Abstract
To understand the molecular basis for the different self-association and lipoprotein preferences of apolipoprotein (apo) E isoforms, we compared the effects of progressive truncation of the C-terminal domain in human apoE3 and apoE4 on their lipid-free structure and lipid-binding properties. A VLDL/HDL distribution assay demonstrated that apoE3 binds much better than apoE4 to HDL3 whereas both isoforms bind similarly to VLDL. Removal of the C-terminal helical regions spanning residues 273–299 reduced the ability of both isoforms to bind to lipoproteins; this led to the elimination of the isoform lipoprotein preference, indicating that the C-terminal helices mediate the lipoprotein selectivity of apoE3 and apoE4 isoforms. Gel filtration chromatography experiments demonstrated that the monomer-tetramer distribution is different for the two isoforms with apoE4 being more monomeric than apoE3 and that removal of the C-terminal helices favors the monomeric state in both isoforms. Consistent with this, fluorescence measurements of Trp-264 in single Trp mutants revealed that the C-terminal domain in apoE4 is less organized and more exposed to the aqueous environment compared to apoE3. In addition, the solubilization of dimyristoylphosphatidylcholine multilamellar vesicles is more rapid with apoE4 than with apoE3; removal of the C-terminal helices significantly affected solubilization rates with both isoforms. Taken together, these results indicate that the C-terminal domain is organized differently in apoE3 and apoE4 so that apoE4 self-associates less and binds less than apoE3 to HDL surfaces; these alterations may lead to the pathological sequelae for cardiovascular and neurodegenerative diseases.
Apolipoprotein (apo) E is a protein of major biological and clinical importance, regulating lipid transport and cholesterol homeostasis in the cardiovascular and central nervous systems (1–5). In humans, apoE is a polymorphic protein with three major isoforms, apoE2, apoE3, and apoE4, each differing by a single amino acid substitution. ApoE3, the most common isoform, contains cysteine at position 112 and arginine at position 158, whereas less common apoE2 and apoE4 contain cysteine and arginine at both sites, respectively (6). ApoE2 displays defective binding to the low density lipoprotein (LDL) receptor and is associated with type III hyperlipoproteinemia (7). Besides being a risk factor for atherosclerosis, apoE4 is a major genetic risk factor for Alzheimer’s disease (AD), accounting for 40–60% of the genetic variation in the disease (8, 9). ApoE4 is also associated with neuronal damage, including poor recovery from traumatic brain injury (10) and other central nervous system stresses (11). ApoE is thought to play a key role in neuronal repair and remodeling, with apoE4 being much less effective in these processes than apoE2 or apoE3 (12).
ApoE contains two independently folded functional domains; a 22-kDa N-terminal domain (residues 1–191) and a 10-kDa C-terminal domain (residues 216–299) linked by a hinge region (2). The N-terminal domain is folded into a four-helix bundle of amphipathic α-helices (13) and contains the region (residues 136–150) that binds to the LDL receptor superfamily (2, 6). The C-terminal domain contains amphipathic α-helices that are involved in binding to lipoproteins with high affinity (14 – 16). Lipid-free apoE exists as oligomer in solution (14, 17, 18) and the C-terminal domain is responsible for this self-association through the intermolecular coiled-coil formation (19). These helix-helix contacts are converted to helix-lipid interaction upon binding of apoE to lipoprotein surfaces, with the N-terminal four-helix bundle being in either an open or a closed conformation depending upon the surface area availability (16, 20).
Polymorphism in apoE dictates different structural and biophysical properties that result in functional differences. For example, apoE3 binds preferentially to high density lipoprotein (HDL), whereas apoE4 prefers very low density lipoprotein (VLDL) (21). This lipoprotein preference may contribute, by an unknown mechanism, to the increased plasma concentrations of cholesterol and LDL associated with the presence of apoE4 (22). The presence of the positive charge from Arg-112 causes a rearrangement of the Arg-61 side chain in the N-terminal domain of apoE4, allowing it to interact with Glu-255 in the C-terminal domain (22, 23). This domain interaction appears to alter the amphipathic α-helix organization in the C-terminal domain (24 – 26), thereby promoting preferential binding of apoE4 to VLDL. Indeed, our previous fluorescence study showed that the C-terminal domain of apoE4 has more exposed hydrophobic surface, contributing to its high affinity binding to VLDL (24). However, the mechanism by which the domain interaction in apoE4 modulates the conformation of the molecule is still unknown.
Using progressive C-terminal-truncated mutants of apoE4, we recently reported that the truncation of a hydrophobic C-terminal α-helical segment (residues 273–299) generates a monomeric protein, with the C-terminal domain being less organized and available for hydrophobic interaction (27). This implies that the conformational organization of the C-terminal α-helices is critical for the self-association and lipid interaction of apoE4. In the present study, we compared the effects of progressive truncation in the C-terminal domain of apoE3 and apoE4 on their lipid-free structure and lipoprotein distributions to elucidate the role of the C-terminal α-helices in controlling the self-association and lipoprotein preference of apoE isoforms. The results show that the C-terminal α-helices in apoE4 are organized differently than in apoE3 so that apoE4 self-associates less and binds differently to lipoprotein surfaces compared to apoE3.
EXPERIMENTAL PROCEDURES
Expression and Purification of Proteins
The full-length human apoE3, apoE4, apoE4(E255A) and their 22-kDa (residues 1–191), 12-kDa (residues 192–299), and 10-kDa (residues 222–299) fragments were expressed and purified as described (16, 24). The mutations in apoE to create the truncated forms (Δ251–299, Δ261–299 and Δ273–299) were made using PCR methods and the single tryptophan point mutation (W@264) was made using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described (27). The apoE preparations were at least 95% pure as assessed by SDS-PAGE. In all experiments, the apoE sample was freshly dialyzed from 6 M GdnHCl and 1% β-mercaptoethanol (or 10 mM DTT) solution into buffer solution before use.
VLDL/HDL distribution assay
VLDL and HDL3 were isolated by sequential density ultracentrifugation from a pool of fresh-frozen human plasma (similar results were obtained when lipoproteins from fresh plasma were utilized). The various apoE preparations were trace-labeled with either 3H or 14C by reductive methylation (16, 28, 29) and incubated at 4 °C for 30 min with a mixture of human HDL3 and VLDL. 5 µg of each of the pair of 3H- and 14C-labeled proteins to be compared was mixed and incubated with 0.45 mg VLDL protein and 0.9 mg HDL3 protein (these concentrations gave approximately equal total VLDL and HDL3 particle surface areas available for apoE binding) in a total volume of 1 ml of Tris buffer (10 mM Tris-HCl, 150 mM NaCl, 0.02% NaN3, 1 mM EDTA, pH 7.4). The VLDL, HDL3 and unbound protein were separated by sequential density gradient ultracentrifugation and the amounts and ratios of 3H/14C radioactivity in each fraction were determined by liquid scintillation counting. Similar results were obtained when the lipoproteins were isolated by gel filtration chromatography (data not shown).
Circular Dichroism (CD) Spectroscopy
Far-UV CD spectra were recorded from 185 to 260 nm at 25 °C using a Jasco J-600 or Aviv 62DS spectropolarimeter. After dialysis, the apoE sample was diluted to 25–50 µg/ml in 10 mM phosphate buffer (pH 7.4) for obtaining the CD spectrum. The results were corrected by subtracting the buffer baseline or a blank sample containing an identical concentration of GdnHCl. The α-helix content was derived from the molar ellipticity at 222 nm, [θ]222, according to the following equation (30, 31):
The thermal denaturation was monitored by the change in molar ellipticity at 222 nm over the temperature range 20−90 °C, as described (27, 31). The cooperativity index, n, describing the sigmoidicity of the thermal denaturation curve was calculated as described (27). The equilibrium constants of denaturation, KD, at each temperature were derived from the measured molar ellipticity at each temperature and the molar ellipticities of the native and denatured forms of the protein (27, 32). The van’t Hoff enthalpy, ΔHv, was calculated from the slope of the line fitted by linear regression to the equation, ln KD = −(ΔHv/R) 1/T + constant, where R is the gas constant and T is temperature. For monitoring chemical denaturation, proteins at concentration of 50 µg/ml were incubated overnight at 4 °C with GdnHCl at various concentrations. KD at a given GdnHCl concentration were calculated from the ellipticity values and, the free energy of denaturation, ΔGD°, the midpoint of denaturation, D1/2, and m value which reflects the cooperativity of denaturation in the transition region, were determined by the linear equation, ΔGD = ΔGD° − m[GdnHCl], where ΔGD = − RT ln KD (27, 32).
Fluorescence Measurements
Fluorescence measurements were carried out with a Hitachi F-7000 or F-4500 fluorescence spectrophotometer at 25 °C in Tris buffer (pH 7.4). Trp emission fluorescence of proteins at concentrations of 25–100 µg/ml were recorded from 300 to 400 nm using a 295 nm excitation wavelength to avoid tyrosine fluorescence. For monitoring chemical denaturation, KD at a given GdnHCl concentration were derived from the change in fluorescence intensity at 335 nm (27, 33). ΔGD°, D1/2, m value, and cooperative index were determined as described above. In fluorescence quenching experiments, the Trp emission spectra of proteins were recorded at increasing concentrations of KI (0-0.2 M) using a 5 M stock solution containing 1 mM NaS2O3 to prevent the formation of iodine. After correction for dilution, the integrated fluorescence intensities were plotted according to the Stern-Volmer equation, F0/F = 1 + Ksv[KI], where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively, and Ksv is the Stern-Volmer constant. Quenching parameters were obtained by fitting to the modified Stern-Volmer equation, F0/(F0 − F) = 1/fa + 1/faKsv[KI], where fa is the fraction of Trp residues accessible to the quencher. Steady-state anisotropy of Trp fluorescence was measured with excitation at 295 nm and emission at 340 nm, as described (34). ANS fluorescence spectra were collected from 400 to 600 nm at an excitation wavelength of 395 nm in the presence of 50 µg/ml protein and an excess of ANS (250 µM) (35).
Gel Filtration Chromatography
ApoE samples trace-labeled with 14C at a concentration of 5 µg/ml were subjected to gel filtration on a calibrated Superdex 200 column (60 × 1.6 cm) using an Akta FPLC system (27, 36). The sample was eluted with Tris buffer at a flow rate of 1 ml/min and 2 ml fractions were collected, and radioactivity was determined by liquid scintillation counting. Similar elution profiles were observed when higher concentrations of either labeled or unlabeled apoE were used so that the absorbance at 280nm could be monitored. The elution volumes of the monomer and tetramer were established by using covalently cross-linked samples of apoE containing monomer and tetramers.
DMPC clearance assay
The kinetics of solubilization of DMPC multilamellar vesicles by the apoE variants were measured by monitoring the time-dependent decrease in turbidity (35, 37). DMPC vesicles extruded through a 200-nm filter were mixed with apoE samples to a final volume of 600 µl in a cuvette, and incubated for 15 min at 24.6 °C. Sample light scattering intensity was monitored at 325 nm on a Shimadzu UV-2450 spectrophotometer.
RESULTS
VLDL/HDL distributions of apoE variants
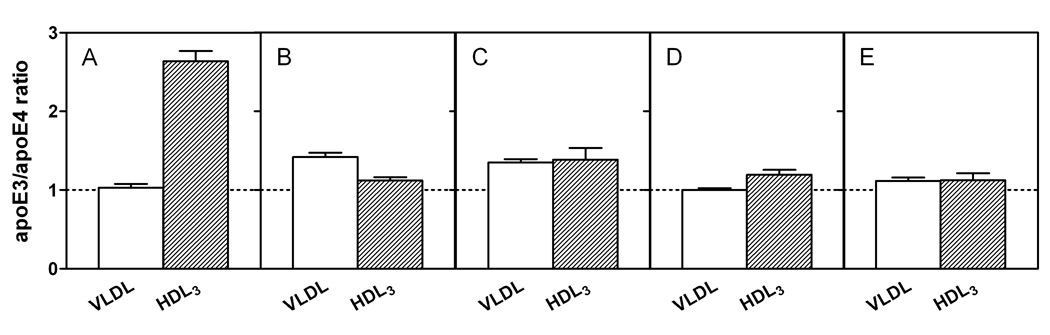
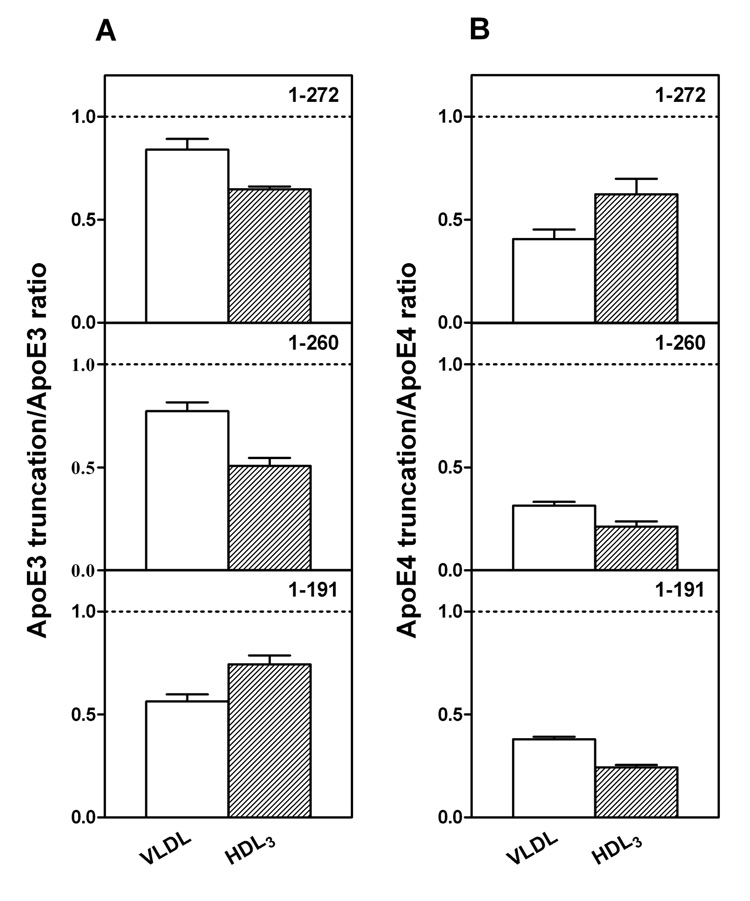
It is known that apoE3 and apoE4 partition differently between VLDL and HDL when added to human plasma (21 – 23). The data in Figure 1A show that apoE3 and apoE4 bind similarly to VLDL (cf. (24)) when added to a mixture of the two lipoproteins whereas apoE3 binds markedly better than apoE4 to HDL3. The large increase in the apoE3/apoE4 ratio in HDL3, as compared to VLDL, is consistent with prior results where the lipoprotein distribution of apoE3 and apoE4 was examined after each protein was added separately to human plasma (23). However, in our competitive binding assay, where the lipoprotein distribution is different to that in human plasma, the ability of apoE4 to bind better than apoE3 to VLDL is not apparent. The isolated apoE3 and apoE4 N-terminal fragments (residues 1–191) showed no discrimination between VLDL and HDL3 (Figure 1E), further confirming that the C-terminal domain mediates the preferential HDL3 binding of the intact apoE3 molecule and that interactions between the N- and C-terminal domains modulate this lipoprotein selectivity (22, 23). Removal of the C-terminal residues (273–299, 261–299, or 251–299) reduced the overall lipoprotein binding ability (more protein appeared in the unbound fraction (22)) and largely eliminated the differential binding of apoE3 and apoE4 to HDL3 (Figures 1B–1D), indicating that the region spanning residues 273–299 is critical to the lipoprotein preferences of apoE3 and apoE4. Figure 2 shows that the truncation of the C-terminal residues has different effects on the lipoprotein binding of the apoE3 and apoE4 isoforms. Removal of either residues 273–299 or 261–299 greatly reduced the binding of apoE4 to VLDL while having a relatively small effect on the binding of apoE3 to VLDL. Loss of these residues reduced the ability of both isoforms to bind to HDL3 with the effect of residues 261–272 being somewhat greater for apoE4. Removal of the region of the C-terminal domain spanning residues 192–260 did not lead to any further major changes in VLDL or HDL3 binding for either isoform. Overall, the effect of C-terminal truncation on reducing the ability to bind to the lipoproteins is larger for apoE4 than for apoE3.
Fig. 1. Effects of C-terminal truncation of apoE3 and apoE4 on the relative distributions between VLDL and HDL3.
The apoE variants were incubated at 4 °C for 30 min with a mixture of human HDL3 and VLDL. For ease of comparison, the ratios for the initial mixtures are normalized to one in each case. (A) apoE3/apoE4 (B) apoE3 (1–272)/apoE4 (1–272) (C) apoE3 (1–260)/apoE4 (1–260) (D) apoE3 (1–250)/apoE4 (1–250) (E) apoE3 (1–191)/apoE4 (1–191). The results from 2–5 independent experiments, each done in triplicate, are reported as mean ± SD.
Fig. 2. Effects of C-terminal truncations on the ability of apoE3 and apoE4 to bind to VLDL and HDL3.
(A) apoE3 C-terminal truncation mutants/apoE3. (B) apoE4 C-terminal truncation mutants/apoE4. The results from 2–5 independent experiments, each done in triplicate, are reported as mean ± SD.
Far-UV CD Analysis
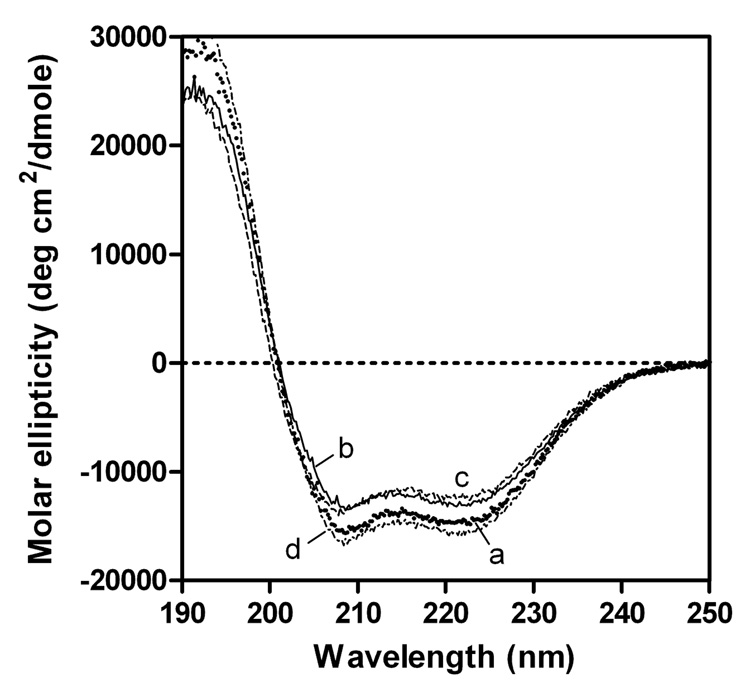
The effect of the C-terminal truncation on the secondary structure of apoE3 was evaluated by CD spectroscopy. The far-UV CD spectra of full-length and some C-terminal-truncated variants of apoE3 are shown in Figure 3, and the α-helix contents estimated from the molar ellipticity at 222 nm are listed in Table 1. Based on the similar measurements for the C-terminal-truncated apoE4, we previously reported that residues 261–299 in apoE4 form α-helical structure whereas residues 192–260 are largely nonhelical (27). Comparison of the number of α-helical residues estimated from CD and the number of residues removed in the apoE3 variants indicated that residues 261–299 in apoE3 also largely form α-helical structure, whereas residues 192–260 have almost random coil structure.
Fig. 3. Far-UV CD spectra of apoE3 C-terminal-truncated mutants.
apoE3 (a), apoE3 (1–272) (b), apoE3 (1–260) (c), and apoE3 22-kDa (d). The protein concentration was 50 µg/ml.
Table 1.
α-helix content and thermal denaturation parameters of apoE3 C-terminal truncated mutants
| apoE3 variant | α-helixa | number of residues in α-helix | Tmb | Cooperativity index | ΔHvc |
|---|---|---|---|---|---|
| % | amino acids | °C | kcal/mol | ||
| apoE3 | 45 ± 3 | 131 | 47 | 2.6 | 22 |
| apoE3 (1–272) | 40 ± 2 | 109 | 58 | N.D.d | N.D.d |
| apoE3 (1–260) | 38 ± 2 | 101 | 57 | 7.5 | 33 |
| apoE3 (1–250) | 40 ± 3 | 101 | 60 | 13 | 49 |
| apoE3 22-kDa | 47 ± 4 | 91 | 51 | 3.6 | 24 |
Mean ± S.D. from at least three measurements.
The reproducibility in Tm is ± 1.5 °C.
Estimated error is ± 2 kcal/mol.
Thermal denaturation was not cooperative.
Table 1 also summarizes parameters for thermal unfolding of the C-terminal-truncated variants of apoE3. Progressive truncations in the C-terminal domain of apoE3 tend to increase the midpoint and cooperativity of unfolding, consistent with the previous results for apoE4 (27). Interestingly, apoE3 (1–250) exhibited much greater stability and cooperativity of unfolding than the apoE3 22-kDa fragment although residues 192–250 are predicted to have a less organized structure than the N-terminal domain (2).
GdnHCl Denaturation Studies
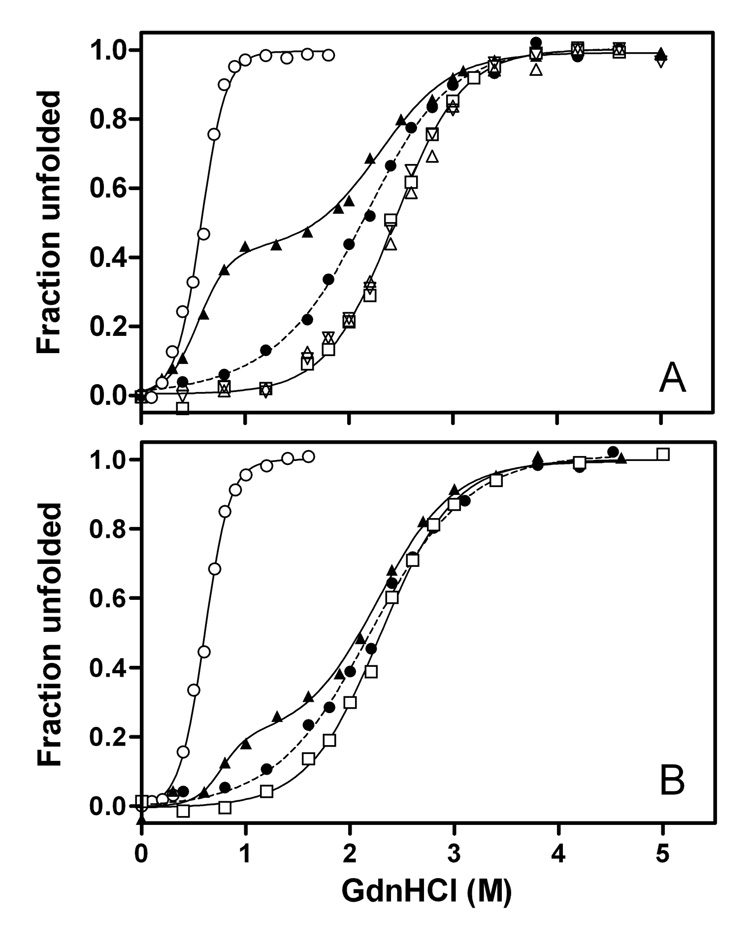
To characterize the structural stability of the C-terminal-truncated apoE3 variants, GdnHCl denaturation experiments were performed using CD and Trp fluorescence. Human apoE possesses seven Trp residues, located at positions 20, 26, 34, and 39 in the N-terminal domain, 210 in the hinge region, and 264 and 276 in the C-terminal domain, allowing tertiary structural changes to be probed by monitoring Trp fluorescence spectra (27, 38). Figure 4 compares GdnHCl denaturation curves for the C-terminal-truncated apoE3 variants. For full-length apoE3, both denaturation curves obtained by Trp fluorescence (A) and CD (B) displayed biphasic behavior, in which the first and the second phases correspond to denaturation of the 10-kDa C-terminal and the 22-kDa N-terminal domains, respectively (38, 39). The midpoints of denaturation for the C- and N-terminal domains were 0.6–0.8 M and 2.3–2.4 M GdnHCl, respectively. Removal of the C-terminal α-helical residues exhibited drastic effects on the denaturation curves. As shown in Figure 4A, apoE3 (1–250) and apoE3 (1–260) displayed identical denaturation curves to the 22-kDa fragment, indicating that residues 192–260 in these C-terminal-truncated apoE3 have a negligible effect on the structural stability of the molecule. Interestingly, apoE3 (1–272) denatured at a significantly lower GdnHCl concentration than the other C-terminal-truncated mutants (midpoint = 2.13± 0.03M compared to 2.44 ± 0.02M), although there was no difference in the denaturation of the apoE4 counterparts (27). This unique denaturation behavior was also observed when monitored by CD (Figure 4B).
Fig. 4. GdnHCl denaturation of full-length apoE3, 22-kDa and 10-kDa fragments of apoE3, and C-terminal-truncated apoE3 monitored by Trp fluorescence (A) and CD (B).
Full-length apoE3 (▲), apoE3 (1–272) (●), apoE3 (1–260) (△), apoE3 (1–250) (▽), apoE3 22-kDa (□), and apoE 10-kDa (○).
ANS Binding
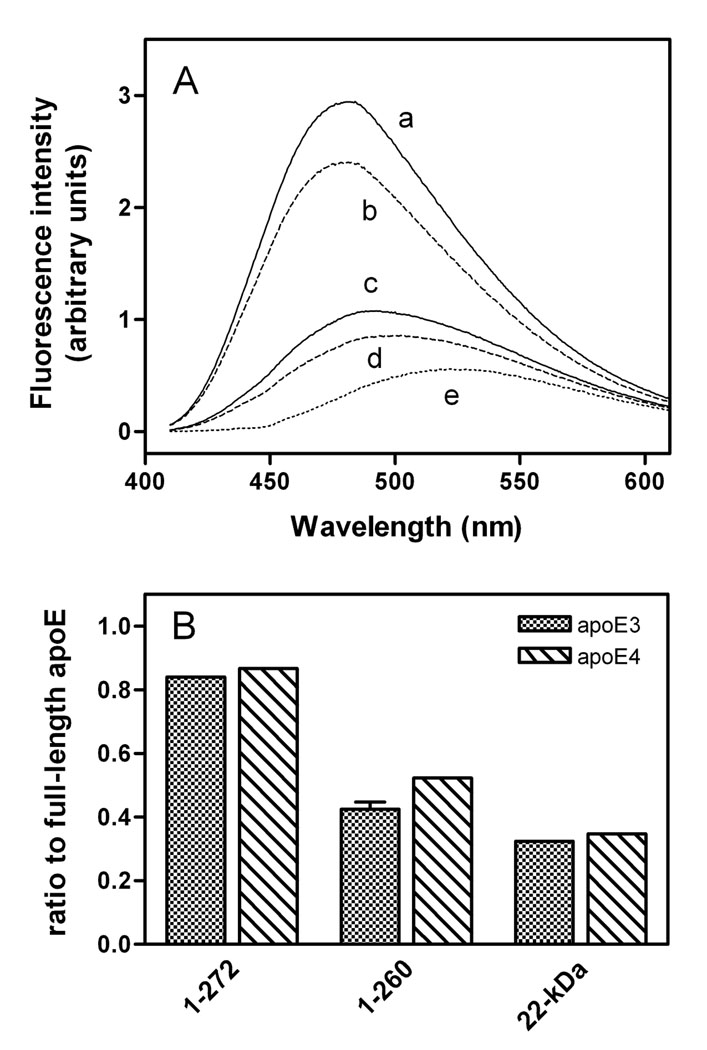
We next performed ANS binding experiments to monitor the change in the tertiary structure of the C-terminal-truncated apoE isoforms. The fluorescence intensity of ANS significantly increases when bound to the accessible hydrophobic sites in intact apoE, mostly created by the C-terminal domain (24, 27). Figure 5A shows that removal of residues 273–299 caused a small decrease in ANS fluorescence intensity, whereas the major decrease was observed for the further truncation of residues 261–272. Similar effects were also seen with the apoE4 counterparts (Figure 5B), indicating that residues 261–272 create an exposed hydrophobic site in the C-terminal domain of apoE3 and apoE4 in a similar manner.
Fig. 5. ANS fluorescence for apoE3 C-terminal-truncated mutants.
(A) Emission spectra of ANS in the presence of apoE3 (a), apoE3 (1–272) (b), apoE3 (1–260) (c), apoE3 22-kDa (d), and buffer (e). (B) ANS fluorescence intensity ratio of the C-terminal-truncated mutants to full-length apoE. The results from at least 2 independent experiments, each done in triplicate, are shown.
Gel Filtration Chromatography
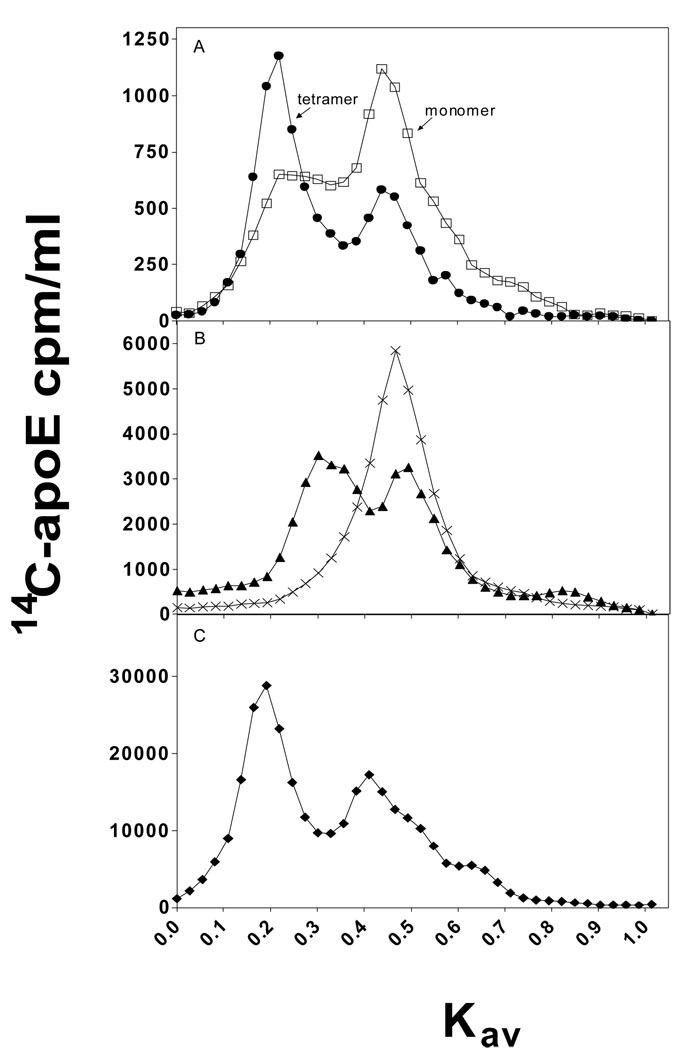
To compare the contributions of the C-terminal α-helical residues to the self-association of apoE3 and apoE4 in solution, gel filtration chromatography was employed. The data in Figure 6A demonstrate that full-length apoE3 and apoE4 exist as both monomeric and tetrameric forms, but the monomer-tetramer distribution is different for the two isoforms, with apoE4 being more monomeric. The tetrameric and monomeric forms are in equilibrium because re-passage of the tetramer peak through the gel filtration column gives rise to both tetramer and monomer peaks. Removal of residues 273–299 favors the monomeric state over the tetrameric state for both isoforms; apoE4 (1–272) exists completely as a monomer (27) whereas apoE3 (1–272) is significantly tetramerized (Figure 6B). In contrast, removal of residues 261–299 caused both proteins to exist completely in the monomeric state (data not shown). These results indicate that the C-terminal α-helical regions modulate the self-association differently for the two isoforms. The monomer-tetramer distribution of apoE4(E255A) was examined to explore the influence of interactions between the N- and C-terminal domains of the apoE molecule on self-association. The elution profile in Figure 6C reveals that the apoE4 variant self-associates more than wild-type apo E4 and behaves similarly to wild-type apoE3.
Fig. 6. Elution profiles upon gel filtration of apoE3 and apoE4 variants on a Superdex 200 column.
Proteins at a concentration of 5 µg/ml (Panel A and B) or 50 µg/ml (Panel C) were subjected to gel filtration. (A) Full-length apoE3 (●), full-length apoE4 (□); (B) apoE3 (1–272) (▲), apoE4 (1–272) (×); (C) apoE4(E255A) (◆). Representative profiles from at least 3 independent experiments using different batches of protein are shown.
Fluorescence Studies of Single Trp Mutants
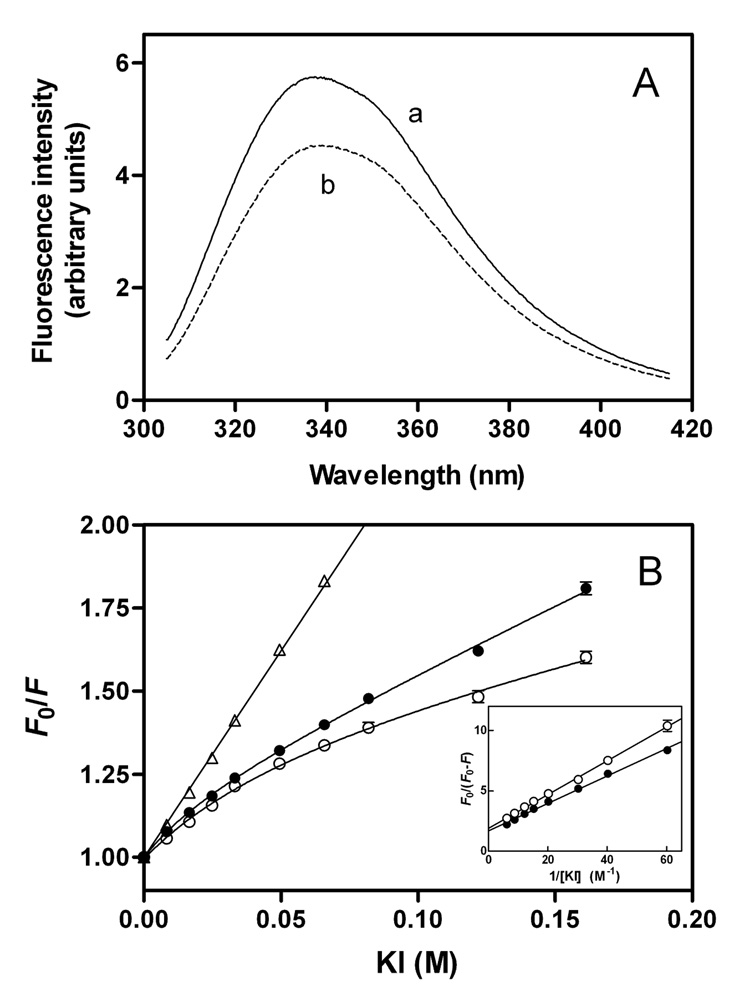
To obtain site-specific information about the C-terminal domain in apoE isoforms, we prepared single Trp mutants of apoE3 and apoE4, in which all the Trp residues except that at position 264 were mutated to Phe (27, 40). Figure 7A shows Trp fluorescence emission spectra from these W@264 mutants for full-length apoE3 and apoE4. Although the fluorescence intensity observed with apoE3 W@264 was increased compared to apoE4 W@264, the wavelength of maximum fluorescence was similar (337–338nm). Fluorescence quenching results in Figure 7B demonstrated that W264 in apoE4 is quenched by the aqueous quencher KI more than apoE3, indicating that this residue in apoE4 is relatively more exposed to the aqueous phase compared to apoE3. Quenching parameters for full-length and the C-terminal-truncated apoE single Trp mutants are shown in Table 2. Compared to the apoE3 counterparts, larger values in fa for apoE4 W@264 and in Ksv for apoE4 (1–272) W@264 indicate that W264 in both full-length and the C-terminal-truncated apoE4 is more exposed to the aqueous environment.
Fig. 7. Fluorescence spectroscopy of apoE3 and apoE4 single Trp mutants.
Solutions of the proteins (100 µg/ml) were excited at 295 nm to obtain the emission fluorescence from W264. (A) Fluorescence spectra for apoE3 W@264 (a) and apoE4 W@264 (b). (B) Stern-Volmer plot comparing quenching by KI of W264 fluorescence from apoE3 (○) and apoE4 (●). The data for N-acetyltryptophanamide are also shown for comparison (△). The inset shows a modified Stern-Volmer plot. The results from 2–4 independent experiments, each done in triplicate, are shown.
Table 2.
Parameters of GdnHCl-induced denaturation, KI quenching, and fluorescence anisotropy of apoE single Trp mutants at 25 °Ca
| apoE variant | Thermodynamic parameters of denaturation |
Quenching by KIb |
Flurorescene anisotropy | |||
|---|---|---|---|---|---|---|
| ΔGD° | D1/2 | m | fa | Ksv | ||
| kcal/mol | M | kcal/mol apoE/mol GdnHCl | M−1 | |||
| apoE3 W@264 | 1.4 ± 0.1 | 0.53 ± 0.02 | 2.7 ± 0.3 | 0.52 ± 0.03 | 13.6 ± 1.3 | 0.084 ± 0.005 |
| apoE4 W@264 | 1.3 ± 0.1 | 0.52 ± 0.02 | 2.5 ± 0.2 | 0.59 ± 0.02 | 14.9 ± 0.7 | 0.078 ± 0.004 |
| apoE3 (1–272) W@264 | 0.6 ± 0.1 | 0.38 ± 0.03 | 1.6 ± 0.3 | 0.62 ± 0.02 | 7.8 ± 0.4 | 0.058 ± 0.003 |
| apoE4 (1–272) W@264 | 0.6 ± 0.1 | 0.31 ± 0.04 | 1.8 ± 0.4 | 0.58 ± 0.05 | 13.1 ± 1.6 | 0.064 ± 0.004 |
Values are drived from duplicate or triplicate measurements from at least two independent preparations of proteins.
Parameters of Trp fluorescence quenching: fa, fraction of Trp residue accessible to I−; Ksv, Stern-Volmer constant.
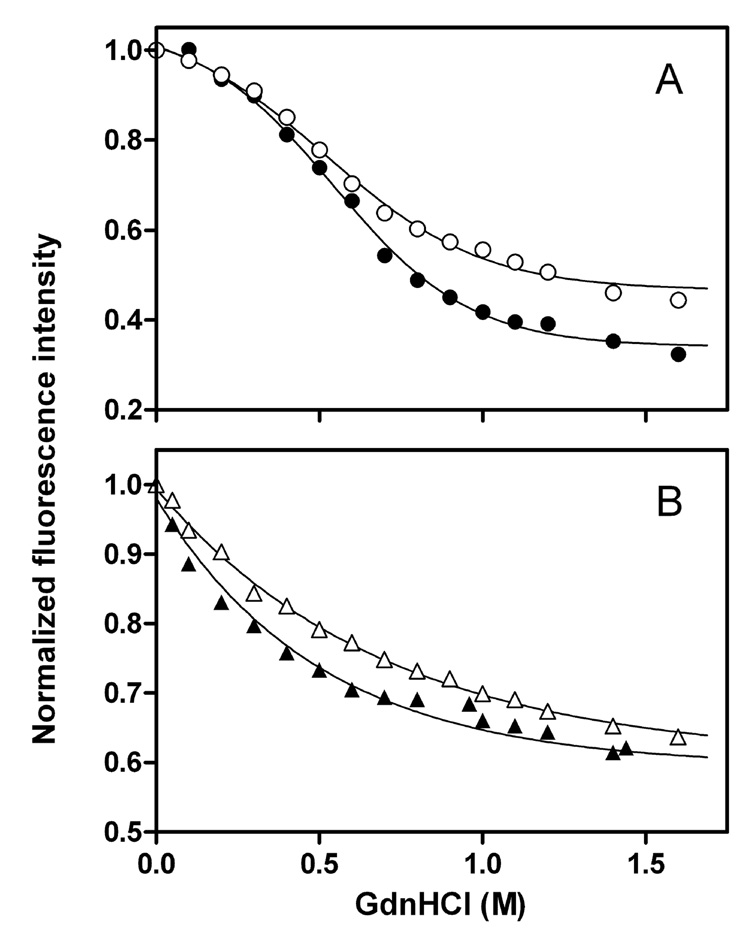
To further examine the structural organization of the C-terminal domain, GdnHCl denaturation was employed. Figure 8 shows GdnHCl denaturation curves of single Trp mutants for full-length and the C-terminal-truncated apoE3 and apoE4 monitored by the change in Trp fluorescence intensity (27). Table 2 summarizes the conformational stability, ΔGD°, the midpoint of denaturation, D1/2, and m value which reflects the cooperativity of denaturation. Comparison of full-length and the C-terminal-truncated forms indicates that removal of α-helical residues 273–299 significantly reduces the structural stability and cooperativity of the C-terminal domain in apoE3 and apoE4. No significant difference in denaturation parameters was observed between the two isoforms in both full-length and 1–272 apoE. Consistent with these results, fluorescence anisotropy values reflecting the motional restriction of W264 in the full-length proteins were higher than those for the C-terminal-truncated forms although there was no significant difference between the two isoforms (Table 2).
Fig. 8. GdnHCl denaturation of single Trp mutants of apoE3 and apoE4 monitored by Trp fluorescence.
Normalized fluorescence intensity at 335 nm was plotted as a function of GdnHCl concentration. (A) ApoE3 W@264 (○), ApoE4 W@264 (●), (B) apoE3 (1–272) W@264 (△), and apoE4 (1–272) W@264 (▲). The results from 2–3 independent experiments, each done in duplicate, are shown.
DMPC Clearance Assay
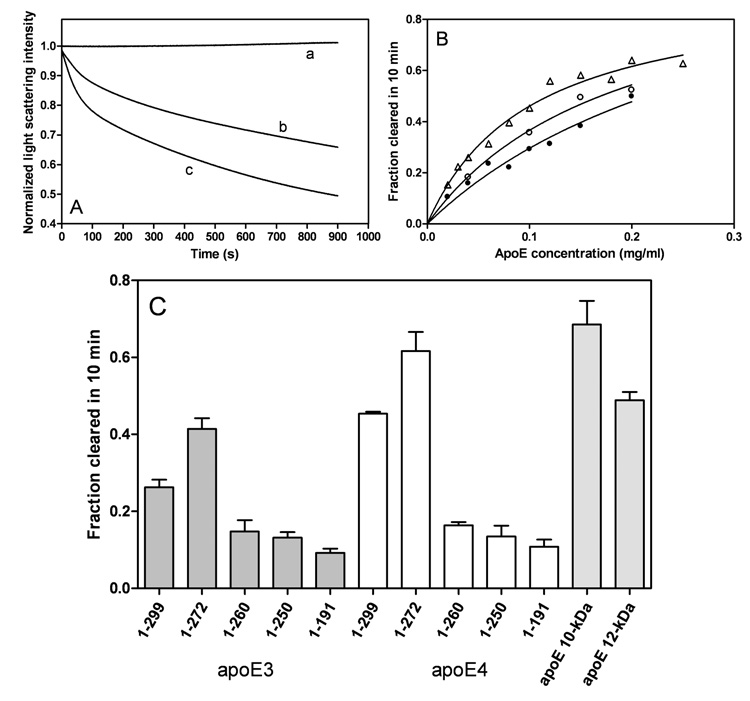
To assess the effects of the C-terminal truncations on the lipid-binding abilities of the apoE variants, we used a DMPC clearance assay. As shown in Figure 9A, full-length apoE3 and apoE4 at a concentration of 0.1 mg/ml displayed a time-dependent decrease in light scattering intensity with apoE4 giving a faster rate than apoE3. Such faster clearance rates for apoE4 than apoE3 were seen over a wide range of apoE concentrations (Figure 9B), demonstrating that apoE4 has a stronger ability to solubilize DMPC vesicles than apoE3 (37, 41, 42). This stronger ability of apoE4 to solubilize DMPC vesicles was reduced by introduction of the mutation E255A (Figure 9B). Removal of residues 273–299 in both apoE3 and apoE4 enhanced the clearance activities to the similar level for the isolated C-terminal fragments, whereas the further truncated mutants 1–260 and 1–250 displayed greatly reduced clearance activities comparable to the 22-kDa fragments (residues 1–191) (Figure 9C). These results indicate that α-helical residues 261–272 play a critical role in the efficient lipid interaction of apoE.
Fig. 9. Solubilization of DMPC multilamellar vesicles by apoE C-terminal-truncated mutants.
A: Time courses for turbidity clearance. DMPC alone (a), apoE3 (b), and apoE4 (c). The DMPC and protein concentrations were 0.25 and 0.1 mg/ml, respectively. B: Effect of protein concentration on fraction cleared in 10 min. ApoE3 (●), apoE4 (△), and apo E4(E255A) (○). C: Comparison of DMPC solubilization efficiency. Protein concentration was 0.1 mg/ml. The results from 3–5 independent experiments using different batches of protein are shown.
DISCUSSION
Polymorphism in apoE leads to the different interactions between the N- and C-terminal domains, contributing to the association of apoE isoforms with cardiovascular diseases as well as various neurodegenerative diseases, including AD (5, 9, 22). The domain interaction in apoE4 was shown to cause a closer spatial proximity of the two domains compared to apoE3 (25, 26), affecting the organization of the C-terminal domain to create an additional exposed hydrophobic site in apoE4 (24). However, the molecular mechanism underlying such structural effects in the C-terminal domain remains to be established.
The VLDL/HDL distribution assay we have developed gave quantitative comparisons of the distributions of the C-terminal-truncated apoE isoforms between these lipoprotein particles. The results in Figure 1 clearly demonstrate that while apoE3 and apoE4 bind similarly to VLDL under the competitive binding assay conditions employed, apoE3 partitions more than apoE4 to HDL3, and that the C-terminal α-helical region (residues 273–299) is largely responsible for such lipoprotein preferences. As far as the intact apoE3 and apoE4 molecules are concerned, examination of the distributions of protein between the VLDL, HDL3 and unbound fractions indicates that both proteins bind well to the lipoproteins in this assay system (≤ 5% apoE in the unbound fraction) where the total surface areas of the VLDL and HDL3 particles present are similar. ApoE3 distributes approximately equally between the VLDL and HDL3 fractions whereas apoE4 distributes much more to VLDL than to HDL3 (VLDL/HDL3 ratio ~ 3). It follows that, since apoE3 and apoE4 bind similarly to VLDL (Fig. 1A), the reason for the apoE3/apoE4 ratio in HDL3 being higher than the ratio for VLDL (Fig. 1A) is that apoE4 binds poorly to the small HDL3 particle. A contributing factor to this difference is perhaps that the unstable N-terminal helix bundle domain in apoE4 tends to bind in an open conformation (16) requiring more interfacial area than is readily available on a small HDL3 particle. In contrast, apoE3 can bind with only its C-terminal domain exerting a small interfacial footprint and its more stable helix bundle (38) remaining detached from the HDL particle surface.
As far as the contribution of the C-terminal 27 residues is concerned, the data in Fig. 1B show that the VLDL/HDL3 apoE3/apoE4 discrimination is lost largely because of the reduction in the ratio of apoE3 (1–272) to apoE4 (1–272) bound to HDL3. To determine the reason for this reduced ratio, we compared the distributions of apoE3 (1–272) and apoE4 (1–272) between the VLDL, HDL3 and unbound protein fractions, as well as the equivalent distributions of intact apoE3 and apoE4. The truncation reduces the binding of apoE4 somewhat more than that of apoE3 for VLDL (also see Fig. 2) so that the VLDL apoE3/apoE4 ratio is slightly increased (cf. Fig. 1A and B). The reduction in binding ability causes significant increases in the amounts of both of the apoE (1–272) variants in the unbound fraction (~ 50% versus ~ 5% for intact apoE). In the case of the HDL3 fraction, the truncation reduces the ability of both isoforms to bind to HDL3 (Fig. 2), but direct comparison of the HDL3 distribution of apoE3 (1–272) versus apoE4 (1–272) indicates that the truncation causes a bigger decrease in the amount of the former variant bound than of the latter (as mentioned above, the amount of apoE4 bound to HDL3 is already low). Thus, the loss of the apoE isoform VLDL/HDL3 distribution specificity caused by deletion of the C-terminal 27 residues occurs because, unlike intact apoE3, the apoE3 (1–272) variant cannot bind effectively to HDL3. These findings are apparently inconsistent with a prior examination of the distribution of apoE truncation mutants between lipoproteins in plasma where the deletion of the C-terminal 27 residues had little effect on lipoprotein binding (23). The reasons for this discrepancy are presumably associated with the different assay systems employed where binding of apoE likely saturates under different conditions. However, our results in Fig. 1 and 2 showing that more extensive C-terminal truncations also eliminate the apoE3/apoE4 VLDL/HDL3 preference and reduce the overall ability to bind to lipoproteins are consistent with the earlier study. Like apoE3 (1–272), the apoE3 variants (1–260), (1–250) and (1–191) (Fig. 1 C–E) all show reduced ability to bind to HDL3; this effect explains the loss of the HDL3 binding selectivity that is characteristic of intact apoE3 (Fig. 1A). Consistent with this, residues 260–272 in apoE4 are particularly important in mediating the small amount of binding to HDL3 that does occur because their removal significantly reduces the binding (Figure 2B); the greater impact of residues 260–272 in apoE4 compared to apoE3 is consistent with the greater exposure to the aqueous phase of this region in apoE4.
Physicochemical characterizations of the C-terminal-truncated apoE3 and apoE4 in the present and previous (27) studies give important insights into the structural organization of the C-terminal domain in the two isoforms. Based on CD measurements, both lipid-free apoE3 and apoE4 appear to have a relatively random coil-rich structure in the C-terminal domain, in which there are some α-helical segments in the extreme C-terminus (residues 261–299). Gel filtration chromatography demonstrated that apoE3 and apoE4 exist as both monomeric and tetrameric forms, but the monomer-tetramer distribution is different for the two isoforms (Figure 6). ApoE3 favors a tetrameric form whereas apoE4 tends to be more monomeric, contrary to previous reports showing that apoE4 has a greater propensity to self-associate than apoE3 (17, 43). This discrepancy perhaps arises because the self-association of apoE depends on the apoE concentration; we used a much lower concentration of apoE (5 µg/ml) for gel filtration compared to the experimental conditions used by others. The truncation of α-helical residues 261–299 or 273–299 has different effects on the self-association of the two isoforms; the self-association of apoE4 (1–272) is completely prevented whereas apoE3 (1–272) is significantly tetramerized (Figure 6B). This indicates that the C-terminal α-helical segments modulate the tetramerization of apoE in solution (14) qualitatively similarly but quantitatively differently for the two isoforms. Protein engineering studies of apoE demonstrated that four hydrophobic residues (F257, W264, L279, and V287) in the C-terminal region are critical for the oligomerization of apoE (44, 45). These residues are likely to participate in hydrophobic interactions which stabilize tetramer through an intermolecular coiled-coil formation in the C-terminal domain (19). Therefore, it is possible that the different domain interactions in apoE isoforms lead to the different organization of the C-terminal α-helical regions, affecting the quaternary oligomerization state mediated by intermolecular helix-helix interaction. This concept is supported by the finding that removal of the interaction between R61 in the N-terminal domain and E255 in the C-terminal domain by introducing the mutation E255A into apoE4 (22) enhances the ability of this variant apoE4 molecule to self-associate so that it behaves more like apoE3 (Figure 6C).
The idea of different organization of the C-terminal domain in apoE3 and apoE4 is supported by the fluorescence studies of single Trp mutants. The results in Figure 7 and Table 2 indicate that the C-terminal domain in apoE4 is organized with more exposure to the aqueous environment than apoE3, consistent with the less organized, less self-associated state of apoE4. Removal of α-helical residues 273–299 leads to a much less organized and cooperative structure in the C-terminal domain for the two isoforms (Figure 8 and Table 2). Using the C-terminal-truncated mutants of apoE4, we previously showed that residues 192–272 in apoE4 (1–272), predominantly forming a random coil structure, do not contribute to the structural stability of the protein (27). In contrast, apoE3 (1–272) displayed different GdnHCl denaturation characteristics to the other C-terminal-truncated mutants of apoE3 (Figure 4). Since the N- and C-terminal domains in apoE are folded independently (38), it is likely that apoE3 (1–272) has a different organization of the C-terminal domain from apoE4 (1–272) due to the self-association in the former case (Figure 6B), contributing to the protein stability to some extent.
The different organization of the C-terminal domain for the apoE isoforms also affects their lipid-binding ability. The results from the DMPC clearance assay (Figure 9) demonstrate that apoE4 has a better ability to solubilize DMPC vesicles than apoE3 (37, 41, 42). Solubilization of large DMPC vesicles by apoE to form smaller discoidal complexes involves a structural reorganization of the apoE molecule (37). Therefore, apoE3 having a greater tendency to form oligomer seems to display slower solubilization kinetics than apoE4 which favors the monomeric state. Consistent with the presence of the monomeric form of apoE enhancing DMPC clearance, the E255A mutation in apoE4 increases its self-association (Figure 6C) and decreases the ability to solubilize DMPC vesicles (Figure 9B); this observation indicates that domain-domain interactions can modulate the ability of apoE to interact with lipid surfaces. The importance of the physical state of the apoE molecule is further demonstrated by the fact that monomeric apoE3 mutants generated by protein engineering display faster DMPC clearance rates than wild-type apoE3 (45). Removal of α-helical residues 273–299 favors the monomeric form in apoE isoforms, thereby leading to the enhanced clearance activities for apoE (1–272) truncation mutants (46). The results of ANS binding (Figure 5) indicate that these truncation mutants retain a significant exposed hydrophobic site created by α-helical residues 261–272, being available for lipid binding (27). Supporting this, further truncation of residues 261–272 largely decreases ANS fluorescence intensity (Figure 5) and DMPC clearance activity (Figure 9) (41) for both isoforms.
We have previously proposed that the increased exposed hydrophobic surface in the C-terminal domain of apoE4 has the potential to increase the lipid affinity of this isoform (24). The hydrophobic segments in the C-terminal domain are known to be responsible for many physiological functions of apoE. Thus, the C-terminal residues 261–299 in apoE are important for the efficient stimulation of ABCA1-dependent cholesterol efflux from J774 macrophages (47). In the C-terminal-truncated fragments of apoE4 found in AD brains, the presence of a lipid-binding region (residues 244–272) is critical for their neurotoxic effects in vivo (48). The lipid-binding region in the C-terminal domain is also involved in the binding of apoE to amyloid β peptide (49, 50). In addition, the hydrophobic amino acids between residues 261–269 account for the contribution of the C-terminal domain to the apoE-induced hypertriglyceridemia (51) and dyslipidemia (52). Thus, the less organized, more exposed conformation of the C-terminal domain in apoE4 shown in this study may lead to the pathological sequelae for cardiovascular and various neurodegenerative diseases.
Acknowledgments
This work was supported by NIH grant HL56083, Grant-in-aid for Scientific Research from JSPS (19590048), the Naito Foundation, and NOVARTIS Foundation (Japan) for the Promotion of Science.
The abbreviations used are
- AD
Alzheimer’s disease
- ANS
8-anilino-1- naphthalenesulfonic acid
- apoE
apolipoprotein E
- CD
circular dichroism
- DMPC
dimyristoylphosphatidylcholine
- GdnHCl
guanidine hydrochloride
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- VLDL
very low density lipoprotein
REFERENCES
- 1.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 3.Mahley RW, Huang Y. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 1999;10:207–217. doi: 10.1097/00041433-199906000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Davignon J. Apolipoprotein E and atherosclerosis: beyond lipid effect. Arterioscler Thromb Vasc Biol. 2005;25:267–269. doi: 10.1161/01.ATV.0000154570.50696.2c. [DOI] [PubMed] [Google Scholar]
- 5.Mahley RW, Huang Y. Apolipoprotein (apo) E4 and Alzheimer's disease: unique conformational and biophysical properties of apoE4 can modulate neuropathology. Acta Neurol Scand Suppl. 2006;185:8–14. doi: 10.1111/j.1600-0404.2006.00679.x. [DOI] [PubMed] [Google Scholar]
- 6.Mahley RW, Rall SC., Jr. Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 7.Mahley RW, Huang Y, Rall SC., Jr. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J. Lipid Res. 1999;40:1933–1949. [PubMed] [Google Scholar]
- 8.Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4725–4727. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisgraber KH, Mahley RW. Human apolipoprotein E: the Alzheimer's disease connection. FASEB J. 1996;10:1485–1494. doi: 10.1096/fasebj.10.13.8940294. [DOI] [PubMed] [Google Scholar]
- 10.Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda B, Groswasser Z. Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology. 1999;52:244–248. doi: 10.1212/wnl.52.2.244. [DOI] [PubMed] [Google Scholar]
- 11.Roses AD. Apolipoprotein E and Alzheimer's disease. The tip of the susceptibility iceberg. Ann. N. Y. Acad. Sci. 1998;855:738–743. doi: 10.1111/j.1749-6632.1998.tb10653.x. [DOI] [PubMed] [Google Scholar]
- 12.Raffai RL, Weisgraber KH. Cholesterol: from heart attacks to Alzheimer's disease. J. Lipid Res. 2003;44:1423–1430. doi: 10.1194/jlr.R300007-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 14.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 15.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 16.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Lund-Katz S, Phillips MC. Lipid binding-induced conformational change in human apolipoprotein E. Evidence for two lipid-bound states on spherical particles. J. Biol. Chem. 2001;276:40949–40954. doi: 10.1074/jbc.M106337200. [DOI] [PubMed] [Google Scholar]
- 17.Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J. Biol. Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama S, Kawai Y, Tajima S, Yamamoto A. Behavior of human apolipoprotein E in aqueous solutions and at interfaces. J. Biol. Chem. 1985;260:16375–16382. [PubMed] [Google Scholar]
- 19.Choy N, Raussens V, Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 2003;334:527–539. doi: 10.1016/j.jmb.2003.09.059. [DOI] [PubMed] [Google Scholar]
- 20.Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 21.Weisgraber KH. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990;31:1503–1511. [PubMed] [Google Scholar]
- 22.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 23.Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 24.Saito H, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC, Lund-Katz S. Effects of polymorphism on the lipid interaction of human apolipoprotein E. J. Biol. Chem. 2003;278:40723–40729. doi: 10.1074/jbc.M304814200. [DOI] [PubMed] [Google Scholar]
- 25.Drury J, Narayanaswami V. Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence. J. Biol. Chem. 2005;280:14605–14610. doi: 10.1074/jbc.M414019200. [DOI] [PubMed] [Google Scholar]
- 26.Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of apolipoprotein E structure by domain interaction: differences in lipid-bound and lipid-free forms. J. Biol. Chem. 2005;280:34288–34295. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 2006;45:4240–4247. doi: 10.1021/bi060023b. [DOI] [PubMed] [Google Scholar]
- 28.Lund-Katz S, Weisgraber KH, Mahley RW, Phillips MC. Conformation of apolipoprotein E in lipoproteins. J. Biol. Chem. 1993;268:23008–23015. [PubMed] [Google Scholar]
- 29.Lund-Katz S, Zaiou M, Wehrli S, Dhanasekaran P, Baldwin F, Weisgraber KH, Phillips MC. Effects of lipid interaction on the lysine microenvironments in apolipoprotein E. J. Biol. Chem. 2000;275:34459–34464. doi: 10.1074/jbc.M005265200. [DOI] [PubMed] [Google Scholar]
- 30.Morrisett JD, David JS, Pownall HJ, Gotto AM., Jr. Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12:1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 31.Acharya P, Segall ML, Zaiou M, Morrow J, Weisgraber KH, Phillips MC, Lund-Katz S, Snow J. Comparison of the stabilities and unfolding pathways of human apolipoprotein E isoforms by differential scanning calorimetry and circular dichroism. Biochim. Biophys. Acta. 2002;1584:9–19. doi: 10.1016/s1388-1981(02)00263-9. [DOI] [PubMed] [Google Scholar]
- 32.Sparks DL, Lund-Katz S, Phillips MC. The charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. J. Biol. Chem. 1992;267:25839–25847. [PubMed] [Google Scholar]
- 33.Eftink MR. Use of fluorescence spectroscopy as thermodynamics tool. Methods Enzymol. 2000;323:459–473. doi: 10.1016/s0076-6879(00)23378-8. [DOI] [PubMed] [Google Scholar]
- 34.Saito H, Minamida T, Arimoto I, Handa T, Miyajima K. Physical states of surface and core lipids in lipid emulsions and apolipoprotein binding to the emulsion surface. J. Biol. Chem. 1996;271:15515–15520. doi: 10.1074/jbc.271.26.15515. [DOI] [PubMed] [Google Scholar]
- 35.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Bortnick AE, Nickel M, Dhanasekaran P, Subbaiah PV, Lund-Katz S, Rothblat GH, Phillips MC. Effects of apolipoprotein A-I on ATP-binding cassette transporter A1-mediated efflux of macrophage phospholipid and cholesterol: formation of nascent high density lipoprotein particles. J. Biol. Chem. 2003;278:42976–42984. doi: 10.1074/jbc.M308420200. [DOI] [PubMed] [Google Scholar]
- 37.Segall ML, Dhanasekaran P, Baldwin F, Anantharamaiah GM, Weisgraber KH, Phillips MC, Lund-Katz S. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- 38.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 39.Wetterau JR, Aggerbeck LP, Rall SC, Jr., Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 40.Narayanaswami V, Szeto SS, Ryan RO. Lipid association-induced N- and C-terminal domain reorganization in human apolipoprotein E3. J. Biol. Chem. 2001;276:37853–37860. doi: 10.1074/jbc.M102953200. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Kypreos K, Zanni EE, Zannis V. Domains of apoE required for binding to apoE receptor 2 and to phospholipids: implications for the functions of apoE in the brain. Biochemistry. 2003;42:10406–10417. doi: 10.1021/bi027093c. [DOI] [PubMed] [Google Scholar]
- 42.Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, Mahley RW. Reactivity of Apolipoprotein E4 and Amyloid β Peptide: Lysosomal Stability and Neurodegeneration. J. Biol. Chem. 2006;281:2683–2692. doi: 10.1074/jbc.M506646200. [DOI] [PubMed] [Google Scholar]
- 43.Chou CY, Lin YL, Huang YC, Sheu SY, Lin TH, Tsay HJ, Chang GG, Shiao MS. Structural variation in human apolipoprotein E3 and E4: secondary structure, tertiary structure, and size distribution. Biophys. J. 2005;88:455–466. doi: 10.1529/biophysj.104.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan D, Li Q, Korando L, Jerome WG, Wang J. A monomeric human apolipoprotein E carboxyl-terminal domain. Biochemistry. 2004;43:5055–5064. doi: 10.1021/bi035958w. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A Monomeric, Biologically Active, Full-Length Human Apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 46.Chou CY, Jen WP, Hsieh YH, Shiao MS, Chang GG. Structural and functional variations in human apolipoprotein E3 and E4. J Biol Chem. 2006;281:13333–13344. doi: 10.1074/jbc.M511077200. [DOI] [PubMed] [Google Scholar]
- 47.Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, Bielicki JK. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46:2583–2593. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]
- 48.Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, Weisgraber KH, Mucke L, Mahley RW, Huang Y. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pillot T, Goethals M, Najib J, Labeur C, Lins L, Chambaz J, Brasseur R, Vandekerckhove J, Rosseneu M. β-Amyloid peptide interacts specifically with the carboxy-terminal domain of human apolipoprotein E: relevance to Alzheimer's disease. J. Neurochem. 1999;72:230–237. doi: 10.1046/j.1471-4159.1999.0720230.x. [DOI] [PubMed] [Google Scholar]
- 50.Phu MJ, Hawbecker SK, Narayanaswami V. Fluorescence resonance energy transfer analysis of apolipoprotein E C-terminal domain and amyloid beta peptide (1–42) interaction. J. Neurosci. Res. 2005;80:877–886. doi: 10.1002/jnr.20503. [DOI] [PubMed] [Google Scholar]
- 51.Kypreos KE, van Dijk KW, Havekes LM, Zannis VI. Generation of a recombinant apolipoprotein E variant with improved biological functions: hydrophobic residues (LEU-261, TRP-264, PHE-265, LEU-268, VAL-269) of apoE can account for the apoE-induced hypertriglyceridemia. J. Biol. Chem. 2005;280:6276–6284. doi: 10.1074/jbc.M413458200. [DOI] [PubMed] [Google Scholar]
- 52.Drosatos K, Kypreos KE, Zannis VI. Residues Leu261, Trp264, and Phe265 Account for Apolipoprotein E-Induced Dyslipidemia and Affect the Formation of Apolipoprotein E-Containing High-Density Lipoprotein. Biochemistry. 2007;46:9645–9653. doi: 10.1021/bi700232g. [DOI] [PMC free article] [PubMed] [Google Scholar]