Abstract
Worldwide, breast cancer is the most frequently occurring malignancy in women. Early age at full term pregnancy has a protective effect against breast cancer. Evidence coming from a rat breast cancer model suggests a possible role for the pregnancy hormone hCG, a ligand of the LH receptor, as a mediator for this effect. In a previous study, we found that a common polymorphism in the LH receptor associates with tumor progression in premenopausal breast cancer patients, as carriers of the variant receptor showed a shorter disease free survival compared to non-carriers.
How hCG and its receptor exert their effects on breast cancer, however, is unclear. One possibility is that these effects take place through LH receptors present in the ovaries, thereby influencing steroid hormone production. Another possibility is that the effects take place through LH receptors present in breast tumor cells itself, as some studies have detected the receptor in both normal and neoplastic breast tissues and in breast cancer cell lines.
To investigate whether a direct effect of LH signalling in breast cancer is likely, we measured LH receptor mRNA expression levels in 1551 breast tumors and 42 different human breast cancer cell lines using a qRT-PCR with a wide dynamic range. In addition, associations between LH receptor expression and clinicopathologic factors were investigated.
Assay validation showed that as little as ∼10 copies per reaction volume of LH receptor cDNA could still be detected by our assay. We show that LH receptors are undetectable in 62% of breast tumor samples and 41 of 42 breast cancer cell lines. For the remaining samples we found expression levels to be very low. Although low, expression of the LH receptor appears to be associated with normal breast cells, favorable tumor characteristics and low tumor percentage.
Since expression of the LH receptor in breast cancer cells is very low, it is almost excludes the possibility of direct signaling effects. We therefore conclude that signaling effects of the LH receptor on breast cancer most likely take place by an indirect pathway through the ovaries.
Keywords: breast cancer, LH receptor, prognosis, qRT-PCR
Introduction
Breast cancer is the most common malignancy in women around the world, with an estimated number of 1.5 million new cases to occur in the year 2010 [1]. While incidence rates are highest in western industrialized countries, an increase in incidence rates is seen in many developing countries, most likely as a consequence of the adoption of western lifestyles [2].
Several risk factors for developing breast cancer have been identified and established over the years. Gender, older age and carriership of high risk susceptibility genes (i.e. BRCA1 or BRCA2) each give rise to a more than ten fold increase in risk [3,4]. In addition to these, factors related to hormone exposure, especially estrogens, and several reproductive factors are also highly associated with the disease [3,5].
A reproductive factor that has frequently been associated with breast cancer risk is a woman's age at birth of her first child [3,6-8]. It was shown that a linear association exist between age at first full term pregnancy and breast cancer risk, a younger age having a protective effect against the disease [8]. However, the mechanism by which this protective effect takes place is largely unknown. It has been postulated that the protective effect of pregnancy on breast cancer is mediated by the hormone hCG [9,10]. hCG is a glycoprotein hormone, produced by the placenta during early pregnancy. Structurally and functionally, hCG is very similar to LH and both hormones exert their effects through the common LH/CG receptor [11].
Evidence for a protective effect of hCG on breast cancer was found by Russo and coworkers [9,10]. In a rat model for breast cancer, tumor induction with the chemical 7,12-dimethylbenz[α]anthracene (DMBA) was more effective in nulliparous than parous animals. Notwithstanding the fact that CG does not naturally occur in rats, Russo and coworkers found that a similar protective effect could be obtained when nonparous animals were pretreated with hCG for 21 days, a period equivalent to full-term pregnancy in rats [9,10]. These results suggest that both pregnancy and treatment with hCG have a protective effect against mammary tumors and tumor progression when given after tumor induction [9,10].
In addition to the findings that hCG has protective effects on breast cancer progression, evidence that the LH receptor is involved in tumor progression as well has came from studies performed by our research group [12-14]. In two independent cohorts, breast cancer patients that are carrier of a common polymorphism in the LH receptor, the insLQ variant, show a significantly shorter disease free survival compared to non-carriers [13,14]. Interestingly, the adverse effect of the insLQ variant on disease free survival was observed in premenopausal women only [14]. In addition to this, it was shown that the insLQ variant is a functional polymorphism, as transfection studies revealed that the variant receptor shows both a higher surface expression and a slight increase in receptor sensitivity after stimulation with LH or hCG when compared to the more common nonLQ variant [13].
These apparent contradictory results indicate that hCG and its target receptor play a role in breast cancer progression. However, the mechanisms of these effects remain unclear. A possibility would be that the effects are mediated by LH receptors present in the ovaries, thereby influencing the production of ovarian hormones, subsequently leading to either a protective or adverse effect on breast cancer progression. However, an effect on disease progression of LH/hCG through LH receptors present in breast cancer tissue cannot be excluded.
Although LH receptors have long been considered to be expressed exclusively in ovarian and testicular tissues, over the last decade several studies have been published that claim their presence also in both normal and neoplastic breast tissues [15-17], and in breast cancer cell lines [15,18-23]. In these studies, LH receptor expression was determined using qualitative techniques such as immunohistochemistry and non-quantitative RT-PCR. In one study, on 160 breast cancer specimens, this qualitative immunohistochemical assessed LH receptor expression was associated with younger age at diagnosis, premenopausal status, lower grade tumors, lobular invasive carcinomas and the presence of estrogen receptors [16].
In the present study we investigated whether a direct effect of LH receptor signaling in breast cancer is probable. In order to do this, we have determined LH receptor mRNA expression levels using a specific and quantitative method on a large cohort of 1551 breast tumors and on a panel of 42 different breast cancer cell lines. We found that expression levels are in most cases undetectable or extremely low. If present, intriguingly, these low LH receptor expression levels are associated with favorable tumor characteristics.
Materials and Methods
Breast tumor samples
The study design was approved by the Medical Ethics Committee of the Erasmus MC (Rotterdam, The Netherlands; MEC 02.953). It included breast tumor tissue specimens of 1,683 female patients with primary operable breast cancer. Frozen tumor samples were processed from patients with breast cancer who entered the clinic between 1979 and 1996. Details on follow-up and analyses are given in [24]. Tumors with specimens of poor RNA quality were excluded from analysis [25] and as a consequence tumor specimens of 1551 patients (92%) were analyzed for LH receptor expression. The cut-off point to classify primary breast tumors as ER and/or progesterone receptor (PgR) –positive was 10 fmol / mg cytosolic protein.
Breast cancer cell lines
cDNA samples derived from 42 different human breast cancer cell lines were a kind gift of Dr. Mieke Schutte (Department of Internal Oncology, Erasmus MC, Rotterdam, the Netherlands). Detailed information about the origin of these cell lines has been described by Elstrodt et al. [26].
TRex-hLHR cell line
TRex-hLHR cells were used to provide a stable cell line expressing the LH receptor and were established as described previously [27]. To induce expression of the LH receptor, cells were plated in Costar six-well plates (Corning Inc., Corning, NY) and treated overnight with 15 ng / ml tetracycline (Sigma, St. Louis, MO), after which RNA was isolated.
LH receptor plasmid DNA
Construction of the expression plasmid pSG5-LHRWT is described by Kraaij et al. [28].
RNA isolation and cDNA synthesis
RNA isolation and cDNA synthesis from breast tumor samples and breast cancer cell lines were performed as described previously by Sieuwerts et al [25].
For the TRex-hLHR and testis samples, total RNA was extracted with Trizol (Invitrogen, Breda, the Netherlands) according to the manufacturers protocol and treated for 30 minutes at 37 °C with DNase (Promega, Leiden, the Netherlands). Thereafter, for the TRex-hLHR samples, reverse transcription was performed using 1.5 μg of total RNA aliquots in a final volume of 40 μl and subsequent RNase H-treatment was performed. For the testis samples, reverse transcription was performed using 1 μg of total RNA aliquots in a final volume of 25 μl without subsequent RNase H- treatment.
qRT-PCR
qRT-PCR reactions were performed in 25-μl reaction volume on an MX 3000P Stratagene System, in accordance with the manufacturer's protocol. A commercially available Assay-on-Demand kit (Applied Biosystems) was used for the LH receptor (Hs00174885_m1) on all samples. This Assay-on-Demand kit targets the exons 7 and 8 of the LH receptor mRNA transcripts. Although the method does not detect alternatively spliced LH receptor transcripts not containing exons 7 and 8, such transcripts are not relevant to the present study, since the amino acids encode by exons 7 and 8 are essential to LH receptor protein function [11]. For the breast tumor samples and testis samples, three reference genes (ie, porphobilinogen deaminase (PBGD), hypoxanthine-guanine phospho-ribosyltransferase (HPRT), and β-2-microglobulin (β2M)) were determined to allow for correction for differences in total RNA content between samples. Primer sequences and amplification protocol for these reference genes have been previously described by Sieuwerts et al [25].
Forty-five rounds of amplification were performed according to the supplier's protocol, and at the end of every round of amplification fluorescent signals of the TaqMan probes (Applied Biosystems) were acquired and used to generate cycle threshold (Ct) values from which mRNA expression levels were calculated when applicable.
Measurements were performed in triplicate for both the TRex-hLHR cDNA and LH receptor plasmid dilution series. For the breast tumor and testis samples, expression levels of the LH receptor were determined in duplicate and triplicate respectively, while expression levels of each of the three reference genes were determined only once. A relative copy number of the LH receptor, normalized against the three reference genes, was calculated as follows: relative copy number = 2(mean Ct Ref − mean Ct LHR). We were able to reliably measure LH receptor expression up to 37 amplification rounds using a dilution series of LH receptor cDNA derived from the TRex-hLHR cell line. We therefore used a relative copy number of 2-16, approximating a Ct value of 37, as detection threshold for the LH receptor. Whenever LH receptor quantification rounds exceeded this value, quantities were considered to be undetectable and measurements were considered to be invalid. In case that two valid measurements were obtained for a breast tumor, the tumor was classified as having ‘detectable’ expression. In case that only one valid measurement was obtained for a tumor, expression was classified as ‘inconclusive’ and a relative copy number was computed as described above using only the valid measurement. In case that two invalid measurements were obtained for a tumor, no relative expression level could be calculated and expression was classified as ‘not detectable’. For the breast cancer cell lines, crude Ct values were used for analysis.
Data analysis and statistics
Graphpad Prism version 3.02 for Windows (Graphpad Software, Inc., San Diego, CA) was used to generate linear fits through the data points obtained from the qRT-PCR on the dilution series of the TRex-hLHR cDNA and LHR plasmid DNA. Slopes were held constant to −1/log2, corresponding to the expected decrease of one Ct per two-fold increase in DNA content.
Associations between LH receptor expression status and the various patient and tumor characteristics were investigated using Pearson's χ2 test. All tests were two-tailed, and statistical significance was assumed at P ≤ 0.05. Statistical analyses were performed with the STATA statistical package, release 9.2 (STATA Corp, College Station, TX).
Results
Assay Validation
The range of Ct values was defined within which LH receptor expression levels can be adequately determined by our qRT-PCR. For this purpose a 4-fold dilution series was made of cDNA derived from TRex-hLHR cells stably transfected with a wild type human LH receptor expression construct and qRT-PCR was performed (Figure 1A). A straight line with slope held constant to -1/log 2 (corresponding to a decrease of 1 Ct value per 2-fold increase in DNA content) was fitted through the data points. We concluded that the assay runs linear up to a Ct value of approximately 37, which was subsequently defined as the detection threshold of our assay.
Figure 1.
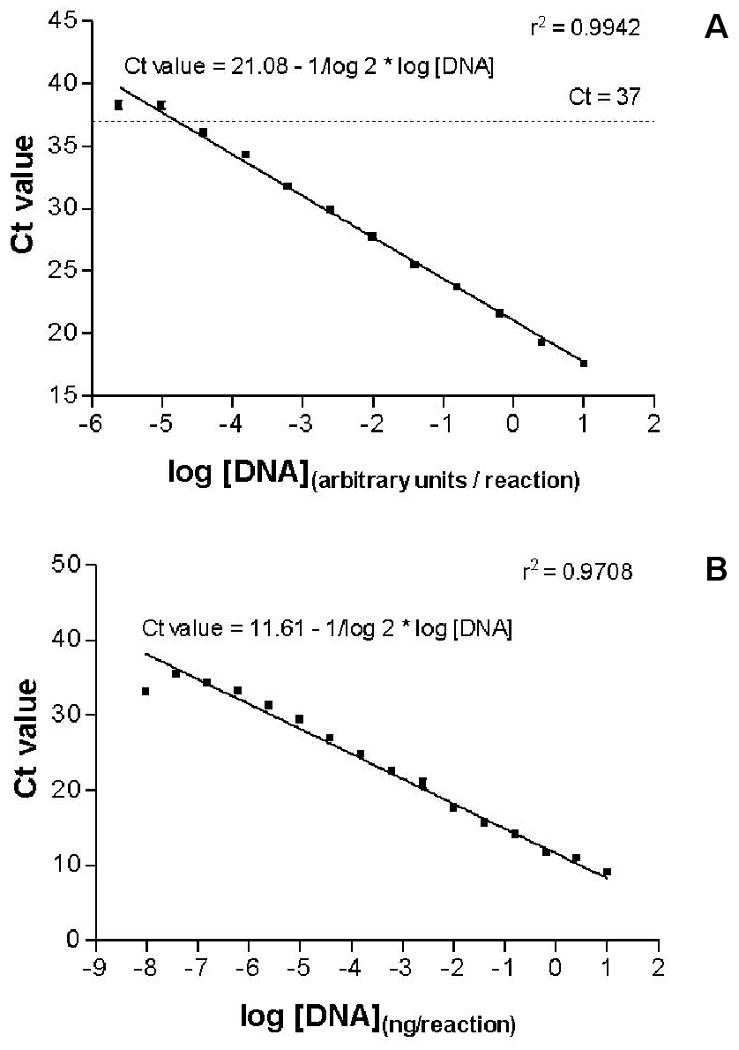
Cycle threshold values obtained by performing the LH receptor qRT-PCR on a dilution series of cDNA derived from the LH receptor expressing TRex-hLHR cell line (panel A) and a dilution series of plasmid DNA containing cDNA encoding the LH receptor (panel B). Values are presented as means ± SE. A straight line with slope held constant to −1/log2 (corresponding to a decrease of one Ct per two-fold increase in DNA content) was fitted through the data points.
To study the minimum number of LH receptor cDNA copies that can still be adequately detected by the LH receptor qRT-PCR, a dilution series was made of plasmid DNA encoding the LH receptor. A straight line fit as described above showed that the assay runs linear largely across the whole dilution series and that as little as ∼10 copies (∼6 fg) of plasmid DNA per reaction could still be detected by the qRT-PCR (Figure 1B).
For comparison expression of LH receptor mRNA in a human testis sample was determined using the identical qRT-PCR technique. A reference-corrected copy number of 5.64·10-3 for the LH receptor in the testis tissue sample was found (Figure 2)
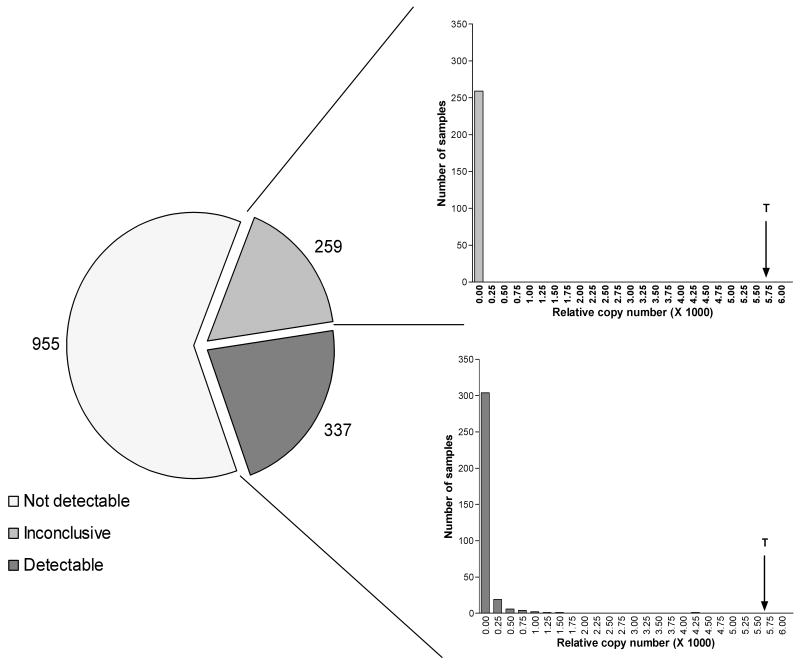
Figure 2.
LH receptor expression levels in 1551 human breast tumors. Pie chart shows the number of tumors within each category of expression (Panel A). Histograms show the expression levels for tumors in the ‘inconclusive’ (Panel B) and ‘detectable’ (Panel C) expression category. For comparison, the level of LH receptor expression as determined on a human testis specimen has been indicated by an arrow; marked with a ‘T’.
Expression of the LH receptor in breast cancer cell lines
Of the 42 breast cancer cell lines investigated, only five cell lines generated a Ct value after the maximum number of 45 amplification rounds. However, only one cell line, MDA-MB-415, was positive since it generated a Ct value that did not exceed the detection threshold of 37 cycles (Table 1).
Table 1.
Cycle threshold values as a measure for LH receptor mRNA expression in a panel of 42 different breast cancer cell lines.
| Cell line | Ct value | Cell line | Ct value |
|---|---|---|---|
| BT20 | ND | MPE600 | ND |
| BT474 | ND | OCUB-F | ND |
| BT483 | ND | OCUB-M | ND |
| BT549 | ND | SK-BR-3 | ND |
| CAMA-1 | ND | SK-BR-5 | ND |
| DU4475 | ND | SK-BR-7 | ND |
| EVSA-T | ND | SUM44PE | ND |
| HCC1937 | ND | SUM52PE | ND |
| HS578T | inconclusivea | SUM102PT | ND |
| MCF-7 | ND | SUM149PT | inconclusivea |
| MDA-MB-134VI | ND | SUM159PT | ND |
| MDA-MB-157 | inconclusivea | SUM185PE | ND |
| MDA-MB-175VII | ND | SUM190PT | ND |
| MDA-MB-231 | inconclusivea | SUM225CWN | ND |
| MDA-MB-330 | ND | SUM229PE | ND |
| MDA-MB-361 | ND | SUM1315MO2 | ND |
| MDA-MB-415 | 35.89 | T47D | ND |
| MDA-MB-435S | ND | UACC812 | ND |
| MDA-MB-436 | ND | UACC893 | ND |
| MDA-MB-453 | ND | ZR75-1 | ND |
| MDA-MB-468 | ND | ZR75-30 | ND |
Measurement exceeds the detection threshold of 37 but was below 45 cycles, and expression should therefore be considered inconclusive. Cell lines that did not generate a Ct value after 45 rounds of amplification are indicated with ‘Not detectable’ (ND).
LH receptor expression in breast tumors
LH receptor expression levels were determined in 1551 breast tumor samples by qRT-PCR (Figure 2). In 955 tumors (62%), LH receptor cDNA could not be detected. For 259 tumors (17%), only one of the two measurements was considered valid. Values found in the latter tumors fall in the lowest copy number category (0-0.025), indicating that LH receptor expression is very low in these tumors (Figure 2B). Moreover, 90% of the 337 remaining tumors with detectable expression also fall in the lowest expression category (Figure 2C). Only 33 tumors (approximately 2% of all breast cancer specimens studied) show a higher copy number. Remarkably, one tumor shows a relatively high expression of the LH receptor in comparison to the other tumors with a relative copy number of 4.78·10-3 (Figure 2C).
LH receptor status in relation to patient and tumor characteristics
Associations between receptor status and clinico-pathologic factors are shown in Table 2. Patients were grouped based on LH receptor expression and included 1446 tumors of which LH receptor status and all clinico-pathologic data were available for further analysis. LH receptor expression is significantly associated with older age as well as postmenopausal status.
Table 2.
LH receptor expression in relation to patient and tumor characteristics.
| Number of patients (%) | |||||
|---|---|---|---|---|---|
| LH receptor expression | |||||
| Characteristics | Total population | Not detectable | Inconclusive | Detectable | P valuea |
| Total | 1446 (100) | 886 (61.3) | 245 (16.9) | 315 (21.8) | - |
| Slice tumor percentage | |||||
| 0-29% | 36 (2.5) | 14 (38.9) | 6 (16.7) | 16 (44.4) | |
| 30-70% | 1044 (72.2) | 628 (60.2) | 186 (17.8) | 230 (22.0) | |
| >70% | 366 (25.3) | 244 (66.7) | 53 (14.5) | 69 (18.9) | 0.002 |
| Surgery | |||||
| Lumpectomy | 626 (43.3) | 369 (58.9) | 109 (17.4) | 148 (23.6) | |
| Ablation | 820 (56.7) | 517 (63.0) | 136 (16.6) | 167 (20.4) | 0.238 |
| Tumor size | |||||
| ≤ 2 cm | 469 (32.4) | 247 (52.7) | 96 (20.5) | 126 (26.9) | |
| > 2 cm | 977 (67.6) | 639 (65.4) | 149 (15.3) | 189 (19.3) | 0.000 |
| Lymph nodes involved | |||||
| 0 | 753 (52.1) | 463 (61.5) | 126 (16.7) | 164 (21.8) | |
| 1 to 3 | 314 (21.7) | 184 (58.6) | 60 (19.1) | 70 (22.3) | |
| > 3 | 379 (26.2) | 239 (63.1) | 59 (15.6) | 81 (21.4) | 0.745 |
| Grade | |||||
| Poor | 795 (55.0) | 505 (63.5) | 129 (16.2) | 161 (20.3) | |
| Good/moderate | 210 (14.5) | 117 (55.7) | 41 (19.5) | 52 (24.8) | |
| Unknown | 441 (30.5) | 264 (59.9) | 75 (17.0) | 102 (23.1) | 0.287 |
| Age in categories, years | |||||
| < 41 | 198 (13.7) | 149 (75.3) | 28 (14.1) | 21 (10.6) | |
| 41-55 | 551 (38.1) | 338 (61.3) | 95 (17.2) | 118 (21.4) | |
| 56-70 | 468 (32.4) | 265 (56.6) | 89 (19.0) | 114 (24.4) | |
| > 70 | 229 (15.8) | 134 (58.5) | 33 (14.4) | 62 (27.1) | 0.000 |
| Menopausal status | |||||
| Premenopausal | 504 (34.9) | 347 (68.8) | 72 (14.3) | 85 (16.9) | |
| Perimenopausal | 58 (4.0) | 25 (43.1) | 18 (31.0) | 15 (25.9) | |
| Postmenopausal | 794 (54.9) | 455 (57.3) | 140 (17.6) | 199 (25.1) | |
| Unknown | 90 (6.2) | 59 (65.6) | 15 (16.7) | 16 (17.8) | 0.000 |
| ER statusb,c | |||||
| Negative | 394 (27.6) | 292 (74.1) | 48 (12.2) | 54 (13.7) | |
| Positive | 1032 (72.4) | 584 (56.6) | 193 (18.7) | 255 (24.7) | 0.000 |
| PgR statusb,d | |||||
| Negative | 491 (36.5) | 339 (69.0) | 64 (13.0) | 88 (17.9) | |
| Positive | 854 (63.5) | 482 (56.4) | 164 (19.2) | 208 (24.4) | 0.000 |
Associations were tested using Pearson's χ2 test.
Cut-off value used was 10 fmol/mg cytosolic protein.
ER status was not known for 20 patients.
PgR status was not known for 101 patients.
No associations were found between expression status and surgical procedure, the number of lymph nodes involved or tumor grade. LH receptors are more frequently expressed by tumors that also express the estrogen and/or the progesterone receptor. Furthermore, expression of the LH receptor was significantly more often found in smaller tumors and tumor sections with lower tumor content.
Discussion
To our knowledge this is the first study that addresses expression of the LH receptor in breast cancer specimens on such a large cohort, using 1551 breast tumor samples and 42 different human breast cancer cell lines. Moreover, it is the first to measure expression levels of LH receptor mRNA in breast tumors using a sensitive quantitative real time-PCR with a wide dynamic range.
The sensitivity of this assay to detect even low levels of LH receptor cDNA content was demonstrated by several experiments. First of all, we have shown that LH receptor cDNA levels could be adequately determined on a dilution series of cDNA derived from the TRex-hLHR cell line up to a Ct value of 37, corresponding to an approximately 8.5·106 fold dilution of cDNA obtained from the cDNA synthesis reaction. Secondly, the assay was able to adequately detect physiological levels of the LH receptor in a human testis sample. It should be noted that testicular tissue has a highly heterogeneous composition and that LH receptor-containing Leydig cells only contribute a small fraction of the total number of cells present in the human testis. Finally, using a dilution series made of plasmid DNA containing the LH receptor, we show that the assay is able to detect a low number of approximately 10 plasmid copies per reaction volume.
These results indicate that this assay is highly sensitive and is able to detect very low levels of LH receptor mRNA. Nevertheless, we find that in about two third of the investigated breast tumors LH receptors cannot be detected at all. For the remaining tumors we find very low expression levels, generally more than 20-fold lower than the expression level measured in the testis sample. Of the 42 breast cancer cell lines investigated, only one cell line, MDA-MB-415, generated a Ct value just below the maximum number of 37 amplification rounds. This means that expression is detectable, but very low in this single cell line. The results obtained for the breast cancer cell lines support our findings in the breast tumors, meaning that expression is lost, or very low, in breast cancer specimens.
Few studies have been published over the last decade in which LH receptors were reported to be present both in human breast tumors [15-17] and in breast cancer cell lines MCF-7, MBA-MB-231, T47D and ZR-75-1 [15,18-23,29]. It is possible that in the studies that were able to detect the LH receptor, expression levels in cell lines were low, but detectable, whereas in the cell lines we investigated expression had dropped to levels that are not detectable anymore. Cell lines as they are kept in culture in different laboratories may have evolved and lost or acquired expression of certain genes, such as the LH receptor gene. Indeed, Zhang et al. [29] found low but detectable expression levels of the LH receptor in the MCF-7 cell line using qRT-PCR. The discrepancy between our results and those reported in the literature cannot be explained by limitations in sensitivity. In most studies, less sensitive techniques than the qRT-PCR method employed in the present study were used to detect the LH receptor in breast cancer cell lines, such as conventional RT-PCR, northern and western blotting or immunostaining techniques.
Two studies have been published on the presence of LH receptor in human female breast tumors [15,16], using immunohistochemical methods. In a pilot study [15] conducted on only 19 malignant breast tumors, nine benign breast lesions and three normal breast tissue specimens, using the monoclonal LHR29 antibody for immunostaining, the majority of breast tumors and all benign breast lesions but also normal breast tissue samples were found to be positive for the LH receptor. Remarkably, expression levels were found to be highest in the normal breast tissue samples rather than in the malignant breast tumor specimens. In the follow-up study of Meduri [16], 160 malignant breast tumors, approximately 10% of the number of tumors in our study, were investigated using the same antibody. Based on the staining pattern and an arbitrary cut-off value (18% labeled cells), 72% of the breast tumors were found to be positive for the LH receptor [16]. The rationale for this cut-off value was not further specified in this publication. This high prevalence is in contrast with our findings that LH receptor mRNA can be detected in maximally 38% of breast tumors. However, with mRNA levels as low as we observed, it seems highly unlikely that any specific staining of LH receptor proteins could be observed in these tumor samples. We should therefore take into consideration the possibility that in the studies performed by Meduri et al. [16], most of the staining observed in breast tumors is due to nonspecific staining instead of staining of LH receptor proteins present at the cellular membranes.
In the present study we found that expression of the LH receptor appears to be related to a lower percentage of tumor cells present in the tumor sections. In addition, we found that presence of the LH receptor is associated with smaller tumors. This leads us to hypothesize that the little LH receptor expression that we find in tumors may occur for the greatest part, or even completely, in the surrounding non-tumorous tissue, instead of in the tumor cells. This is in line with, Meduri et al. who found expression levels to be highest in normal breast tissue samples compared to breast tumors [15] and our findings using immunohistochemical analyses that also show higher expression in normal cells (data not shown). Another possibility would be that expression of the LH receptor by tumors reflects in some way the grade of differentiation of the tumor. In accordance with this is the finding that LH receptors are more often found in tumors that also express estrogen and/or progesterone receptors, features of good differentiation and less aggressive behavior of the tumor. Furthermore, we observe a trend between expression of the LH receptor and good/moderate tumor grade in this study. If expression of the LH receptor is indeed associated with less aggressive tumor behavior, then this could in turn explain the association we found between expression and smaller tumor size and low percentage of tumor cells. Further research is needed to find a definite explanation for these observations.
Preclinical findings have led researchers to the idea that hCG may be used, either alone or in conjugate with a toxic compound, as a therapeutic agent that is capable to specifically target breast cancer cells [21,22,30,31]. In addition to finding a cytotoxic effect of the Hecate-CGβ conjugate on MCF-7 and MDA-MB-435S cells in vitro, Leuschner et al. [30,31] demonstrated this compound to be effective in vivo as well. After establishing MDA-MB-435S xenografts in athymic nude mice, they found that Hecate-CGβ could not only inhibit tumor growth [30], but was even able to cause tumor regression [31]. This resulted in one clinical trial in which hCG alone was given as a treatment to breast cancer patients [32]. According to the authors, treatment with hCG decreased the proliferative index (Ki67) of tumors (n=25) and was capable to slow down tumor progression in some patients with metastatic disease (n=13). This seems to be in contrast with the overt clinical effect of LHRH agonists for treatment of premenopausal breast cancer, which for a large part acts by suppression of LHR signaling. Furthermore, in the era of evidence-based medicine and of targeted treatment the authors failed to measure the LH receptor expression in the breast tumor specimens.
Although these results may argue for an effect of hCG, either alone or in conjugated form, taking into consideration the expression levels of LH receptors in tumors as low as we find, it seems highly unlikely that any of these observed effects could be explained by the binding of hCG to the LH receptor. We must therefore consider the possibility that another factor is present at the cellular membranes of breast tumor cells that is also capable to bind hCG. Indeed, the study Zaleska et al. [33] underscores our view. They observed in a rat model with DES/DMBA induced tumors that the Hecate-CGβ conjugate inhibited tumor growth to a greater extent than treatment with Hecate alone. However, similar to our present data, they could only detect low levels of LH receptor expression in 17% of the tumors using a nested RT-PCR method. Based on the observed almost absence of LHR on breast tumor specimens, targeted treatment of breast cancer using hCG-derivatives are unjustified. On the other hand, a possible protective effect of hCG –high during the superphysiological condition of pregnancy-, on normal breast cells might make this a chemopreventive agent for breast cancer. Yet, elaborate studies are needed before final conclusions can be drawn.
Lastly, previous work of our group indicated that premenopausal breast cancer patients that are carrier of a common and functional polymorphism in the LH receptor, the insLQ variant, show a significant shorter disease free survival compared to non-carriers [13,14]. It was postulated that the variant receptor may exert its effects by an indirect effect through the ovaries, affecting steroid hormone synthesis, or by a direct effect on breast cancer tissue itself. With expression levels in breast tumor tissues from the same cohort as low as we currently present, however, we may exclude the possibility of a direct effect of the LH receptor gene variant on breast tumor behavior. Thus, the insLQ polymorphism, causing a change in the expression level of the LH receptor in the ovary[13,14], may exposure of the breast tissue to ovary-derived estrogens. The ovarian origin of the effect of the polymorphism is underlined by the observation that an association between breast cancer disease and the insLQ polymorphism is only seen in premenopausal women.
Acknowledgments
The authors would like to thank Dr Mieke Schutte for providing the cDNA samples from the breast cancer cell lines and Drs John Foekens, Anieta Sieuwerts and Maxime Look for their contribution to this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Fernandez LM. Use of statistics to assess the global burden of breast cancer. Breast J. 2006;12 1:S70–80. doi: 10.1111/j.1075-122X.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 3.Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–85. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 4.Nelson HD, Huffman LH, Fu R, Harris EL. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: systematic evidence review for the U.S. Preventive Services Task Force. Ann Intern Med. 2005;143:362–79. doi: 10.7326/0003-4819-143-5-200509060-00012. [DOI] [PubMed] [Google Scholar]
- 5.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 6.Brekelmans CT. Risk factors and risk reduction of breast and ovarian cancer. Curr Opin Obstet Gynecol. 2003;15:63–8. doi: 10.1097/00001703-200302000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Dumitrescu RG, Cotarla I. Understanding breast cancer risk -- where do we stand in 2005? J Cell Mol Med. 2005;9:208–21. doi: 10.1111/j.1582-4934.2005.tb00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacMahon B. Epidemiology and the causes of breast cancer. Int J Cancer. 2006;118:2373–8. doi: 10.1002/ijc.21404. [DOI] [PubMed] [Google Scholar]
- 9.Russo IH, Russo J. Hormonal approach to breast cancer prevention. J Cell Biochem Suppl. 2000;34:1–6. [PubMed] [Google Scholar]
- 10.Rao CV. Does full-term pregnancy at a young age protect women against breast cancer through hCG? Obstet Gynecol. 2000;96:783–6. doi: 10.1016/s0029-7844(00)00983-2. [DOI] [PubMed] [Google Scholar]
- 11.Themmen APN, Huhtaniemi IT. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev. 2000;21:551–83. doi: 10.1210/edrv.21.5.0409. [DOI] [PubMed] [Google Scholar]
- 12.Powell BL, Piersma D, Kevenaar ME, van Staveren IL, Themmen AP, Iacopetta BJ, Berns EM. Luteinizing hormone signaling and breast cancer: polymorphisms and age of onset. J Clin Endocrinol Metab. 2003;88:1653–7. doi: 10.1210/jc.2002-021585. [DOI] [PubMed] [Google Scholar]
- 13.Piersma D, Berns EM, Verhoef-Post M, Uitterlinden AG, Braakman I, Pols HA, Themmen AP. A common polymorphism renders the luteinizing hormone receptor protein more active by improving signal peptide function and predicts adverse outcome in breast cancer patients. J Clin Endocrinol Metab. 2006;91:1470–6. doi: 10.1210/jc.2005-2156. [DOI] [PubMed] [Google Scholar]
- 14.Piersma D, Themmen AP, Look MP, Klijn JG, Foekens JA, Uitterlinden AG, Pols HA, Berns EM. GnRH and LHR gene variants predict adverse outcome in premenopausal breast cancer patients. Breast Cancer Res. 2007;9:R51. doi: 10.1186/bcr1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meduri G, Charnaux N, Loosfelt H, Jolivet A, Spyratos F, Brailly S, Milgrom E. Luteinizing hormone/human chorionic gonadotropin receptors in breast cancer. Cancer Res. 1997;57:857–64. [PubMed] [Google Scholar]
- 16.Meduri G, Charnaux N, Spyratos F, Hacene K, Loosfelt H, Milgrom E. Luteinizing hormone receptor status and clinical, pathologic, and prognostic features in patients with breast carcinomas. Cancer. 2003;97:1810–6. doi: 10.1002/cncr.11294. [DOI] [PubMed] [Google Scholar]
- 17.Carlson HE, Kane P, Lei ZM, Li X, Rao CV. Presence of luteinizing hormone/human chorionic gonadotropin receptors in male breast tissues. J Clin Endocrinol Metab. 2004;89:4119–23. doi: 10.1210/jc.2003-031882. [DOI] [PubMed] [Google Scholar]
- 18.Lojun S, Bao S, Lei ZM, Rao CV. Presence of functional luteinizing hormone/chorionic gonadotropin (hCG) receptors in human breast cell lines: implications supporting the premise that hCG protects women against breast cancer. Biol Reprod. 1997;57:1202–10. doi: 10.1095/biolreprod57.5.1202. [DOI] [PubMed] [Google Scholar]
- 19.Hu YL, Lei ZM, Huang ZH, Rao CV. Determinants of Transcription of the Chorionic Gonadotropin /Luteinizing Hormone Receptor Gene in Human Breast Cells. Breast J. 1999;5:186–193. doi: 10.1046/j.1524-4741.1999.98067.x. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X, Russo IH, Russo J. Alternately spliced luteinizing hormone/human chorionic gonadotropin receptor mRNA in human breast epithelial cells. Int J Oncol. 2002;20:735–8. [PubMed] [Google Scholar]
- 21.Gebauer G, Fehm T, Beck EP, Berkholz A, Licht P, Jager W. Cytotoxic effect of conjugates of doxorubicin and human chorionic gonadotropin (hCG) in breast cancer cells. Breast Cancer Res Treat. 2003;77:125–31. doi: 10.1023/a:1021301001208. [DOI] [PubMed] [Google Scholar]
- 22.Bodek G, Rahman NA, Zaleska M, Soliymani R, Lankinen H, Hansel W, Huhtaniemi I, Ziecik AJ. A novel approach of targeted ablation of mammary carcinoma cells through luteinizing hormone receptors using Hecate-CGbeta conjugate. Breast Cancer Res Treat. 2003;79:1–10. doi: 10.1023/a:1023351819956. [DOI] [PubMed] [Google Scholar]
- 23.Rao Ch V, Li X, Manna SK, Lei ZM, Aggarwal BB. Human chorionic gonadotropin decreases proliferation and invasion of breast cancer MCF-7 cells by inhibiting NF-kappaB and AP-1 activation. J Biol Chem. 2004;279:25503–10. doi: 10.1074/jbc.M400683200. [DOI] [PubMed] [Google Scholar]
- 24.Foekens JA, Peters HA, Grebenchtchikov N, Look MP, Meijer-van Gelder ME, Geurts-Moespot A, van der Kwast TH, Sweep CG, Klijn JG. High tumor levels of vascular endothelial growth factor predict poor response to systemic therapy in advanced breast cancer. Cancer Res. 2001;61:5407–14. [PubMed] [Google Scholar]
- 25.Sieuwerts AM, Meijer-van Gelder ME, Timmermans M, Trapman AM, Garcia RR, Arnold M, Goedheer AJ, Portengen H, Klijn JG, Foekens JA. How ADAM-9 and ADAM-11 differentially from estrogen receptor predict response to tamoxifen treatment in patients with recurrent breast cancer: a retrospective study. Clin Cancer Res. 2005;11:7311–21. doi: 10.1158/1078-0432.CCR-05-0560. [DOI] [PubMed] [Google Scholar]
- 26.Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, Merajver SD, Ethier SP, Schutte M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–5. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 27.Richter-Unruh A, Verhoef-Post M, Malak S, Homoki J, Hauffa BP, Themmen AP. Leydig cell hypoplasia: absent luteinizing hormone receptor cell surface expression caused by a novel homozygous mutation in the extracellular domain. J Clin Endocrinol Metab. 2004;89:5161–7. doi: 10.1210/jc.2004-0298. [DOI] [PubMed] [Google Scholar]
- 28.Kraaij R, Post M, Kremer H, Milgrom E, Epping W, Brunner HG, Grootegoed JA, Themmen AP. A missense mutation in the second transmembrane segment of the luteinizing hormone receptor causes familial male-limited precocious puberty. J Clin Endocrinol Metab. 1995;80:3168–72. doi: 10.1210/jcem.80.11.7593421. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Fatima N, Dufau ML. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol Cell Biol. 2005;25:7929–39. doi: 10.1128/MCB.25.18.7929-7939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leuschner C, Enright FM, Gawronska B, Hansel W. Membrane disrupting lytic peptide conjugates destroy hormone dependent and independent breast cancer cells in vitro and in vivo. Breast Cancer Res Treat. 2003;78:17–27. doi: 10.1023/a:1022169525521. [DOI] [PubMed] [Google Scholar]
- 31.Hansel W, Enright F, Leuschner C. Destruction of breast cancers and their metastases by lytic peptide conjugates in vitro and in vivo. Mol Cell Endocrinol. 2007;260-262:183–9. doi: 10.1016/j.mce.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 32.Janssens JP, Russo J, Russo I, Michiels L, Donders G, Verjans M, Riphagen I, Van den Bossche T, Deleu M, Sieprath P. Human chorionic gonadotropin (hCG) and prevention of breast cancer. Mol Cell Endocrinol. 2007;269:93–8. doi: 10.1016/j.mce.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Zaleska M, Waclawik A, Bodek G, Zezula-Szpyra A, Li X, Janowski T, Hansel WH, Rahman NA, Ziecik AJ. Growth repression in diethylstilbestrol/dimethylbenz[a]anthracene-induced rat mammary gland tumor using Hecate-CGbeta conjugate. Exp Biol Med (Maywood) 2004;229:335–44. doi: 10.1177/153537020422900408. [DOI] [PubMed] [Google Scholar]