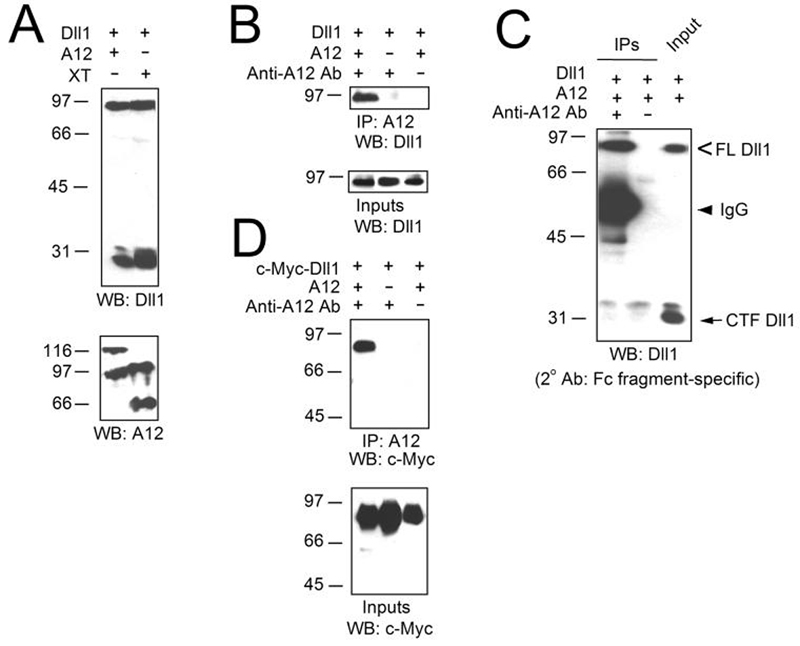
FIG. 4. Cytoplasmic domain of ADAM12 is dispensable for Dll1 cleavage (A).
COS-7 cells were co-transfected with Dll1 and either the full length ADAM12 (lane 1) or ADAM12 containing a 169-amino acid deletion at the C-terminus (XT; lane 2). Total cell lysates were analyzed by Western blotting using antibody specific for the C-terminus of Dll1 (top) or the disintegrin domain of ADAM12 (bottom). The upper bands detected with anti-ADAM12 antibody in ADAM12- and XT-transfected cells represent the nascent forms of ADAM12 or XT, respectively, the lower bands represent the mature forms lacking the pro-domain. ADAM12 forms a complex with the full length Dll1, but not with NTF or CTF (B-D). B, Cells were co-transfected with Dll1 and ADAM12 (lanes 1 and 3) or with Dll1 and empty ADAM12 expression vector (lane 2), cell extracts were subjected to immunoprecipitation with antibody specific for the C-terminus of ADAM12 (lanes 1 and 2) or were incubated without antibody (lane 3), the immunoprecipitates (IPs) and inputs were analyzed by Western blotting using anti-Dll1 antibody. C, COS-7 cells were co-transfected with Dll1 and ADAM12, cell extracts were subjected to immunoprecipitation with (lane 1) or without (lane 2) anti-ADAM12 antibody; immunoprecipitates (IPs) and inputs were analyzed by immunoblotting with anti-Dll1 antibody. The secondary antibody was anti-rabbit IgG, Fc fragment-specific, and detected only the heavy chain of anti-ADAM12 antibody used for immunoprecipitation. Notice that the relative abundance of CTF Dll1 was much higher in the input than in the immunoprecipitate. D, Experiment was performed as in B using the Dll1 construct with c-Myc tag in the N-terminal domain. The immunoprecipitates (IPs) and inputs were analyzed by Western blotting using anti-c-Myc antibody.
Metabolic labeling and immunoprecipitation
To immunoprecipitate the N-terminal fragment of Dll1 from culture media, 24 h after co-transfection with c-Myc-Dll1 and ADAM12, COS-7 cells were transferred to DMEM: methionine/cysteine-free DMEM (1:9), containing 35S EasyTag Express protein labeling mix (267 μCi/ml, Perkin Elmer). After 16 h, medium was collected, cell debris was removed by centrifugation, and supernatant was used for immunoprecipitation with 9E10 antibody (5 μg/ml). To immunoprecipitate c-Myc-tagged ADAMs or un-tagged ADAM12, 36 h after transfection cells were lyzed with extraction buffer, cell extracts were centrifuged at 21,000g for 15 min, supernatants were used for immunoprecipitation with 9E10 antibody (5 μg/ml) or with anti-ADAM12 cytoplasmic peptide antibody (1:250), respectively. To immunoprecipitate Dll1 from transfected MEFs or the endogenous Dll1 from primary myoblasts, anti-Dll1 antibody (H-265, 5 μg/ml) was used. After pre-clearing, supernatants were incubated with antibodies and Protein G Sepharose 4 Fast Flow (Amersham Biosciences), the beads were washed three times with extraction buffer, immuno-complexes were eluted with SDS-PAGE sample buffer and analyzed by electrophoresis and autoradiography or Western blotting.
CBF1 reporter assay
NIH3T3 cells in 6-well plates were transfected at 50% confluence with 0.5 μg Notch1, 0.5 μg CBF1 firefly luciferase reporter, 0.05 μg Renilla luciferase (pRL-TK), 0.5 μg ADAM12 or empty pcDNA3.1 vector, and 0.5 μg Dll1 or empty pIRES-puro vector. Twenty four hours after transfection, CHO-K1 cells stably transfected with c-Myc-Dll1 or with empty vector were added (106 cells/well) and co-cultured for additional 24 h. Firefly and Renilla luciferase activities were determined using the Dual-Luciferase reporter assay system (Promega). The activity of Renilla luciferase was used as an internal control for transfection efficiency.
RESULTS
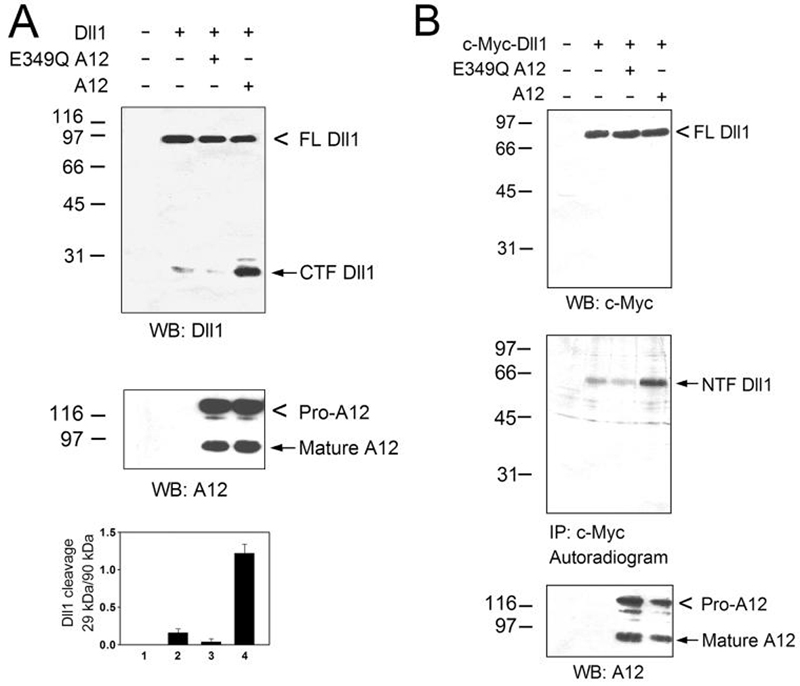
First, we asked whether ADAM12 is capable of processing Dll1. When untagged Dll1 was transfected into COS-7 cells, antibody specific for the C-terminus of Dll1 detected the full length protein of ∼90 kDa and a low level of the C-terminal Dll1 fragment (CTF) of ∼29 kDa in cell lysates (Fig. 1A). Co-transfection of wild-type mouse ADAM12, but not the catalytically-inactive E349Q mutant, strongly increased the abundance of CTF (Fig. 1A), suggesting that ADAM12 cleaved Dll1. To follow the fate of the N-terminal portion of Dll1, we introduced c-Myc tag between amino acids 46 and 47 of Dll1 (Fig. 1B). After metabolic labeling of transfected cells with [35S]methionine+cysteine, we immunoprecipitated the ∼60-kDa soluble N-terminal fragment (NTF) of Dll1 from culture medium (Fig. 1B, middle panel). The amount of detected NTF was much higher when cells were co-transfected with wild-type ADAM12, indicating that Dll1 cleavage took place in intact cells and that NTF was released to medium.
FIG. 1. ADAM12 cleaves Dll1 in intact cells.
A, COS-7 cells were transiently transfected with two empty vectors (lane 1), with mouse Delta-like 1 and empty ADAM12 expression vector (Dll1; lane 2), co-transfected with Dll1 and catalytically inactive mutant form of ADAM12 (E349Q A12; lane 3), or co-transfected with Dll1 and wild-type ADAM12 (A12; lane 4). Total cell lysates were analyzed by Western blotting using antibody specific for the C-terminus of Dll1 (top panel) or for the C-terminal domain of ADAM12 (middle panel). Full length (FL, 90-kDa band) and the C-terminal fragment (CTF, 29-kDa band) of Dll1 are indicated; the nascent pro-form of ADAM12 and the mature form of ADAM12 lacking the pro-domain are shown. The extent of Dll1 cleavage was determined as the ratio of intensities of the 29-kDa and 90-kDa Dll1 bands (mean ± S.E., n=5, bottom panel). B, Cells were transfected as in A, Dll1 construct contained c-Myc tag in the N-terminal domain. Cells were metabolically labeled with [35S]methionine+cysteine, cell lysates were analyzed by Western blotting using anti-c-Myc antibody (top) or anti-ADAM12 antibody (bottom), the N-terminal fragment (NTF) of Dll1 was immunoprecipitated from medium using anti-c-Myc antibody and visualized by SDS-PAGE and autoradiography (middle).
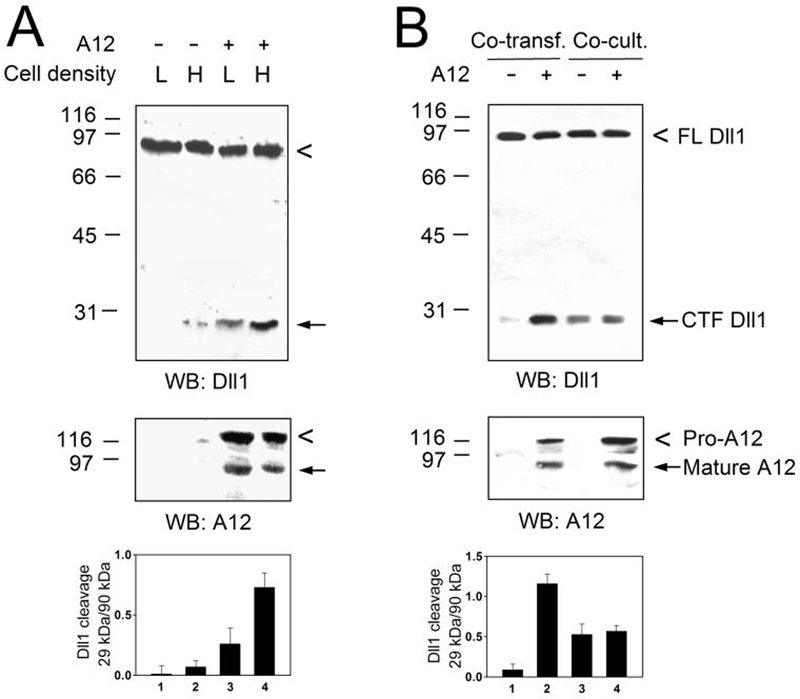
ADAM12-catalyzed processing of Dll1 (as well as the processing mediated by endogenous ADAMs present in COS-7 cells) was more efficient at high cell density (∼90% confluence at the time of assaying) than at low cell density (∼50% confluence; Fig. 2A). This result suggested that the cleavage might have taken place in trans orientation. However, co-transfection of ADAM12 and Dll1 yielded significantly more CTF than co-culture of cells that were singly-transfected with Dll1 or with ADAM12 (Fig. 2B), indicating that the cleavage occurred in cis. Thus, cell density-dependence of Dll1 cleavage suggests that interactions of Dll1 or ADAM12 with proteins present on the surface of neighboring cells may be required for the efficient processing of Dll1.
FIG. 2. Dll1 cleavage by ADAM12 is cell density-dependent and occurs in cis.
A, COS-7 cells were co-transfected with Dll1 and either empty ADAM12 expression vector (lanes 1, 2) or with vector containing ADAM12 cDNA (lanes 3,4). Cells were ∼50% confluent (low density; L) or ∼90% confluent (high density; H) at the time of harvesting. B, COS-7 cells (∼90% confluent) were co-transfected with Dll1 and empty vector (lane 1) or Dll1 and ADAM12 (lane 2). Alternatively, cells (∼50% confluent) were transfected with Dll1 alone and co-cultured with vector-transfected (lane 3) or ADAM12-transfected cells (lane 4). In A and B, total cell lysates were analyzed by Western blotting using antibodies specific for the C-termini of Dll1 or ADAM12 (top and middle panels, respectively). The extent of Dll1 cleavage was determined as the ratio of intensities of the 29-kDa and 90-kDa Dll1 bands (mean ± S.E., n=3, bottom).
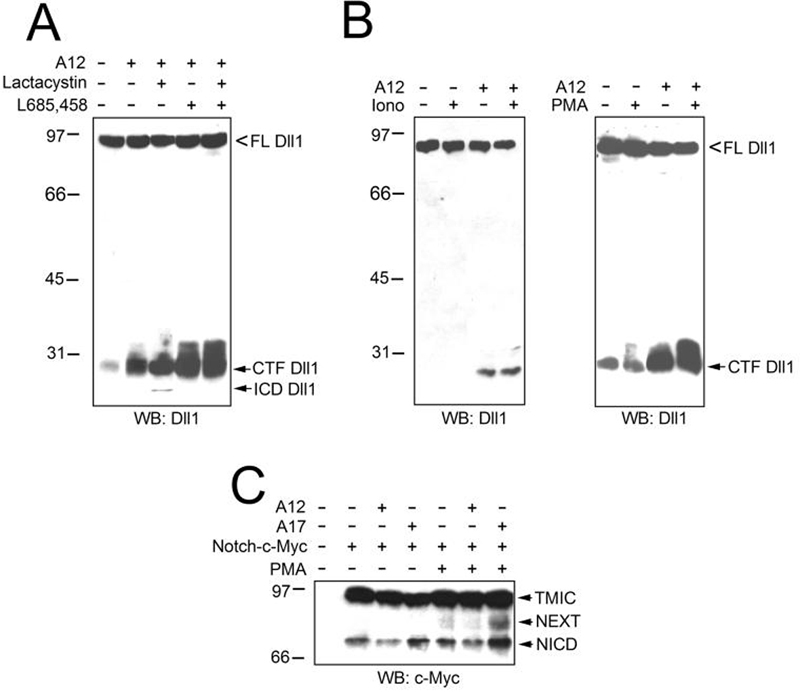
It was reported that removal of the extracellular domain of Dll1 was followed by cleavage of CTF by γ-secretase (12-14). In our experiments, the product of γ-secretase (ICD, intracellular domain of Dll1, ∼3 kDa smaller than CTF) was poorly detectable in immunoblots, but it was augmented in cells treated with lactacystin, a proteasomal inhibitor (Fig. 3A). Incubation of cells with L685,458, a γ-secretase inhibitor, resulted in increased accumulation of CTF (Fig. 3A). These results suggest that at least part of ADAM12-produced CTF Dll1 is further processed by γ-secretase to produce ICD Dll1, and the ICD fragment is then subject to proteasomal degradation.
FIG. 3. ADAM12-mediated cleavage of Dll1 is followed by γ-secretase cleavage (A), is not stimulated by ionomycin, and is weakly enhanced by PMA (B).
A, COS-7 cells co-transfected with Dll1 and empty vector (lane 1) or ADAM12 (lanes 2-5) were incubated for 3 h with 5 μM lactacystin (a proteasomal inhibitor, lanes 3 and 5) or for 6 h with 1 μM L685,458 (a γ-secretase inhibitor, lanes 4 and 5). Cell lysates were analyzed by Western blotting using anti-Dll1 antibody. ICD, intracellular domain of Dll1, is the product of γ-secretase cleavage of CTF. B, COS-7 cells were co-transfected with Dll1 and either empty vector (lanes 1 and 2) or ADAM12 (lanes 3 and 4). Twenty four hours after transfection, cells were incubated for 5 h with 1 μM L685,458 and for additional 1 h with or without 5 μM ionomycin (left) or 25 ng/ml of PMA (right), as indicated. Cells were serum-starved for 30 min prior to and during the PMA treatment. Evidence for a role of ADAM17, but not ADAM12, in cleaving Notch1 (C). COS-7 cells were transfected with Notch1 containing C-terminal c-Myc tag (lanes 2-7, Notch-c-Myc) and either empty vector (lanes 1, 2, 5), ADAM12 (lanes 3 and 6), or ADAM17 (lanes 4 and 7). Two days after transfection, confluent cells were serum-starved for 30 min and then incubated for 1 h without (lanes 1-4) or with 25 ng/ml PMA (lanes 5-7), followed by analysis of Notch cleavage by Western blotting using anti-c-Myc antibody. TMIC, the S1 cleavage product; NEXT, Notch extracellular truncation, the S2 cleavage product; NICD, Notch intracellular domain, the S3 cleavage product.
ADAM10- or ADAM17-mediated cleavage of several substrate proteins is increased by ionomycin or phorbol esters, respectively (16, 26-28). Here, ADAM12-mediated processing of Dll1 was not increased by ionomycin and was only weakly stimulated by phorbol myristate acetate (PMA, Fig. 3B). This result indicates that ADAM12 is capable of constitutive, rather than stimulated, cleavage of Dll1.
To test whether ADAM12 can also cleave Notch, a receptor for Dll1, we co-transfected COS-7 cells with ADAM12 and Notch1, and cultured cells at high density. As positive control, we used ADAM17, which acts as α-secretase for Notch and, upon ligand binding, cleaves Notch at the S2 site (4). Following stimulation of cells with PMA, the S2 cleavage product, “NEXT”, was clearly detected in ADAM17-cotransfected cells (Fig. 3C). In contrast, the S2 product was not detected in ADAM12-transfected cells, suggesting that ADAM12 lacks α-secretase activity towards Notch1.
ADAM12-mediated Dll1 processing did not require an intact cytoplasmic tail of ADAM12, as the full length ADAM12 and the XT fragment containing a 169-amino acid deletion at the C-terminus (29) processed Dll1 equally well (Fig. 4A). Dll1 and ADAM12 co-immunoprecipitated together by an antibody specific to the C-terminus of ADAM12 (Fig. 4B). Interestingly, the immuno-complexes contained the full length Dll1, but not the 29-kDa CTF (Fig. 4C). The N-terminal extracellular fragment of Dll1 generated after ADAM12 cleavage was, however, also excluded from the ADAM12-Dll1 immuno-complexes (Fig. 4D), suggesting that only the full length Dll1 was capable of interacting with ADAM12. The lack of 60-kDa NTF Dll1 in ADAM12 immunoprecipitates is consistent with the result shown in Fig. 1B, in which the N-terminal fragment of Dll1 was released to medium and was not found in association with cells.
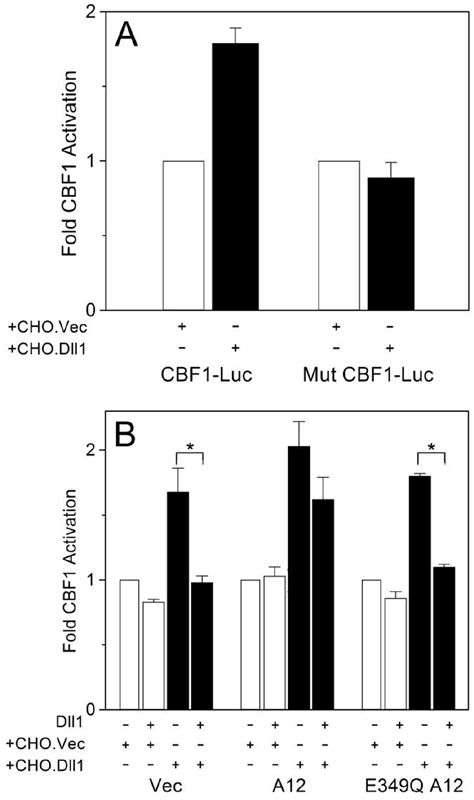
Having established that Dll1, a Notch ligand, but not Notch itself, is cleaved by ADAM12, we examined the effect of Dll1 cleavage on Notch signaling. To monitor the activation status of the Notch pathway, we utilized CBF1-luc reporters containing four (30) or eight binding sites (31) for CBF1, a CSL transcription factor activated by Notch (11). NIH3T3 cells were transiently co-transfected with mouse Notch1 and a Notch reporter, and co-cultured with CHO cells stably transfected with Dll1 (CHO.Dll1) or with empty vector (CHO.Vec). Co-culture with CHO.Dll1 cells led to higher activity of the reporter than co-culture with CHO.Vec cells (Fig. 5A), suggesting that the exogenous Dll1 expressed in CHO.Dll1 cells activated Notch. The extent of this activation was more modest than reported previously in several other studies, in which quail QT6 or mouse L fibroblasts were employed to present Notch ligands to Notch-expressing cells (21, 32). One of the reasons of a modest increase in Notch activity in our system may be that co-culture with control CHO.Vec cells had already stimulated Notch 4-5-fold when compared to the conditions without CHO.Vec cells (result not shown). This was most likely due to high expression levels of the endogenous Notch ligands in CHO cells that were capable of Notch activation. In such case, further increase of the amount of ligand by transfecting exogenous Dll1 might have had a limited effect and, understandably, might not have produced additional strong increase in Notch activity. Nonetheless, increased activity of the Notch reporter induced by co-culture with CHO.Dll1 versus CHO.Vec cells was observed for the reporter containing intact, but not mutated, CBF1 binding sites (Fig. 5A), which validated our co-culture assay to measure Notch activation. When Dll1 was further co-expressed with Notch in NIH3T3 cells, the activation of Notch was abolished (Fig. 5B), due to formation of Notch/Dll1 complexes in cis (19-22). Most importantly, further co-transfection of Dll1-processing ADAM12 resulted in re-activation of the Notch reporter (Fig. 5B). The catalytically inactive mutant of ADAM12, E349Q, which did not process Dll1 (Fig. 1), did not activate Notch either (Fig. 5B). This result suggested that when Notch and Dll1 were expressed in the same cell, proteolytic processing of Dll1 increased the ability of Notch to receive signals and to activate its downstream signaling pathway.
FIG. 5. Proteolytic processing of Dll1 by ADAM12 increases Notch signaling in a cell-autonomous manner.
A, NIH3T3 cells were co-transfected with mouse Notch1 and either a CBF1 reporter (CBF1-Luc; pJH23A) or the same reporter in which CBF1 binding sites were mutated (Mut CBF1-Luc; pJH25A). After 24 h, cells were co-cultured with CHO cells stably transfected with vector only (CHO.Vec; white bars) or with CHO-cells stably transfected with Dll1 (CHO.Dll1; black bars). The activities of firefly luciferase, normalized to Renilla luciferase as internal control, were assayed 24 h later. B, NIH3T3 cells were transiently co-transfected with Notch1, a CBF1 reporter (pJT123A) and Dll1, wild type ADAM12 (A12), or catalytically inactive ADAM12 (E349Q A12), as indicated. After 24 h, cells were co-cultured for additional 24 h with CHO.Vec (white bars) or CHO.Dll1 cells (black bars), followed by measurement of luciferase activity. Notice that co-expression of Dll1 with Notch in the same cell inhibits Notch signaling induced by CHO.Dll1 (black bars) in the absence of A12 or in the presence of the E349Q mutant (*, p<0.05). In the presence of catalytically active A12, the inhibition was diminished and was not statistically significant. In A and B, error bars represent standard error of the mean (n=3).
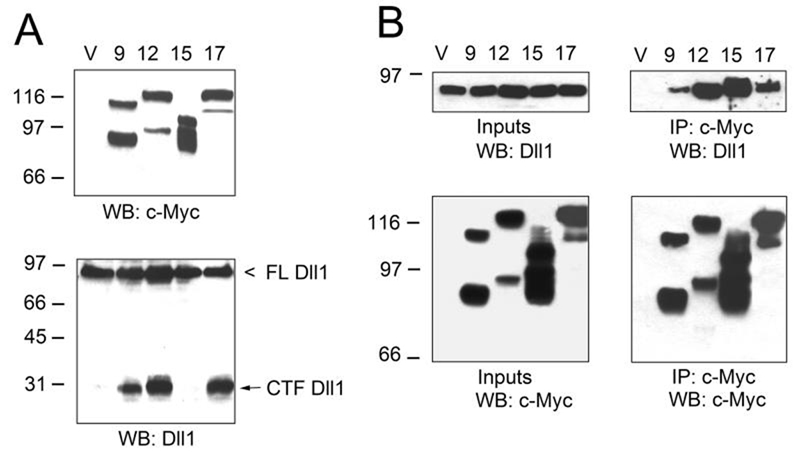
We next tested whether Dll1 can serve as a substrate for three other ADAMs that have not been previously implicated in Dll1 cleavage, ADAM9, 15, and 17. All these ADAMs, as well as ADAM12, were engineered to contain a C-terminal c-Myc tag for a common detection with anti-c-Myc antibody (Fig. 6). Co-transfection with Dll1 demonstrated that ADAM9 and 17, similarly to ADAM12, had catalytic activity towards Dll1, but ADAM15 was not able to process Dll1 (Fig. 6A). After comparing the amounts of the 29-kDa fragment of Dll1 and the amounts of mature, catalytically active forms of each ADAM, we concluded that the cleavage of Dll1 occurred with the following potency order: ADAM17>ADAM12>ADAM9. Interestingly, all ADAM proteins used in this study formed stable complexes with Dll1, as assessed by co-immunoprecipitation with anti-c-Myc antibody (Fig. 6B). Thus, ADAM15 interacted with Dll1, but was unable to cleave it. Importantly, ADAM15 contains a consensus sequence of the catalytic site of zinc-dependent metalloproteases and appears catalytically active (33).
FIG. 6. Dll1 is a substrate for ADAM9, 12, and 17, but not for ADAM15.
A, COS-7 cells were co-transfected with Dll1 and empty vector (V), c-Myc-tagged ADAM9, 12, 15 or 17. Cell extracts were analyzed by Western blotting using anti-c-Myc tag antibody (top) or anti-Dll1 antibody (bottom). Upper bands detected with anti-c-Myc antibody represent nascent full length ADAM proteins, lower bands correspond to the catalytically active forms lacking pro-domains. B, COS-7 cells were co-transfected with Dll1 and empty vector (V), c-Myc-tagged ADAM9, 12, 15 or 17. ADAM proteins were immunoprecipitated with anti-c-Myc tag antibody. The inputs (left) and immunoprecipitates (IPs; right) were analyzed by anti-Dll1 (upper panels) and anti-c-Myc (lower panels) antibodies.
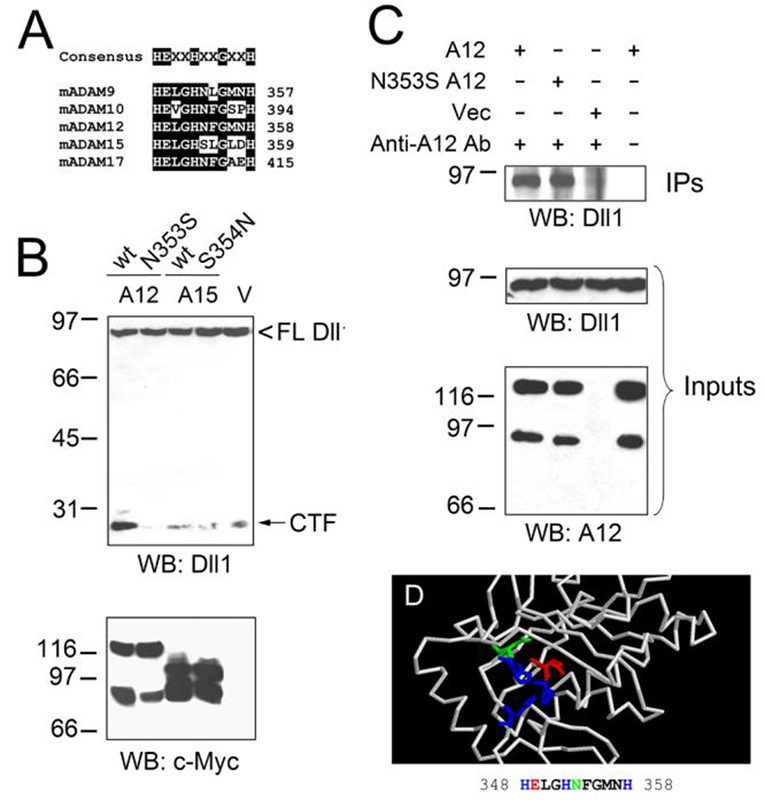
To better understand the molecular features that are required for Dll1 cleavage, we compared the sequences of the catalytic site of Dll1-processing ADAMs and of ADAM15. In addition to the newly identified Dll1-processing enzymes ADAM9, 12, and 17, we included the sequence of the catalytic site of ADAM10, a well-established Dll1-cutter (13, 14). The consensus motif for ADAM proteases is HEXXHXXGXXH (34), with three His residues binding a zinc ion at the active site and Glu being the catalytic residue (Fig. 7A). We observed that all Dll1-cleaving ADAMs contained an Asn after the second His. In contrast, ADAM15 contained a Ser (Ser354) at this position. We thus asked whether the identity of the amino acid following the second His was important for Dll1 cleavage. We generated two mutants, N353S ADAM12 and S354N ADAM15. When expressed in COS-7 cells, both mutants were processed correctly and both their nascent and mature forms lacking the pro-domains were detected (Fig. 7B). While the N353S ADAM12 mutant completely lost its ability to process Dll1, the S354N ADAM15 mutant did not gain catalytic activity towards Dll1 (Fig. 7B). The lack of Dll1 processing by N353S ADAM12 was not due to its lost capacity to interact with Dll1, as demonstrated in the co-immunoprecipitation experiment (Fig. 7C). We thus conclude that Asn353 in ADAM12 is necessary for Dll1 cleavage, but the presence of Asn at the corresponding position in ADAM15 is not sufficient for the cleavage. Fig. 7D shows a predicted structure of the metalloprotease domain of ADAM12, obtained by homology modeling using the structure of ADAM33 metalloprotease as a template. It is apparent that the side chain of Asn353 faces the catalytic cleft, where it might participate in the interactions with Dll1.
FIG. 7. Asn353 in ADAM12 is necessary for Dll1 cleavage.
A, Alignment of the amino acid sequences of the catalytic sites of mouse ADAM9, 10, 12, 15, and 17. The consensus sequence of the catalytic site of ADAM proteases is shown on the top. B, COS-7 cells were co-transfected with Dll1 and either wild-type or N353S ADAM12 mutant, wild-type or S354N ADAM15 mutant, or empty vector. All ADAM constructs contained a C-terminal c-Myc tag. Cell extracts were analyzed by Western blotting using anti-Dll1 antibody (upper panel) or anti-c-Myc antibody (bottom panel). C, COS-7 cells were co-transfected with Dll1 and wild-type ADAM12, the N353S mutant of ADAM12, or empty vector, as indicated. Cell extracts were immunoprecipitated using anti-ADAM12 antibody (lanes 1-3) or were incubated without antibody (lane 4). Immunoprecipitates (IPs) and inputs were analyzed by Western blotting with anti-Dll1 antibody, the expression of ADAM12 was verified by Western blotting of the inputs with anti-ADAM12 antibody. D, The predicted structure of the metalloprotease domain of ADAM12, obtained by threading mouse ADAM12 sequence (amino acids 212-414) onto mouse ADAM33 (PDB structure 1R55) using the Swiss-PdbViewer software, and then submitted to the SWISS-MODEL server for refinement. Side chains of His348, His352, and His358 coordinating Zn2+ are shown in blue, catalytic E349 is shown in red, and N353 is depicted in green.
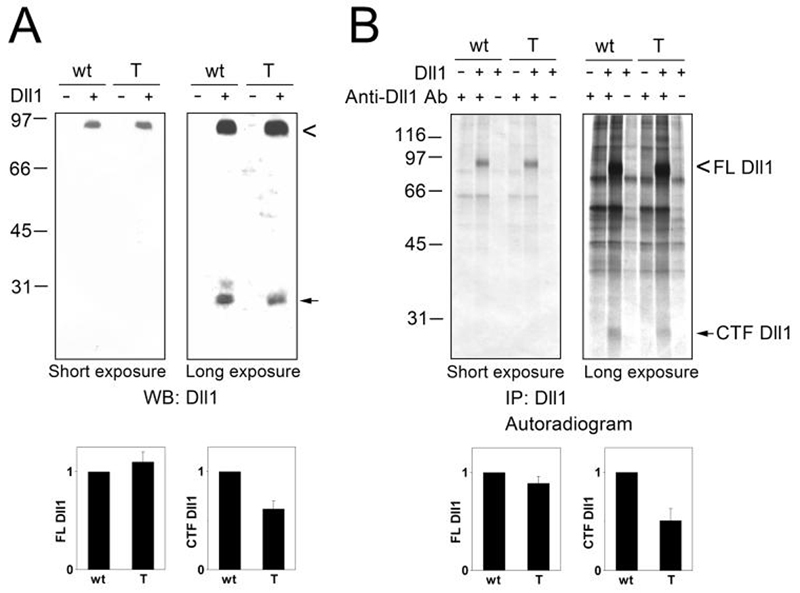
To determine whether ADAM proteases studied in this work can process Dll1 when they are expressed at the endogenous levels, we compared the amount of Dll1 cleavage in wild type (wt) and ADAM9/12/15−/− triple knockout (T) mouse embryonic fibroblasts (MEFs). Dll1 was transfected into wt-MEFs or T-MEFs, and the extent of Dll1 cleavage was examined by subjecting total cell lysates to immunoblotting with anti-Dll1 antibody (Fig. 8A) or by immunoprecipitation of FL Dll1 and CTF Dll1 from [35S]-labeled cells (Fig. 8B). The level of Dll1 cleavage mediated by endogenous proteases in MEFs was significantly lower than the cleavage observed in COS-7 overexpressing ADAM9, 12 or 17 (compare Fig. 8A and Fig. 6A). Significant differences in the intensities of the bands corresponding to FL Dll1 and CTF Dll1 in Fig. 8A precluded an accurate quantification of the extent of cleavage (the CTF Dll1/FL Dll1 ratio). These differences were even more pronounced in Fig. 8B, because the number of cysteine and methionine residues in FL Dll1 is ∼10 higher than in CTF Dll1 (13). Nevertheless, while the amounts of FL Dll1 in wt-MEFs and T-MEFs were similar, the level of CTF Dll1 in T-MEFs corresponded to ∼60% of the level in wt-MEFs (Fig. 8A,B). Since ADAM15 is not capable of cleaving Dll1 (Fig. 6A), this result suggested that ADAM9 and/or ADAM12 contributed to Dll1 processing in wt-MEFs.
FIG. 8. Endogenous ADAM9 and/or ADAM12 contribute to Dll1 cleavage in mouse embryonic fibroblasts (MEFs).
SV40-immortalized MEFs isolated from wild type mice (wt) or from ADAM9/12/15−/− triple knockout mice (T) were transiently transfected to express Dll1, as indicated. A, After 48 h, Dll1 processing was analyzed by Western blotting with anti-Dll1 antibody. The intensities of the bands corresponding to FL Dll1 were determined after short exposure times (left), the intensities of the bands representing CTF Dll1 were measured after long exposure times (right). The amounts of FL Dll1 and CTF Dll1 in T-MEFs were normalized to the amounts in wt-MEFs; the results (mean from three different experiments, ± S.E) are shown below each Western blot. B, Twenty four hours after transfection, cells were metabolically labeled with [35S]methionine+cysteine, 16 h later cell lysates were subjected to immunoprecipitation using anti-Dll1 antibody, as indicated. The immunoprecipitates were resolved by SDS-PAGE and analyzed by autoradiography. The intensities of the bands corresponding to FL Dll1 and CTF Dll1 were determined after short exposure times (left) and long exposure times (right), respectively, and the results were plotted as in A (mean ± S.E., n=2). In A and B, cells were incubated for 6 h in the presence of 1 μM γ-secretase inhibitor L685,458 prior to harvesting.
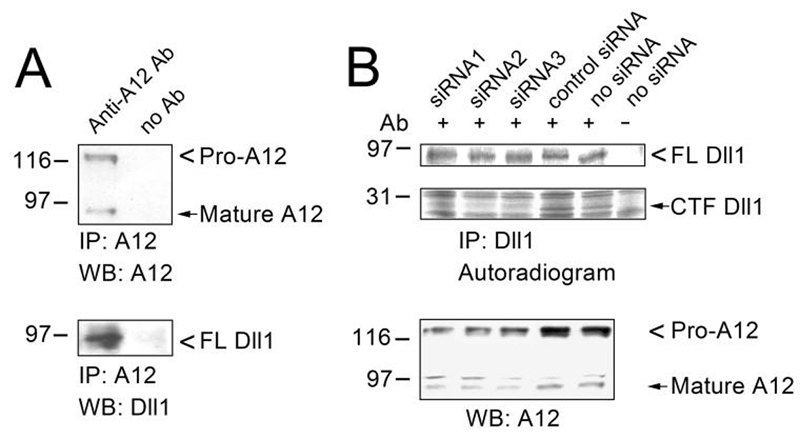
Activation of the Notch pathway inhibits myogenic differentiation in vitro and in vivo (35-39), suggesting that this pathway may act to maintain the myogenic cells in an undifferentiated proliferating state (40-43). Here, we asked whether the endogenous Dll1 present in myogenic cells interacts with ADAM12, whether it undergoes proteolytic processing, and what role ADAM12 might have in this processing. To address these questions, we used mouse primary myoblasts, which express the endogenous Dll1 and ADAM12 and in which the expression of both proteins increases during the first few days after transfer to differentiation medium (42, 44, 45). As shown in Fig. 9A, the endogenous ADAM12 co-immunoprecipitated with the endogenous Dll1 from primary myoblasts. Metabolic labeling of cells with [35S]methionine+cysteine, followed by immunoprecipitation with anti-Dll1 antibody, revealed the presence of antibody-specific bands of 90 kDa and 29 kDa, corresponding to the FL Dll1 and CTF Dll1, respectively. Transfection of cells with three different small interfering RNAs (siRNAs) targeting ADAM12, but not with a control siRNA, decreased ADAM12 protein levels by ∼50-60% and reduced the amount of CTF Dll1 by more than 50% (Fig. 9B). Thus, we conclude that the endogenous Dll1 is subject to proteolytic processing in confluent, differentiating myoblast cultures, giving rise to the 29-kDa fragment, and that the endogenous ADAM12 contributes to this processing.
FIG. 9. Endogenous ADAM12 co-immunoprecipitates with Dll1 and contributes to Dll1 cleavage in primary mouse myoblasts.
A, Primary mouse myoblasts grown to confluence and incubated for one day in differentiation medium were subjected to immunoprecipitation with anti-ADAM12 antibody. The immunoprecipitates were analyzed by immunoblotting with anti-ADAM12 (top) or anti-Dll1 antibody (bottom). B, Primary mouse myoblasts were transfected with one of three ADAM12 siRNAs or control siRNA, as described in Experimental Procedures. One day after transfection, cells reached confluence and were transferred to differentiation medium. After 24 h, cells were metabolically labeled with [35S]methionine+cysteine for 10 h, 1 μM γ-secretase inhibitor L685,458 was added, and the labeling continued for next 6 h. The level of endogenous ADAM12 was analyzed by Western blotting after partial purification on concanavalin A agarose (bottom). In parallel, the endogenous Dll1 was immunoprecipitated with anti-Dll1 antibody, the immunoprecipitates were analyzed by SDS-PAGE and direct autoradiography (top). The exposure time of the region in the autoradiogram containing the FL Dll1 was 5 times longer that the exposure time of the region containing CTF Dll1. Notice that the abundance of CTF Dll1 is diminished in cells transfected with ADAM12 siRNAs.
DISCUSSION
Notch ligands undergo proteolytic processing by ADAMs, but the identities of ADAMs that can mediate cleavage, as well as the consequences of such processing are not clear. In Drosophila, the best characterized Notch ligand is Delta. Co-transfection experiments in Drosophila S2 cells demonstrated that three ADAMs could cleave Delta: Kuzbanian (a Drosophila homolog of ADAM10), Kuzbanian-like (Kul), and DTACE (a homolog of ADAM17) (17, 23). Interestingly, DMeltrin, a Drosophila homolog of ADAM12, clearly lacked the ability to process Delta (23).
Mammalian Dll1 expressed in HEK293, COS, CHO, N2a, or NIH3T3 cells was cleaved by endogenous ADAMs present in these cells (12-14). Transfection of Dll1 into ADAM10−/− or ADAM10+/+ MEFs demonstrated that the cleavage was reduced by ∼50% in the absence of ADAM10 (13). This result suggested that ADAM10 was only partially responsible for Dll1 cleavage and that other ADAMs catalyzed the remaining cleavage of Dll1 in ADAM10−/− cells. MEFs express several catalytically active ADAMs, including ADAM9, 12, 15, and 17 (16), and we focused our study on these proteases. ADAM17 is the closest relative of ADAM10 and its activity towards Dll1 has been somewhat expected (13). Dll1 processing in CHO cells was not affected, however, by the ADAM17 inhibitors batimastat or TAPI-1, raising questions about the ability of ADAM17 to cleave Dll1. Processing of Dll1 by ADAM9, 12 or 15 has not been tested previously.
Here, we show that ADAM 9, 12, and 17, but not ADAM15, when co-transfected with Dll1 into COS-7 cells, are capable of Dll1 cleavage. Although we have not determined the exact cleavage sites in Dll1, each of these ADAMs generated a CTF of a similar size, suggesting that the cleavage sites were either identical or located very close to each other. In the case of ADAM12, we provided evidence that Dll1 processing was accompanied by the release of the N-terminal fragment of Dll1 to medium, it was cell density-dependent, occurred in cis, and it was weakly stimulated by PMA. Furthermore, processing of Dll1 transfected into ADAM9/12/15−/− MEFs was diminished when compared with processing in wild type MEFs. This result suggested that Dll1 was also subject to processing by the endogenous ADAM9 or ADAM12 or both (since ADAM15 was not capable of cleaving Dll1 even when overexpressed in COS-7 cells, most likely it did not contribute to Dll1 processing in MEFs at the endogenous level). Although the reduction of Dll1 cleavage in ADAM9/12/15−/− MEFs was rather modest (∼40%), it was consistent with ∼50% inhibition of Dll1 processing observed in ADAM10−/− MEFs (13) and with the remaining activity of ADAM17, which might have also contributed to the processing.
Our studies show that Asn353 in ADAM12 is required for Dll1 cleavage. Replacement of Asn353 with a Ser residue found in ADAM15 completely abolished ADAM12 activity towards Dll1. Consistently, other ADAMs capable of cleaving Dll1, namely ADAM9 and 17 (this study) and ADAM10 (13) also contain an Asn residue at the corresponding position, suggesting that this may be a general feature of Dll1-processing ADAMs. Currently, high resolution structures of metalloprotease domains are available only for ADAM17 and ADAM33. Analysis of the architecture of the catalytic site in ADAM17 and in the predicted structure of ADAM12 modeled onto the closely related ADAM33 (this work) indicates that the side chain of Asn410 in ADAM17 and of Asn353 in ADAM12 are facing the catalytic cleft and thus might be involved in the interaction with Dll1. Replacement of Ser354 in ADAM15 with Asn, however, did not increase Dll1 cleavage by ADAM15, indicating that the presence of Asn at that position is not sufficient for Dll1 processing and that other molecular events in ADAMs are required for efficient cleavage of the Dll1 substrate.
The ability of ADAM12 to cleave Dll1 and the apparent lack of activity towards Notch allowed us to study the effect of ligand cleavage on Notch signaling. While shedding of the extracellular domain of Notch ligand limits the ligand presentation in trans and terminates Notch signaling in a neighboring cell (17, 18), cell-autonomous effects of ligand cleavage are less clear. Notch ligands form complexes in cis with Notch, which leads to sequestration of Notch receptors, reduction of Notch receptivity to signals from outside, and attenuation of Notch signaling (19-22). Shedding of the extracellular domain of the ligand could relieve this inhibitory effect and activate Notch, a scenario that is supported by results of in vivo experiments in flies. Overexpression of Kul in juxta-marginal cells in Drosophila wing discs (which are characterized by high levels of Delta and low levels of Notch signaling) resulted in increased expression of Notch-target genes (23). Alternatively, the C-terminal fragment of a ligand could compete with Notch S2 cleavage product for γ-secretase and thus interfere with Notch signaling. Indeed, overexpression of an ectodomain-truncated Jagged 1 in COS-7 cells, together with an ectomain-truncated Notch, led to inhibition of the Notch function (14). However, expression of a truncated Jagged that mimicked an already cleaved Jagged, did not allow evaluation of the actual effect of the removal of the N-terminal portion of Jagged by ADAMs. In our studies, both Dll1 and Notch constructs represented the intact proteins, and the relative contribution of Dll1 ectodomain shedding (a stimulatory effect) and competition of Dll1 and Notch C-terminal fragments for γ-secretase (an inhibitory effect) were addressed. Our results demonstrate that ADAM12, capable of mediating a constitutive cleavage of Dll1 but not of Notch, activated Notch signaling in a cell autonomous manner, whereas the catalytically inactive ADAM12 mutant did not process Dll1 and did not activate Notch. Thus, alleviation of the inhibition of Notch mediated by Dll1 appears to outweigh the generation of a Dll1 fragment that can compete for γ-secretase.
The essence of Notch signaling is the amplification of small differences in the levels of Notch receptors and their ligands between adjacent cells (1-3). Cells that are initially equivalent but at one point develop a bias towards receptors or ligands will eventually become signal-receiving and signal-sending cells, respectively. Amplification of the differences in receptor/ligand levels is mainly achieved by a transcriptional feedback mechanism (1-3). We propose that proteolytic cleavage of Dll1 may represent another mechanism for reinforcing small differences in the signaling capacities of different cells and for establishing the uni-directionality in Notch signaling.
During myogenic differentiation in vitro, a pool of seemingly equivalent cells assume different fates that coincide with different levels of Notch signaling (42). In proliferating cells and then in cell cycle-arrested but undifferentiated “reserve cells”, the Notch activity is high, whereas in differentiated myotubes the level of Notch signaling is low (42). While Notch signaling is generally regulated at multiple levels, one of the most direct mechanisms involves regulation of expression and membrane localization of Notch ligands. In Xenopus, expression of XDelta-1 is positively regulated at the transcriptional level by MyoD (46). In Drosophila, expression of Delta has been recently shown to be negatively regulated by dmiR-1 (47), a muscle-specific microRNA (48). Our results suggest that in mouse myogenic cells, the Dll1 protein may be additionally regulated at the post-translational level by cell surface proteolysis and that ADAM12 is one of the ADAM proteases mediating this processing (Fig. 9). Interestingly, gene profiling experiments suggest that several ADAMs capable of cleaving Dll1, including ADAM9, 10, and 12, are strongly up-regulated during muscle regeneration in vivo (ref. 49, data available at the Children's National Medical Center web site http://perp.cnmcresearch.org/), and several reports suggested a role for ADAM12 in myogenesis (50,51). Whether Dll1 processing by ADAMs indeed represents a mechanism of regulation of Notch signaling during myogenic differentiation associated with muscle regeneration remains to be determined.
ACKNOWLEDGMENTS
We thank Drs. Raphael Kopan for the Notch1 plasmid and Diane Hayward and Paul D. Ling for the CBF1 reporters. We also thank Dr. Elena Tasheva for help in studying cleavage of Notch1.
Footnotes
This work was supported by NIH grants GM065528 to AZ and GM64750 to CPB. This is contribution 06-334-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Lai EC. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 2.Kadesch T. Curr. Opin. Genet. Dev. 2004;14:506–512. doi: 10.1016/j.gde.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Louvi A, Artavanis-Tsakonas S. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 4.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. Mol. Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 5.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena IA, von Figura K, Saftig P. Hum. Mol. Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 7.Blobel CP. Nat. Rev. Mol. Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 8.Huovila AP, Turner AJ, Pelto-Huikko M, Karkkainen I, Ortiz RM. Trends Biochem. Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Mumm JS, Kopan R. Dev. Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 10.Selkoe D, Kopan R. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 11.Hayward SD. Semin. Cancer Biol. 2004;14:387–396. doi: 10.1016/j.semcancer.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Ikeuchi T, Sisodia SS. J. Biol. Chem. 2003;278:7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- 13.Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israël A, Logeat F. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaVoie MJ, Selkoe DJ. J. Biol. Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 15.Bland CE, Kimberly P, Rand MD. J. Biol. Chem. 2003;278:13607–13610. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- 16.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. J. Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 18.Mishra-Gorur K, Rand MD, Perez-Villamil B, Artavanis-Tsakonas S. J. Cell Biol. 2002;159:313–324. doi: 10.1083/jcb.200203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto K, Ohara O, Takagi M, Takeda S, Katsube K. Dev. Biol. 2002;241:313–326. doi: 10.1006/dbio.2001.0517. [DOI] [PubMed] [Google Scholar]
- 20.Katsube K, Sakamoto K. Int. J. Dev. Biol. 2005;49:369–374. doi: 10.1387/ijdb.041950kk. [DOI] [PubMed] [Google Scholar]
- 21.Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. J. Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franklin JL, Berechid BE, Cutting FB, Presente A, Chambers CB, Foltz DR, Ferreira A, Nye JS. Curr. Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- 23.Sapir A, Assa-Kunik E, Tsruya R, Schejter E, Shilo BZ. Development. 2005;132:123–132. doi: 10.1242/dev.01546. [DOI] [PubMed] [Google Scholar]
- 24.Rando TA, Blau HM. J. Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Y, Kang Q, Zhao Z, Zolkiewska A. J. Biol. Chem. 2002;277:26403–26411. doi: 10.1074/jbc.M110814200. [DOI] [PubMed] [Google Scholar]
- 26.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano O, Murakami D, Hartmann D, de Strooper B, Saftig P, Iwatsubo T, Nakajima M, Shinohara M, Saya H. J. Cell Biol. 2004;165:893–902. doi: 10.1083/jcb.200310024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi H, Gruszczynska-Biegala J, Wood D, Zhao Z, Zolkiewska A. J. Biol. Chem. 2005;280:23475–23483. doi: 10.1074/jbc.M413550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Mol. Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng R, Gordadze AV, Fuentes Panana EM, Wang F, Zong J, Hayward GS, Tan J, Ling PD. J. Virol. 2000;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarriault S, Le Bail O, Hirsinger E, Pourquié O, Logeat F, Strong SF, Brou C, Seidah NG, Israël A. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin J, Eynstone LV, Davies M, Williams JD, Steadman R. J. Biol. Chem. 2002;277:33683–33689. doi: 10.1074/jbc.M200988200. [DOI] [PubMed] [Google Scholar]
- 34.Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX, McKay DB, Bode W. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopan R, Nye JS, Weintraub H. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 36.Shawber C, Nofziger D, Hsieh JJ, Lindsell C, Bogler O, Hayward D, Weinmaster G. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 37.Nofziger D, Miyamoto A, Lyons KM, Weinmaster G. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 38.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. J. Biol. Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 39.Delfini MC, Hirsinger E, Pourquie O, Duprez D. Development. 2000;127:5213–5224. doi: 10.1242/dev.127.23.5213. [DOI] [PubMed] [Google Scholar]
- 40.Conboy IM, Rando TA. Dev. Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 41.Luo D, Renault VM, Rando TA. Semin. Cell Dev. Biol. 2005;16:612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Kitzmann M, Bonnieu A, Duret C, Vernus B, Barro M, Laoudj-Chenivesse D, Verdi JM, Carnac G. J. Cell Physiol. 2006;208:538–548. doi: 10.1002/jcp.20688. [DOI] [PubMed] [Google Scholar]
- 43.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Nat. Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Zhao Z, Gruszczynska-Biegala J, Zolkiewska A. Mol. Cell Biol. 2003;23:6725–6738. doi: 10.1128/MCB.23.19.6725-6738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, Lendahl U. Development. 2003;130:6089–6099. doi: 10.1242/dev.00834. [DOI] [PubMed] [Google Scholar]
- 46.Wittenberger T, Steinbach OC, Authaler A, Kopan R, Rupp RA. EMBO J. 1999;18:1915–1922. doi: 10.1093/emboj/18.7.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon C, Han Z, Olson EN, Srivastava D. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao P, Hoffman EP. Dev. Dyn. 2004;229:380–392. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- 50.Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- 51.Kurisaki T, Masuda A, Sudo K, Sakagami J, Higashiyama S, Matsuda Y, Nagabukuro A, Tsuji A, Nabeshima Y, Asano M, Iwakura Y, Sehara-Fujisawa A. Mol. Cell. Biol. 2003;23:55–61. doi: 10.1128/MCB.23.1.55-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]