Abstract
BRMS1 and SUDS3 are related members of SIN3-HDAC chromatin remodeling complexes. We hypothesized that they might have overlapping functions and that SUDS3 over-expression could compensate for BRMS1 deficiency. SUDS3 expression was ubiquitous in seven breast cell lines, regardless of metastatic potential. SUDS3 over-expression in BRMS1-non-expressing metastatic cells did not suppress metastasis, motility, osteopontin secretion nor EGF receptor expression, phenotypes associated with BRMS1-mediated metastasis suppression. This study demonstrates functional differences for BRMS1 family members and highlights how the composition of SIN3-HDAC (BRMS1/SUDS3) complexes uniquely affects protein expression and biological behaviors.
Keywords: SUDS3, BRMS1, metastasis suppression, motility
1. Introduction
Breast Cancer Metastasis Suppressor 1 (BRMS1) is a functionally validated metastasis suppressor, defined by blockage of metastasis without preventing orthotopic tumor growth, in both human and murine breast cancer and melanoma cells lines, as well as ovarian cancer cells (reviewed in [1]). The mechanism(s) by which BRMS1 suppresses metastasis are complex and varied, but appear to be dependent upon transcriptional regulation through interaction with SWI-independent 3 (SIN3)-histone deacetylase (HDAC) chromatin remodelling complexes [2-4].
BRMS1 re-expression in metastatic cancerous cells restores homotypic and heterotypic gap junctional intercellular communication [5;6], increases the sensitivity of cells to anoikis [7;8], and significantly decreases motility [9]. BRMS1 also alters transcription and expression of multiple genes [10-12]; in particular BRMS1 decreases expression of several tumor promoting and metastasis activating genes including epidermal growth factor receptor (EGFR; [13]), osteopontin (OPN; [14;15]), urokinase-type plasminogen activator [16], and fascin [17].
Suppressor of Defective Silencing 3 (SUDS3, formerly SDS3) was first identified in yeast when mutation restored silencing at the HMR locus [18]. Co-immunoprecipitation studies showed it to be an integral part of orthologous SIN3-HDAC chromatin remodeling complexes in yeast, mouse, and human cells [19-22]. Moreover Sds3, the yeast ortholog of SUDS3, promoted Sin3 complex integrity and was essential for histone deacetylase activity [19]. Mammalian studies demonstrated that SUDS3 is essential for embryonic development [23] and implicated a role for SUDS3 in cancer [24].
In addition to being involved in many of the same chromatin remodeling complexes [3;4;25-27], BRMS1 shares 23% identity and 49% similarity [3] with the entirety of the SUDS3 protein. Amino acids 69-110 of BRMS1 and 63-104 of SUDS3 were originally designated as the Sds3 domain; but recently, the domain was redefined to include amino acids 52-223 of BRMS1 and 58-229 of SUDS3 [28]. The Sds3 domain is common to all BRMS1 family members including BRMS1, SUDS3, BRMS1-like (p40), as well as the yeast protein Dep1 [28]. Based on sequence homology, we hypothesized that these two proteins might share overlapping functions. Of the phenotypes examined, SUDS3 over-expression in BRMS1 non-expressing cells did not mimic BRMS1 re-expression. Therefore, we conclude that, despite being related, the two proteins are functionally distinct.
2. Materials and Methods
2.1. Cell lines and cell culture
MDA-MB-231 and MDA-MB-435 are human estrogen receptor- and progesterone receptor-negative cell lines derived from metastatic infiltrating ductal breast carcinomas [29;30]. While there is some controversy as to the origin of MDA-MB-435 [31;32], the findings presented here are not dependent upon cellular origin. MDA-MB-436 and MDA-MB-468 cells are human mammary adenocarcinoma cells. These cells were cultured in a mixture (1:1, v/v) of Dulbecco's modified Eagle's medium and Ham's F12 medium (DMEM/F12) with 5% fetal bovine serum, 2mM L-glutamine (Invitrogen, Carlsbad, CA), and 0.02 mM non-essential amino acids (Mediatech, Herndon, VA) without antibiotics or antimycotics. Cells were grown on 100-mm tissue culture dishes (Corning, Corning, NY) at 37°C with 5% CO2 in a humidified atmosphere. Cultures were passaged upon reaching 80-90% confluency using a solution of 0.05% Trypsin/2 mM EDTA (Invitrogen) and were confirmed negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa, Shiga, Japan).
MCF7 cells are tumorigenic human mammary cells. These cells were cultured in minimal essential medium (MEM) with L-glutamine and Earle's salts supplemented with 10% fetal bovine serum (Invitrogen), 0.1 mM non-essential amino acids, 1 mM sodium pyruvate (Mediatech), and 10 mg/mL insulin (Sigma-Aldrich, St. Louis, MO).
MCF10 and derived cell lines model cancer progression and originated from benign fibrocystic breast tissue [33-37]. They include MCF10A (immortalized but nontumorigenic epithelial cells), MCF10AT (mutant ras-expressing, premalignant, mildly tumorigenic epithelial cells), and MCF10CA1a.1 and MCF10CA1d.1α (form invasive orthotopic tumors that metastasize to lung and regional lymph nodes). These cell lines were cultured as described above with the substitution of 5% horse serum for fetal bovine serum. MCF10A and MCF10AT growth medium was supplemented with 10 ng/mL EGF, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin, and 10 μg/mL insulin (Sigma-Aldrich).
Stably transfected cells were selected using 500 μg/mL active G418 (Mediatech) and maintained in 100 μg/mL G418.
2.2. Constructs and transfection
pcDNA3.1-V5/His-SUDS3, pcDNA3.1-V5/His, and pcDNA3 (Invitrogen) plasmids were transfected into MDA-MB-435 and MDA-MB-231 cells using Lipofectamine 2000 (Invitrogen). Vector controls were kept as a mixed population while SUDS3 transfectants were single cell cloned and screened for expression of SUDS3 by immunoblotting.
2.3. Antibodies and immunoblots
A polyclonal rabbit anti-SUDS3 antibody was previously described [22]. The antibody was generated against a peptide corresponding to amino acids 83-328 of the SUDS3 protein. Other antibodies were purchased as indicated: mouse-anti-β-actin (A2228; Sigma-Aldrich), rabbit-anti-OPN (WH0006696M1; Sigma Aldrich), mouse-anti-V5 (R962-25; Invitrogen), anti-EGFR (2232; Cell Signaling Technology, Danvers, MA), mouse-mab-anti-GAPDH (ab9482; Abcam, Cambridge, MA), anti-mouse secondary antibody conjugated to horseradish peroxidase (NXA931; Amersham-Pharmacia, Biotech, Buckinghamshire, UK) and anti-rabbit secondary antibody conjugated to horseradish peroxidase (NA934; Amersham-Pharmacia).
Cells were grown to 80-90% confluence and then lysed in either RIPA Buffer (Millipore, Billerica, MA) or a 0.1% Triton X-100 lysis buffer as previously described [7]. Both lysis buffers were supplemented with 1 μL/mL protease inhibitor cocktail (P8340; Sigma-Aldrich). To evaluate OPN secretion, media were collected from cells that had been washed three times with ice-cold PBS and serum starved for 12 -24 hours in 5 mL of media/10-cm dish. Equal protein loading was determined by using the BCA assay for whole cell lysates (Pierce, Rockford, IL) or by cell count for media loading.
2.4. In vitro growth, wound healing, and motility assay
Cells at 80–90% confluence were detached and seeded at a density of 50,000 cells per well in a 6-well tissue culture dish (Corning) in triplicate. The growth of cells was monitored for 14-16 days. Cell number and viability were determined using a hemacytometer.
Cells at 80–90% confluence were detached and seeded in triplicate at a density of 100,000 cells per well in a 6-well tissue culture dish. A scratch was made in the shape of an octothorpe (#) using 10 μL pipette tips. Cells were kept in serum-free media to minimize effects of proliferation. Phase contrast photomicrographs were taken using a Nikon Eclipse inverted microscope (Nikon, Chiyoda-ku, Tokyo) equipped with the QICAM Mono capture device (Media Cybernetics, Inc., Bethesda, MD). Four images were taken, one from each intersection, at 0 and 8 hr for MDA-MB-231 cell lines. Initial and final distances were measured using QCapture Pro software (Media Cybernetics, Inc., Bethesda, MD). Parent and vector control cell lines and SUDS3-transfected clones were compared using Dunn's comparison test. Calculations were performed using SigmaStat statistical analysis software (SPSS Inc., Chicago, IL). Statistical significance was defined as a probability p ≤ 0.05.
2.5. Experimental metastasis assay
Experimental metastasis assays were performed as previously described [38]. Ten mice per experimental group were initially injected with 5 × 105 cells. Animals were maintained under the guidelines of the National Institutes of Health and the University of Alabama at Birmingham. All protocols were approved by the Institutional Animal Care and Use Committee. Food and water were provided ad libitum. ANOVA calculations were performed using SigmaStat statistical analysis software (SPSS Inc., Chicago, IL). Statistical significance was defined as a probability p ≤ 0.05.
3. Results
3.1. SUDS3 expression does not correlate with metastasis
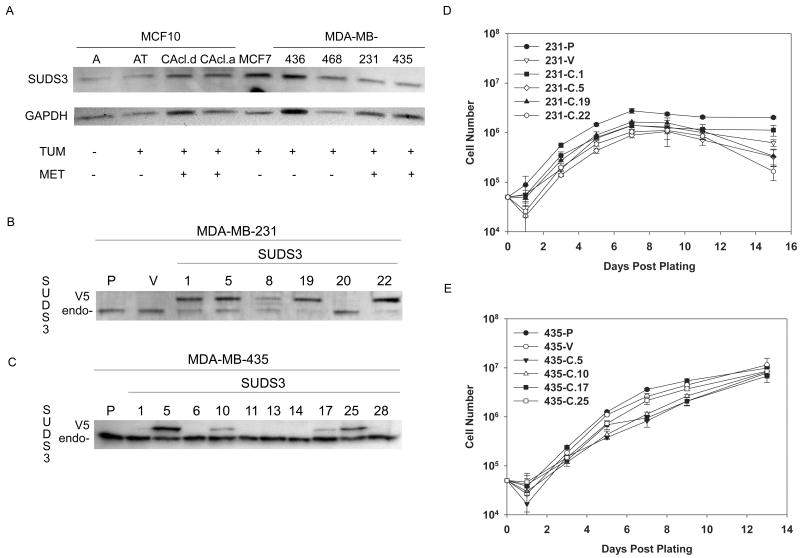
Previous studies have shown that BRMS1 re-expression in metastatic cells blocks metastasis without blocking orthotopic tumor growth [4;7;39]. Immunohistochemical analysis showed that BRMS1 expression was inversely correlated with prognosis and metastasis in a subset of human breast cancers [40]. Based primarily upon relatedness of BRMS1 and SUDS3, we hypothesized that SUDS3 shared BRMS1 metastasis suppressor as well as other functions. Levels of SUDS3 were measured in multiple human breast cell lines using a polyclonal antibody generated specifically against SUDS3. SUDS3 (45 kDa) was present in all of the cell lines examined, regardless of their tumorigenicity or metastatic potential (Fig. 1A). Although all lanes were loaded with equal amounts of whole cell lysate protein, probing with antibodies directed against housekeeping proteins (i.e., GAPDH; β-actin, α-tubulin) exhibited variability among cell lines, suggesting that we have not yet identified a consistent loading control. Nonetheless, it is apparent that no gross trend in expression levels was observed with tumor progression.
Fig. 1.
SUDS3 expression does not correlate with tumor progression or metastatic potential (A). A panel of human breast cell lines were probed with anti-SUDS3 antiserum. Endogenous SUDS3 was ubiquitously present. Levels varied from experiment to experiment, but did not correlate with metastatic potential. Blots were re-probed with GAPDH to verify equal loading. MDA-MB-231 (B) and -435 (C) cells were stably transfected to express a SUDS3-V5 fusion protein. Western blots with SUDS3 specific antiserum shows an endogenous (endo-) 45 kDa band in all clones. A second band at 50 kDa corresponding to the fusion protein (V5) was verified with anti-V5 antibody (data not shown). Ectopic over-expression of SUDS3 did not affect the in vitro growth of MDA-MB-231 cells (D) or -435 (E) cells when compared to parental and vector controls.
3.2. Ectopic expression of SUDS3 in MDA-MB-231 and MDA-MB-435 cells does not affect proliferation
To examine whether ectopic of SUDS3 affected in vitro cell growth or phenotypes associated with BRMS1 metastasis suppression, stable MDA-MB-231 and -435 breast cancer cell lines were generated to ectopically express a SUDS3-V5/His fusion protein. Several clones were isolated and SUDS3 expression was evaluated by immunoblot. Endogenous SUDS3 (45 kDa) and SUDS3-V5 (∼ 50 kDa) were detected with anti-SUDS3 (Fig. 1B & 1C). Since all of the clones were derived from the same parental population, endogenous SUDS3 was used as a loading control to assess ectopic SUDS3 expression. The identity of the 50 kDa band was verified using an anti-V5 antibody (data not shown). Ectopic SUDS3 expression did not affect in vitro growth rates or saturation densities (Fig. 1D & 1E). Similarly, no gross differences in morphology were observed (data not shown). Several clones of each cell line were selected to represent varying levels of ectopic SUDS3 expression in MDA-MB-435 (clones 5, 10, 17, and 25) and -231 (clones 1, 5, and 22) cells.
3.3. Ectopic expression of SUDS3 does not affect motility of MDA-MB-231 cells
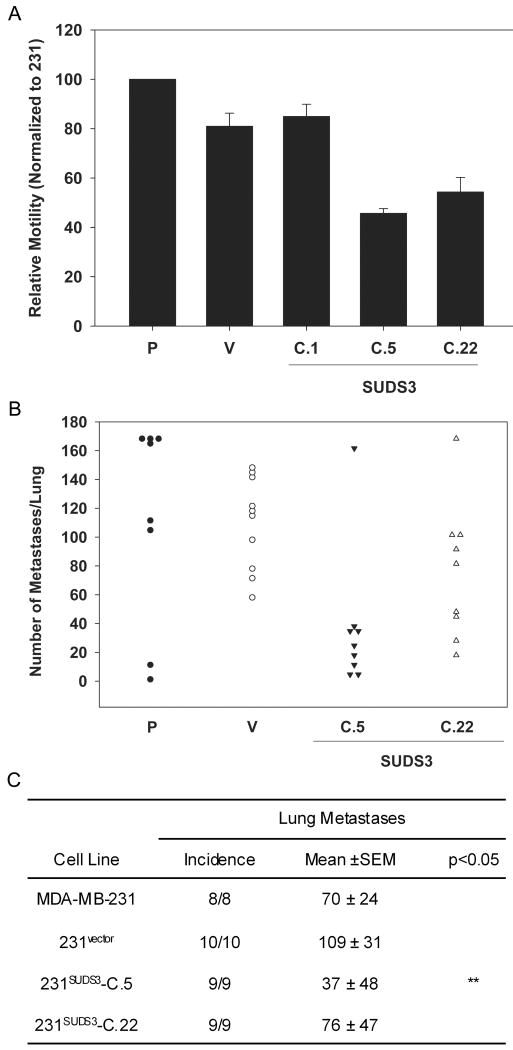
Motility is required for tumor cell invasion and metastasis. 231BRMS1 cells showed a modest, but significant inhibition (∼60%) of motility as measured using an in vitro wound healing assay [41]. To determine whether ectopic expression of SUDS3 affected in vitro motility in MDA-MB-231 cells, a similar in vitro wounding/motility assay was performed. SUDS3 did not alter motility compared to parental or vector controls (Fig. 2A).
Fig. 2.
SUDS3 does not significantly nor consistently suppress motility or metastatic behavior of MDA-MB-231 breast carcinoma cells. (A) Motility was measured using an in vitro scratch/wound healing assay. Confluent MDA-MB-231 monolayers were scratched and distances from edge to edge were measured at 0 and 8 hr. Relative motility is normalized to parental MDA-MB-231 (P) cells. Vector control cells (V) and three selected 231SUDS3 clones are shown. Data are cumulative for 3 independent experiments with replicate wells. (B) Lung colonization of MDA-MB-231 (P), vector control (V), and 231SUDS3 transfectants (C.5 and C.22) was measured following i.v. of 5 × 105 cells into athymic mice. Each symbol represents the number of surface lung metastases (maximum counted = 170) per mouse. (C) Summary statistics of experimental metastasis for data represented in panel B. One-way analysis of variance with Dunn's post-test was used to determine differences among groups (** = p<0.05).
3.4. Ectopic expression of SUDS3 does not consistently suppress metastasis of MDA-MB-231 cells
To examine whether over-expression of SUDS3 could suppress metastasis, representative clones of 231SUDS3 were injected into the lateral tail vein of athymic mice. Formation of macroscopic lung metastases was assessed as described previously [4;7;42]. 231SUDS3 clones 5 and 22 produced an average of 37 and 76 lung colonies/lung compared to 70-109 lung colonies in parental and vector controls (Fig. 2B & 2C). The size of the lung colonies were approximately equal for all of the cells. The equivalence of clone 22 to parental and vector cells demonstrates that, in this model, over-expression of SUDS3 does not suppress metastasis.
3.5. Ectopic expression of SUDS3 does not reduce OPN or EGFR
OPN is a secreted glycoprotein that can bind cell surface receptors to promote cell adhesion and migration. High expression of OPN generally correlates with aggressive tumor cell behavior and poor prognosis [43]. There exist various forms of endogenous osteopontin due to differential RNA splicing, protein modification, and susceptibility to proteases [44;45]. Ectopic expression of BRMS1 in MDA-MB-435 cells decreased OPN mRNA and protein by 90- 95% [4;14;15]. OPN down-regulation is crucial to BRMS1 metastasis suppression since restoration of OPN in 435BRMS1 cells resulted in increased incidence of spontaneous metastasis to lymph nodes and lungs [15].
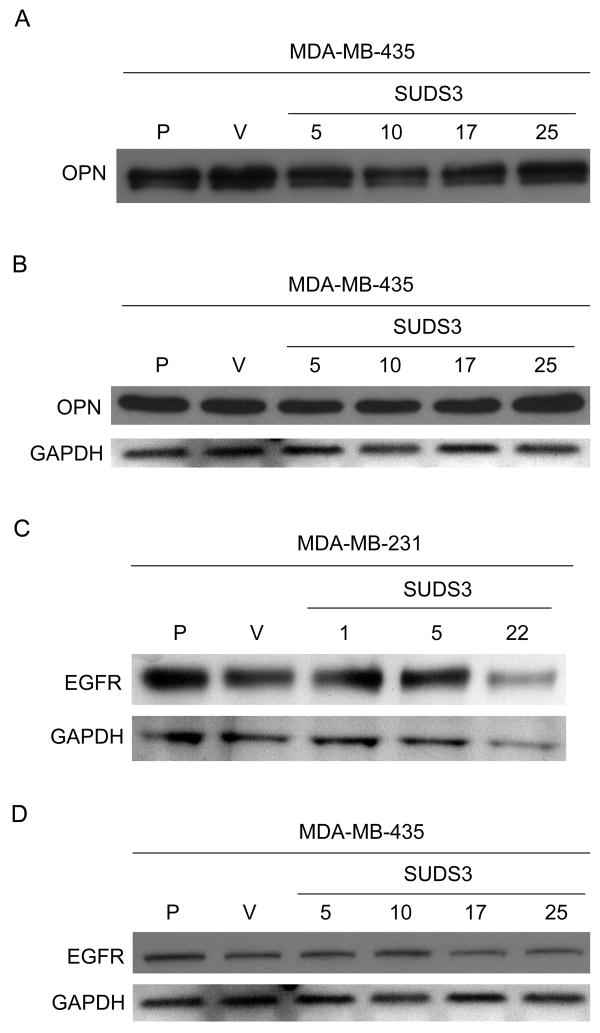
To assess whether OPN was similarly affected by SUDS3, 435SUDS3 cell-conditioned media and whole cell lysate was collected from serum-starved cells. Immunoblots with anti-OPN identified a single band at 50 kDa in the whole cell lysate and two bands between 40 and 70 kDa in conditioned media as previously described [15]. Ectopic expression of SUDS3 did not decrease either intracellular or secreted levels of OPN (Fig. 3A & 3B).
Fig. 3.
SUDS3 does not affect levels of secreted (A) or intracellular (B) OPN or EGFR in MDA-MB-231 (C) or -435 (A, B, D) cells. GAPDH was evaluated as a control for equal loading (B, C, D), while secreted OPN measurements were normalized by cell number (A).
The EGF receptor tyrosine kinase activates a variety of signaling transduction pathways that affect cell proliferation, differentiation, adhesion, migration, and apoptosis. EGFR is expressed in patients with breast cancer and activated EGFR is often associated with poor patient survival in invasive breast cancer [46]. BRMS1 significantly decreased EGFR in MDA-MB-231 and -435 cells by ∼50 to 100%, respectively [13]. However, ectopic over-expression of SUDS3 did not change EGFR when comparing parental, vector, and SUDS3 over-expressing MDA-MB-231 and -435cells (Fig. 3C).
4. Discussion
Functional and transcriptional compensation among protein families is a well-characterized phenomenon, as clearly demonstrated by the Rb family of proteins for example [47]. The studies reported here were undertaken, in part, because of the strong homology between SUDS3 and BRMS1 and the involvement of both proteins in SIN3-HDAC chromatin remodeling complexes. These facts suggested that perhaps SUDS3 and BRMS1 share some functional redundancy and that SUDS3 could compensate for BRMS1 deficiency. Not only did SUDS3 not suppress metastasis like BRMS1, but their apparent functions and/or regulation of chromatin remodeling-based transcriptional regulation differed as well.
We chose to isolate clones from the transfected MDA-MB-231 population in order to address phenotypic changes related to tumor heterogeneity and dose-dependency. That SUDS3 transfectant clones were not universally low (compared to parental MDA-MB-231) for metastatic ability or motility allows the conclusion that SUDS3 is not a metastasis suppressor. Differences among clones are most likely attributable to genetic instability and tumor cell heterogeneity. Taken together, these experiments clearly show that BRMS1 and SUDS3, while sharing the Sds3 domain and participating in similar (and sometimes even the same) SIN3-HDAC complexes, have distinct functions in the breast cancer cell lines examined.
Among the many proteins that comprise the SIN3-HDAC core complexes, there are several that have high sequence similarity for which both overlapping and distinct functions have already been, and are continually being, discovered. These proteins include SIN3A/B, HDAC1/2, ARID4A/B, and Rbbp4/7 [48;49]. Similarly, although BRMS1 and SUDS3 did not share overlapping functions for all of the phenotypes examined, there may be other shared characteristics. Nonetheless, our findings emphasize BRMS1 and SUDS3 distinctness and further emphasize that the diversity and mix-and-match nature of chromatin remodeling machinery remains relatively ill-defined. Further defining the binding partners and functions of the myriad proteins in the larger SIN3-HDAC complexes is essential.
While SUDS3 is necessary for normal somatic cell survival [23], there have been no reports to date on levels of endogenous SUDS3 in breast cell lines. SUDS3 is expressed in every breast cell line examined. SUDS3 expression did not appear to change to compensate for loss of BRMS1 expression in the MDA-MB-231 and -435 cell lines. Not shown here, we frequently observed a second SUDS3 band in MCF10-derived cells (data not shown). Thus, modified variants may exist within these cells, however, additional experimentation is required in order to identify and characterize this band.
A possible explanation for the apparent differences between SUDS3 and BRMS1 relates to protein-protein interactions. A part of the originally defined Sds3 domain has been shown in separate studies to be necessary for the homodimerization of murine SUDS [21]. SUDS3 directly interacts with BRMS1 as shown in yeast two-hybrid genetic screens [3]. Studies are currently underway to determine whether the Sds3 domain is responsible for direct binding of BRMS1 to SUDS3 or whether BRMS1-SUDS3 interactions occur at other portions of each molecule. Although the data presented here do not support our original hypothesis - that SUDS3 is a metastasis suppressor - the collective findings begin to address the role(s) that the Sds3 domain plays in these individual proteins and in the formation/regulation of the SIN3-HDAC complexes.
Acknowledgments
This study was supported by funding from the National Institute of Health: CA87728, (D.R.W.), F32CA113037, (D.R.H.), and the National Foundation for Cancer Research Center for Metastasis Research. Special thanks to members of the Welch lab for assistance in preparation of this manuscript. This work is submitted in partial fulfillment of the requirements of the UAB Molecular and Cellular Graduate Program (A.C.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meehan WJ, Welch DR. Breast cancer metastasis suppressor 1: Update. Clin Exptl Metastasis. 2003;20:45–50. doi: 10.1023/a:1022542519586. [DOI] [PubMed] [Google Scholar]
- 2.Nikolaev AY, Papanikolaou NA, Li M, Qin J, Gu W. Identification of a novel BRMS1-homologue protein p40 as a component of the mSin3A/p33(ING1b)/HDAC1 deacetylase complex. Biochem Biophys Res Comm. 2004;323:1216–1222. doi: 10.1016/j.bbrc.2004.08.227. [DOI] [PubMed] [Google Scholar]
- 3.Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 4.Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR. Alterations of breast cancer metastasis suppressor 1:at rich interactive domain 4a interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–7444. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapoor P, Saunders MM, Li Z, Zhou Z, Schaeffer N, Kunze EL, Samant RS, Welch DR, Donahue HJ. Breast cancer metastatic potential: Correlation with increased heterotypic gap junctional intercellular communication between breast cancer cells and osteoblastic cells. Int J Cancer. 2004;111:693–697. doi: 10.1002/ijc.20318. [DOI] [PubMed] [Google Scholar]
- 6.Saunders MM, Seraj MJ, Li ZY, Zhou ZY, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- 7.Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809–817. doi: 10.2353/ajpath.2008.070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Molec Cell Biol. 2006;26:8683–8696. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exptl Metastasis. 2001;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- 10.Champine PJ, Michaelson J, Weimer B, Welch DR, DeWald DB. Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exptl Metastasis. 2007;24:551–565. doi: 10.1007/s10585-007-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–4018. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 12.Cicek M, Samant RS, Kinter M, Welch DR, Casey G. Identification of metastasis-associated proteins through protein analysis of metastatic MDA-MB-435 and metastasis-suppressed BRMS1 transfected-MDA-MB-435 cells. Clin Exptl Metastasis. 2004;21:149–157. doi: 10.1023/b:clin.0000024729.19084.f0. [DOI] [PubMed] [Google Scholar]
- 13.Vaidya KS, Harihar S, Stafford LJ, Hurst DR, Hicks DG, Casey G, DeWald DB, Welch DR. Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J Biol Chem. 2008 doi: 10.1074/jbc M710068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedley BD, Welch DR, Allan AL, Al-Katib W, Dales DW, Postenka CO, Casey G, MacDonald IC, Chambers AF. Downregulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int J Cancer. 2008;123:526–534. doi: 10.1002/ijc.23542. [DOI] [PubMed] [Google Scholar]
- 16.Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-κB activity. Cancer Res. 2005;65:3586–3595. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer. 2006;16:522–531. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 18.Vannier D, Balderes D, Shore D. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics. 1996;144:1343–1353. doi: 10.1093/genetics/144.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lechner T, Carrozza MJ, Yu Y, Grant PA, Eberharter A, Vannier D, Brosch G, Stillman DJ, Shore D, Workman JL. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3*Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J Biol Chem. 2000;275:40961–40966. doi: 10.1074/jbc.M005730200. [DOI] [PubMed] [Google Scholar]
- 20.Vannier D, Damay P, Shore D. A role for Sds3p, a component of the Rpd3p/Sin3p deacetylase complex, in maintaining cellular integrity in Saccharomyces cerevisiae. Mol Genet Genom. 2001;265:560–568. doi: 10.1007/s004380100447. [DOI] [PubMed] [Google Scholar]
- 21.Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Molec Cell Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Molec Cell Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David G, Dannenberg JH, Simpson N, Finnerty PM, Miao L, Turner GM, Ding Z, Carrasco R, DePinho RA. Haploinsufficiency of the mSds3 chromatin regulator promotes chromosomal instability and cancer only upon complete neutralization of p53. Oncogene. 2006;25:7354–7360. doi: 10.1038/sj.onc.1209734. [DOI] [PubMed] [Google Scholar]
- 25.Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Comm. 2006;348:1429–1435. doi: 10.1016/j.bbrc.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiio Y, Rose DW, Aur R, Donohoe S, Aebersold R, Eisenman RN. Identification and characterization of SAP25, a novel component of the mSin3 corepressor complex. Mol Cell Biol. 2006;26:1386–1397. doi: 10.1128/MCB.26.4.1386-1397.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Guezennec X, Vermeulen M, Stunnenberg HG. Molecular characterization of Sin3 PAH-domain interactor specificity and identification of PAH partners. Nucleic Acids Res. 2006;34:3929–3937. doi: 10.1093/nar/gkl537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cailleau R, Young R, Olive M, Reeves WJ. Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;53:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cailleau R, Olive M, Cruciger QVJ, Cailleau R, Olive M, Cruciger QVJ. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14:911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 31.Sellappan S, Grijalva R, Zhou XY, Yang WT, BarEli M, Mills GB, Yu DH. Lineage infidelity of MDA-MB-435 cells: Expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- 32.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2006;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 33.Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 34.Shekhar MP, Tait L, Pauley RJ, Wu GS, Santner SJ, Nangia-Makker P, Shekhar V, Nassar H, Visscher DW, Heppner GH, Miller FR. Comedo-ductal carcinoma in situ: A paradoxical role for programmed cell death. Cancer Biol Ther. 2008;7 doi: 10.4161/cbt.7.11.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauley RJ, Soule HD, Tait L, Miller FR, Wolman SR, Dawson PJ, Heppner GH. The MCF10 family of spontaneously immortalized human breast epithelial cell lines: models of neoplastic progression. Eur J Cancer Prev. 1993;2 3:67–76. [PubMed] [Google Scholar]
- 36.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 37.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:1185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 38.Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exptl Metastasis. 1997;15:272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- 39.Locke D, Perusinghe N, Newman T, Jayatilake H, Evans WH, Monaghan P. Developmental expression and assembly of connexins into homomeric and heteromeric gap junction hemichannels in the mouse mammary gland. J Cell Physiol. 2000;183:228–237. doi: 10.1002/(SICI)1097-4652(200005)183:2<228::AID-JCP9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri T, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G. Loss of BRMS1 protein expression predicts reduced disease-free survival in hormone receptor negative and HER2 positive subsets of breast cancer. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weng LP, Smith WM, Dahia PLM, Ziebold U, Gil E, Lees JA, Eng C. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59:5808–5814. [PubMed] [Google Scholar]
- 42.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 43.Tuck AB, Chambers AF, Allan AL. Osteopontin overexpression in breast cancer: Knowledge gained and possible implications for clinical management. J Cell Biochem. 2007;102:859–868. doi: 10.1002/jcb.21520. [DOI] [PubMed] [Google Scholar]
- 44.Denhardt DT, Guo X. Osteopontin: a protein with diverse functions. FASEB J. 1993;7:1475–1482. [PubMed] [Google Scholar]
- 45.Patarca R, Saavedra RA, Cantor H. Molecular and cellular basis of genetic resistance to bacterial infection: the role of the early T-lymphocyte activation-1/osteopontin gene. Crit Rev Immunol. 1993;13:225–246. [PubMed] [Google Scholar]
- 46.Magkou C, Nakopoulou L, Zoubouli C, Karali K, Theohari I, Bakarakos P, Giannopoulou I. Expression of the epidermal growth factor receptor (EGFR) and the phosphorylated EGFR in invasive breast carcinomas. Breast Cancer Res. 2008;10:R49. doi: 10.1186/bcr2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14–36. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 49.McDonel P, Costello I, Hendrich B. Keeping things quiet: Roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol. 2008 doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]