Abstract
We report a streamlined procedure to efficiently carry samples from chromatin to qPCR-compatible DNA in as little as 4 hours. We use this streamlined ChIP to quantify histone H3 modifications at active (cad) and repressed (T early alpha) promoters in a Rag1-deficient pro-T cell line after 1–2 hour IP. We further show that the protocol is equally effective when usingreadily quantified histone modifications in chromatin from as few as 104 Rag-deficient DN thymocytes. Taken together, these data outline a simple, cost-effective procedure for efficient ChIP analysis.
Keywords: Chromatin immunoprecipitation, histone, epigenetics, cad, TEA, thymocyte, Rag
1. Introduction
The fast-growing fields of epigenetics and epigenomics have necessitated development of assays that probe interactions between DNA, structural histone proteins, and regulatory transcription factors. One of the primary tools used to unravel the so-called “histone code” has been chromatin immunoprecipitation, or ChIP (Dedon et al., 1991; O’Neill and Turner, 1996; Kuo and Allis, 1999). Indeed, ChIP, coupled with realtime PCR (qPCR) has become the gold standard assay for chromatin organization (Jenuwein and Allis, 2001), and is increasingly used to demonstrate differential transcription factor recruitment to various promoters (Morshead et al., 2003; O’Neill et al., 2006). Despite such widespread use, the complexity, lengthiness, and scale of the standard ChIP protocol, which can take up to 3 days and require ≥106 cells per reaction, make it extremely sensitive to experimenter-induced variability and to contamination, and limit its utility for scarce cell populations, such as those found in select compartments of the immune system.
A number of groups have now proposed modifications to the standard ChIP protocol (Nelson et al., 2006; O’Neill et al., 2006; Attema et al., 2007; Dahl and Collas, 2007; Dahl and Collas, 2008). The newer protocols have demonstrate that ChIP is amenable to variations in virtually every aspect of the assay from the size of chromatin input, the time dedicated to immunoprecipitation, washing, elution, and crosslink reversal, to Proteinase K treatment regiments. By replacing agarose or sepharose beads with Protein A- or Protein G-coupled paramagnetic beads, newer approaches minimize the need to preclear input chromatin of antibody-independent bead binding activities. At the same time, the ability to easily and quantitatively capture magnetic bead complexes eliminates the need for centrifugation, which both reduces the time required for each of the many washes in the ChIP protocol, and reduces the potential for sample loss or contamination during wash aspiration.
We have designed a streamlined protocol that incorporates improvements offered by the Q2-ChIP (Dahl and Collas, 2007), FastChIP (Nelson et al., 2006), ChIP-IT Express (ActiveMotif), and miniChIP (Attema et al., 2007) into a simplified format. We find this streamlined protocol is easily mastered, rapid, and highly reproducible. Demonstrating its adaptability to reduced sample size, our streamlined ChIP readily quantitated histone modifications and even binding of the RAG1 component of V(D)J recombinase in as few as 104 CD4/CD8 double negative thymocytes harvested from a Rag-2 deficient mouse. Though we find each of the existing ChIP protocols can be highly effective, we propose our streamlined protocol as a simplified approach for those new to ChIP or for higher throughput ChIP screens.
2. Materials and Methods
2.1 Cells
The RAG1−/−, p53−/− pro-T cell line, P5424, has been previously described (Mombaerts et al., 1995). P5424 cells were cultured at 37°C/5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, 0.01% penicillin/streptomycin, and 50 μM β-mercaptoethanol. Thymii were isolated from 4–8 wk old Rag2−/− mice, crushed and filtered to yield a single cell suspension, and red blood cells were removed by hypotonic lysis. The mouse studies described here were reviewed and approved by the institutional animal care and use committee at North Carolina State University.
2.2 Antibodies
Rabbit polyclonal antisera to acetylated H3K9 (06–599) and dimethylated H3K4 (07–030), along with rabbit control IgG (12–370) were purchased from Upstate. Rabbit polyclonal antisera to dimethylated H3K9 (ab1772) and trimethylated H3K4 (ab8580), and mouse monoclonal antibody to RNA polymerase II (ab5408) were purchased from Abcam.
2.3 Chromatin Preparation
Protein:DNA complexes in 4 × 106 P5424 or freshly isolated thymocyte suspension cells were cross-linked using formaldehyde (1% final) in either tissue culture dishes or conical centrifuge tubes with gentle shaking for 10 min. at room temperature. Crosslinking was stopped by drop-wise addition of glycine (125 mM final concentration) and gentle shaking for 5 min. at room temperature. Cells were pelleted, washed in 1X PBS (5 ml). Pelleted cells were resuspended in 500 μl lysis buffer (10 mM Tris-HCl, pH7.5, 10 mM NaCl, 3 mM MgCl2, and 0.5% NP-40) supplemented with 1 mM PMSF and 1X Protease Inhibitor Cocktail (PIC, (Roche), and incubated for 30 min on ice. Nuclei from the lysed cells were pelleted by microcentrifugation (5000 rpm for 10 min. @ 4°C). Nuclei were resuspended in 100 μl MNase reaction buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 4% NP-40) supplemented with 1 mM PMSF and 1X PIC, and chromatin was sheared with the addition of 1–2 U MNase (Sigma) for 10 min at 37°C. Digestion was stopped with the addition of EDTA (10 mM final), and the resultant chromatin was stored @ −80°C.
2.4 Chromatin Immunoprecipitation
For ChIP, paramagnetic Dynabeads (Dynal) separately coupled to Protein A (10 μl/IP) and Protein G (10 μl/IP) were combined in a 1.5 ml microcentrifuge tube, captured by placing the tube against a strong magnet, and the suspension medium was immediately aspirated while the beads were held in place by the magnet. Beads were washed as described (Dahl and Collas, 2007) with the addition of 100 μl/IP 1X RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% NaDeoxycholate) containing BSA (50 mg/ml) and sheared salmon sperm DNA (0.5 mg/ml), denoted as RBD hereafter. Beads were vortexed (3 sec.), and recaptured. Wash solution was aspirated, and the wash was repeated. Beads were resuspended in RBD (50 μl/IP) containing PMSF and PIC, aliquoted into sterile 200 μl PCR strip tubes containing 1–5 μg antibody, and rotated 1 hr at 4°C. Conjugated antibody:bead complexes were washed 2X in RBD as described above, and resuspended in 75 μl/IP volumes of RBD supplemented with PMSF and PIC.
For each IP, antibody:bead suspensions were mixed with 25 μl undiluted chromatin (106 cell equivalents) or chromatin diluted in RBD containing PMSF and PIC as indicated (Fig. 3). Protein-DNA complexes were immunoprecipitated for 2 hours (except Fig. 1) at 4°C with rotation. Bead complexes were captured by placing the PCR strip tubes on a horizontal bar magnet (attached to an empty 200 μl micropipettor tip rack), IP solution was aspirated, and beads were resuspended in 180 μl RBD. To wash the bead complexes, the strip tubes were rapidly moved back and forth across the magnet 5 times (sequentially forcing the beads from one side of the tube to the other). Beads were recaptured and the wash was repeated 3X. Following RBD wash, bead complexes were washed 2X in 180 μl TE, pH 8.0.
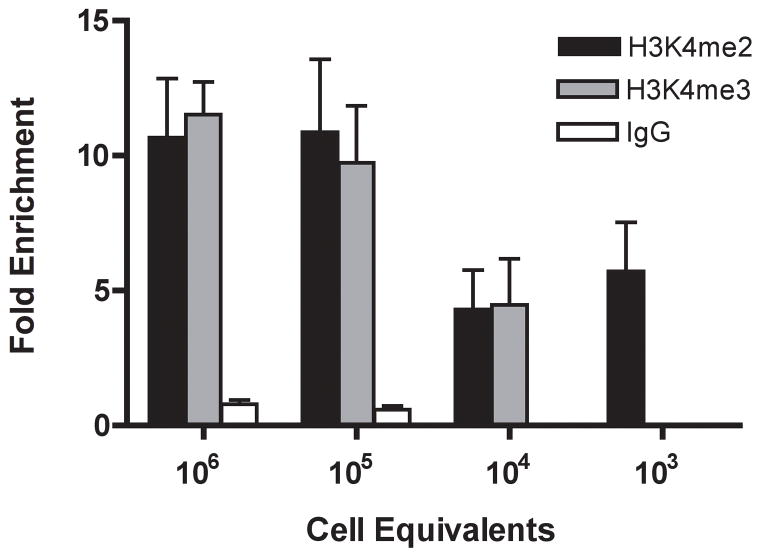
Figure 3.
Sensitivity of ChIP to reduction in the amount of input chromatin. Prior to IP, chromatin was prepared from 106 P5424 cells, and serially diluted with RIPA buffer to the indicated cell equivalents. Bead complexes with antibodies to H3K4me2 and H3K4me3, and control IgG were incubated for 2 hours with each chromatin dilution, and fold enrichment of cad promoter DNA in the IP samples relative to input is calculated as in Fig. 1.
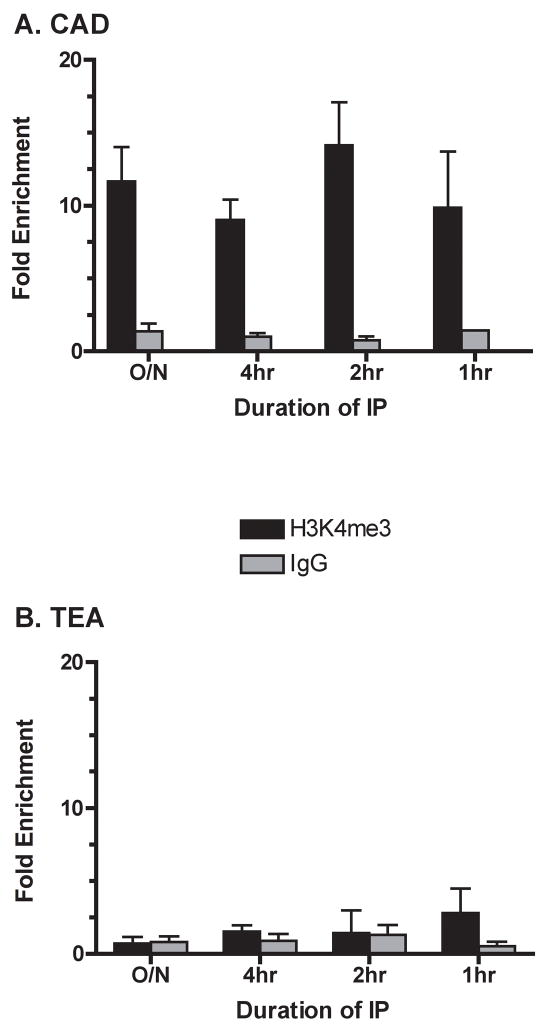
Figure 1.
Sensitivity of ChIP to reduction in the duration of IP incubation. Chromatin from the Rag1−/−p53−/− mouse thymocyte cell line, P5424, was immunoprecipitated for the indicated times with paramagnetic beads complexed with either anti-H3K4me3 (black) or control IgG (grey). Fold enrichment of cad (A) or TEA (B) DNA following IP with each antibody is shown relative to an input sample. Bars indicate means (± SD) of triplicate Q-PCRs, and are representative of 2 experiments with independent chromatin preparations.
To elute protein-DNA complexes, beads were resuspended in 100 mM NaHCO3 (100 μl), strip tubes were resealed with fresh strip caps, and were gently vortexed 15 min @ room temperature. Tubes were returned to the magnet, and protein-DNA eluates were transferred to fresh PCR strip tubes containing 4 μl 5M NaCl. To prepare matched input samples, 10 μl of input chromatin was diluted in 90 μl 100 mM NaHCO3, transferred to a strip tube containing 4 μl 5M NaCl, and carried along with the IP samples for all subsequent steps. Crosslinks were reversed @ 95°C for 15 min., tubes were cooled to room temperature, and then incubated with Proteinase K (10 μg/ml final) 1 hr @ 45°C. Digestion was stopped with addition of PMSF (2 mM final), DNA was purified using Qiaquick nucleotide removal columns (Qiagen) according to the manufacturer’s instructions, and eluted in 100 μl Qiagen elution buffer.
2.5 Q-PCR and data analysis
For realtime PCR, bound (3 μl) and input (3 μl of 1:100 dilution) samples were amplified in a MyIQ thermal cycler (Bio-RAD) using 1X SensiMix Plus (Quantace) and primers specific for the cad promoter (forward: 5′-GTCTGCGTGCTTGCCCTGTCTCAGC-3′; reverse: 5′-CGGGCTTGCTTACCCACTTCCCCAGC-3′), or TEA (McMurry and Krangel, 2000; Sikes et al., 2002), or 57prime;PDβ2 promoter (forward: 5′-GTTTCTGAGGCATGTGTCTCTGCG-3′; reverse: 5′-TCCTCTTTGTCACAGTGCCCACC-3′). Cycling parameters for 20 μl reactions were 95°C 10 min., followed by 45 cycles of 95°C, 30 sec.; 57°C (TEA) or 72°C (cad), 30 sec.; 72°C, 30 sec, followed by melt curve analysis. Fold enrichment in the bound fractions relative to input was calculated from as previously described (Ciccone et al., 2004), and the average enrichment for triplicate amplifications was reported.
3. Results
3.1 Synthesis of a streamlined ChIP protocol from existing methods
Although extremely effective, the standard ChIP protocol as described by Kuo and Allis (Kuo and Allis, 1999) remains time-intensive and difficult. By contrast, we recently found that multiple modified ChIP protocols including the Q2ChIP (Dahl and Collas, 2007) and ChIP-IT Express (ActiveMotif) were readily mastered by less experienced lab members. Noting the remarkable differences between each of these effective protocols, we sought to synthesize a single method that would minimize tradeoffs for cost, ease of use, speed, and reproducibility.
For our studies, we used input chromatin prepared essentially as described by Ciccone et al. (Ciccone et al., 2004), in which sonication-based shearing is replaced with micrococcal nuclease digestion. We favored the enzymatic digestion as a means to minimize technique-dependent variability between users and laboratories. Additionally, crosslinking and glycine treatments were performed in conical centrifuge tubes rather than culture dishes. However, this alteration necessitated that we eliminate a PBS wash between the formaldehyde and glycine treatments. Consequently, reactions were stopped by dropwise addition of glycine.
Our antibody:bead conjugation and IP conditions essentially followed the Q2ChIP protocol (Dahl and Collas, 2007). Consequently, preblocking the beads and preclearing chromatin to prevent nonspecific DNA binding proved unnecessary. BSA (50 mg/ml) and sheared salmon sperm DNA (0.5 mg/ml) were added, and antibody:bead conjugates were thoroughly washed to block nonspecific antibody:DNA interactions and reduce ChIP variability. Finally, to increase the uniformity of bead conjugation with different antibody preparations, we incubated 1:1 mixes of Protein A- and Protein G-coupled beads with each antibody for 1 hr, though the manufacturer’s instructions and our own tests suggest that conjugation can be reduced to as little as 15 minutes (data not shown).
Conjugates were mixed with varying concentrations of chromatin at 4°C, and immune complexes were allowed to form over a 2 hr window. We adapted the post-IP sample processing procedures used in ActiveMotif’s ChIP-IT Express kit, in which the crosslink reversal times are dramatically shortened. We used the Q2ChIP buffers (RIPA and TE) to wash immune complexes 4X and 2X respectively. In each case, washes were performed by repeatedly passing PCR strip tubes containing resuspended immune complexes across an immobilized bar magnet. In this way, all 6 washes for as many as 16 IP reactions could be completed in as little as 15 min. We observed no benefit from incubating complexes in the wash buffers prior to aspiration.
After washing, captured immune complexes were eluted from the beads in 100 mM NaHCO3 by affixing the strip tubes to a benchtop vortexer set at 50% speed for 15 min. Crosslink reversal was similarly limited to 15 min. For this, NaCl (250 mM final) was added, and eluates were immediately recovered from the beads. Crosslinks in the eluates were then reversed in a thermal cycler for 15 min. at 95°C, Proteinase K was added, and eluates were incubated for 1 hr at 45°C. To minimize handling and experimental variability, we stopped protease digestion by adding PMSF, and purified sample DNAs using Qiaquick Nucleotide Removal columns (Qiagen). However, we found that phenol:chloroform extraction and ethanol precipitation were equally effective at yielding Q-PCR-competent DNA.
3.2 Increased IP duration does not enhance specific signal
To determine if the 2 hr IP time could be reduced, we set up ChIP reactions using chromatin from 106 Rag1−/−p53−/− P5424 cells (a murine CD4/CD8 double negative pro-T cell line) and antibodies to histone H3 carrying a trimethylation of lysine 4 (H3K4me3). To validate our IP results, we measured Q-PCR signals at 2 distinct promoters, cad, which has previously been shown in P5424 to bear histone modifications consistent with open chromatin and transcriptional activity (Morshead et al., 2003), and TEA, which exists in a repressive chromatin state in Rag2-deficient thymocytes (McMurry and Krangel, 2000). Two hr IP with anti-H3K4me3 specifically enriched an amplicon within the cad gene promoter >14 fold above untreated input, while IP with nonspecific IgG showed no enrichment (Fig. 1A). Increasing IP duration to 4 or 16 hours did not increase fold enrichment of the anti-H3K4me3 samples relative to input or IgG controls. In fact, enrichment was equally effective after only a single hr IP. In contrast to our cad findings, H3K4me3 levels at TEA were not significantly enriched above control IgG, even after overnight IP (Fig. 1B). We did not determine if IP reactions shorter than 1 hr would be equally effective, though other protocols have limited IP to 45 min. (Nelson et al., 2006).
3.3 Streamlined ChIP effectively distinguishes between accessible and repressed gene targets
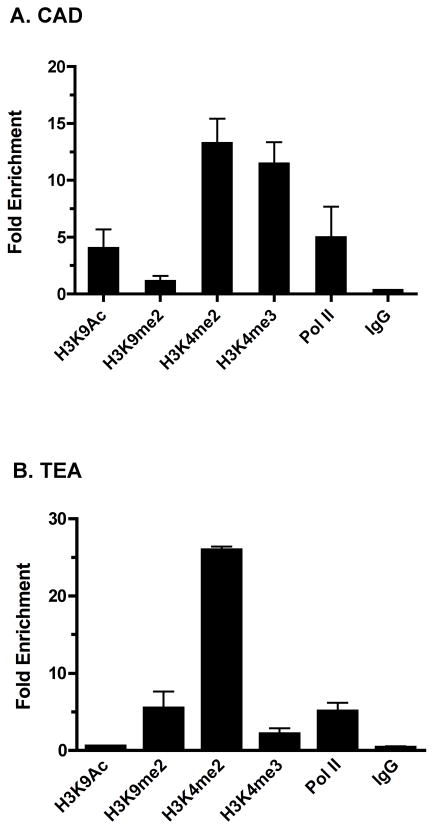
To extend our validation, we next screened cad and TEA chromatin with a panel of activation state-specific antibodies (Fig. 2). Two hr IP with antibodies to either dimethylated or trimethylated H3K4 showed equivalent enrichment over input control at the cad promoter (Fig. 2A; 13.25 ± 2.17 and 11.44 ± 1.92, respectively). Enrichment of H3K4me2 was similarly observed at TEA, though H3K4me3 levels were significantly reduced (Fig. 2B; 25.93 ± 0.45 and 2.15 ± 0.70, respectively). We next measured levels of opposing acetylation and dimethylation at H3K9 and recruitment of RNA polymerase II. The cad promoter was enriched for both H3K9 acetylation and RNA Pol II recruitment, both markers of accessible chromatin, but not for the repressive chromatin marker H3K9me2 (Fig. 2A). Conversely, H3K9me2 levels but not H3K9ac levels were elevated at TEA (Fig. 2B), consistent with a repressed chromatin state. When we assessed Pol II binding at TEA, we again saw enrichment equivalent to that at cad. RNA Pol II stalling prior to transcription elongation has been recently shown across the human genome (Muse et al., 2007). Likewise, the differential H3K4 methylation status (me2+/me3−) of TEA is mirrors that observed for developmentally poised genes (Santos-Rosa et al., 2002; Mikkelsen et al., 2007; Orford et al., 2008). Taken together, our observations suggest that TEA exists in a poised state in P5424 DN cells.
Figure 2.
Analysis of histone H3 modifications at differentially accessible promoters in P5424 chromatin. Bead complexes with each of the indicated antibodies were incubated with P5424 chromatin for 2 hours. Resultant fold enrichments of cad (A) and TEA(B) promoter sequences in the IP samples relative to input are calculated as in Fig. 1.
3.4 Streamlined ChIP remains effective with reduced chromatin input
The Q2ChIP is reported to remain effective despite dramatic reduction of input chromatin levels (Dahl and Collas, 2007). To determine if our modified protocol was equally effective in the presence of reduced input, we measured enrichment of H3K4me2 and H3K4me3 at cad after IP of serially diluted P5424 chromatin (Fig. 3). In each case, antibody and bead concentrations were held constant to isolate the impact of chromatin reduction. We found that 104 cell equivalency was the minimum amount of chromatin that would still yield reproducible Q-PCR signal for both H3K4 antibodies. AntibodyWhen normalized to input antibody-dependent enrichment when using chromatin from 104 cells showed roughly a 2-fold reduction relative to more concentrated input levels. However, PCR signals in control IgG samples failed to cross threshold, suggesting that antibody-specific enrichment was still considerable. Chromatin dilution to 103 cell equivalents yielded reproducible Q-PCR signal only for H3K4me2 enrichment, and subsequent dilutions failed to amplify altogether, a marked difference from the Q2-ChIP which showed effective ChIP for ≤102 cells (Dahl and Collas, 2007).
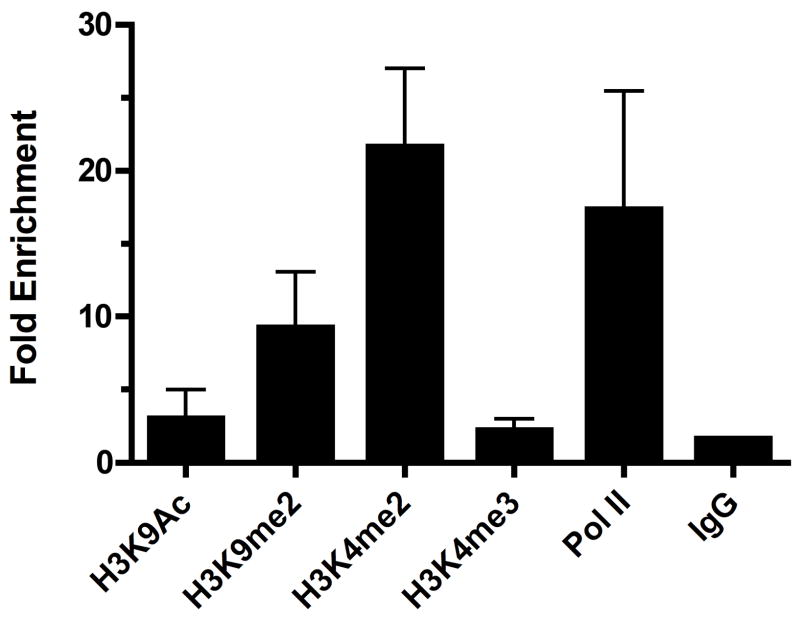
Although dilution of chromatin prepared from large-scale P5424 cultures suggested that our ChIP protocol was effective when chromatin was limiting, we wanted to demonstrate its application to analysis of rare cell populations in vivo. Thymii from Rag-deficient mice contain approximately 106 thymocytes, all prevented from progressing beyond DN development by their inability to rearrange the TCRβ gene locus (Mombaerts et al., 1992; Shinkai et al., 1992). To test the utility of our streamlined ChIP protocol for limiting cell populations, we measured histone modifications at TEA in chromatin from 104 freshly isolated Rag2−/− thymocytes (Fig. 4). Consistent with our data using 106 P5424 cells (Fig. 2B), ChIP of the TEA promoter in Rag2-deficient thymocytes showed elevated H3K9 methylation (9.27 ± 3.79), H3K4me2 (21.65 ± 5.36), and RNA Pol II loading (17.35 ± 8.10), despite moderately increased background levels obtained with control IgG (1.61 ± 0.01). By contrast, enrichments using antibodies to H3K9ac and H3K4me3 (3.01 ± 1.97 and 2.21 ± 0.77, respectively) were similar to IgG. Taken together with our analysis in P5424, these data strongly suggest that TEA exists in a poised chromatin state during DN thymocyte development prior to β selection and the activation of Tcra recombination.
Figure 4.
Chromatin modifications at the TEA promoter of Rag2-deficient thymocytes. Bead complexes with each of the indicated antibodies were incubated for 2 hrs with chromatin prepared from 104 freshly isolated Rag2−/− thymocytes. Fold enrichment of TEA in each IP sample relative to input is calculated as in Fig. 1.
4. Discussion
Although enormously powerful, the conventional methodology for ChIP is a somewhat daunting procedure that requires large amounts of chromatin input and stretches across several days. A series of recent studies have shown that individual steps of the standard ChIP protocol can be extensively modified. We have synthesized the various improvements to the conventional ChIP protocol to develop a simplified method that is significantly shorter, uses a minimum amount of costly reagents and equipment, minimizes potential for sample loss or contamination, and is compatible with reduced chromatin input. Most importantly, IP, post-IP washes, and crosslink reversal times are all dramatically reduced in the streamlined protocol without compromising the ability to quantify gene-specific differences in histone modification and binding site occupancy.
Whereas recent protocols have reported successful ChIP with chomatin prepared from as few as 100 cells (Dahl and Collas, 2007), we found that amplification signals declined when chromatin was IPed from fewer than 105 cells (Fig. 3). Despite reduced signal, we had no difficulty quantifying epigenetic alterations in chromatin from 104 freshly isolated RAG2-deficient thymocytes (Fig. 4). Though apparently less sensitive than Q2-ChIP, the practical impact of reduced chromatin demand in our protocol allows us to run as many as 100 IP reactions with chromatin isolated from a single RAG knockout animal, a marked difference with standard protocols that would require between 1 and 10 thymii per IP. Quantitation of purified DNA using a NanoDrop spectrophotometer prior to Q-PCR showed that IPs with 104 cell equivalent inputs typically yielded between 50 and 100 ng DNA, of which 1 to 3 ng were amplified per reaction. With this amount of input DNA, threshold cycles in both the TEA and cad PCRs ranged from 33 to 36 cycles. Although IP reactions may remain quite effective using even less chromatin, we expect that 104 cell equivalents reflects the limit below which our lab cannot effectively amplify available targets.
In developing the streamlined ChIP protocol, we uncovered a previously unreported aspect of thymocyte biology; namely the poised state of TEA in DN cells prior to Tcra recombination. The observed dichotomy between dimethylated and trimethylated H3K4 at TEA has been recently shown to mark poised developmentally regulated promoters at a variety of hematopoietic genes (Orford et al., 2008) as well as at promoters across the genome (Mikkelsen et al., 2007). In addition to selective H3K4 dimethylation, enrichment of RNA pol II at TEA strongly argues that in the DN3 developmental state of P5424 the TEA promoter is repressed but primed for rapid activation in later staged cells.
In summary, the streamlined ChIP protocol outlined here provides a rapid, sensitive, and efficient alternative to the standard protocol. Its ease of use should allow the protocol’s ready adoption by programs not currently using ChIP, and its reduced dependence on input chromatin should facilitate extension of current epigenetic studies. Indeed, we are currently using our streamlined ChIP protocol to pursue imposition of a poised state at the TEA promoter during individual stages of T lineage development.
Acknowledgments
We gratefully thank Drs Hosni Hassan and Jose Bruno Barcena for use of their Q-PCR equipment.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases (R56AI070848–01A1).
Abbreviations used in this paper
- ChIP
chomatin immunoprecipitation
- H3
histone 3
- Pol II
RNA polymerase 2
- Ab
antibody
- cad
carbamoylphosphate synthetase/aspartate carbamyltransferase/dihydroorotase gene
- TEA
T early alpha promoter
- Rag
Recombination activating gene
- TCR
T cell receptor
- DN
double negative
Footnotes
Disclosures
The authors have no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci U S A. 2007;104:12371–6. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Morshead KB, Oettinger MA. Chromatin immunoprecipitation in the analysis of large chromatin domains across murine antigen receptor loci. Methods Enzymol. 2004;376:334–48. doi: 10.1016/S0076-6879(03)76022-4. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–46. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–33. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–8. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–77. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Terhorst C, Jacks T, Tonegawa S, Sancho J. Characterization of immature thymocyte lines derived from T-cell receptor or recombination activating gene 1 and p53 double mutant mice. Proc Natl Acad Sci U S A. 1995;92:7420–4. doi: 10.1073/pnas.92.16.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–82. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Immunoprecipitation of chromatin. Methods Enzymol. 1996;274:189–97. doi: 10.1016/s0076-6879(96)74017-x. [DOI] [PubMed] [Google Scholar]
- O’Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–41. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–67. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Sikes ML, Meade A, Tripathi R, Krangel MS, Oltz EM. Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc Natl Acad Sci U S A. 2002;99:12309–14. doi: 10.1073/pnas.182166699. [DOI] [PMC free article] [PubMed] [Google Scholar]