Abstract
Chimpanzees in west central Africa (Pan troglodytes troglodytes) are known to harbor simian immunodeficiency viruses (SIVcpzPtt) that represent the closest relatives of human immunodeficiency virus type 1 (HIV-1); however, the number of SIVcpzPtt strains that have been fully characterized is still limited. Here, we report the complete nucleotide sequence of SIVcpzGAB2, a virus originally identified in 1989 in a chimpanzee (P. t. troglodytes) from Gabon. Analysis of this sequence reveals that SIVcpzGAB2 is a member of the SIVcpzPtt group of viruses, but that it differs from other SIVcpzPtt strains by exhibiting a highly divergent Env V3 loop with an unusual crown (NLSPGTT) containing a canonical N-linked glycosylation site, an unpaired cysteine residue in Env V4, and two late (L) domain motifs (PTAP and YPSL) in Gag p6. Moreover, phylogenetic analyses indicate evidence of recombination during the early divergence of SIVcpzPtt strains; in particular, part of the pol gene sequence of SIVcpzGAB2 appears to be derived from a previously unidentified SIVcpz lineage ancestral to HIV-1 group O. These data indicate extensive diversity among naturally occurring SIVcpzPtt strains and provide new insight into the origin of HIV-1 group O.
Simian immunodeficiency viruses infecting chimpanzees (SIVcpz) were first described in 1989 1 and soon identified as representing the closest relatives of human immunodeficiency virus type 1 (HIV-1).2 However, it was not until 1999 that SIVcpz, and in particular strains of the virus infecting the subspecies Pan troglodytes troglodytes in west central Africa, were recognized as the most likely source of three cross-species transmissions that gave rise to HIV-1 groups M, N, and O.3 One reason for this was the very small number of SIVcpz strains that had been molecularly characterized; even today, only seven strains of SIVcpz have been completely sequenced. Five of these represent SIVcpzPtt from the central subspecies P. t. troglodytes,2–6 while the other two are strains of (the more distantly related) SIVcpzPts from the eastern subspecies P. t. schweinfurthii.7,8 None of these SIVcpz infections is known to be pathogenic for their natural hosts, despite the close genetic similarity of humans and chimpanzees. Moreover, this lack of pathogenicity is unlikely to be the result of an ancient virus–host relationship since chimpanzees have acquired their infection relatively more recently from Cercopithecid monkeys, which represent a major SIV reservoir in sub-Saharan Africa.9 Understanding the evolutionary history of SIVcpz is thus of fundamental importance, not only because it may reveal why HIV-1 is pathogenic, but also because it may yield new insight into the mechanisms that govern cross-species transmission and the emergence of new human pathogens.
The first two SIVcpz-infected apes (GAB1 and GAB2) were identified among 50 chimpanzees wild-caught in Gabon that were screened for HIV-1 cross-reactive antibodies.1 To further characterize their infections, peripheral blood mononuclear cells (PBMCs) of these chimpanzees were cocultivated with normal human donor PBMCs to recover replicating virus. This yielded a virus isolate (SIVcpzGAB1) for one of the chimpanzees (GAB1), which was molecularly cloned and sequenced.2 The second chimpanzee (GAB2), a 2-year-old infant from northeastern Gabon, died shortly after capture from wounds inflicted by hunters. Only a single blood sample was taken in April 1988 and attempts to isolate virus from this sample remained unsuccessful. SIVcpz infection was subsequently verified by polymerase chain reaction (PCR) amplification of a small subgenomic pol fragment (280 bp) from uncultured PBMC DNA.10 Both GAB1 and GAB2 were confirmed to be members of the P. t. troglodytes subspecies by mitochondrial DNA analysis.3
To characterize SIVcpzGAB2 in greater detail, we used long PCR approaches to amplify a complete genomic equivalent from uncultured PBMC DNA. This was done by using primer combinations that targeted circular unintegrated viral DNA intermediates as described previously.11 Briefly, two short fragments, located at the 5′ and 3′ ends of the pol gene, were first obtained using SIV consensus primers. SIVcpzGAB2–specific primers were then designed from these amplicons and used, in combination with SIV consensus primers, to amplify the remainder of the genome in four overlapping fragments. Amplification products were gel purified, cloned, and sequenced. In regions of overlap, the 5′ sequence was arbitrarily selected for compilation of the final genomic sequence. The concatenated SIVcpzGAB2 sequence (R-U5-gag-pol-env-U3) was determined to be 9354 bp in length.
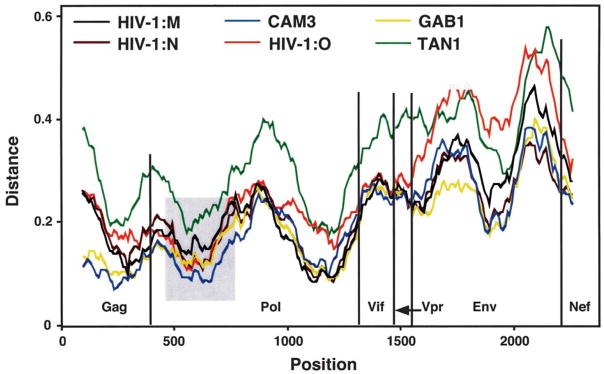
Inspection of the SIVcpzGAB2 sequence identified complete open reading frames for all nine genes characteristic of SIVcpz and HIV-1 strains, with no obvious mutations in any protein or RNA motifs of known functional importance. To compare SIVcpzGAB2 to previously described members of the SIVcpz/HIV-1 group of viruses, we first performed diversity plot analyses of the major proteins (Fig. 1). Most plots revealed roughly parallel shifts in the extent of divergence along the proteome, with consistently greater similarity of SIVcpzGAB2 to other SIVcpzPtt strains (CAM3, GAB1) or HIV-1 (group M) than to SIVcpzPts (TAN1). However, the plot for SIVcpzGAB2 versus HIV-1 group O was exceptional in showing surprisingly high similarity across the first third of Pol relative to the values across Gag, Env, and the rest of Pol. Such nonparallel behavior of diversity plots reflects either gene-specific variations in evolutionary rates or recombination during the divergence of strains.
FIG. 1.
Diversity plots illustrating the extent of amino acid sequence difference between SIVcpzGAB2 and SIVcpzPtt (GAB1, CAM3), SIVcpzPts (TAN1), and representative HIV-1 group M, N, and O strains. Plots for SIVcpz strains US and CAM13 followed similar trajectories to that for GAB1; those for strains CAM5 and ANT were similar to those for CAM3 and TAN1, respectively. The HIV-1 strains used were U455 (M), YBF30 (N), and ANT70 (O). Predicted protein sequences were aligned using Clustal W16 with minor manual adjustment, then concatenated (C-terminal overlaps were removed); sites with ambiguous alignment, or a gap in any sequence, were excluded. Differences were calculated for windows of 200 amino acids, moved in steps of 20 amino acids along the alignment (0.1 = 10% difference); values are plotted at the midpoint of the window. The gray area shows the region within which the distance of SIVcpz GAB2 to HIV-1 group O (red line) is reduced.
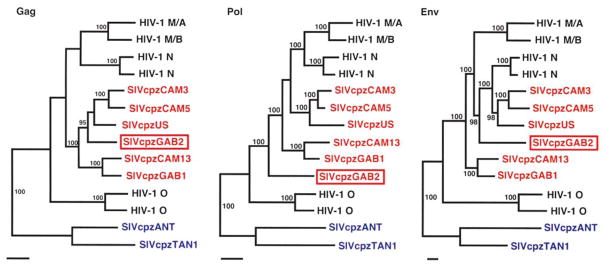
To examine the evolutionary relationships of SIVcpzGAB2 to other strains in more detail, we performed phylogenetic analyses of Gag, Pol, and Env proteins (Fig. 2). As expected, SIVcpzGAB2 grouped within the SIVcpzPtt/HIV-1 lineage rather than the SIVcpzPts lineage, but its position varied among the three trees. In trees derived from Gag and Env proteins, SIVcpzGAB2 joined the CAM3/CAM5/US clade of SIVcpzPtt, with strong statistical support (note that the insertion of HIV-1 group N within this clade in the Env tree, but not in the Gag or Pol trees, reflects the previously inferred recombinant ancestry of that group3). In contrast, in the Pol tree, SIVcpzGAB2 was an outlier to the entire clade composed of other SIVcpzPtt and HIV-1 groups M and N.
FIG. 2.
Phylogenetic trees depicting the relationship of SIVcpzGAB2 (boxed) to other SIVcpz and HIV-1 isolates. SIVcpz strains from P. t. troglodytes and P. t. schweinfurthii are shown in red and blue, respectively. Protein sequences were aligned using Clustal W16 with minor manual adjustment. Maximum likelihood trees were estimated by the Bayesian method (implemented in MRBAYES),17 using the JTT matrix of amino acid replacement and a gamma distribution of rates across sites, then midpoint rooted. Numbers on internal branches indicate estimated posterior probabilities (as percentages); only values of 95% or higher are shown. The scale bars indicate 5% replacement per site. Representative HIV-1 sequences used: group M subtypes A (strain U455) and B (LAI), group N strains YBF30 and YBF106, and group O strains ANT70 and MVP5180.
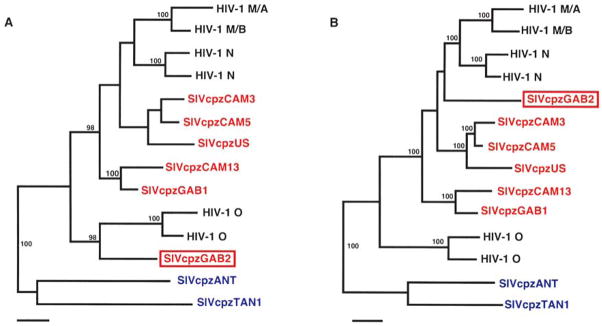
The position of SIVcpzGAB2 in the Pol tree could be an artifact if the pol gene comprised regions with different evolutionary histories due to recombination, as suggested by the diversity plots. To investigate this, we estimated phylogenies for two regions of Pol separately (Fig. 3). Region A was initially selected on the basis of the diversity plot, and subsequently optimized by repeated phylogenetic analyses with redefined boundaries; the final version comprised 300 residues in the gap-stripped alignment, covering sites 129–430 in the SIVcpzGAB2 Pol sequence. Region B comprised the remainder of Pol, downstream of region A. In Pol region A, SIVcpzGAB2 was found to cluster with HIV-1 group O, with the branch supporting this estimated to have an a posteriori probability of 98%. In contrast, for Pol region B SIVcpzGAB2 was found to fall within the clade composed of SIVcpzPtt and HIV-1 groups M and N, although no grouping with any specific viral lineage within this clade was statistically supported. These results thus provide evidence that recombination occurred during the early divergence of the SIVcpzPtt lineages, including those that gave rise to HIV-1 groups M and N, implying coinfection of chimpanzees with divergent strains of SIVcpzPtt.
FIG. 3.
Phylogenetic analyses of regions of the Pol protein. Region A comprised 300 residues (in the gap-stripped alignment) from the amino-terminal half of the protein (see Fig. 1); region B comprised the remaining 554 residues between region A and the carboxyl-terminus. See Figure 2 for methods and details of presentation.
The clustering of SIVcpzGAB2 with HIV-1 group O in Pol region A is particularly interesting; while this result must be viewed with some caution, given the tendency of the Bayesian phylogenetic methodology to overestimate the strength of support for clades, 12 it suggests some insight into the origin of HIV-1 group O. Group O is clearly more closely related to SIVcpzPtt than to SIVcpzPts, and most cases of HIV-1 group O infection have been reported in Cameroon and neighboring countries, in an area overlapping the range of P. t. troglodytes. However, no strains of SIVcpzPtt that are specifically closely related to group O have been described. This could be due to the as yet limited sampling of SIVcpzPtt strains. However, it is worth noting that a third subspecies of chimpanzee, P. t. vellerosus, inhabits Cameroon north of the Sanaga River; while this subspecies is not known to harbor SIVcpz infection, only about 50 individuals have been tested.6 Thus, SIVcpz relatives of HIV-1 group O could yet be found among P. t. vellerosus, but the clustering of SIVcpzGAB2 with HIV-1 group O in the 5′ pol region suggests that the ancestors of HIV-1 group O infected P. t. troglodytes.
Detailed comparisons of Gag and Env sequences revealed a number of protein signatures that are unique for SIVcpzGAB2. For example, alignment of SIVcpz/HIV-1 Gag p6 protein sequences identified an unusual arrangement of late (L) domain sequences involved in primate lentivirus budding.13 All known SIVcpz and HIV-1 strains contain a PT/SAP motif that interacts with Tsg101, a cellular protein that facilitates budding of vesicles into late endosomes. The Gag p6 sequence of SIVcpz-GAB2 has this motif, but also contains a YPSL motif known to bind to AIP1, a second host protein involved in endosomal sorting. The presence of both of these motifs within Gag p6 has previously been observed only in SIVs infecting certain Cercopithecus monkey species.11
The V3 loop of the HIV-1 gp120 glycoprotein is involved in coreceptor binding, yet is extremely variable, apparently in response to immune pressure due to its exposure on the protein surface.14 A notable feature of previously characterized SIVcpz sequences has been a relative lack of diversity in this region.5,7,15 In a phenetic analysis of V3 loop sequence similarity (Fig. 4), the SIVcpzGAB2 V3 sequence was the most divergent of all the SIVcpz, including SIVcpzPts (note that while SIVcpz strains from P. t. troglodytes and P. t. schweinfurthii form two very distinct clades in phylogenetic analyses of Env and other proteins, their V3 loop sequences do not, even when SIVcpzGAB2 is excluded). The V3 loop of SIVcpzGAB2 differs from other SIVcpzPtt strains at 17–19 (out of 35) sites, whereas the other SIVcpzPtt strains differ from each other at only 6–12 residues. At 13 sites, the SIVcpzGAB2 sequence contained residues not found in any of the other SIVcpz strains, including four sites (shown in red) where the amino acid was conserved among all seven other SIVcpz sequences (Fig. 4). Most notably, the first Gly in the GPG motif found at the crown of the loop was replaced by a Ser in SIVcpzGAB2, which together with the preceding Glu to Asn change generated a highly unusual predicted N-linked glycosylation site near the tip of the loop. To ensure that the observed V3 loop sequence did not represent a minor SIVcpzGAB2 variant, we PCR amplified and sequenced additional env fragments. None of these exhibited changes in their V3 loop sequences (not shown). Moreover, we constructed full-length SIVcpzGAB2 clones and tested their infectivity and replication potential in vitro. Several of these clones yielded SIVcpzGAB2 virions that were infectious and replicated in primary human PBMC cultures (not shown). Although the biological significance of the divergent SIVcpz-GAB2 V3 loop remains to be determined, it is clear that the predominant viral species that infected the chimpanzee GAB2 encoded this unusual envelope domain.
FIG. 4.

Comparison of gp120 V3 loop sequences among SIVcpz and representative HIV-1 strains. Sites conserved among all SIVcpz strains except SIVczpGAB2 are highlighted in the SIVcpzGAB2 sequence. Predicted N-linked glycosylation sites are underlined. The dendrogram at the left represents the phenetic similarity among sequences, inferred by the UPGMA method applied to fractional differences among pairs of sequences. SIVcpzPtt and SIVcpzPts strains are indicated by cpz/t (in red) and cpz/s (in blue), respectively. Other sequences are HIV-1 strains: for example, M/A:UG037 denotes group M, subtype A, strain UG037.
Finally, the SIVcpzGAB2 gp120 glycoprotein contains an additional cysteine residue in a region corresponding to the V4 loop of HIV-1. SIVcpz and HIV-1 group M strains typically have no Cys residues within the V4 loop, but HIV-1 group O strains have a CX4C motif. SIVcpzCAM5 also has two extra Cys residues in the V4 region, but in a CX5C motif. Cys residues in HIV or SIV envelope glycoprotein surface subunits are known to form disulfide bonds that determine the tertiary structure and folding of the gp120 subunit and are essential for envelope function. Whether the unpaired cysteine residue in the SIVcpzGAB2 V4 loop has structural consequences important for in vivo envelope function is not known; however, it should be noted that the replication-competent SIVcpzGAB2 clones mentioned above also contain this unpaired cysteine residue.
In conclusion, the full-length sequences of six SIVcpzPtt strains have now been determined, including three from Cameroon, two from Gabon, and one of unknown west central African origin. It is interesting to note that the two Gabonese viruses are not particularly closely related (Fig. 2). The SIVcpz-GAB2 sequence reported here shows several novel features, adding significantly to our knowledge of SIVcpzPtt diversity, but also raising questions concerning the historical, geographical, and functional nature of that diversity. Thus, given the importance of this group of viruses as the recent sources of HIV-1, it is clear that sequences of additional SIVcpzPtt strains will need to be determined.
SEQUENCE DATA
The complete genome sequence of SIVcpzGAB2 has been deposited in the GenBank/EMBL DNA sequence database with the accession number AF382828.
Acknowledgments
We thank John Moore for helpful discussions, Maria Salazar for technical assistance, and W.J. Abbott for artwork and manuscript preparation. This work was supported in part by grants from the National Institutes of Health R01 AI50529, R01 AI 58715, R01 AI 44596, and NO1 AI85338, the Sequencing Core of the UAB Center for AIDS Research (P30 AI 27767), and the Howard Hughes Medical Institute.
References
- 1.Peeters M, Honore C, Huet T, et al. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 3.Gao F, Bailes E, Robertson DL, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 4.Corbet S, Muller-Trutwin MC, Versmisse P, et al. Env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller-Trutwin MC, Corbet S, Souquiere S, et al. SIVcpz from a naturally infected Cameroonian chimpanzee: Biological and genetic comparison with HIV-1 N. J Med Primatol. 2000;29:166–172. doi: 10.1034/j.1600-0684.2000.290310.x. [DOI] [PubMed] [Google Scholar]
- 6.Nerrienet E, Santiago ML, Foupouapouognigni Y, et al. Simian immunodeficiency virus infection in wild-caught chimpanzees from Cameroon. J Virol. 2005 doi: 10.1128/JVI.79.2.1312-1319.2005. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanden Haesevelde MM, Peeters M, Jannes G, et al. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology. 1996;221:346–350. doi: 10.1006/viro.1996.0384. [DOI] [PubMed] [Google Scholar]
- 8.Santiago ML, Bibollet-Ruche F, Bailes E, et al. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J Virol. 2003;77:2233–2242. doi: 10.1128/JVI.77.3.2233-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailes E, Gao F, Bibollet-Ruche F, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 10.Janssens W, Fransen K, Peeters M, et al. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpzGAB2 from a wild-captured chimpanzee from Gabon. AIDS Res Hum Retroviruses. 1994;10:1191–1192. doi: 10.1089/aid.1994.10.1191. [DOI] [PubMed] [Google Scholar]
- 11.Bibollet-Ruche F, Bailes E, Gao F, et al. New simian immunodeficiency virus infecting De Brazza’s monkeys (Cercopithecus neglectus): Evidence for a Cercopithecus monkey virus clade. J Virol. 2004;78:7748–7762. doi: 10.1128/JVI.78.14.7748-7762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki Y, Glazko GV, Nei M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc Natl Acad Sci USA. 2002;99:16138–16143. doi: 10.1073/pnas.212646199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Schwedler U, Stuchell M, Muller B, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 14.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV-1’s switch hitter. AIDS Res Hum Retroviruses. 2005 doi: 10.1089/aid.2005.21.171. (in press) [DOI] [PubMed] [Google Scholar]
- 15.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–617. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic tress. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]