Abstract
We report on a general structure- and NMR- based approach to derive drug-like small molecule inhibitors of protein-protein interactions in a rapid and efficient manner. We demonstrate the utility of the approach by deriving novel and effective SMAC mimetics targeting the anti-apoptotic protein X-Linked Inhibitor of Apoptosis Protein (XIAP). The XIAP baculovirus IAP repeat 3 (Bir3) domain binds directly to the N-terminal of Caspase-9 and thus inhibiting programmed cell death. It has been shown that in the cell this interaction can be displaced by the protein second mitochondrial activator of caspases (SMAC) and that its N-terminal tetrapeptide region (NH2-AVPI, Ala-Val-Pro-Ile) is responsible for this activity. However, due to their limited cell-permeability, synthetic SMAC peptides are inefficient when tested in cultured cells, limiting their use as potential chemical tools or drug candidates against cancer cells. Hence, as an application, we report on the derivation of novel, selective, drug-like, cell permeable SMAC mimics with cellular activity.
Introduction
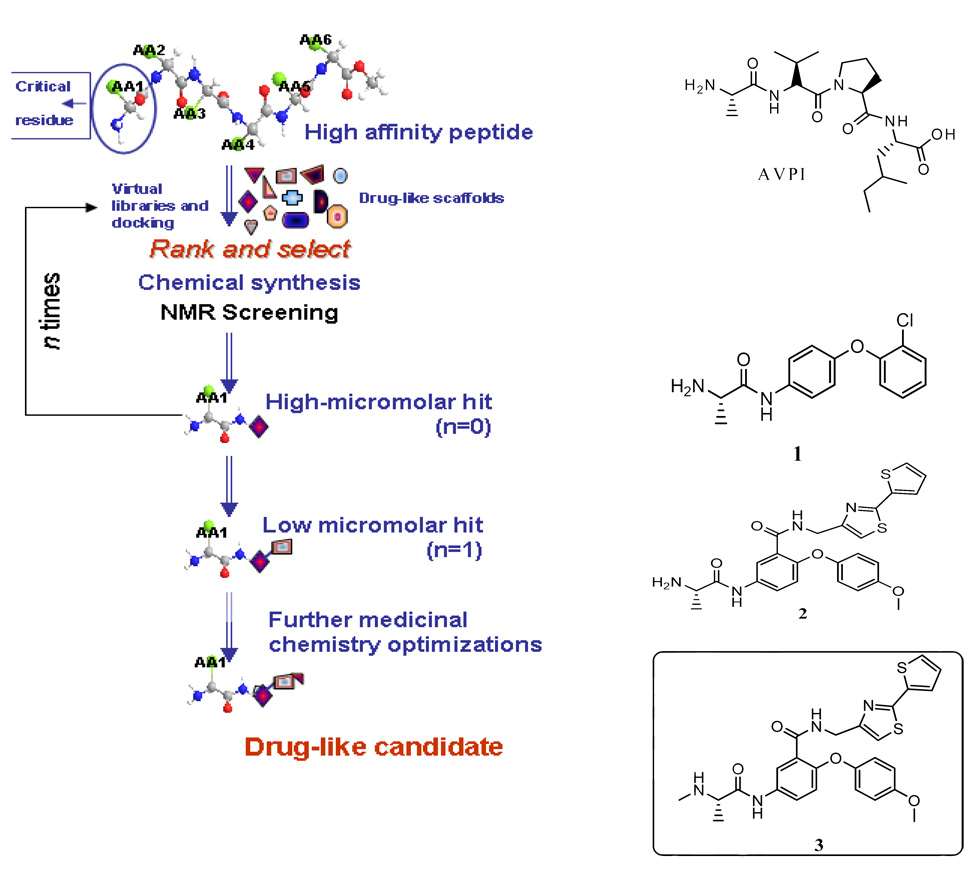
The X-Linked Inhibitor of Apoptosis Protein (XIAP) baculovirus IAP repeat 3 (Bir3) domain binds directly to the N-terminal of Caspase-9 and thus inhibiting programmed cell death.1–3 It has been shown that in the cell this interaction can be displaced by the protein SMAC (second mitochondrial activator of caspases) and that its N-terminal tetrapeptide region (AVPI) is responsible for the binding.3,4 However, the use of synthetic SMAC-derived peptides as therapeutic compounds is hindered due to their limited cell-permeability, proteolytic instability, and poor pharmacokinetics.5–8 A number of research groups have reported the discovery of small-molecule Bir3 inhibitors by various methods,6–14 including peptidomimetic approaches,8,10,14 virtual screening/structure-based design,6,11–13 or screening of natural-product or synthetic libraries.7,9 In this study, we present a simple strategy in which individual amino-acids are replaced in an iterative manner with more drug-like scaffolds (Figure 1). By starting from the single most important amino acid of the template peptide, alanine,8–11,15–16 a first virtual library is obtained by coupling the selected amino-acid with low molecular weight, drug-like, synthetically accessible scaffolds. Subsequently, the library elements are docked against the target in order to select those compounds with the best fit for the binding site. Following chemical synthesis of top scoring compounds, these are experimentally tested by NMR spectroscopy techniques. The use of NMR is pivotal to the approach given that at this stage only high-micromolar binders, at best, are expected. Hit compounds are subsequently used for a second round of in silico derivatizations followed by synthetic chemistry of top scoring compounds. The approach can be repeated until the desired potency is achieved, keeping molecular weight and other drug-like properties in check (Figure 1).
Figure 1.
The Schematic representation of approach used to derive non-peptide SMAC mimics.
Results and Discussion
As an application we report the derivation of drug-like, cell permeable SMAC mimics. Following our strategy, we first designed an initial virtual library of L-Alanine derivatives, a critical amino acid in SMAC peptides, by coupling it with 578 primary and 815 secondary commercially available, low molecular weight, drug-like, amines. After molecular docking studies, a series of 15 selected candidate compounds (structures and Goldscore values are listed in supplementary figure 1A) were synthesized and tested experimentally by NMR for their ability to bind to the Bir3 domain of XIAP (supplementary figure 1B). Through observation the differences of chemical shift perturbations on Bir3 in presence of the selected putative SMAC mimics, compound 1 (BI-75A1, Figure 1) was identified as a weak binder (Kd ~ 200 µM) for the Bir3 domain. Molecular docking studies support that 1 presents several binding characteristics that overlap with these observed with the SMAC peptide, in particular mimicking the interactions provided by the first three amino-acids in AVPI (see also supplementary Figure 1C). From the experimentally derived structure of Bir3 in complex with AVPI, it appears clear that the Ala and Val residues occupy the first of two sub-pockets (P1 and P2 in Figure 2A) on the surface of the domain, while the side chain of the Ile residue occupies the second (Figure 2A). Therefore, structure modifications of 1 at position 2 of the 4-phenoxybenzene scaffold could be proposed in a second iteration, which may result in the selection of an additional scaffold mimicking the interactions provided by the isoleucine residue of AVPI, into the P2 sub-pocket (Figure 2).
Figure 2.
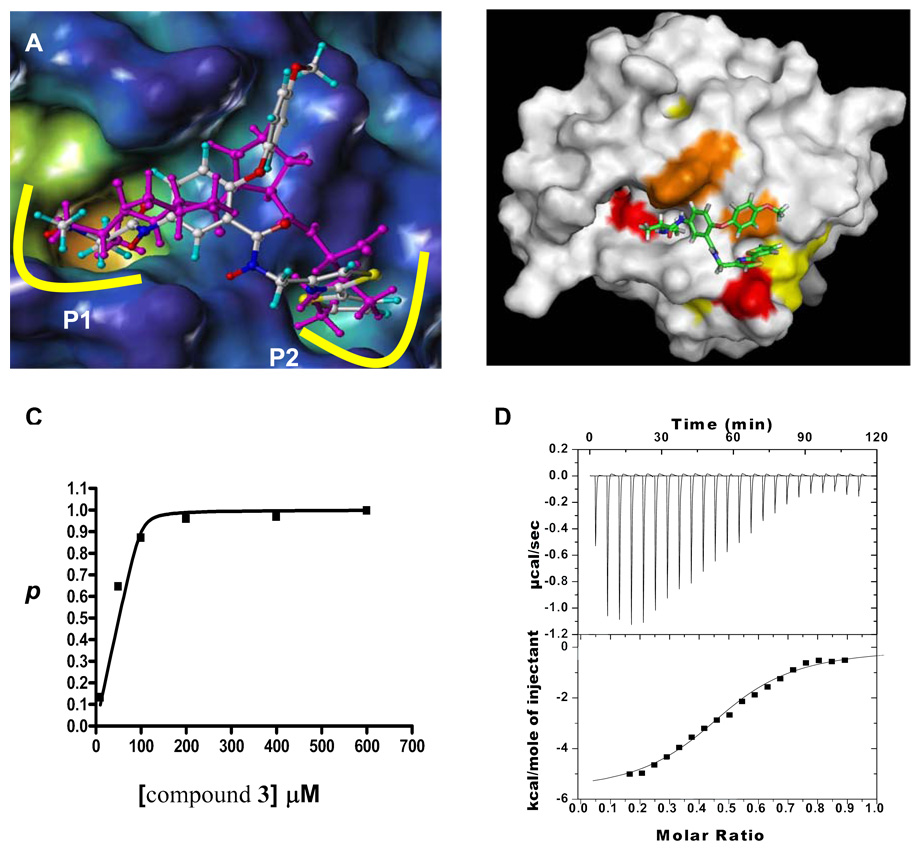
(A) Superposition between the X-ray structure of AVPI (magenta) in complex with the Bir3 domain of XIAP (surface representation) and the docked conformation of compound 3; The Ala and Val residues occupy the first of two sub-pockets (P1 and P2) on the surface of the domain, while the Ile occupies the second. (B) Chemical shift mapping data with 15N Bir3 and compound 3. The surface of the Bir3 domain of XIAP is colored according to the shifts induced by compound 3: red > orange> yellow ≫ white = no shifts). (C) Fractional changes (p) of chemical shifts in Bir3 as a function of ligand concentration. The experimental data were fitted to the non-linear equation as described in the methods resulting in Kd = 1.2 µM. (D) Calorimetric titration of Bir3 with compound 3. The top panel shows raw data in power versus time. The bottom panel shows data after peak integration, subtraction of blank titration data, and concentration normalization. The solid line is the fit to a single binding site model. Kd ~ 2.5 µM.
Based on this hypothesis, a second virtual library of compound 1’s derivatives (about 900 compounds) was similarly designed, compounds rank ordered by using in silico docking, and top scoring compounds, such as compound 2 (BI-75D1; Goldscore is 63.0, Figure 1 and compounds listed in Supplementary Figure 2), were further synthesized and tested by NMR. The molecular docking model of the analogue of 2, compound 3 (BI-75D2) is shown in Figure 2A. As also corroborated by NMR chemical shift mapping data with 15N labeled Bir3 (Figure 2B and supplementary Figure 3A), compound 3 docks on the Bir3 surface by occupying each of the two sub-pockets occupied by the SMAC peptide AVPI. Additional interactions between the domain and the bridging Proline residue in AVPI are mimicked by the peripheral phenyl ring in compound 3 (Figure 2A).
From this second iteration and structure refinement by N-methylation to improve drug-likeness,10,14 compound 3 emerged showing the tightest binding affinity, among the tested compounds, for Bir3, with a Kd value of 1.2 µM (Figure 2C). When tested in fluorescence polarization displacement assay with FITC-labeled Smac peptides, the compound appeared weaker depending on experimental conditions (with inhibition constants of ~ 30 µM). This may be explained by possible non-specific interactions of the FITC-peptide with the protein that cannot be displaced by the compound. Moreover, the binding affinity between Bir3 domain of XIAP and compound 3 was also analyzed by Isothermal Titration Calorimetry (ITC), indicating a binding affinity (Ka) of 6.5 ± 3.3 × 105 (M−1) (Kd ~ 2.5 µM) (Figure 2D).
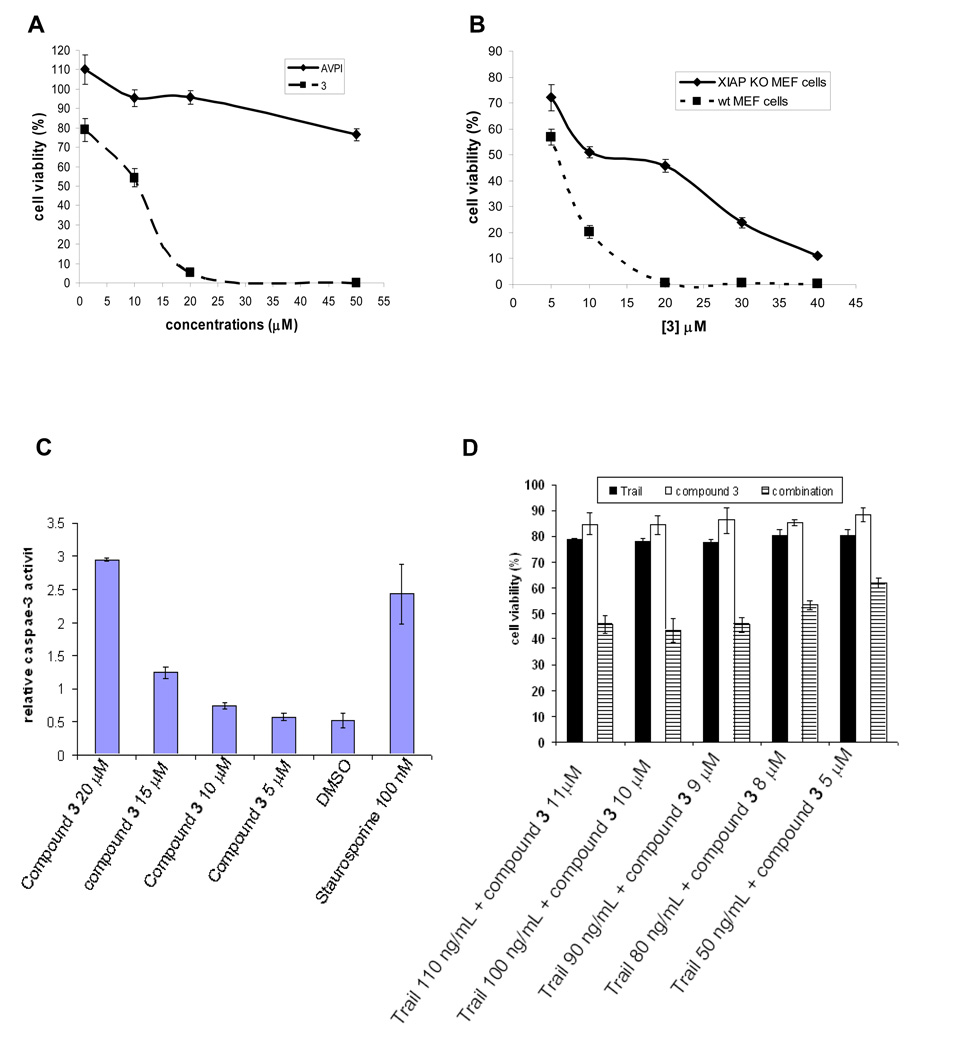
Given the superior drug-like characteristics, better human plasma stability, longer drug metabolic stability, high permeability, and good solubility, of the compound (Table 1) compared to the SMAC N-terminal peptides such as AVPI or AVPF (cell activity with IC50 values > 50 µM), compound 3 exhibited marked cellular activity as apoptotic inducer (IC50 = 16.4 µM) in MDA-MB-231 breast cancer cell lines (Figure 3A). The cell line was chosen since it is one of the most sensitive human cancer lines to the reported Bir3 inhibitors and with a high Inhibitor of Apoptosis Proteins (IAPs) expression levels.10,17 In fact, the IAP induced resistance in these cells resulted in no obvious DNA fragmentation when treated with TRAIL (30 ng/ml) or etoposide (5 µM).17 To confirm whether the pro-apoptotic activity of compound 3 is mainly linked to its interaction with the Bir3 domain of XIAP with concomitant caspase-9 activation, we treated XIAP knockout (−/−) and wild type mouse embryonic fibroblasts (MEF) cells with similar amounts of compound 3. Because the compounds are designed to induce apoptosis by inhibition of the anti-apoptotic protein XIAP, it is expected that XIAP−/− cells would be indifferent to the compound. As expected, compound 3 exhibited much less pro-apoptotic activity in XIAP knockout (−/−) MEF cells rather than in wild type cells (Figure 3B) further confirming that compound 3 down-regulates the XIAP function in MEF cells. It is noteworthy that compound 3 still killed XIAP knockout (−/−) MEF cells although at much higher concentrations. This may be caused by off-target effects at higher concentrations and/or by the possible interaction with of other Bir3 containing proteins. Compound 3 also increases caspase-3 activation in MDA-MB-231 cells (Figure 3C) further confirming that the pro-apoptotic activity of compound 3 in cancer cells occurs through caspase activation. Staurosporine was used as control to validate the assay.
Table 1.
| AVPI | compound 3 | |
|---|---|---|
| Human Plasma Stabilitya,e | 23 mins | >120 mins (93.9 % ± 7.7) |
| Drug Metabolic Stabilityb,e | t1/2 = 11 mins | t1/2 = 14.5 mins (> 45 min)*; CLint = 47.7 |
| Permeability (2 %)c,e | Low (5.2 %) | High (27 %) |
| Solubilityd,e | >100 µg/mL | >100 µg/mL |
| Kd from ITC | 0.73 µM | 2.5 µM |
The test compounds were incubated in human plasma at 37 °C for 15, 60, 120 minutes. Liquid chromatography/tandem mass spectrometry (LC/MS/MS) has been applied in quantitative analysis of drug candidates in plasma. Triple quadrupole mass spectrometer (API 3200 QTrap, Applied Biosystems/MDS SCIEX) was used to conduct mass analysis in multiple reaction monitoring (MRM) mode. The plasma stability expressed as the percentages of the remaining compound to the initial concentration were calculated using Analyst software (version 1.4.2) and Excel template.
To determine the metabolic stability, the compounds in triplicates were incubated with S9 fraction in the presence of NADPH at 37 °C for 30 minutes. Liquid chromatography/tandem mass spectrometry (LC/MS/MS) has been applied in quantitative analysis of drug candidates in biological matrix. Triple quadrupole mass spectrometer (API 3200 QTrap, Applied Biosystems/MDS SCIEX) was used to conduct mass analysis in multiple-reaction-monitoring (MRM) mode. The t1/2 and CLint of a test compound in the liver microsomes were calculated according to “in vitro half-life” approach using Analyst software (version 1.4.1) and Excel template. Intrinsic clearance (CLint) was determined using the following equation based on “well-stirred” model: CLint (µL/(min·mg)) = k (min−1) · Vincubation(µL)/Pincubation(mg) where Vincubation – incubation volume in µL; Pincubation – amount of microsomal protein in the incubation volume.
The permeability was determined by using PAMPA method described in the literature44
The apparent solubility was determined by using nephelometry-based method described in the literature.45 The compounds were tested at 8 concentrations in triplicate. The compound was dissolved in DMSO to 10 mM solution on day 1. The assay was performed on the day one with detection on the day 2.
The assay is performed by ASINEX Ltd. (Winston-Salem, NC).
The major metabolic product of compound 3 is its N de-methylation product (compound 2) which is still active against Bir3.
Figure 3.
(A) Cell based activity of compounds AVPI and compound 3. MDA-MB-231 breast cancer cells were cultured in 10 % FBS DMEM media in 96-well plates overnight and followed by dosed with AVPI, compound 3, or vehicle at varying concentrations in serum free DMEM media. After 48 hours treatment, the effects on cells proliferation were evaluated by the ATPLite assay. Cell survival was expressed as percentage of control ± s.e.m., compared to DMSO control, from two independent experiments, each having fours wells per drug concentration. (B) Similar amounts of wild type and XIAP knockout (−/−) mouse embryonic fibroblasts (MEF) cells were cultured 24 hours in 10 % FBS RMPI-1640 media in white 96-well plate before treated by compound 3 or DMSO (control) at varying concentrations. After 24 hours treatment in 10 % FBS RMPI-1640 media, the effects on cells proliferation were evaluated by the ATPLite assay. Cell survival was expressed as percentage of control ± s.e.m., compared to DMSO control, from three independent experiments, each having fours wells per drug concentration. (C) Caspase-3 activation assay. After cultured in 10 % FBS DMEM media overnight, MDA-MB 231 breast cancer cells were treated by compound 3 at various concentrations, DMSO vehicle, or staurosporine 100 nM. After 24 hours culture, the cells were lysed and caspase-3 activities of the lysates were detected by meso scale discovery (MSD) caspase-3 assay kit detecting cleaved versus total caspase-3. Staurosporine was used to indicate the efficiency of the assay. (D) Growth inhibition of MDAMB-231 cells by compound 3 alone or in combination with TRAIL. Cells were cultured in 10 % FBS DMEM media in white 96-well plates overnight and than treated with TRAIL or compound 3 at indicated concentrations alone for 20 and 24 hours, respectively. When used together, compound 3 was added 4 hours before TRAIL (total incubation time, 24 hours). Cell viability (%) was determined by ATPLite assay and was expressed as percentage of control ± s.e.m., compared to DMSO control, from two independent experiments, each having fours wells per drug concentration.
It has been reported that SMAC-mimic compounds sensitize tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) -induced apoptosis in breast cancer cells,17 hence we tested whether compound 3 has synergistic effects with this agent. MDA-MB-231 cells were treated with compound 3 (Figure 3D) or TRAIL alone for 24 or 20 hours respectively. When used together, compound 3 was added 4 hours earlier than TRAIL. Cell viability was determined by ATPLite assay. The synergy combination index (CI) value18 of the combination between compound 3 and TRAIL is 0.48 at both IC50s (CI<1) indicating that those agents induce apoptosis in MDA-MB-231 cells in a synergistic manner. The synergistic effect is rational since TRAIL is believed to trigger apoptosis trough regulating extrinsic death-receptor pathway and compound 3, a Bir3 inhibitor, should activate caspase-9 to induce intrinsic mitochondrial pathway of apoptosis. TRAIL (Genentech, South San Francisco, CA) has recently enter clinic trail, but showed resistances in certain cancers, such as acute myeloid leukemia (AML).19–23 It has been reported that the resistances of TRAIL can be overcame, at least in part, by down-regulation of XIAP.19,24–27 On the other hand, TRAIL itself shows very little effects to induce tumor cell death at low concentrations (up to 110 ng/mL, Figure 3D). Therefore, the combination of compound 3 and TRAIL may provide a novel approach for TRAIL-based therapeutic strategy, such as lower resistance and lower dose of TRAIL.
Conclusion
In conclusion, we propose a simple and effective strategy that combining in silico docking, fragment-based drug design and NMR-spectroscopy, can be used to rapidly derive peptide mimics with improved drug-like properties. When applied to the design of SMAC mimetics, the approach resulted in the design of compound 3 with demonstrated activity in cell. This novel class of compounds not only can be used to further investigate the role of XIAP in tumorigenesis but also provides a viable starting point for the development of novel anti-cancer agents.
Experimental Section
Protein Expression and Purification
The plasmid of His6-tagged Bir3 were described previously.28 The protein was expressed in the Escherichia coli strain BL21(DE3) and purified using Ni2+ affinity chromatography. The uniformly 15N-lableled Bir3 was produced by growing the bacteria in M9 minimal media containing 15NH4Cl as the sole nitrogen source. The NMR samples were dissolved in 20 µM or 100 µM sodium phosphate buffer (pH 7.0) for 1D or 2D NMR experiments containing 90 %/10 % (H2O/2H2O) or 99.5 % 2H2O.
General Synthetic Procedures
All commercially available starting materials were purchased from Sigma-Aldrich, Acros, or Chembridge and were used without further purification. Column chromatography was performed with silica gel (63–200 Å; Machrey-Nagel GmbH & Co. KG) or RediSep Flash Column (ISCO Ltd.). 1H NMR and 13C NMR for QC analysis were acquired on a Varian Inova 300 MHz or Bruker 600 MHz spectrometer. Chemical shifts are reported in ppm from residual solvent peaks (δ 7.27 or 3.31 for CDCl3 and CD3OD for 1H NMR respectively). High resolution ESI-TOF mass spectra were acquired at the Center for Mass Spectrometry, the Scripps Research Institute, La Jolla, CA. Compounds were all found to be in excess of 95 % pure as established by LC-MS.
HPLC analyses were performed using the following conditions: Waters Symmetry® C18 column (Reverse phase, 4.6×150 mm). A linear gradient using 5 % acetonitrile and 95 % water, to 95 % acetonitrile and 5 % water over 20 minutes. Flow rate was 1 mL/min and UV detection was set to 254 nm. The LC column was maintained at ambient temperature.
(S)-2-amino-N-(4-(2-chlorophenoxy)phenyl)propanamide (1)
N-(tert-Butoxycarbonyl)-L-alanine (0.1925 g; 1.02 mmole) and 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (WSC· HCl; 0.2341 g; 1.22 mmole) were mixed in 5 mL THF in a round-bottomed flask, and then 4-(2-chlorophenoxy)aniline (0.2116 g; 0.96 mmole) in small amount THF was then added dropwise into the flask. The reaction mixture was stirred overnight at room temperature under nitrogen. The mixture was extracted with ethyl acetate and saturated NaHCO3(aq), and dried over Na2SO4. The crude product was purified by the CombiFlash Companion machine (ISCO, Inc. Lincoln, NE) by 4 g RediSep normal-phase flash columns with hexane and ethyl acetate solvent system following the recommend procedures. (S)-tert-butyl 1-(4-(2-chlorophenoxy)phenylamino)-1-oxopropan-2-ylcarbamate was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.2088 g, 73 %).
To de-protect Boc groups, the purified (S)-tert-butyl 1-(4-(2-chlorophenoxy)phenylamino)-1-oxopropan-2-ylcarbamate (0.2088 g) was dissolved in dichloromethane (CH2Cl2) and than added 10 equivalent trifloroacetic acid. The reaction mixture was stirred at room temperature for 2–3 hours after the starting material was all consumed (checked by TLC). After solvent was removed, the reactions were than quenched by saturated Na2CO3(aq) solution. The product was extracted with ethyl acetate and the combined organic phase was adjusted to pH 2.0 with concentrated HCl. The final product (salt form) was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.2341 g, 85 %). HPLC purity >99.9 %, tr = 11.26 min. 1H NMR (600 MHz, CDCl3) δ 9.47 (s, 1 H; -NH), 7.95-8.32 (br, 2 H; -NH2), 7.49 (d, J = 5.40 Hz, 2 H), 7.32 (d, J = 7.24 Hz, 2 H), 7.07 (m, 1 H), 6.98 (m, 1H), 6.79 (d, J = 7.2 Hz, 1H), 6.68 (d, J = 6.00 Hz, 2 H), 4.64 (m, 1 H) 1.87 (d, 3 H). HRMS exact mass of (M + H)+, 291.0895 amu; observed mass of (M + H)+, 291.0900 amu.
(S)-5-(2-aminopropanamido)-2-(4-methoxyphenoxy)-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (2)
To a stirring solution of 2-(4-methoxyphenoxy)-5-nitrobenzoyl chloride (0.5188 g; 1.69 mmole) in DMF (5 mL) at 0 °C was added 4-Dimethylaminopyridine (DMAP; 0.2716 g; 2.22 mmole) in one potion. After 30 minutes, the stirring mixture was added (2-(2-thienyl)-1,3-thiazol-4-yl)methylamine (0.3496 g; 1.78 mmole) in a small amount DMF dropwise. The solution was then stirred overnight at room temperature under nitrogen. The mixture was quenched by saturated NH4Cl(aq), extracted with ethyl acetate, and dried over Na2SO4. The resulting residue was purified by the CombiFlash Companion machine (ISCO, Inc. Lincoln, NE) with 4 g RediSep normal-phase flash columns with hexane and ethyl acetate solvent system to afford 2-(4-methoxyphenoxy)-5-nitro-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (0.5926 g, 75 %).
To hydrogenate nitro group, the product from last step was dissolved in ethyl acetate (8 mL) and the catalytic amount palladium/C (10 wt. %) and a balloon of hydrogen gas were added. The mixture was stirred overnight at room temperature, filtered, and concentrated under reduced pressure. The resulting residue was used for next step without further purification.
N-(tert-Butoxycarbonyl)-L-alanine (0.1334 g; 0.71 mmole) and 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (WSC· HCl; 0.2022 g; 1.05 mmole) were mixed in 8 mL THF in a round-bottomed flask, and 5-amino-2-(4-methoxyphenoxy)-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (the hydrogenated product, 0.2238 g; 0.5 mmole) in small amount THF was then added dropwise into the flask. The reaction mixture was stirred overnight at room temperature under nitrogen. The mixture was extracted with ethyl acetate and saturated NaHCO3(aq), and dried over Na2SO4. The crude product was purified by the CombiFlash Companion machine (ISCO, Inc. Lincoln, NE) by 4 g RediSep normal-phase flash columns with hexane and ethyl acetate solvent system following the recommend procedures. (S)-tert-butyl 1-(4-(4-methoxyphenoxy)-3-((2-(thiophen-2-yl)thiazol-4-yl)methylcarbamoyl)phenylamino)-1-oxopropan-2-ylcarbamate was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.2070 g, 68 %).
To de-protect Boc groups, the purified (S)-tert-butyl 1-(4-(4-methoxyphenoxy)-3-((2-(thiophen-2-yl)thiazol-4-yl)methylcarbamoyl)phenylamino)-1-oxopropan-2-ylcarbamate (0.2070 g) was dissolved in dichloromethane (CH2Cl2) and than added 10 equivalent trifloroacetic acid. The reaction mixture was stirred at room temperature for 2–3 hours after the starting material was all consumed (checked by TLC). After solvent was removed, the reactions were than quenched by saturated Na2CO3(aqe) solution. The product was extracted with ethyl acetate and the combined organic phase was adjusted to pH 2.0 with concentrated HCl. The final product (salt form) was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.1435 g, 83 %). HPLC purity 98.11 %, tr = 12.0 min. 1H NMR (600 MHz, CD3OD) δ 8.16 (s, 1 H), 7.72 (d, J = 6.60 Hz, 1 H), 7.61 (s, 2 H), 7.25 (s, 1 H), 7.15 (s, 1 H), 7.01 (d, J = 7.20 Hz, 2 H), 6.92 (d, J = 7.80 Hz, 2H), 6.88 (d, J = 8.40 Hz, 1 H), 4.71 (s, 2 H), 4.09 (m, 1 H), 3.86 (s, 3 H), 1.62 (d, J = 6.00 Hz, 3 H). HRMS exact mass of (M + H)+, 509.1312 amu; observed mass of (M + H)+, 509.1326 amu.
(S)-2-(4-methoxyphenoxy)-5-(2-(methylamino)propanamido)-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (3)
To a stirring solution of 2-(4-methoxyphenoxy)-5-nitrobenzoyl chloride (1.4882 g; 4.84 mmole) in DMF (10 mL) at 0 °C was added 4-Dimethylaminopyridine (DMAP; 0.7115 g; 5.85 mmole) in one potion. After 30 minutes, the stirring mixture was added (2-(2-thienyl)-1,3-thiazol-4-yl)methylamine (1.002 g; 5.11 mmole) in a small amount DMF dropwise. The solution was then stirred overnight at room temperature under nitrogen. The mixture was quenched by saturated NH4Cl(aq), extracted with ethyl acetate, and dried over Na2SO4. The resulting residue was purified by the CombiFlash Companion machine (ISCO, Inc. Lincoln, NE) with 12 g RediSep normal-phase flash columns with hexane and ethyl acetate solvent system to afford 2-(4-methoxyphenoxy)-5-nitro-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (1.4708 g, 65 %).
To hydrogenate nitro group, the product from last step was dissolved in dichloromethane (CH2Cl2, 10 mL) and the catalytic amount palladium/C (10 wt. %) and a balloon of hydrogen gas were added. The mixture was stirred 5 hours at room temperature, filtered, and concentrated under reduced pressure. The resulting residue was used for next step without further purification.
Boc-N-methyl-L-alanine (0.1222 g; 0.60 mmole) and 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (WSC· HCl; 0.1435 g; 0.75 mmole) were mixed in 8 mL THF in a round-bottomed flask, and 5-amino-2-(4-methoxyphenoxy)-N-((2-(thiophen-2-yl)thiazol-4-yl)methyl)benzamide (the hydrogenated product, 0.2238 g; 0.5 mmole) in small amount THF was then added dropwise into the flask. The reaction mixture was stirred overnight at room temperature under nitrogen. The mixture was extracted with ethyl acetate and saturated NaHCO3(aq), and dried over Na2SO4. The crude product was purified by the CombiFlash Companion machine (ISCO, Inc. Lincoln, NE) by 4 g RediSep normal-phase flash columns with hexane and ethyl acetate solvent system following the recommend procedures. (S)-tert-butyl 1-(4-(4-methoxyphenoxy)-3-((2-(thiophen-2-yl)thiazol-4-yl)methylcarbamoyl)phenylamino)-1-oxopropan-2-yl(methyl)carbamate was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.2460 g, 79 %).
To de-protect Boc groups, the purified (S)-tert-butyl 1-(4-(4-methoxyphenoxy)-3-((2-(thiophen-2-yl)thiazol-4-yl)methylcarbamoyl)phenylamino)-1-oxopropan-2-yl(methyl)carbamate was dissolved in dichloromethane (CH2Cl2) and than added 10 equivalent trifloroacetic acid. The reaction mixture was stirred at room temperature for 2–3 hours after the starting material was all consumed (checked by TLC). After solvent was removed, the reactions were than quenched by saturated Na2CO3(aqe) solution. The product was extracted with ethyl acetate and the combined organic phase was adjusted to pH 2.0 with concentrated HCl. The final product (salt form) was obtained after drying the samples in an evaporator and high-pressure vacuum system (0.1817 g, 88 %, HCl salt form). HPLC purity 99.14 %, tr = 12.28 min. 1H NMR (600 MHz, CD3OD) δ 8.19 (s, 1 H), 7.72 (d, J = 7.20 Hz, 1 H), 7.59 (s, 2 H), 7.25 (s, 1 H), 7.14 (s, 1 H), 7.00 (d, J = 7.20 Hz, 2 H), 6.91 (d, J = 7.80 Hz, 2 H), 6.88 (d, J = 8.40 Hz, 1 H), 4.61 (s, 2 H), 4.11 (m, 1 H), 3.90 (s, 3 H), 2.88 (s, 3 H), 1.74 (d, J = 6.0 Hz, 3 H). HRMS exact mass of (M + H)+, 523.1468 amu; observed mass of (M + H)+, 523.1485 amu.
NMR spectroscopy
Spectra were acquired on a 600 MHz Bruker Avance spectrometer equipped with TXI probe and z-shielded gradient coils or on a 600 MHz Bruker Avance equipped with TCI cryoprobe.
Dissociation equilibrium constants (Kd) of ligands were determined by monitoring the protein chemical shift changes as a function of ligand concentration. Data were collected for a set of resolved Bir3 (concentration was 100 µM) 1H NMR resonances and fitted to a single binding site model according to the equation:29
To and Lo are total concentration of target protein and ligand, respectively. The parameter p represents the fractional population of bound versus free species at equilibrium, which for fast exchanging ligands is measured as:
δobs is the observed receptor chemical shift during the titration, and δfree and δsat are the chemical shifts for the receptor in the unbound and fully bound (saturated) states, respectively.
Compounds with estimated Kd values of 100 µM or less are subsequently determined more accurately by a more complete NMR titration. At least six data points were collected for different concentration of ligand and Kd can then be determined by a non-linear least squares fit of p versus [Lo].
All NMR data were processed and analyzed using TOPSPIN2.0 (Bruker BioSpin Corp., Billerica, MA) and SPARKY.30 For chemical shift mapping the NMR samples contained 0.1 mM 15N-labeled Bir3, 100 mM potassium phosphate buffer (pH 7.0), 5 % DMSO and 7 % D2O. 2D [15N, 1H]-HSQC experiments were acquired using 32 scans with 2048 and 128 complex data points in the 1H and 15N dimensions at 300 K.
Isothermal Titration Calorimetry (ITC)
Purified Bir3 domain of XIAP and the 3 compounds were dissolved in PBS buffer (pH = 7.4) and then degassed for 10 min prior to sample loading. Titrations were performed using a VP-ITC calorimeter from Microcal (Northampton, MA). The solution containing Bir3 (100 µM) was loaded into the sample cell, whereas the compound 3’s solution (1 mM) was loaded into the injection syringe. The calorimeter was first equilibrated at 25 °C, and the baseline was monitored during equilibration. The total observed heat effects were corrected for these small contributions. All titration data were analyzed using Microcal Origin software provided by the ITC manufacturer. (MicroCal, LLC). The association constant (Ka) through the equation: ΔG = −RTln Ka, where R was the gas constant and T was the absolute temperature in Kelvin. Equilibrium dissociation constant (Kd) was calculated as the reciprocal of Ka.
Molecular Modeling
Some commercial available alanine-containing compounds are downloaded from ZINC databases (total 2,062,906 drug-like compounds from RYAN, CHEMBRIDGE, CHEMDIV, SIGMA ALDRICH, SPECS, COMGENEX, OTAVA, ASINEX, NCI, TIMTEC, ENAMINE, MAYBRIDGE, AMBINTER, INTERBIOSCREEN, LIFECHEMICALS, KEY ORGANICS, MICROSOURCE, NANOSYN, PHARMEKS, PUBCHEM, INTERCHIM databases).31 The libraries of commercial available amines were modified by CombiChem into a virtual alanine-containing library with 1,400 compounds.32–34 The 3D structures of the 1,400 compounds were built with CONCORD35 and energy minimized with Sybyl (Tripos, St. Louis, MO).36 The X-ray coordinates of Smac-XIAP-BIR3 is available (PDB ID: 1G73, 2.0A resolution)37 and was used as the docking host of the virtual compounds. Docking calculations were performed with GOLD version 3.0 (Cambridge Crystallographic Data Centre, Cambridge, U.K.).38,39 For each compound, 10 solutions were generated and subsequently ranked according to their Goldscore.40 The Top 200 compounds predicted by GOLD were further analyzed using SYBYL. The goals are to visually inspect all their docking poses and select compounds whose alanine moieties primarily bind to BIR3 the same mode as that of AVPI observed in the X-ray structure. The Figure was made with SYBYL and the molecular surface was generated with MOLCAD.41 The second virtual library of compound 1’s derivatives (about 900 compounds) was similarly built up by downloading drug-like amines from Sigma-Aldrich, Acros, and Chembridge and further modifying into alanine-containing compounds by Combichem.
Cell Culture
ER-negative MDA-MB 231 breast cancer cells were obtained from the American Type Culture Collection (Manassas, VA) and were maintained in DMEM medium supplemented with 10 % fetal bovine serum (FBS, Gibco) at 37 °C in a humidified incubator containing 5 % CO2. Wild type and XIAP knockout (−/−) mouse embryonic fibroblasts (MEF)2,42 were cultured in RPMI-1640 medium supplemented with 10 % fetal bovine serum (FBS, Gibco) at 37 °C in a humidified incubator containing 5 % CO2.
ATPlite® Cell viability assay
The effect of individual test agents on cell viability was assessed by using the ATPlite 1 step Luminescence Assay System (PerkinElmer, Waltham, MA)43 in four replicates. For cell viability data of compound 3 and AVPI (Figure 3A), MDA-MB-231 cells (8000 cells/well) were seeded and incubated in white 96-well, flat-bottomed plates in DMEM medium with 10 % FBS overnight. The cells were exposed to various concentrations of test agents dissolved in DMSO (final DMSO concentration, 0.1 %) in serum free DMEM medium for 48 hours. To compare the difference of cell growth inhibition of compound 3 between wild type and XIAP knockout (−/−) MEF cells (Figure 3B), the same amount of each type cells (5000 cells/well) were seeded and incubated in white 96-well, flat-bottomed plates in RPMI-1640 medium with 10 % FBS overnight. The cells were exposed to various concentrations of compound 3 dissolved in DMSO (final DMSO concentration, 0.1 %) in 10 % FBS RPMI-1640 medium for 24 hours. For TRAIL combination experiment (Figure 3D), MDA-MB-231 cells (8000 cells/well) were seeded and incubated in white 96-well, flat-bottomed plates in DMEM medium with 10 % FBS overnight. The cells were exposed to various concentrations of compound 3 alone or TRAIL alone or combined in 10 % FBS DMEM medium. The cells were treated by compound 3 for 24 hours and TRAIL for 20 hours. When used together, compound 3 was added into cells four hours before TRAIL. Controls received DMSO vehicle at a concentration equal to that of drug-treated cells. After the indicated length of treatments, the cells in each well were treated with 10 µL ATPlite substrate solution. Dark adapt the plate for 5–10 minutes and measure the luminescence by Wallac 1420 plate reader (PerkinElmer). The curves or graphs were generated using Excel (Microsoft®) and the IC50 was calculated by PRISM (Graphpad®). The Combination index (CI) of 3 and TRAIL was calculated by CalcuSyn (version 2.1, Biosoft®) software.
Caspase-3 activation
MDA-MB-231 cells were cultured in 12 wells plates (100,000 cells/well) with 10 % FBS DMEM media overnight. The cells were treated compound 3 (varying concentrations), DMSO vehicle, or 100 nM Staurosporine and incubated for another 24 hours. The effect of individual test agents on caspase-3 activation was assessed by using the cleaved/total caspase-3 assay (Meso Scale Discovery, Meso Scale Diagnostics, LLC, Gaithersburg, MD) following the recommend buffer and procedures in two replicates. The number of relative caspase-3 activation is calculated by following equation:
Supplementary Material
General synthetic schemes, details about molecular models NMR spectra of the derivatives of compound 1. NMR Bir3 15N-HSQC spectra, and the mapping data of compound 3. Statistical analysis of the synergy data reported n Figure 3. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This work was supported in part by NIH grants U19 CA113318 (to M.P. and J.C.R.), R01 AI059572 (to M.P.) and P01 CA102583 (to M.P.).
Abbreviations
- XIAP
X-Linked Inhibitor of Apoptosis Protein
- Bir3
baculovirus IAP repeat 3
- SMAC
second mitochondrial activator of caspases
- IAPs
Inhibitor of Apoptosis Proteins
- FITC-labled
fluorescein-isothiocyanate-labeled
- ITC
Isothermal Titration Calorimetry
- MEF
mouse embryonic fibroblasts
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- CI
combination index
- AML
acute myeloid leukemia
- AVPI
Alanine Valine Proline Isoleucine.
References
- 1.Fesik SW, Shi Y. Structural biology. Controlling the caspases. Science. 2001;294:1477–1478. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Cuddy M, Samuel T, Welsh K, Schimmer A, Hanaii F, Houghten R, Pinilla C, Reed JC. Cellular, biochemical, and genetic analysis of mechanism of small molecule IAP inhibitors. J. Biol. Chem. 2004;279:48168–48176. doi: 10.1074/jbc.M405022200. [DOI] [PubMed] [Google Scholar]
- 3.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 5.Schimmer AD, Welsh K, Pinilla C, Wang Z, Krajewska M, Bonneau MJ, Pedersen IM, Kitada S, Scott FL, Bailly-Maitre B, Glinsky G, Scudiero D, Sausville E, Salvesen G, Nefzi A, Ostresh JM, Houghten RA, Reed JC. Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 6.Nikolovska-Coleska Z, Xu L, Hu ZJ, Tomita Y, Li P, Roller PP, Wang RX, Fang XL, Guo RB, Zhang MC, Lippman ME, Yang DJ, Wang SM. Discovery of embelin as a cell-permeable, small-molecular weight inhibitor of XLAP through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. J. Med. Chem. 2004;47:2430–2440. doi: 10.1021/jm030420+. [DOI] [PubMed] [Google Scholar]
- 7.Wu TY, Wagner KW, Bursulaya B, Schultz PG, Deveraux QL. Development and characterization of nonpeptidic small molecule inhibitors of the XIAP/caspase-3 interaction. Chem. Biol. 2003;10:759–767. doi: 10.1016/s1074-5521(03)00157-1. [DOI] [PubMed] [Google Scholar]
- 8.Sun HY, Nikolovska-Coleska Z, Yang CY, Xu L, Tomita Y, Krajewski K, Roller PP, Wang SM. Structure-based design, synthesis, and evaluation of conformationally constrained mimetics of the second mitochondria-derived activator of caspase that target the X-linked inhibitor of apoptosis protein/caspase-9 interaction site. J. Med. Chem. 2004;47:4147–4150. doi: 10.1021/jm0499108. [DOI] [PubMed] [Google Scholar]
- 9.Park CM, Sun C, Olejniczak ET, Wilson AE, Meadows RP, Betz SF, Elmore SW, Fesik SW. Non-peptidic small molecule inhibitors of XIAP. Bioorg. Med. Chem. Lett. 2005;15:771–775. doi: 10.1016/j.bmcl.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Oost TK, Sun C, Armstrong RC, Al-Assaad AS, Betz SF, Deckwerth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg SH, Shoemaker AR, Tomaselli KJ, Zou H, Fesik SW. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J. Med. Chem. 2004;47:4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 11.Sun HY, Nikolovska-Coleska Z, Chen JY, Yang CY, Tomita Y, Pan HG, Yoshioka Y, Krajewski K, Roller PP, Wang SM. Structure-based design, synthesis and biochemical testing of novel and potent Smac peptido-mimetics. Bioorg. Med. Chem. Lett. 2005;15:793–797. doi: 10.1016/j.bmcl.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Chen JY, Nikolovska-Coleska Z, Wang GP, Qiu S, Wang SM. Design, synthesis, and characterization of new embelin derivatives as potent inhibitors of X-linked inhibitor of apoptosis protein. Bioorg. Med. Chem. Lett. 2006;16:5805–5808. doi: 10.1016/j.bmcl.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 13.Sun HY, Nikolovska-Coleska Z, Lu JF, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang SM. Design, synthesis, and characterization of a potent, nonpeptide, cell-permeable, bivalent smac mimetic that concurrently targets both the BIR2 and BIR3 domains in XIAP. J. Am. Chem. Soc. 2007;129:15279–15294. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 15.Sharma SK, Straub C, Zawel L. Development of peptidomimetics targeting IAPs. Int. J. Pept. Res. Ther. 2006;12:21–32. doi: 10.1007/s10989-005-9003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney MC, Wang XX, Park J, Liu YS, Pei DH. Determination of the sequence specificity of XIAP BIR domains by screening a combinatorial peptide library. Biochemistry-Us. 2006;45:14740–14748. doi: 10.1021/bi061782x. [DOI] [PubMed] [Google Scholar]
- 17.Bockbrader KM, Tan M, Sun Y. A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene. 2005;24:7381–7388. doi: 10.1038/sj.onc.1208888. [DOI] [PubMed] [Google Scholar]
- 18.Milella M, Estrov Z, Kornblau SM, Carter BZ, Konopleva M, Tari A, Schober WD, Harris D, Leysath CE, Lopez-Berestein G, Huang Z, Andreeff M. Synergistic induction of apoptosis by simultaneous disruption of the Bcl-2 and MEK/MAPK pathways in acute myelogenous leukemia. Blood. 2002;99:3461–3464. doi: 10.1182/blood.v99.9.3461. [DOI] [PubMed] [Google Scholar]
- 19.Carter BZ, Mak DH, Schober WD, Dietrich MF, Pinilla C, Vassilev LT, Reed JC, Andreeff M. Triptolide sensitizes AML cells to TRAIL-induced apoptosis via decrease of XIAP and p53-mediated increase of DR5. Blood. 2008;111:3742–3750. doi: 10.1182/blood-2007-05-091504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 21.Wuchter C, Krappmann D, Cai Z, Ruppert V, Scheidereit C, Dorken B, Ludwig WD, Karawajew L. In vitro susceptibility to TRAIL-induced apoptosis of acute leukemia cells in the context of TRAIL receptor gene expression and constitutive NF-kappa B activity. Leukemia. 2001;15:921–928. doi: 10.1038/sj.leu.2402131. [DOI] [PubMed] [Google Scholar]
- 22.Riccioni R, Pasquini L, Mariani G, Saulle E, Rossini A, Diverio D, Pelosi E, Vitale A, Chierichini A, Cedrone M, Foa R, Lo Coco F, Peschle C, Testa U. TRAIL decoy receptors mediate resistance of acute myeloid leukemia cells to TRAIL. Haematologica. 2005;90:612–624. [PubMed] [Google Scholar]
- 23.Mori T, Doi R, Kida A, Nagai K, Kami K, Ito D, Toyoda E, Kawaguchi Y, Uemoto S. Effect of the XIAP inhibitor Embelin on TRAIL-induced apoptosis of pancreatic cancer cells. J. Surg. Res. 2007;142:281–286. doi: 10.1016/j.jss.2007.03.068. [DOI] [PubMed] [Google Scholar]
- 24.Rosato RR, Dai Y, Almenara JA, Maggio SC, Grant S. Potent antileukemic interactions between flavopiridol and TRAIL/Apo2L involve flavopiridol-mediated XIAP downregulation. Leukemia. 2004;18:1780–1788. doi: 10.1038/sj.leu.2403491. [DOI] [PubMed] [Google Scholar]
- 25.Cummins JM, Kohli M, Rago C, Kinzler KW, Vogelstein B, Bunz F. X-linked inhibitor of apoptosis protein (XIAP) is a nonredundant modulator of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in human cancer cells. Cancer Res. 2004;64:3006–3008. doi: 10.1158/0008-5472.can-04-0046. [DOI] [PubMed] [Google Scholar]
- 26.McManus DC, Lefebvre CA, Cherton-Horvat G, St-Jean M, Kandimalla ER, Agrawal S, Morris SJ, Durkin JP, Lacasse EC. Loss of XIAP protein expression by RNAi and antisense approaches sensitizes cancer cells to functionally diverse chemotherapeutics. Oncogene. 2004;23:8105–8117. doi: 10.1038/sj.onc.1207967. [DOI] [PubMed] [Google Scholar]
- 27.Kim EH, Kim SU, Choi KS. Rottlerin sensitizes glioma cells to TRAIL-induced apoptosis by inhibition of Cdc2 and the subsequent downregulation of survivin and XIAP. Oncogene. 2005;24:838–849. doi: 10.1038/sj.onc.1208241. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 29.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 30.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 31.Irwin JJ, Shoichet BK. ZINC: A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. http://zinc.docking.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies K, Briant C. Combinatorial chemistry library design using pharmacophore diversity. Network Sci. 1996;1 http://www.netsci.org/Science/combichem/feature05.html. [Google Scholar]
- 33.ChemBridgeSoft. Cambridge, MA: Corporation; http://www.cambridgesoft.com/webinars/info/default.aspx?webinarid=236. [Google Scholar]
- 34.Polinsky A. Combichem and cheminformatics. Curr. Opin. Drug Discov. Dev. 1999;2:197–203. [PubMed] [Google Scholar]
- 35.Pearlman RS. CONCORD. 1998 distributed by Tripos (St. Louis) [Google Scholar]
- 36.Pearlman RS. SYBYL 7.0. Tripos Inc.: 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA; 1998. [Google Scholar]
- 37.Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- 38.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 39.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 40.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 41.Teschner M, Henn C, Vollhardt H, Reiling S, Brickmann J. Texture mapping: a new tool for molecular graphics. J. Mol. Graph. 1994;12:98–105. doi: 10.1016/0263-7855(94)80074-x. [DOI] [PubMed] [Google Scholar]
- 42.Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC. An IAP-IAP complex inhibits apoptosis. J. Biol. Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 43.Kangas L, Gronroos M, Nieminen AL. Bioluminescence of cellular ATP: a new method for evaluating cytotoxic agents in vitro. Med. Biol. 1984;62:338–343. [PubMed] [Google Scholar]
- 44.Ko JK, Ma JJ, Chow JY, Ma L, Cho CH. The correlation of the weakening effect on gastric mucosal integrity by 5-HT with neutrophil activation. Free Radic. Biol. Med. 1998;24:1007–1014. doi: 10.1016/s0891-5849(97)00428-0. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd BA, Arif AM, Allred EL. A norbornenyl derivative with through-space ketone pi-interaction. Acta Crystallogr C. 2000;56(Pt 11):1377–1379. doi: 10.1107/s0108270100011185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
General synthetic schemes, details about molecular models NMR spectra of the derivatives of compound 1. NMR Bir3 15N-HSQC spectra, and the mapping data of compound 3. Statistical analysis of the synergy data reported n Figure 3. This material is available free of charge via the Internet at http://pubs.acs.org.