Abstract
Background
Intraductal papillary mucinous neoplasms (IPMNs) are non-invasive precursor lesions of pancreatic cancer. Misexpression of microRNAs (miRNAs) is commonly observed in pancreatic adenocarcinoma. In contrast, miRNA abnormalities in pancreatic cancer precursor lesions have not been documented.
Experimental design
Relative expression levels of a panel of twelve miRNAs upregulated in pancreatic cancers were assessed in 15 non-invasive IPMNs, using quantitative reverse transcription PCR (qRT-PCR). Two significantly overexpressed miRNAs—miR-155 and miR-21—were evaluated by locked nucleic acid in situ hybridization (LNA-ISH) in a panel of 64 archival IPMNs. The expression of miR-155 and miR-21 was also evaluated in pancreatic juice samples obtained from ten patients with surgically resected IPMNs and five patients with non-neoplastic pancreato-biliary disorders (“disease controls”).
Results
Significant overexpression by qRT-PCR of ten of the twelve miRNAs was observed in the 15 IPMNs versus matched controls (p < 0.05), with miR-155 (mean 11.6-fold) and miR-21 (mean 12.1-fold) demonstrating highest relative fold-changes in the precursor lesions. LNA-ISH confirmed the expression of miR-155 in 53 of 64 (83%) IPMNs compared to 4 of 54 (7%) normal ducts, and of miR-21 in 52 of 64 (81%) IPMNs compared to 1 of 54 (2%) normal ducts, respectively (p < 0.0001). Upregulation of miR-155 transcripts by qRT-PCR was observed in 6 of 10 (60%) IPMN-associated pancreatic juice samples compared to 0 of 5 (0%) disease controls.
Conclusions
Aberrant miRNA expression is an early event in the multistage progression of pancreatic cancer, and miR-155 warrants further evaluation as a biomarker for IPMNs in clinical samples.
Keywords: pancreatic cancer, intraductal papillary mucinous neoplasm, microRNA, miR-155
Introduction
Pancreatic cancer is the fourth most common cause of cancer-related mortality in the United States.1 The overwhelming majority of patients present with locally advanced or distant metastatic disease, rendering their malignancy surgically inoperable. Despite advances in chemo-radiation therapies over the last few decades, the dire prognosis of pancreatic cancer has remained essentially unchanged. Early diagnosis of this neoplasm at an early, and hence potentially resectable stage, offers one of the best avenues for cure.2
It is now well established that pancreatic cancers do not arise de novo, but rather represent the culmination of a multistep progression involving non-invasive precursor lesions within exocrine pancreatic ducts.3 The most common precursors are pancreatic intraepithelial neoplasia (PanIN), which are microscopic lesions arising within ducts less than 5 mm in diameter.4 In contrast, IPMNs represent macroscopic precursors of pancreatic adenocarcinoma, typically presenting as cystic lesions within the main pancreatic duct or one of its branches.5 Since their original description in the 1982,6 insights into the histological features and genetics of these cystic precursor lesions have expanded considerably. Akin to PanIN lesions, the lining epithelium of IPMNs can demonstrate varying degrees of histologic atypia, ranging from the innocuous IPMN adenoma, through borderline IPMN to IPMN with carcinoma-in-situ.4 In addition to variable risk of progression associated with degrees of histological atypia, biological heterogeneity is also observed within IPMNs based on the nature of the lining epithelium. Thus, IPMNs with intestinal-type or pancreato-biliary epithelium harbour a greater propensity for progression to invasive neoplasia than those with gastric foveolar type epithelium.7,8 Genetic studies have established that many of the seminal alterations observed in invasive pancreatic cancer, such as mutations of KRAS2, DPC4/SMAD4 and TP53, are also present in a variable proportion of non-invasive IPMNs, further cementing their status as bona fide precursor lesions.3,9 At the same, a subset of molecular changes identified in IPMNs is not commonly observed in PanINs (for example, mutations of PI3KCA and STK11/LKB1 genes, or expression of the cellular apomucin Muc2),10-12 suggesting that in addition to histological digression, there are also two distinct molecular pathways to invasive adenocarcinoma in the pancreas.13,14
MicroRNAs (miRNAs) are a diverse class of 18–24 nucleotide RNA molecules that demonstrate remarkable evolutionary conservation.15 The principal function of these non-coding RNAs is to regulate the stability and translation of nuclear mRNA transcripts. Physiologic regulation of the cellular transcriptome by miRNAs plays a critical role in development and in homeostasis. Aberrant expression of miRNA is widespread, if not ubiquitous, in human cancers, with the identification of both over and underexpressed miRNAs in neoplastic cells compared to their normal counterparts.16 Several recent studies have identified a multitude of misexpressed miRNAs in human pancreatic cancers compared to normal pancreatic tissues.17-20 In contrast, patterns of miRNA abnormalities in the non-invasive precursor lesions of pancreatic adenocarcinoma remain largely unknown. The profiling of miRNAs in IPMNs has several conceivable advantages. In addition to enhancing our understanding of the pathogenesis of early pancreatic neoplasia, these aberrantly expressed miRNAs potentially represent targets for therapy, as well as candidate biomarkers for the diagnosis of pancreatic cancer precursors in clinical specimens. In this study, we demonstrate that miR-155 and miR-21 are significantly upregulated in the majority of non-invasive IPMNs, and their expression correlates with histological features of progression in these neoplasms. Further, our studies provide promising, albeit preliminary evidence, that upregulation of miR-155 in clinical samples like pancreatic juice harbors the potential to emerge as a diagnostic adjunct for IPMNs.
Results
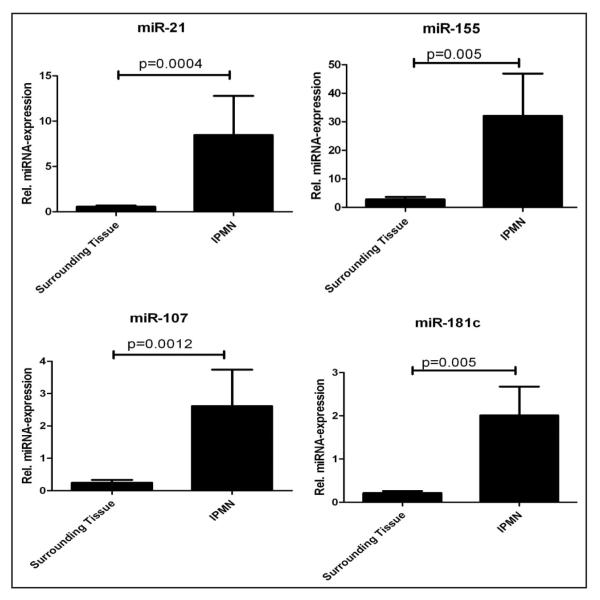
Analysis of miRNA profiles in the 15 microdissected IPMN lesions confirmed significant overexpression of ten of 12 miRNAs, compared to matched non-neoplastic samples (p < 0.05) (Fig. 1 and Table 1); only miR-15a and miR-17-5p did not reach statistically significant differences in expression, although both demonstrated a trend towards upregulation in the neoplastic epithelium. Of the significantly overexpressed miRNAs, miR-21 and miR-155 had the highest relative fold expression levels in IPMNs versus non-neoplastic pancreata (12.1-fold and 11.6-fold, respectively). Given the established association of these two seminal “onco-miRs” with human cancers,25,30 including pancreatic adenocarcinoma,17,20,31 we decided to explore the expression of miR-21 and miR-155 in a larger panel of 64 IPMN lesions by LNA-ISH. Both miRNAs were frequently overexpressed within the neoplastic epithelium of IPMNs, with 53 of 64 (83%) IPMNs expressing miR-155 (Fig. 2A, Suppl. Fig. 2A) and 52 of 64 (81%) expressing miR-21 (Fig. 2B, Suppl. Fig. 2B). Expectedly, the scrambled probes failed to demonstrate any evidence of ISH signal (Fig. 2C, Suppl. Fig. 2C). In contrast, miR-155 expression was observed in only 4 of 54 (7%) matched non-neoplastic pancreata on the TMAs, while miR-21 was expressed in only 1 of 54 (2%) non-neoplastic pancreata (p < 0.001, Chi-square test; Suppl. Fig. 3 and Table 2). We also retrieved archival formalin-fixed paraffin-embedded blocks from five of the 15 cases that were utilized in the qRT-PCR study. LNA-ISH was performed for miR-155 expression in these five cases and confirmed that aberrant miRNA expression was restricted to the neoplastic epithelium in these cases, although a strict correlation with qRT-PCR levels in the corresponding frozen material was not observed (Suppl. Fig. 4).
Figure 1.
Relative fold expression of miRNAs in microdissected IPMNs compared to matched non-neoplastic pancreata. Each panel represents a miRNA assessed by qRT-PCR. The two bars represent either surrounding non-neoplastic tissue, or IPMN epithelium. Each sample was assessed in triplicate, and U6 non-coding RNA was used as housekeeping control. The Y-axis represents the average of the relative fold expression levels for each group (IPMNs versus controls). Error bars represent standard errors of the mean. The significance level is designated on the panel. Only four of the 12 miRNAs examined are shown.
Table 1.
Relative differential expression of twelve pancreatic cancer associated miRNAs in microdissected non-invasive IPMNs
| miRNA | Fold-change | p-value | Pancreatic cancer reference |
|---|---|---|---|
| miR-21 | 12.1 | 0.0004 | (17–20) |
| miR-155 | 11.6 | 0.005 | (17–20) |
| miR-107 | 10.8 | 0.01 | (17, 18, 20) |
| miR-223 | 9.7 | 0.02 | (17–19) |
| miR-181c | 9.5 | 0.005 | (18, 20) |
| miR-181a | 7.7 | 0.01 | (18, 20) |
| miR-221 | 6.6 | 0.01 | (17–20) |
| miR-210 | 5.7 | 0.005 | (18, 19) |
| miR-16 | 4.8 | 0.02 | (20) |
| miR-100 | 3.8 | 0.01 | (20) |
| miR-15a | 3.5 | 0.2 | (18) |
| miR-17-5p | 3.4 | 0.1 | (17) |
Figure 2.
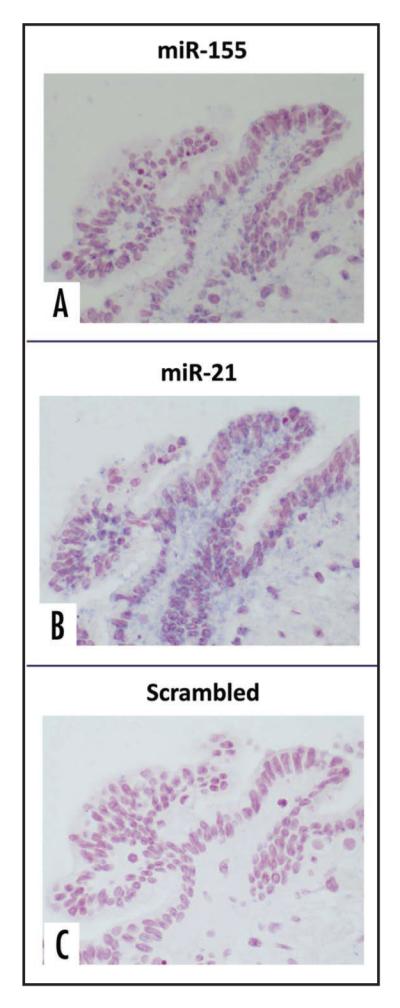
Locked nucleic acid in situ hybridization (LNA-ISH) for miR-155 and miR-21 expression in archival IPMN tissues. High magnification photomicrographs of an archival IPMN with robust expression of DIG-labeled miR-155 probe (A) and miR-21 probe (B) within the neoplastic epithelium, consistent with expression of these miRNAs within the neoplasm. In contrast, the scrambled miRNA probe demonstrates complete absence of expression (C). Magnification 40x.
Table 2.
Frequency of miR-155 and miR-21 expression in IPMNs and non-neoplastic pancreata by in situ hybridization
| miRNA | Expression | ISH-score | Non-neoplastic pancreas |
IPMN |
|---|---|---|---|---|
| miR-155 | Negative | 0 | 50 (93%) | 11 (17%) |
| Positive | 1 | 3 (5%) | 16 (25%) | |
| 2 | 1 (2%) | 37 (58%) | ||
| miR-21 | Negative | 0 | 53 (98%) | 12 (19%) |
| Positive | 1 | 1 (2%) | 39 (61%) | |
| 2 | 0 | 13 (20%) | ||
| Total | 54 | 64 |
Note: None of the ISH-scores reached 2 × 2 = 4.
Upon stratification by the degree of histological atypia in the lining epithelium, a significantly higher proportion of IPMNs with carcinoma-in-situ expressed both miRNAs, compared to IPMN adenomas (Table 3A). Specifically, 20 of 20 (100%) IPMNs with carcinoma-in situ expressed miR-155 compared to 7 of 13 (54%) IPMN adenomas (p = 0.0002), while 19 of 20 (95%) IPMNs with carcinoma-in situ expressed miR-21 compared to 7 of 13 (54%) IPMN adenomas (p = 0.008). However, there were no statistically significant differences between IPMN adenoma and borderline IPMN, or between borderline IPMN and IPMN with carcinoma-insitu, based on either miR-155 or miR-21 expression. When stratified by the histological subtype of the lining epithelium, we did not elicit significant differences in the frequency of miR-155 expression between pancreato-biliary type and intestinal type of IPMNs (Table 3B). In contrast, we found a significantly lower proportion of IPMNs with gastric foveolar type lining expressing miR-155 (4 of 7 cases, 57%), compared to the intestinal type IPMNs (19 of 19 cases 100%; p = 0.01). Admittedly the numbers of gastric foveolar type IPMNs in our series are small; nevertheless contingent upon this caveat, it is worth noting that the intestinal types of IPMNs are usually main-duct lesions with a higher propensity for malignancy than the typically branch duct, gastric foveolar type IPMNs.7,8 Comparable significant differences in expression between the histological variants were, however, not observed in the case of miR-21.
Table 3 (A).
Frequency of miR-155 and miR-21 expression in IPMNs stratified by histologic atypia of the lining epithelium
| miRNA | Expression | ISH score | IPMN adenoma | IPMN borderline | IPMN carcinoma-in-situ | Total |
|---|---|---|---|---|---|---|
| miR-155 | Negative | 0 | 6 (46%) | 5 (16%) | 0 | 11 (17%) |
| Positive | 1 | 4 (31%) | 7 (23%) | 5 (25%) | 16 (25%) | |
| 2 | 3 (23%) | 19 (61%) | 15 (75%) | 37 (58%) | ||
| miR-21 | Negative | 0 | 6 (46%) | 5 (16%) | 1 (5%) | 12 (19%) |
| Positive | 1 | 5 (39%) | 19 (61%) | 15 (75%) | 39 (61%) | |
| 2 | 2 (15%) | 7 (23%) | 4 (20%) | 13 (20%) | ||
| Total | 13 | 31 | 20 | 64 |
Note: None of the ISH-scores reached 2 × 2 = 4.
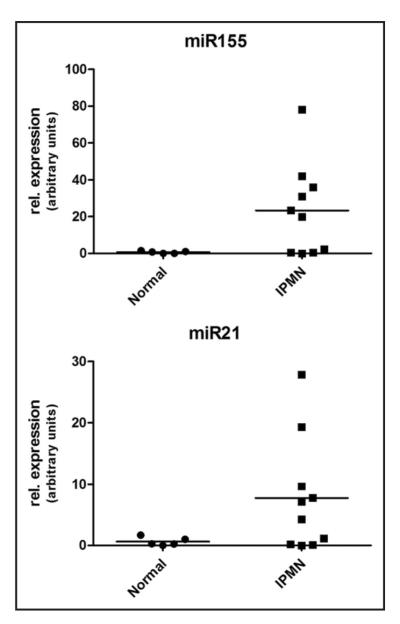
In light of the promising data with both miRNAs in IPMN tissue sections, we next examined the potential of using these targets as biomarkers of IPMNs in pancreatic juice samples. A total of 15 pancreatic juice samples obtained at the time of surgical resection were examined for relative levels of miR-155 and miR-21 by qRT-PCR. The juice samples were obtained from 10 patients with histologically confirmed IPMNs, and five with other pancreato-biliary disorders (“disease controls”). As seen in Figure 3, the mean relative expression level of miR-155 was higher than disease controls in the IPMN juice samples. Specifically, the five disease controls had minimally detectable miR-155 in pancreatic juice, while 6 of 10 (60%) IPMNs had elevated miR-155 in juice samples, with 20-fold or greater relative-fold expression; nevertheless, due likely to the small numbers of samples, the differences did not reach statistical significance (p = 0.1). Similarly the five disease controls had minimal miR-21 expression in the juice samples, although the mean level of miR-21 overexpression in the IPMN juice samples was more attenuated than that observed with miR-155, with only two samples having 20-fold or greater relative expression (Fig. 3).
Figure 3.
Expression of miR-155 and miR-21 in pancreatic juice samples assessed by qRT-PCR. Analysis by qRT-PCR of pancreatic juice samples from ten patients with IPMNs and five disease controls. The X-axis designates the two categories of patients for each miRNA and the Y-axis designates the relative fold expression in each pancreatic juice sample. The horizontal bar represents the group average. Note that the Y-axis values differ in miR-155 and miR-21, reflecting a greater fold elevation in the former.
Discussion
It is estimated that as many as half of individuals over the age of sixty harbor PanIN lesions in their pancreata.32 By contrast, the numbers of IPMNs are likely to be considerably lower in the general population. Nonetheless, due in part to their macroscopic (cystic) nature and the potential for detection upon radiological examination, IPMNs present a unique opportunity for prevention of invasive malignancy in the pancreas.3 With the enhanced usage of CT and other non-invasive scanning techniques in medical practice, increasing numbers of patients with incidental cysts of the pancreas (“incidentalomas”) are being identified.33 While in many instances these cysts are innocuous in their biological potential (for example, serous cystic neoplasms), others represent bona fide precursors of pancreatic adenocarcinomas, such as IPMNs or mucinous cystic neoplasms (MCNs).34,35 Identification of molecular aberrations in IPMNs thus attains considerable clinical significance, not only in terms of understanding the biology of early pancreatic neoplasia, but also as a fertile seedbed for generating therapeutic and diagnostic markers, which might stem the progression to a lethal invasive cancer.2
Several studies have reported miRNA abnormalities in pancreatic cancers, using either array-based platforms or by qRT-PCR.17-20 For a subset of these miRNAs, the underlying mechanism for misexpression has been elucidated. For example, we have demonstrated that loss of p53 function in pancreatic cancer is associated with decreased expression of its transcriptional target miR-34a in pancreatic cancers;23 conversely, aberrant Myc activity in pancreatic and other human cancers results in elevated expression of the miR-17-92 polycistron.36 In most instances however, the mechanism(s) underlying miRNA misexpression in pancreatic cancer remain unknown. In the current study, we profiled a series of non-invasive IPMNs for miRNA abnormalities using the prior published reports in pancreatic cancer as a guide. Specifically, we selected a panel of 12 miRNAs that have been reported as overexpressed in this malignancy. The miRNAs were selected based on multiple criteria, including their identification in more than one pancreatic cancer miRNA profiling study, the relative fold-elevation compared to normal pancreas, and the putative cancer-associated function of the miRNA. Although this selective approach is less optimal than an unbiased “forward genetics” strategy, our prior experience with genomic and transcriptomic analyses of IPMNs suggests that these precursors often harbor many of the genetic alterations observed in pancreatic adenocarcinomas.21,37,38 Indeed, we found that as many as 10 of the 12 miRNAs were significantly overexpressed in the IPMNs compared to non-neoplastic pancreata, establishing that miRNA abnormalities are an early event in the multistep progression of pancreatic cancer.
We focused our attention on further tissue-based validation of two of the highest differentially expressed miRNAs—miR-21 and miR-155. Both miRNAs have been reported as significantly overexpressed in invasive pancreatic cancer in all four prior studies,17-20 furthering underscoring their relevance to this malignancy. The primary transcript for miR-21 is expressed from chromosome 17q23.2 and the resulting mature transcript was one of the first oncogenic miRNAs (onco-miR) identified in human cancer.17,30,39 Upregulation of miR-21 in cancer cells is associated with apoptosis inhibition and acquisition of invasive properties.40,41 Several of the coding genes translationally repressed by miR-21 have now been identified, including the tumor suppressor phosphatase and tensin homolog (PTEN), downregulation of which results in activation of the Akt signaling pathway,42 and programmed cell death 4 (PDCD4), loss of function of which promotes cellular transformation and metastases.43 On the same lines, miR-155 is co-expressed in conjunction with the non-coding transcript BIC, from chromosome 21q21.3.44 Although initially identified as an overexpressed miRNA in hematological malignancies, miR-155 has now been reported as upregulated in several solid tumors, including pancreatic cancer.39,45 Of note, a recent study has shown that misexpressed miR-155 in pancreatic cancer appears to repress the function of tumor protein 53 induced nuclear protein 1 (TP53INP1); the latter is a pro-apoptotic, p53-induced protein, and its miR-155-mediated downregulation in pancreatic cancer enhances tumorigenicity in vivo.31
Our LNA-ISH data confirms that abnormal expression of both miR-155 and miR-21 is commonly observed in IPMNs, while these transcripts are absent in the overwhelming majority of normal pancreatic ductal epithelia. As is true for many of the molecular alterations observed in pancreatic cancer precursors,1 there is a gradation in the frequency of miRNA misexpression along the histological continuum of atypia, with a significantly greater proportion of IPMNs with carcinoma-in-situ expressing either miRNA compared to IPMN adenomas. Further, when stratified by the histological subtype of lining epithelium, we detected an increased frequency of miR-155 expression in IPMNs with an intestinal type or pancreato-biliary type epithelium versus those with gastric foveolar type lining (this difference was statistically significant in the intestinal type IPMNs). Given the greater propensity for the intestinal or pancreato-biliary type IPMNs to progress to invasive adenocarcinomas compared with gastric foveolar IPMNs,7,8 there appears to be a correlation between the proportion of miR-155 expressing cases and intrinsic biological potential of a particular IPMN subtype. Nonetheless, the discrimination was not as unequivocally dichotomous as has been previously reported for certain cellular apomucin expression profiles (specifically, MUC1, MUC2 and MUC5AC).7,12
In addition to profiling IPMN tissues for miRNA abnormalities, we were also interested in determining whether detection of misexpressed miRNAs in pancreatic juice samples might be a feasible biomarker development strategy. To the best of our knowledge, detection of aberrantly expressed miRNAs in pancreatic juice has not been studied to date, although this possibility has been suggested based on the current state of knowledge on miRNA abnormalities in pancreatic cancer.2 In particular, there is a great need for developing adjunct diagnostic strategies for cystic neoplasms of the pancreas such as IPMNs and MCNs. In contrast to most pancreatic adenocarcinomas that tend to be solid lesions,46 “conventional” modalities like endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) present considerable challenges in the diagnosis of cystic entities.47 Typically, the aspirated material tends to be paucicellular, thus impeding accuracy of diagnosis. Of the available biomarker assays on cyst fluid, only CEA appears to be of modest benefit, with an accuracy of 79% in distinguishing mucinous precursor lesions (IPMNs and MCNs) from other cystic entities in the pancreas.48 Recent studies have elaborated upon the use of DNA-based molecular assays like microsatellite analysis and somatic mutations in aiding the differential diagnosis of pancreatic cysts.49,50 Our results provide encouraging, albeit admittedly preliminary, validation that miRNA profiling of pancreatic juice by qRT-PCR is feasible, and further, suggest that such profiling may provide discrimination between pancreato-biliary disease controls and patients harboring IPMNs. Notably, patients with non-neoplastic pancreato-biliary disorders like chronic pancreatitis have minimally detectable miR-155 in their pancreatic juice, while at least 60% of IPMN samples have elevated miR-155 levels (20 fold or greater relative fold expression). It is our intention to build further upon these preliminary studies and examine a larger cohort of patients, factoring in additional standards such as cyst fluid CA19-9 and CEA levels.
In conclusion, we have performed the first miRNA profiling of non-invasive IPMNs and confirmed that these macroscopic precursors share many of the miRNA abnormalities observed in invasive neoplasia. Two candidate miRNAs—miR-155 and miR-21—are commonly expressed in the neoplastic epithelium of IPMNs, and demonstrate correlation with histological features of progression (carcinoma-in-situ) and more ominous biological potential. Finally, elevated miR-155 demonstrates promise as a candidate biomarker for IPMNs in pancreatic juice specimens, warranting further study.
Material and Methods
IPMN specimens and other clinical samples
Cryostat embedded sections of 15 non-invasive IPMN specimens were obtained from the surgical pathology archives of Johns Hopkins Hospital. In each case, the patient had undergone surgical resection for removal of a pancreatic cystic neoplasm, and the histology of the lesion was confirmed by one of the authors (AM), who is an expert on pancreatic pathology.4,9 The IPMN specimens were snap-frozen in liquid nitrogen, embedded in Tissue-Tek OCT compound medium (Sakura FineTek USA, Torrance, CA) and stored at -80°C. The samples were subsequently embedded onto UV-treated PALM membrane slides (Carl Zeiss MicroImaging, Inc., Thornwood, NY) for the purpose of microdissection. Locked nucleic acid in situ hybridization (LNA-ISH) was performed on a panel of 64 archival IPMNs arrayed on duplicate 1.4 mm cores in tissue microarray (TMA) format, as we have previously described.10,21 The 64 samples were comprised of 13 IPMN adenomas, 31 borderline IPMNs, and 20 IPMNs with carcinoma-in-situ, on the basis of histological atypia in their lining epithelium.4 Similarly, when classified by the nature of the lining epithelium,7,8 the 64 samples were comprised of 35 pancreato-biliary type, 19 intestinal type, 7 gastric foveolar type, and one oncocytic IPMNs; in two cases the epithelium could not be accurately classified and these were labeled as “unclassified”. In addition, tissue cores from matched non-neoplastic pancreas were available for evaluation in 54 of the 64 IPMNs. We also examined pancreatic juice samples obtained at the time of surgery from 15 patients, including ten patients with histologically documented IPMNs and five patients with other pancreato-biliary disorders including chronic pancreatitis or bile duct stones (“disease controls”). The pancreatic juice was immediately mixed with Trizol reagent and stored at -80°C for miRNA extraction.
Laser microdissection and miRNA extraction
The IPMN cryostat sections were fixed in cold methanol and stained with hematoxylin and eosin (H&E) prior to microdissection using a PALM MicroBeam (Carl Zeiss MicroImaging, Inc., Thornwood, NY). The neoplastic epithelium was outlined and selectively microdissected as described previously22 (Suppl. Fig. 1). Subsequently, non-neoplastic pancreatic parenchyma from each case was microdissected, and “catapulted” into a different tube. The microdissected tissue samples were then subjected to RNA extraction using the mirVana™ miRNA Isolation kit (Ambion/Applied Biosystems, Austin, TX), according to the manufacturer’s protocol. For the pancreatic juice samples preserved in TriZol, total RNA isolation was performed by ethanol precipitation, as we have previously described.23
Quantitative reverse transcription PCR (qRT-PCR) for miRNA
Quantitative analyses of miRNA levels in microdissected IPMNs and in pancreatic juice samples were performed using pre-designed TaqMan® miRNA assays (Applied Biosystems, Foster City, CA). The TaqMan® miRNA assays are a two-step protocol, involving reverse transcription with human mature miRNA specific primers, followed by real time PCR with TaqMan® probes. These assays target the mature miRNA sequence only, and the precursors are not detected. For the current study, we selected a panel of 12 miRNAs previously described as significantly overexpressed in invasive pancreatic cancers (Table 1):17-20 miR-15a, miR-16, miR-17-5p, miR-21, miR-100, miR-107, miR-155, miR-181a, miR-181c, miR-210, miR-221 and miR-223. The non-coding RNU6B (U6 control) was used as housekeeping control. The 15 microdissected IPMNs were compared against matched normal pancreata, and each sample was assessed in triplicate for any given miRNA. Relative fold expression was calculated using the 2-ΔΔCt method, as described previously.24 On the same lines, qRT-PCR was performed on RNA from the 21 pancreatic juice samples for miR-21 and miR-155.
Locked nucleic acid in situ hybridization (LNA ISH)
LNA-ISH was performed using LNA™ probes against miR-21 and miR-155 (Exiqon, Vedbaek, Denmark), respectively, on archival IPMN tissue microarrays, as per the manufacturer’s protocol. The use of LNA™ probes for the successful cataloging of altered miRNAs in archival human cancer tissues has been recently described.25-27 Briefly, after deparaffinization, the slides were blocked for two hours, and then incubated with hybridization buffer containing the digoxigenin (DIG) labeled LNA™ probe in a hybridization oven, overnight. A parallel set of TMAs was hybridized with a “scrambled” miRNA probe from Exiqon, as previously described,25 as a measure of probe specificity. After several washes for ensuring stringency, the slides were incubated with anti-digoxigenin Fab fragment (1:2000) overnight in a humid chamber at 4°C. The colorimetric detection reaction was performed using NBT/BNI ReadyMix for 48 hours. The slides were then mounted with Cytoseal 60 (Richard-Allan-Scientific). The TMAs were scored on a multi-headed microscope by three of the authors on the panel (J-BK, AM and S-MH). The LNA-ISH results were scored based on intensity of staining as 0 (negative), 1 (weak) or 2 (strong), and based on the percentage of positive epithelial cells as 0 (<1%), 1 (focal, 1–50%) or 2 (diffuse, >50%), respectively. A “ISH-score” was generated as the product of intensity times area, similar to what we have previously described with immunohistochemical analyses on TMAs.28,29 The “ISH-score” was then binned into a two-tier classification of “negative” (score 0), and “positive” (score ≥ 1).
Data processing and statistical analysis
Statistical analyses were performed using SPSS version 11 (SPSS Inc., Chicago, IL). Associations between categorical variables were examined using the Pearson’s chi-square and Fisher’s exact tests. A p-value <0.05 was considered statistically significant. The depiction of miRNA differential expression was plotted using GraphPad Prism.
Supplementary Material
Table 3 (B).
Frequency of miR-155 and miR-21 expression in IPMNs stratified by histologic subtype of the lining epithelium
| miRNA | Expression | Gastric | Pancreato-biliary | Intestinal | Oncocytic | Unclassified | Total |
|---|---|---|---|---|---|---|---|
| miR-155 | negative | 3 (43%) | 7 (20%) | 0 | 1 (100%) | 0 | 11 |
| positive | 4 (57%) | 28 (80%) | 19 (100%) | 0 | 2 (100%) | 53 | |
| miR-21 | negative | 2 (29%) | 7 (20%) | 3 (16%) | 0 | 0 | 12 |
| positive | 5 (71%) | 28 (80%) | 16 (84%) | 1 (100%) | 2 (100%) | 52 | |
| Total | 7 | 35 | 19 | 1 | 2 | 64 |
Acknowledgements
Supported by the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation for Pancreatic Cancer Research, and the NIH SPORE in GI Cancers P50CA062924.
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/HabbeCBT8-4-Sup.pdf
References
- 1.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goggins M. Identifying molecular markers for the early detection of pancreatic neoplasia. Semin Oncol. 2007;34:303–10. doi: 10.1053/j.seminoncol.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hruban RH, Maitra A, Kern SE, Goggins M. Precursors to pancreatic cancer. Gastroenterol Clin North Am. 2007;36:831–49. doi: 10.1016/j.gtc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–87. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 5.Adsay NV. Cystic neoplasia of the pancreas: Pathology and biology. J Gastrointest Surg. 2008;12:401–4. doi: 10.1007/s11605-007-0348-z. [DOI] [PubMed] [Google Scholar]
- 6.Ohhashi K, Murakami F, Maruyama M. Four cases of mucous secreting pancreatic cancer. Prog Dig Endosc. 1982;203:348–51. [Google Scholar]
- 7.Adsay NV, Merati K, Basturk O, Iacobuzio-Donahue C, Levi E, Cheng JD, et al. Pathologically and biologically distinct types of epithelium in intraductal papillary mucinous neoplasms: Delineation of an “intestinal” pathway of carcinogenesis in the pancreas. Am J Surg Pathol. 2004;28:839–48. doi: 10.1097/00000478-200407000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa T, Klöppel G, Adsay N Volkan, Albores-Saavedra J, Fukushima N, Horii A, et al. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: A consensus study. Virchows Arch. 2005;447:794–9. doi: 10.1007/s00428-005-0039-7. [DOI] [PubMed] [Google Scholar]
- 9.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 10.Sahin F, Maitra A, Argani P, Sato N, Maehara N, Montgomery E, et al. Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod Pathol. 2003;16:686–91. doi: 10.1097/01.MP.0000075645.97329.86. [DOI] [PubMed] [Google Scholar]
- 11.Schönleben F, Qiu W, Ciau NT, Ho DJ, Li X, Allendorf JD, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12:3851–5. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levi E, Klimstra DS, Andea A, Basturk O, Adsay NV. MUC1 and MUC2 in pancreatic neoplasia. J Clin Pathol. 2004;57:456–62. doi: 10.1136/jcp.2003.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adsay NV, Merati K, Andea A, Sarkar F, Hruban RH, Wilentz RE, et al. The dichotomy in the preinvasive neoplasia to invasive carcinoma sequence in the pancreas: Differential expression of MUC1 and MUC2 supports the existence of two separate pathways of carcinogenesis. Mod Pathol. 2002;15:1087–95. doi: 10.1097/01.MP.0000028647.98725.8B. [DOI] [PubMed] [Google Scholar]
- 14.Adsay NV, Basturk O, Cheng JD, Andea AA. Ductal neoplasia of the pancreas: Nosologic, clinicopathologic and biologic aspects. Semin Radiat Oncol. 2005;15:254–64. doi: 10.1016/j.semradonc.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–39. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 16.Croce CM, Calin GA. miRNAs, cancer and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 19.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 20.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue CA, van Heek NT, Cameron JL, et al. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004;164:903–14. doi: 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maitra A, Gazdar AF. Tissue microdissection and processing. Cancer Treat Res. 2001;106:63–84. doi: 10.1007/978-1-4615-1657-6_3. [DOI] [PubMed] [Google Scholar]
- 23.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68:1152–64. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 28.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–12. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 29.Swierczynski SL, Maitra A, Abraham SC, Iacobuzio-Donahue CA, Ashfaq R, Cameron JL, et al. Analysis of novel tumor markers in pancreatic and biliary carcinomas using tissue microarrays. Hum Pathol. 2004;35:357–66. doi: 10.1016/j.humpath.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Cho WC. OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cubilla AL, Fitzgerald PJ. Morphological lesions associated with human primary invasive nonendocrine pancreas cancer. Cancer Res. 1976;36:2690–8. [PubMed] [Google Scholar]
- 33.Edirimanne S, Connor SJ. Incidental pancreatic cystic lesions. World J Surg. 2008;32:2028–37. doi: 10.1007/s00268-008-9633-6. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez-del Castillo C, Targarona J, Thayer SP, Rattner DW, Brugge WR, Warshaw AL. Incidental pancreatic cysts: Clinicopathologic characteristics and comparison with symptomatic patients. Arch Surg. 2003;138:427–3. doi: 10.1001/archsurg.138.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter JM, Cameron JL, Lillemoe KD, Campbell KA, Chang D, Riall TS, et al. Periampullary and pancreatic incidentaloma: A single institution’s experience with an increasingly common diagnosis. Ann Surg. 2006;243:673–80. doi: 10.1097/01.sla.0000216763.27673.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato N, Rosty C, Jansen M, Fukushima N, Ueki T, Yeo CJ, et al. STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. Am J Pathol. 2001;159:2017–22. doi: 10.1016/S0002-9440(10)63053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, Cameron JL, et al. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2002;123:365–72. doi: 10.1053/gast.2002.34160. [DOI] [PubMed] [Google Scholar]
- 39.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 40.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 41.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 42.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 43.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. Bio Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 44.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 46.Eloubeidi MA, Jhala D, Chhieng DC, Chen VK, Eltoum I, Vickers S, et al. Yield of endoscopic ultrasound-guided fine-needle aspiration biopsy in patients with suspected pancreatic carcinoma. Cancer. 2003;99:285–92. doi: 10.1002/cncr.11643. [DOI] [PubMed] [Google Scholar]
- 47.Jhala NC, Jhala DN, Chhieng DC, Eloubeidi MA, Eltoum IA. Endoscopic ultrasound-guided fine needle aspiration: A cytopathologist’s perspective. Am J Clin Pathol. 2003;120:351–67. doi: 10.1309/MFRF-J0XY-JLN8-NVDP. [DOI] [PubMed] [Google Scholar]
- 48.Brugge WR, Lewandrowski K, Lee-Lewandrowski E, Centeno BA, Szydlo T, Regan S, et al. Diagnosis of pancreatic cystic neoplasms: A report of the cooperative pancreatic cyst study. Gastroenterology. 2004;126:1330–6. doi: 10.1053/j.gastro.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Schoedel KE, Finkelstein SD, Ohori NP. K-Ras and microsatellite marker analysis of fine-needle aspirates from intraductal papillary mucinous neoplasms of the pancreas. Diagn Cytopathol. 2006;34:605–8. doi: 10.1002/dc.20511. [DOI] [PubMed] [Google Scholar]
- 50.Khalid A, McGrath KM, Zahid M, Wilson M, Brody D, Swalsky P, et al. The role of pancreatic cyst fluid molecular analysis in predicting cyst pathology. Clin Gastroenterol Hepatol. 2005;3:967–73. doi: 10.1016/s1542-3565(05)00409-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.