Abstract
The leukemia cells of unrelated patients with chronic lymphocytic leukemia (CLL) display a restricted repertoire of immunoglobulin (Ig) gene rearrangements with preferential usage of certain Ig gene segments. We developed a computational method to rigorously quantify biases in Ig sequence similarity in large patient databases and to identify groups of patients with unusual levels of sequence similarity. We applied our method to sequences from 1577 CLL patients through the CLL Research Consortium (CRC), and identified 67 similarity groups into which roughly 20% of all patients could be assigned. Immunoglobulin light chain class was highly correlated within all groups and light chain gene usage was similar within sets. Surprisingly, over 40% of the identified groups were composed of somatically mutated genes. This study significantly expands the evidence that antigen selection shapes the Ig repertoire in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the monoclonal expansion of CD5+ B cells that expresses functional, rearranged immunoglobulin genes. The immunoglobulin (Ig) heavy chain variable region (IGHV) genes of roughly half of all CLL patients contain abundant somatic mutations, while the Ig used by leukemia cells of the other half of patients have minimal or no somatic mutations1. The latter group of patients are more likely to have CLL cells that express high levels of CD38 and the zeta-associated protein of 70 kD (ZAP-70)2–4 and to have a more adverse clinical outcome. The underlying biology explaining these differences is unclear, but the observation has provoked extensive efforts by many groups to sequence the IGHV genes from many CLL patients. As these sequence collections have grown, additional features of the IGHV repertoire in CLL have become apparent. Significant gene use biases were confirmed5, and the spectrum of mutations has been analyzed and found to be consistent with conventional somatic hypermutation6.
Recently, several groups have identified sets of CLL patients whose IGHV genes are remarkably similar. This similarity is manifested in terms of V, D, and J gene usage, CDR3 amino acid composition, and light chain usage. Initial examples of this phenomenon included sets of patients with: (i) the 51p1 allele of VH1-69 joined with D3-3 or D3-10, JH6 and distinct long HCDR3 sequence 7; (ii) the VH3-21 gene and the Vlambda2-14 light chain and a distinct, short HCDR3 sequence 8,9; and (iii) a set of patients expressing class switched VH4-39 with D6-13, and JH5 and a defined HCDR3 motif coupled with VκO12/2 light chain10. These observations were followed by several larger studies that identified additional sets of patients 11–14. The patients that have IGHV genes belong to the sets utilizing VH1-69 and its alleles or the VH3-21 gene have been found to have relatively more adverse clinical outcomes, which, in the case of VH3-21, might be independent of whether or not there are IGHV somatic mutations. More recently, a set of patients with CLL cells that use the VH3-72 gene with a distinct HCDR3 motif has been identified that apparently have a relatively good clinical prognosis15.
Collectively, the studies described above have advanced the hypothesis that particular antigen receptors play a significant role in the progress towards neoplasia16. The remarkable similarity of HCDR3 regions within these sets of patients strongly supports the view that particular antigen reactivity is the selective force. Such a hypothesis places antigen stimulation as a key event in the development or perpetuation of the leukemic cells. However, to date, these antigens have not been identified.
The sets of patients discovered thus far have not been identified through rigorous statistical approaches. Since the process of heavy chain V-D-J recombination allows for roughly seven thousand unique combinations, observing the same usage of V, D, and J genes is expected to be rare. However, both normal and leukemic B cells can have non-random use of certain V, D, and J genes. In addition, junctional processes during VDJ recombination diversify the sequences where the genes are joined. Thus, it is not clear what collection of sequences would give an appropriate null model for testing the hypothesis that the Ig repertoire in CLL is distinct because of selection by antigen(s) that contributes to leukemogenesis. The problem is compounded by the fact that Ig sequence collections from normal B cells are limited and it is not clear which type or class of B cell would be the proper “normal” equivalent. As such, it has been difficult to conclude much about the number of sets that may exist among the CLL population or the extent to which these may represent a distinct biological phenomenon.
In the present study, we address these issues by introducing a computational approach that includes a rigorous definition of sequence similarity in the HCDR3 and allows us to screen large datasets for patients whose IGHV sequences are sufficiently similar that they can be considered to be archetypes. Our approach relies on the creation of random datasets of IGHV sequences that preserve certain sequence biases present in CLL patients and allow us to define a level of similarity that is surprising. We applied our method to analyze the largest available collection of IGHV CLL sequences: 1577 evaluated by the CRC17. Our method identified the previously reported sets as well as many new sets with the CRC collection. Furthermore, combining our robust definition of clusters with estimation techniques borrowed from ecology allows us to predict the fraction of patients that belong to a set for both the mutated and unmutated classes. These calculations indicate that set membership at the sequence level is a phenomenon restricted to ~28% of unmutated cases and ~12% of mutated cases.
Additionally, we examined the sequences of the Ig light chains in a subset of the patients in our identified sets, and found that members of sets have highly correlated lambda and kappa chain usage. However, we did not find strong correlations between set membership and several critical clinical parameters.
Materials and Methods
Patient samples and VH sequencing
Blood was collected from consenting patients who satisfied diagnostic criteria for CLL and who presented for evaluation at the referral centers of the CLL Research Consortium (CRC). The samples were prepared and the IGHV sequences were determined as previously described 11.
Immunoglobulin sequence annotation
1577 IGHV sequences from CLL patients were obtained from the CRC collection. A set of Perl scripts were written to perform batch queries of IMGT/VQUEST 18. The alignments returned by IMGT were used to determine the CDR3 region and the percent mutation of the V region. Additional annotations included identification of the origin of each nucleotide: V, D, J or “junctional” from IMGT/JunctionAnalysis. Subsequently, each amino acid was assigned an origin according to table translating the various codon triplets into symbols. The percent mutation from germline sequence for the V gene was calculated and included in the annotations for each sequence for use in creating mutated and unmutated datasets.
Sequence alignments and clustering
We extracted the CDR3 region from each sequence from the IMGT/JunctionAnalysis reports along with the annotations of the source of the encoding nucleotides (V, D, J or junctional process). We performed an all versus all pairwise alignment of the CDR3 sequences using the allversusall program from the EMBOSS 3.0.0 package19 with the BLOSUM62 scoring matrix. The pairwise alignment score S(i, j) between sequences i and j was converted into a distance d(i, j) using the transformation d(i, j) = S(i, i)+S(j, j)−2S(i, j). We set d(i, j) equal to infinity if sequences i and j contained different V genes. This criterion enforced the requirement that CDR3 sequences were deemed similar only if they originate from the same V genes. We performed average linkage hierarchical clustering on the distance matrix d(i, j) using the software of Hoon and colleagues20 to identify clusters of sequences.
The creation of sets of similar patients requires the definition of a similarity or distance threshold. Namely, how close must sequences be to be considered part of the same set? We answered this question using a permutation test. In this test, we compare the clusters we obtain in the CLL sequences to clusters we obtain in a synthetic dataset created by shuffling the components of each CLL sequence. The goal was to determine thresholds that would be unlikely to recover clusters by random chance. To create a synthetic dataset, we first annotated each CLL sequence according to the VH gene used, and the CDR3 residues were split into five segments: (i) encoded by the V gene exclusively; (ii) arising from a V-D junctional event; (iii) encoded by the D gene exclusively; (iv) arising from a the D-J junctional event, or (v) encoded by the J gene. A permuted sequence dataset was generated by randomly shuffling these five segments from all CLL sequences. These permuted datasets preserved IGHV gene usage and sequence biases present in the CLL IGHV sequence collection. We created 100 random datasets by permuting the CLL sequences to determine a threshold for the average distance permitted between sequences in a one given cluster.
Using the random datasets, we estimated the false discovery rate (FDR) for a distance threshold as the ratio of the fraction of random sequences in clusters (any sequence that is contained in a cluster of size 2 or more) divided by the fraction of CRC sequences in clusters. This computation was repeated to find distance thresholds for 1%, 5%, 10%, 15%, 20%, 25% and 30% FDR separately for all CRC sequences. A 5% FDR was obtained for a distance threshold of 58.
Species Estimator Statistics
To estimate the number of unseen patient clusters, we applied our clustering procedure to random subsets of the CLL sequences. We computed a species accumulation curve for the CRC dataset by re-sampling from the CRC sequences. For a fixed subset size K, we extract 100 samples of size K from the CRC sequences. We clustered each of these 100 samples separately and computed the average over the 100 samples of the fraction of sequences contained in clusters. We fit a two-phase exponential association equation Y(x) = Ymax1 (1 − Exp[−k1 x]) + Ymax2 (1 − Exp[−k2 x]) to the species accumulation curve using nonlinear regression. Under the assumptions: (i) only a fraction of patients are contained in clusters; and (ii) the patients that are not members of clusters in the current dataset are not members of IGHV clusters, we estimated the number of unseen clusters by applying statistical methods commonly used to estimate the number of unseen species in an ecological survey 21,22
Results
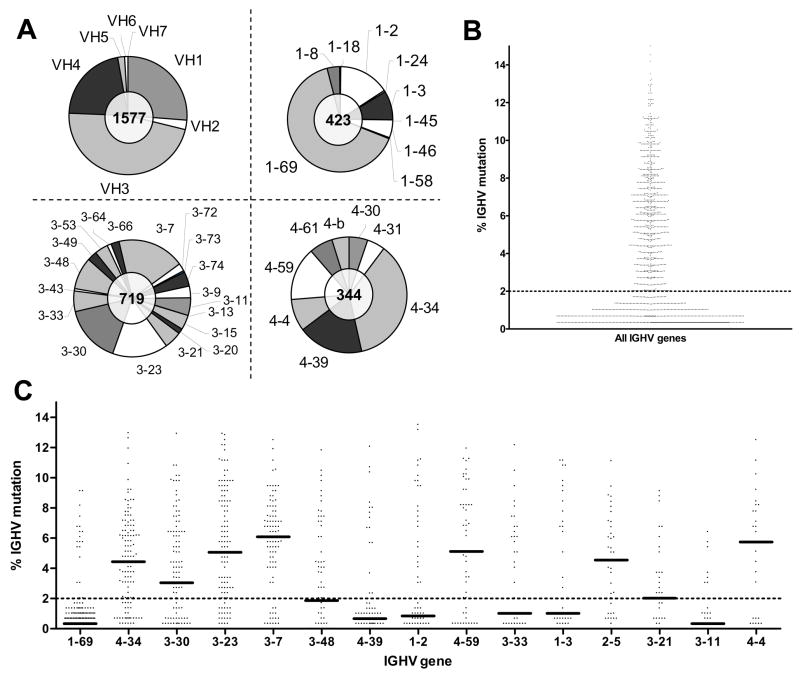
Ig heavy chain variable regions were sequenced from the CLL cells of 1577 patients, evaluated by members of CLL Research Consortium. The sequences were analyzed for V, D, and J gene use and percent mutation in the VH sequence using IMGT/JunctionAnalysis18. The distribution of IGHV genes appearing in the patient collection was similar to other reported collections, with VH3 family genes the most common6 (Figure 1A). The most common IGHV genes were 1–69, 4–34, 3–30, 3–23, and 3–7. The distribution of mutational frequencies was also similar to previously described CLL sequence collection, with 49% of the sequences having more than 2% mutation (Figure 1B). The distributions of mutation among sequences using particular IGHV genes were highly skewed. 1–69 and 4–39 utilizing sequences were almost all unmutated, whereas 4–34 and 3–07 utilizing sequences were predominantly mutated. Figure 1C plots the percent mutation as a function of IGHV sequenced used for the 15 most frequently observed IGHV genes. A bimodal distribution centered on the 2% threshold is clearly evident for some IGHV genes, such as 3–7, 1–2, 1–3, and 4–4, whereas others are more continuous (e.g. 3–23).
Figure 1. Distribution of IGHV genes and mutation frequencies within the dataset.
A. The distribution of IGHV gene families among the data and the distribution of individual genes for the VH1, 3, and 4 families are shown. The total number of cases depicted is indicated in the center of each pie chart. B. Scatterplot of the % mutation for all cases analyzed. The dashed line at 2% reflects the threshold for considering a case “mutated”. C. The % mutation for cases using the 15 most frequent IGHV genes. The bars indicate the median value.
Supplemental Table I summarizes the significant associations found between IGHV, IGD, and IGHJ gene use. The frequency of IGHV/IGD, IGHV/IGHJ, and IGD/IGHJ associations are graphically depicted in Supplemental Figure 1. Many of the statistically significant correlations correspond to previously identified groups of similar sequences, such as the association of IGHV1-69 with IGD 2-2, 3-3, and 3-16, but broadly biased associations were not observed. Therefore, a strategy to identify clusters of patients with similar IGHV gene sequences was employed.
A major concern in clustering IGHV gene sequences is distinguishing true clustering from coincidental similarities that would be found in a random dataset of the same size. In order to identify groups of similar patients it a rigorous manner, it was necessary to identify a threshold of similarity between IGHV sequences that is surprising compared to what would be expected by chance. Ideally, one might define the similarity threshold through examination of normal (non-CLL) sequences, but the normal cell counterpart of CLL is not known. Thus, we decided not to infer CDR3 sequence characteristics or IGHV gene frequency from the publicly available normal B cell derived sequence collections. Instead we employed a permutation approach to determine the background frequency of similar sets while avoiding a need for assumptions about the cellular origin of CLL or the mechanistic biases of VDJ recombination. This approach shuffled the constituent subparts of the IGHV sequences: namely the V, D, and J genes and the junctional residues. This shuffled procedure produced “random” sequences that preserve the VH bias and bias in junctional residues inherent in the CLL sequences but removes any correlations between the subparts. These random sequences allow us to determine a similarity threshold that reveals the biologically meaningful combinations of V, D, and J present in the CLL sequences apart from biases in their constituent parts (Fig. 2).
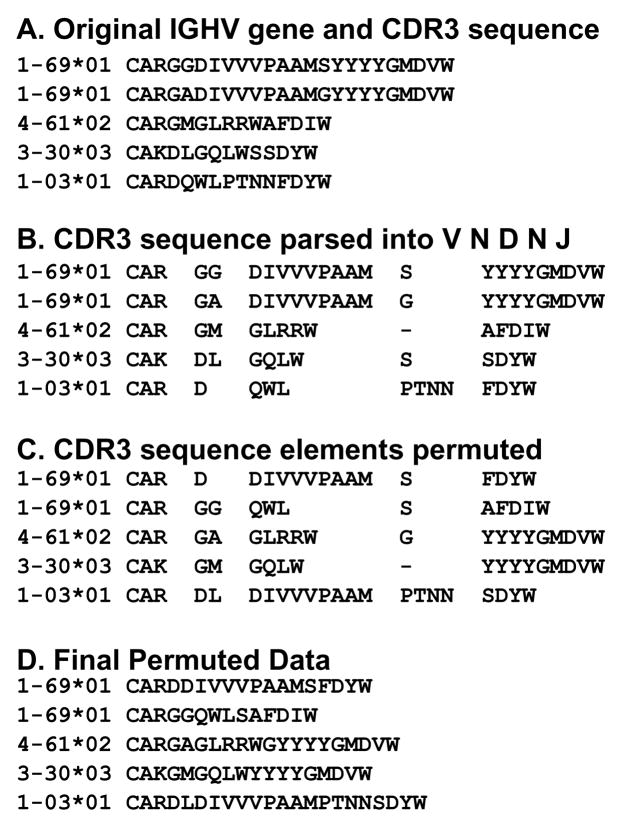
Figure 2. CDR3 permutation scheme.
Permuted datasets were created from the original CLL IGHV sequences in the manner of the example shown. The original CDR3 amino acid sequences (A) were parsed into the sequences derived from V, D, or J genes or junctional processes as annotated by IMGT/JunctionAnalysis (B). Amino acids were categorized as junctional if any of the nucleotides encoding an amino acid were attributable to a junctional event. These sequence elements and the assigned IGHV genes were randomly scrambled (C) to create the final permuted datasets (D).
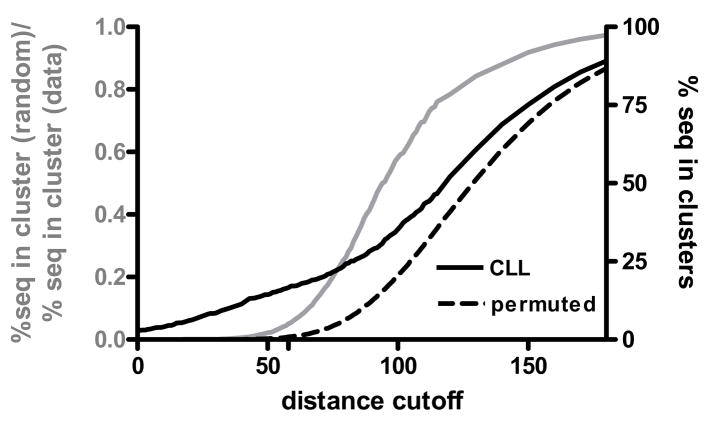
The CLL sequences and permuted sequences were aligned and clustered, and the fraction of sequences that were assigned to a cluster consisting of two or more sequences (cluster fraction) was calculated for both sets of sequences. The cluster fraction from the permuted dataset is essentially an estimate of the false discovery rate (FDR), a measure of random chance of obtaining clusters at a given threshold. We selected a similarity threshold at which the ratio of the cluster fraction from the permuted dataset was 5% of the cluster fraction in the real data (Figure 3), analogous to an FDR or p-value of 0.05. This threshold was used for the subsequent analyses.
Figure 3. Determination of clustering threshold.
The CLL and permuted CDR3 datasets were clustered as described in Methods. The percentage of sequences assigned to a cluster as a function of the distance cutoff used in the clustering algorithm was determined for both datasets (black lines, right y axis). The fraction of sequences in clusters among the permuted data divided by the fraction of sequences in clusters among actual data is shown in gray along the left y axis. At a distance cutoff of 58 the fraction of sequences in clusters among the permuted data was 5% of the value among the actual data.
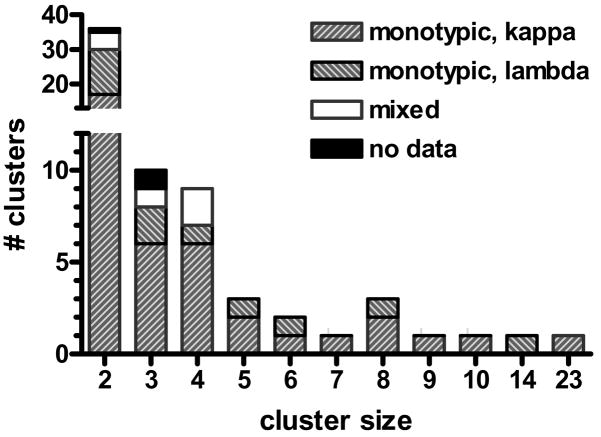
A total of sixty-seven clusters were identified using this procedure. The distribution of cluster sizes is shown in Figure 4. There were several large clusters that consisted of sequences containing previously identified CDR3 archetypes. Almost half of the clusters consisted of pairs of similar sequences and there were 10 clusters with 3 members each. The light chain isotypes had been previously determined for most of the cases studied here as part of the CLL Research Consortium tissue bank program and are indicated in Table 1. Significantly, 30 out of the 36 pairs were monotypic for light chain isotype (Figure 4). Eight out of the nine clusters with three members for which data was available were monotypic as well, and the larger clusters were all monotypic (Table 1).
Figure 4. Cluster size and light chain isotype distribution.
67 clusters were identified at the 5% FDR cutoff value. Flow cytometric data for light chain expression was available for most of the cases. The vast majority of clusters were monotypic for light chain expression, consisting of only kappa or lambda. Only five of the 36 clusters composed of two sequences were of mixed light chain isotype.
Table I.
Summary of IGHV sequence archetypes
| Consensus V gene |
Light chain |
IGHV mutation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cluster | n | IGHV | IGD | IGHJ | representative CDR3 | κ | λ | κλ | M | U | %mut |
| 1 | 23 | 1–69 | 3–16 | 3 | CARGGDYDYIWGSYRSNDAFDIW | 22 | - | - | - | 23 | 0.4 |
| 2 | 14 | 3–21 | 6 | CARDANGMDVW | 1 | 12 | - | 10 | 4 | 2.5 | |
| 3 | 10 | 1–3 | 5–12 | 4 | CAREQWLPSLNFDYW | 9 | - | 1 | - | 10 | 0.3 |
| 4 | 9 | 1–69 | 2–2 | 6 | CARMRPDIVVVPAAISYYYGMDVW | 9 | - | - | - | 9 | 0.3 |
| 5 | 8 | 4–59 | 6–19 | 4 | CARGPDISGWLGLAYW | - | 7 | 1 | 8 | - | 9.1 |
| 6 | 8 | 3–48 | 2–2 | 6 | CARDSPLVVPAAIFYYYYGMDVW | 8 | - | - | - | 8 | 0.0 |
| 7 | 8 | 1–2 | 1–26 | 6 | CARLLSGSYYYYYYGMDVW | 7 | - | - | - | 8 | 1.0 |
| 8 | 7 | 1–2 | 6–19 | 4 | CARGQWLPQDYFDYW | 7 | - | - | - | 7 | 0.2 |
| 9 | 6 | 1–69 | 3–3 | 6 | CARDGSNYDFWSGYYPNYYYYGMDVW | - | 4 | - | - | 6 | 0.4 |
| 10 | 6 | 1–69 | 2–2 | 6 | CAREVPDIVVVPAVYYYYGMDVW | 6 | - | - | - | 6 | 0.2 |
| 11 | 5 | 4–34 | 2–2 | 3 | CARGLRQVGYCSSTSCYVYYYYYYMDVW | 4 | - | - | - | 5 | 0.1 |
| 12 | 5 | 1–69 | 2–2 | 6 | CARGGDIVVVPAAMSYYYYGMDVW | 5 | - | - | - | 5 | 0.6 |
| 13 | 5 | 4–39 | 2–2 | 6 | CARHRLGYCSSTSCYYYYYGMDVW | - | 4 | - | - | 5 | 0.0 |
| 14 | 4 | 4–34 | 3 | CAKDIEVAVPDAFDIW | 4 | - | - | 4 | - | 6.8 | |
| 15 | 4 | 1–69 | 3–22 | 1 | CARGPYSSDYYYAYW | 4 | - | - | 4 | - | 5.6 |
| 16 | 4 | 3–7 | 4 | CARGPWW | 3 | 1 | - | 4 | - | 8.0 | |
| 17 | 4 | 1–69 | 3–3 | 4 | CARADDFWSGYFHW | 4 | - | - | - | 4 | 0.6 |
| 18 | 4 | 3–30 | 1–1 | 4 | CVKDRSATWSFDYW | - | 2 | 1 | 4 | - | 7.2 |
| 19 | 4 | 3–7 | 4–17 | 6 | CAGGYGDYVYYYYYYGMDVW | 2 | - | - | - | 4 | 1.0 |
| 20 | 4 | 3–48 | 3–3 | 6 | CARTYDFWSGYFSYYYYYGMDVW | - | 2 | - | - | 4 | 0.3 |
| 21 | 4 | 3–11 | 3–10 | 5 | CARDRVLYYGSGSYYNWFDPW | 4 | - | - | - | 4 | 0.1 |
| 22 | 4 | 4–34 | 3–9 | 6 | CARLLAGAYYYYYYGMDVW | 3 | - | - | - | 4 | 0.8 |
| 23 | 3 | 3–15 | 5 | CTTDSFIW | - | 3 | - | 1 | 2 | 4.0 | |
| 24 | 3 | 1–69 | 3–10 | 6 | CARAMVQGVIGVDYYYYYMDVW | - | 3 | - | - | 3 | 0.6 |
| 25 | 3 | 1–69 | 3–10 | 6 | CARGDLWFGELLTYYYYYGMDVW | 3 | - | - | - | 3 | 0.5 |
| 26 | 3 | 3–30 | 1–20 | 3 | CAKVWWNDVGDAFDIW | 3 | - | - | 3 | - | 2.9 |
| 27 | 3 | 1–3 | 6 | CARMLTGRNYYYYYGMDVW | 3 | - | - | - | 3 | 0.7 | |
| 28 | 3 | 1–69 | 5–24 | 3 | CARGGEMAAILGRGAFDIW | 2 | - | - | 2 | 1 | 4.5 |
| 29 | 3 | 4-b | 6–19 | 4 | CARVRYSSDWYDYFDFW | - | - | - | 2 | 1 | 2.3 |
| 30 | 3 | 4–59 | 3–22 | 6 | CARARGDYYDSSGYLYYYYGMDVW | 3 | - | - | - | 3 | 0.3 |
| 31 | 3 | 1–46 | 3–22 | 4 | CARDQYYYDSSGYYSGYFDYW | 3 | - | - | - | 3 | 0.1 |
| 32 | 3 | 3–21 | 6–13 | 4 | CARDPSFYSSSWTLFDYW | 1 | 2 | - | 1 | 2 | 0.7 |
| 33 | 2 | 3–53 | 4 | CARGNAFDYW | - | 2 | - | 1 | 1 | 2.4 | |
| 34 | 2 | 4–34 | 4 | CARRPETWDILTGDGFDSW | - | 2 | - | 2 | - | 7.8 | |
| 35 | 2 | 3–9 | 3–16 | 6 | CAKDRSYDYIWGSYRTGPFYYYYYGMDVW | 1 | 1 | - | - | 2 | 0.3 |
| 36 | 2 | 3–74 | 4 | CARDLSAADYW | - | 2 | - | 1 | 1 | 4.2 | |
| 37 | 2 | 1–2 | 6–19 | 4 | CARVQWLGLAHFDYW | 2 | - | - | - | 2 | 0.5 |
| 38 | 2 | 3–72 | 2–2 | 3 | CTRVRICSSTTCRNAFDIW | 1 | 1 | - | 2 | - | 3.5 |
| 39 | 2 | 3–23 | 4–17 | 3 | CAKGDGDSLDAFDIW | - | 2 | - | 2 | - | 3.9 |
| 40 | 2 | 3–7 | 2–2 | 4 | CARIYCSSSSCYSDRHFDYW | 2 | - | - | 2 | - | 4.9 |
| 41 | 2 | 1–69 | 3–3 | 6 | CARRVGRYDFWSGYQTTSYYGMDVW | 2 | - | - | - | 2 | 0.7 |
| 42 | 2 | 6–1 | 6 | CARDYYYGMDVW | 1 | 1 | - | 1 | 1 | 2.3 | |
| 43 | 2 | 1–69 | 3–9 | 6 | CARDCYDILTGWSLYYYYGMDVW | 2 | - | - | - | 2 | 0.0 |
| 44 | 2 | 3–23 | 3–3 | 4 | CAKWFRGYDFWSGYSINYFDYW | 2 | - | - | - | 2 | 0.7 |
| 45 | 2 | 1–2 | 5 | CARDEELRWSQGWFDPW | 1 | - | 1 | 2 | - | 11.7 | |
| 46 | 2 | 1–3 | 3 | CARGVRTGTYYGDDAFDIW | 2 | - | - | 2 | - | 9.0 | |
| 47 | 2 | 4–34 | 3–22 | 4 | CVRGFSHYYDSSGYLTLFDYW | 2 | - | - | 2 | - | 3.2 |
| 48 | 2 | 4–34 | 2–2 | 6 | CARGPCIVVVPAAYYDYYYGMDVW | 2 | - | - | - | 2 | 0.5 |
| 49 | 2 | 3–15 | 4 | CTSGGGTGDYW | 1 | 1 | - | 2 | - | 6.1 | |
| 50 | 2 | 1–69 | 3–10 | 6 | CARGVVQGVINVLYYYYGMDVW | - | 2 | - | - | 2 | 0.7 |
| 51 | 2 | 4–59 | 4 | CARGGSGSPEPFDYW | - | 2 | - | 2 | - | 2.2 | |
| 52 | 2 | 1–69 | 3–3 | 6 | CATRDITIFGVVIIKGYYYYGMDVW | 2 | - | - | - | 2 | 1.0 |
| 53 | 2 | 1–69 | 2–2 | 6 | CARTARYVVVPAAMLYYYGMDVW | 2 | - | - | - | 2 | 1.2 |
| 54 | 2 | 5–51 | 6 | CARPSLTGLNYGMDVW | 1 | 1 | - | 2 | - | 4.1 | |
| 55 | 2 | 4–4 | 6–19 | 4 | CVRGADNSGWNPFDYW | - | 2 | - | 2 | - | 7.4 |
| 56 | 2 | 3–53 | 3–3 | 6 | CARDSNSPYYDFWSGYYTDYYYGMDVW | - | 2 | - | - | 2 | 0.0 |
| 57 | 2 | 4–34 | 6 | CARDSLLVVPAAIYYYYYGMDVW | 2 | - | - | - | 2 | 0.2 | |
| 58 | 2 | 1–69 | 3–3 | 6 | CARGNKGDLIFGVVINYYYYYGMDVW | 2 | - | - | - | 2 | 0.3 |
| 59 | 2 | 3–7 | 1–26 | 4 | CAREARIVGATTIDYW | - | 2 | - | 2 | - | 7.4 |
| 60 | 2 | 1–69 | 3–22 | 4 | CARPYYYDSSGYYYPGYW | - | 2 | - | - | 2 | 1.0 |
| 61 | 2 | 3–48 | 1 | CARDEVGAPYW | - | 2 | - | 1 | 1 | 1.5 | |
| 62 | 2 | 4–30 | 3–10 | 6 | CVRDGQGVW | 2 | - | - | 2 | - | 7.5 |
| 63 | 2 | 2–5 | 1–1 | 4 | CAHRRGGYNWNDGHFDYW | - | 2 | - | 2 | - | 8.6 |
| 64 | 2 | 3–30 | 3–3 | 6 | CARATPTYYDFWSGYYPNYYYYGMDVW | - | 2 | - | - | 2 | 0.3 |
| 65 | 2 | 1–69 | 3 | CASGVLLWFGELLYPLDYW | 2 | - | - | - | 2 | 0.5 | |
| 66 | 2 | 1–69 | 5–5 | 5 | CAREGGIQLWGYNWFDPW | 2 | - | - | - | 2 | 0.2 |
| 67 | 2 | 3–30 | 1–26 | 4 | CARGIVGTTDGIFDYW | 2 | - | - | 2 | - | 5.4 |
We determined the Ig light chain sequences for the patients in 8 sets to test the assumption that the restriction in use of light chain isotype by any one set reflected similarity in the primary light chain sequences of the Ig within the set. All but one set were found to have Ig light chains that were encoded by the same IGLV or IGKV gene, though the J segments were not always the same (Sup Table II). This striking light chain restriction was not part of the cluster determination method and is therefore an independent confirmation that the method for selecting the clusters is identifying biologically relevant clusters and not mere fortuitous similarities.
Table II.
Species estimator non-linear curve fitting parameters
| name | equation | R2 | Runs test p value | Ymax (Ymax1, Ymax2) |
|---|---|---|---|---|
| Two phase exponential association | Y=Ymax1*(1−exp(−K1*X)) + Ymax2*(1−exp(−K2*X)) | 0.9983 | 0.6359 | 0.192 (0.136, 0.056) |
| One phase exponential association | Y=Ymax*(1−exp(−K*X)) | 0.9872 | < 0.0001 | 0.168 |
| Clench | Y=a*x/(1+b*x) | 0.9971 | < 0.0001 | 0.214 |
| linear dependence | Y=(Top/K)*(1−exp(−K*X)) | 0.9872 | < 0.0001 | 0.168 |
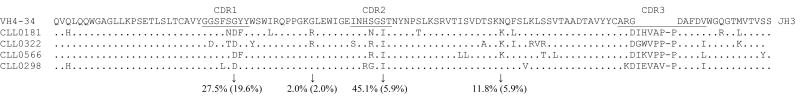
Most of the clusters contained either all unmutated (n=38) or all mutated (n=22) sequences, where unmutated is defined as less than 2% mutation in the IGHV gene. The larger clusters were mostly unmutated, but the distribution among the smaller clusters was more equal. Member sequences of mutated clusters frequently contained common mutations throughout the IGHV gene. Cluster 14, which contained 4 members using the 4–34 gene, is shown as an example in Figure 5. Not only were there several common mutations, but these mutations were also relatively rare among the other CLL sequences that used the 4–34 gene and contained mutations.
Figure 5. IGHV sequence alignment of cluster 14 reveals shared rare mutations.
Cluster 14 was composed of four cases that all used mutated IGHV4-34 and IGHJ3 genes. These are shown aligned with the germline genes with the CDR regions underlined. A (.) indicates identity with the germline amino acid. Several shared mutations were evident. The values indicated below are the frequency with which those amino acids were mutated in all IGHV4-34 using CLL cases (n=100) and the value in parentheses is the frequency with which the mutation resulted in the same amino acid as for this cluster. For example, mutation of the serine in CDR2 was observed in 45.1% of IGHV4-34 CLL cases, but in only 5.9% was the mutation replacement by isoleucine.
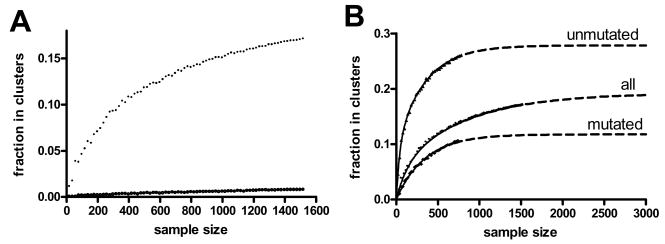
It is apparent that as larger CLL sequence collections are analyzed, additional patient clusters might be found. We used a re-sampling approach to estimate the number of unseen clusters. In this approach, we performed clustering on random subsets of the CLL sequences of various sizes and plotted the number of clusters and the fraction of sequences assignable to a cluster (Figure 6). As expected, both of these quantities grow as more sequences are considered. However, both quantities also appear to be approaching an asymptote. Using this plot, we can estimate the number of clusters and in fraction of patients in clusters as the size of the sequence dataset grows. This problem is analogous to the problem of estimating the total number of species in an ecosystem based on an ecological survey. A number of “species estimator” equations21,22 have been developed for this purpose and we fit several of these formulas to random subsets of the CLL sequences. The fitting parameters and results are summarized in Table II. A two-phase exponential association equation fit the data extremely well and did not show any significant deviation from the model as determined by a Runs test. The best fit parameters of this equation indicated an asymptote at 0.192 with a 95% confidence interval from 0.173 to 0.211. When the data were re-analyzed using less stringent clustering parameters, the asymptote shifted upwards, but the curve fitting quality was similar (data not shown).
Figure 6. Non-linear curve fitting of species estimator models indicates a limited fraction of CLL cases are cluster members.
The fraction of sequences in clusters was determined for random samples of various sizes between the CLL and permuted data. The fraction of sequences assigned to a cluster grew as the sample size grew, but appeared to be approaching an asymptote (A). One or two phase exponential association equations were fit to the cluster fraction data for the entire CLL dataset as well as the mutated and unmutated cases separately. The dashed lines indicate the projections of those fit equations, indicating a clear predicted asymptote for all cases.
Assuming that not all patients can be assigned to one of the defined clusters and that the patients who cannot be currently assigned are not members of unidentified IGHV clusters, we estimate that there are 128 clusters with a 95% confidence interval of 93 to 210 clusters using species estimator statistics 21,22.
Since it was apparent from the clusters identified that there were more clusters among the cases with unmutated Ig genes, the re-sampling and curve fitting was performed independently on cases that used unmutated or mutated Ig genes. The two-phase exponential association curve achieved a good fit for the data from cases that used unmutated Ig genes, but did not provide a confident estimate for the asymptote for the cases that used mutated Ig genes (data not shown). However, the one phase exponential association equation fit the data from these latter cases equally well with a small 95% confidence interval. These are shown in Figure 6. The curve fittings predict a maximum fraction in clusters of 0.278 for the cases with unmutated Ig and 0.116 for the cases with mutated Ig.
Discussion
This study found that the grouping of patients into clusters or sets based on IGHV similarity is a phenomenon restricted to a subset of CLL patients. A similar result was reported by Stamatopoulos et al 14, who also attempted clustering of IGHV sequences using different criteria. However, those and earlier studies that report clustering of IGHV genes in CLL patient relied on ad hoc criterion for defining these clusters. In this study we defined rigorous computational approaches and we found that ~28% of unmutated CLL and ~12% of mutated CLL IGHV sequences can be assigned to a cluster. Furthermore, our species estimator results suggest that while many smaller clusters remain to be found, the current data implies that less than a quarter of all CLL patients can be assigned to a cluster.
The limited light chain data presented here provides evidence that the clustering scheme has identified clusters with biological relevance. The proportion of monotypic light chain clusters far exceeded what would be predicted by chance, and almost all of the clusters (even the pairs) for which light chain sequencing was performed displayed remarkable sequence restriction. Furthermore, the presence of common mutations outside of the CDR3 region among some of the mutated clusters is also highly suggestive of shared biological function.
There are two main caveats to this work. First, while our permutation approach attempts to create a rigorous definition of a cluster that minimizes coincidental similarity, there remains a choice of parameter values in the definition of a cluster. However, the essential result of an asymptotic fraction of sequences in clusters was unaffected by the choice of clustering threshold and the stringent criteria used in this study were selected to avoid accidental clusters. Second, and more significant, is that the analysis done was based solely on IGHV sequences. It is likely that highly similar sequences will share antigen reactivity and the presence of common mutations throughout the IGHV gene in some of the mutated sets is even more suggestive of shared antigen specificity within a cluster, but this is not a formal proof. Ultimately antigen specificity is determined by antibody structure, not sequence, and thus dramatically different IGHV sequences may encode antibodies with identical antigen, or even epitope, specificity 23. Thus the number of functional clusters might be quite different from the number of sequence-defined clusters described here.
Furthermore, the extent to which the antibody response against a given antigen is stereotyped across individuals is not clear. There have been limited studies in mice and humans, primarily with hapten antigens, but there have not been large systematic studies of the antibody sequences generated in many individuals to antigens of diverse type. In fact, the large datasets of IGHV sequences from CLL patients, exceeding many thousands worldwide, represent a unique collection of truly independent origin from many individuals, and may be useful for addressing fundamental issues of the antibody repertoire once the etiology is more clearly established.
This study significantly expands the evidence that antigen selection shapes the repertoire of CLL Ig. It is difficult to conceive an alternative explanation for the extensive presence of sequence clusters among CLL patients. However, it remains unclear at which phase of leukemogenesis antigen selection may play a role and whether ongoing antigen stimulation is relevant at the time of clinical presentation. It is also unclear whether that antigen is necessarily foreign, self, or a class of antigens recognized in a polyspecific manner. Sequence analysis alone cannot shed light on these issue since inferring antibody specificity from primary sequence is difficult and prone to mistake. While antibodies that have highly similar, if not identical, heavy and light chain primary structures can be assumed to have shared specificities for antigen, the converse is not the case. Antibodies with highly disparate primary structures can be found to bind a common antigen 23. As such, it is possible that patients who cannot be assigned to a cluster still could have leukemic cells that express Ig capable of binding a common antigen(s). Recognition of this situation would require methods that can assess for similarities in the tertiary structures of the Ig expressed in this disease24. Further refinement of our ability to understand and analyze the B cell receptor in relation to B cell tumorigenesis is essential in the ultimate dissection of the CLL disease process.
The clinical significance of these sets is unclear. No overall difference was found in time to treatment for patients that belonged to a cluster as compared to those that did not (data not shown). While there have been reports of particular sets having distinct clinical features, both aggressive and stable 15,24, the small number of patients in most of the clusters we identified precludes definitive conclusions. Indeed, large meta-studies from many centers and sites will likely be necessary to achieve sufficient numbers to drawn statistically valid results. Alternatively, if the antigens responsible for these archetypal sequence motifs are identified, cluster membership based on antigen reactivity may ultimately be a better grouping criteria.
Supplementary Material
Supplemental Figure 1. Pairwise association of V, D, and J genes. The number of occurrences of each D-J (A), V-J (B), and V-D (C) pair is shown. The dark bars indicate statistically significant values, as determined by Chi squared statistics.
Acknowledgments
The authors wish to acknowledge the technical assistance of Michael Harabaglia and Ester Avery. This work was supported by a grant from the National Institute of Health (US). BJR was supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. BTM was a Lymphoma Research Foundation Fellow.
Footnotes
Author Contributions
BTM and BJR designed and performed research and wrote the paper, SJA, GFW, and LZR performed research, JGG and NEK provided essential samples and associated data, and TJK designed and supervised the research and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder HW, Jr, Ferrarini M, Chiorazzi N. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, Neuberg DS, Flinn IW, Rai KR, Byrd JC, Kay NE, Greaves A, Weiss A, Kipps TJ. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 4.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 5.Kipps TJ, Tomhave E, Pratt LF, Duffy S, Chen PP, Carson DA. Developmentally restricted immunoglobulin heavy chain variable region gene expressed at high frequency in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1989;86:5913–5917. doi: 10.1073/pnas.86.15.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messmer BT, Albesiano E, Messmer D, Chiorazzi N. The pattern and distribution of immunoglobulin VH gene mutations in chronic lymphocytic leukemia B cells are consistent with the canonical somatic hypermutation process. Blood. 2004;103:3490–3495. doi: 10.1182/blood-2003-10-3407. [DOI] [PubMed] [Google Scholar]
- 7.Widhopf GF, 2nd, Kipps TJ. Normal B cells express 51p1-encoded Ig heavy chains that are distinct from those expressed by chronic lymphocytic leukemia B cells. J Immunol. 2001;166:95–102. doi: 10.4049/jimmunol.166.1.95. [DOI] [PubMed] [Google Scholar]
- 8.Tobin G, Thunberg U, Johnson A, Thorn I, Soderberg O, Hultdin M, Botling J, Enblad G, Sallstrom J, Sundstrom C, Roos G, Rosenquist R. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 2002;99:2262–2264. doi: 10.1182/blood.v99.6.2262. [DOI] [PubMed] [Google Scholar]
- 9.Tobin G, Thunberg U, Johnson A, Eriksson I, Soderberg O, Karlsson K, Merup M, Juliusson G, Vilpo J, Enblad G, Sundstrom C, Roos G, Rosenquist R. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vlambda2-14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood. 2003;101:4952–4957. doi: 10.1182/blood-2002-11-3485. [DOI] [PubMed] [Google Scholar]
- 10.Ghiotto F, Fais F, Valetto A, Albesiano E, Hashimoto S, Dono M, Ikematsu H, Allen SL, Kolitz J, Rai KR, Nardini M, Tramontano A, Ferrarini M, Chiorazzi N. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J Clin Invest. 2004;113:1008–1016. doi: 10.1172/JCI19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004;104:2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 12.Tobin G, Thunberg U, Karlsson K, Murray F, Laurell A, Willander K, Enblad G, Merup M, Vilpo J, Juliusson G, Sundstrom C, Soderberg O, Roos G, Rosenquist R. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104:2879–2885. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 13.Messmer BT, Albesiano E, Efremov DG, Ghiotto F, Allen SL, Kolitz J, Foa R, Damle RN, Fais F, Messmer D, Rai KR, Ferrarini M, Chiorazzi N. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200:519–525. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, Belhoul L, Stella S, Stavroyianni N, Crespo M, Hadzidimitriou A, Sutton L, Bosch F, Laoutaris N, Anagnostopoulos A, Montserrat E, Fassas A, Dighiero G, Caligaris-Cappio F, Merle-Beral H, Ghia P, Davi F. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood. 2007;109:259–270. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 15.Capello D, Guarini A, Berra E, Mauro FR, Rossi D, Ghia E, Cerri M, Logan J, Foa R, Gaidano G. Evidence of biased immunoglobulin variable gene usage in highly stable B-cell chronic lymphocytic leukemia. Leukemia. 2004;18:1941–1947. doi: 10.1038/sj.leu.2403537. [DOI] [PubMed] [Google Scholar]
- 16.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–894. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 17.Greaves AW, Payne PR, Rassenti L, Kipps TJ. CRC Tissue Core Management System (TCMS): integration of basic science and clinical data for translational research. AMIA Annu Symp Proc. 2003:853. [PMC free article] [PubMed] [Google Scholar]
- 18.Giudicelli V, Chaume D, Lefranc MP. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 2004;32:W435–440. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 20.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 21.Chao A. Nonparametric-Estimation of the Number of Classes in a Population. Scandinavian Journal of Statistics. 1984;11:265–270. [Google Scholar]
- 22.Chao A. Estimating the Population-Size for Capture Recapture Data with Unequal Catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- 23.Messmer BT, Sullivan JJ, Chiorazzi N, Rodman TC, Thaler DS. Two human neonatal igm antibodies encoded by different variable-region genes bind the same linear peptide: evidence for a stereotyped repertoire of epitope recognition. J Immunol. 1999;162:2184–2192. [PubMed] [Google Scholar]
- 24.Thorselius M, Krober A, Murray F, Thunberg U, Tobin G, Buhler A, Kienle D, Albesiano E, Maffei R, Dao-Ung LP, Wiley J, Vilpo J, Laurell A, Merup M, Roos G, Karlsson K, Chiorazzi N, Marasca R, Dohner H, Stilgenbauer S, Rosenquist R. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3-21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood. 2006;107:2889–2894. doi: 10.1182/blood-2005-06-2227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Pairwise association of V, D, and J genes. The number of occurrences of each D-J (A), V-J (B), and V-D (C) pair is shown. The dark bars indicate statistically significant values, as determined by Chi squared statistics.