Abstract
The Multi-Vesicular Body (MVB) pathway functions in multiple cellular processes including cell surface receptor down-regulation and viral budding from host cells. An important step in the MVB pathway is the correct sorting of cargo molecules, which requires the assembly and disassembly of Endosomal Sorting Complexes Required for Transport (ESCRTs) on the endosomal membrane. Disassembly of the ESCRTs is catalyzed by AAA-protein (ATPase Associated with various cellular Activities) Vps4. Vps4 contains a single AAA domain and undergoes ATP-dependent quaternary structural change to disassemble the ESCRTs. Structural and biochemical analyses of the Vps4 ATPase reaction cycle are reported here. Crystal structures of S. cerevisiae Vps4 in both the nucleotide-free form and the ADP-bound form provide the first structural view illustrating how nucleotide binding might induce conformational changes within Vps4 that leads to oligomerization and binding to its substrate ESCRT-III subunits. In contrast to previous models, characterization of the Vps4 structure now supports a model where the ground state of Vps4 in the ATPase reaction cycle is predominantly a monomer and the activated state is a dodecamer. Comparison with a previously reported human VPS4B structure suggests that Vps4 functions in the MVB pathway via a highly conserved mechanism supported by similar protein-protein interactions during its ATPase reaction cycle.
Keywords: Crystallography, Membrane Trafficking, Vps4, ESCRT, AAA ATPase
INTRODUCTION
A significant number of activated cell surface receptors are down-regulated through endocytosis and delivery to the hydrolytic organelle the lysosome for degradation1. This process is mediated by a special form of the late endosome called the multi-vesicular body (MVB). During MVB formation, MVB cargoes are sorted into invaginating vesicles which further bud into the lumen of the endosome. Fusion of the MVBs with the lysosome delivers the cargo-carrying intralumenal vesicles to the hydrolytic activity of the lysosome for degradation2–4. Sorting of proteins into the MVB pathway is not limited to endocytic cargoes. Lysosomal resident proteins are sorted directly from the Golgi apparatus into the lysosome without transiting the cell surface. In addition to facilitating delivery of proteins to the lysosome, the MVB machinery also participates in the budding of retroviruses from the host cell5 and more recently has been implicated in abscission, the final stage of cytokinesis6.
MVB cargoes are sorted into the MVB pathway via the action of class E Vps (Vacuolar Protein Sorting) proteins, a majority of which are subunits of three distinct protein complexes called ESCRTs (Endosomal Sorting Complexes Required for Transport, -I, -II and –III)7,8. These complexes are transiently recruited from the cytoplasm to the endosomal membrane where they function sequentially in the sorting of cargo proteins into the MVB pathway and possibly in the formation of MVB vesicles3. After protein sorting is completed, the ESCRTs are disassembled and released from the endosomal membrane for further rounds of action3. Disassembly of the ESCRTs requires the activity of Vps4, a member of the AAA-protein (ATPase Associated with various cellular Activities) family9,10. The functional importance of Vps4 is underscored by the fact that loss of Vps4 activity results in accumulation of the ESCRTs on the endosomal membrane, impairment of cargo sorting and blockage of viral budding11–17.
The ESCRT disassembly activity of Vps4 is tightly coupled to the ATPase reaction cycle of Vps4. Disruption of Vps4 ATPase activity through point mutations resulted in the accumulation of ESCRT on the endosomal membrane9. Upon binding ATP, Vps4 undergoes protein oligomerization which enables its binding to the ESCRTs10. When ATP is hydrolyzed into ADP, the stability of the oligomer decreases and it disassembles into a lower molecular weight species in vitro. In order to understand the molecular mechanism by which Vps4 catalyzes the disassembly of the ESCRTs, it is necessary to define the detailed conformational changes within the Vps4 structure during its ATPase reaction cycle. To this end, we have now determined the crystal structures of S. cerevisiae Vps4 both in the absence of nucleotide and in the presence of ADP. This study provides the first structural view detailing how adenine nucleotide-binding might trigger conformational changes within Vps4 leading to protein oligomerization and ESCRT binding. Given the importance of quaternary structural change in the mechanism of Vps4 action, our structural and biochemical characterization now supports a model of Vps4 monomer as the ground state in the ATPase reaction cycle in contrast to a prior notion that Vps4 is a molecular dimer in the absence of ATP binding. Together with a previously reported nucleotide-free human VPS4B structure18, our study suggests that Vps4 functions in the MVB pathway via a highly conserved mechanism supported by similar protein-protein interactions during its ATPase reaction cycle.
RESULTS
Structure determination
The crystal structure of Vps4 from the budding yeast S. cerevisiae was determined using an ATPase deficient mutant (E233Q) fragment that encompasses residues 83–437 (Supplementary Figure 1). The missing 82 residues are known to fold into an independent domain responsible for binding protein substrates including subunits of ESCRT-III19. Deletion of these residues, however, does not affect the ATPase activity or ATP-dependent oligomerization of Vps4 (Supplementary Figure 1) suggesting that the N-terminal domain is dispensable for the ATPase reaction cycle of Vps4. Two different crystal forms were obtained, one in the absence and the other in the presence of 5 mM ADP. The nucleotide-free form belongs to the P6522 space group with one molecule in the asymmetric unit and the ADP-bound form belongs to the P212121 space group with three molecules in the asymmetric unit. Final structure of the nucleotide-free form was refined to a resolution of 2.9Å with an R-factor of 25.9% and an Rfree of 28.8% and that of the ADP-bound form was refined to a resolution of 3.2Å with an R-factor of 25.5% and an Rfree of 30.5% (Table I). Residues not included in the final structural models have no observable electron density and are presumably disordered.
Table 1.
Crystallographic Data Statistics
| Crystal form I (apo) | Crystal form II (ADP) | ||||||
|---|---|---|---|---|---|---|---|
| Data collection statistics | |||||||
| Native | SeMet | Native | |||||
| Wavelength | 0.9799 Å | 0.9801Å | 0.9499Å | ||||
| Space group | P6522 | P6522 | P212121 | ||||
| Unit cell (Å) | a = b = 86.55 Å c = 236.06 Å |
a = b = 87.20 Å c = 235.59 Å |
a = 75.63 Å, b = 120.64 Å, c = 157.33 Å | ||||
| Resolution (Å) | 2.9 | 3.2 | 3.2 | 3.2 | 3.2 | ||
| Completeness (%) | 98.1 (99.2) | 99.6 (97.8) | 99.4 (95.9) | 99.3 (94.8) | 99.4 (97.3) | ||
| I/σa | 37.5 (3.8) | 38.7 (3.6) | 37.3 (3.0) | 37.0 (2.6) | 26.9 (2.5) | ||
| Rmerge (%)a | 6.3 (60.6) | 8.1 (50.5) | 6.8 (55.9) | 6.8 (50.5) | 4.2 (47.5) | ||
| Redundancy | 13.2 | 13.3 | 13.3 | 13.2 | 3.7 | ||
| Unique reflections | 10,792 | 9,425 | 9,428 | 9,466 | 24,259 | ||
| MAD phasing statistics for Crystal form I | |||||||
| Peak | Edge | Remote | |||||
| Isomorphous | Anomalous | Isomorphous | Anomalous | Isomorphous | Anomalous | ||
| Phasing powerb | |||||||
| Centric | - | - | 0.888 | - | 1.011 | - | |
| Acentric | - | 2.438 | 0.928 | 1.552 | 1.159 | 1.524 | |
| FOMc | |||||||
| Centric | 0.346 | ||||||
| Acentric | 0.507 | ||||||
| FOM after DMd | 0.820 | ||||||
| Refinement statistics (Native data set) | |||||||
| Rwork/Rfree(%)e | 25.8/28.8 | 25.5/30.5 | |||||
| Rmsd, bonds (Å) | 0.009 | 0.008 | |||||
| Rmsd, angles (°) | 1.135 | 1.281 | |||||
| Mean B (Å2) | 61.8 | 120.0 | |||||
| Ramachandran | 86.2%/13.0%/0.4%/0.4% | 73.8%/24.8%/1.4%/0% | |||||
| Molecules/ASU | 1 | 3 | |||||
Values in parentheses are for the highest resolution bin.
Phasing power = < [| Fh(calc) |/phase-integrated lack of closure] >.
Figure of merit.
Figure of merit after solvent flattening in the program DM.
R = Σ (|Fobs| − |Fcalc|)/|Fobs|. Rfree is the R value obtained for a test set of reflections that consisted of a randomly selected 5% subset of the diffraction data used during refinement.
Overall structure
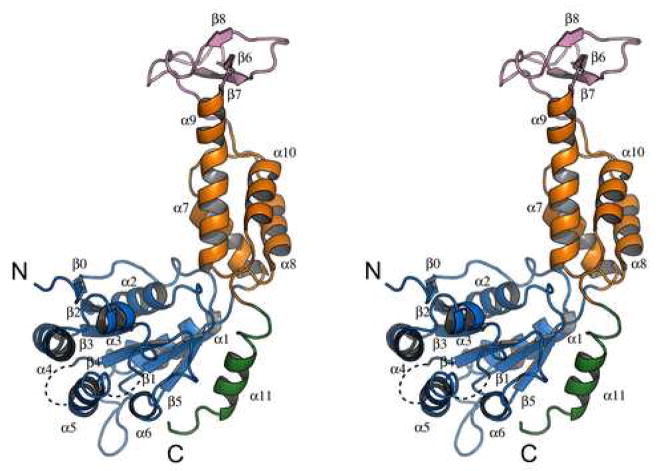
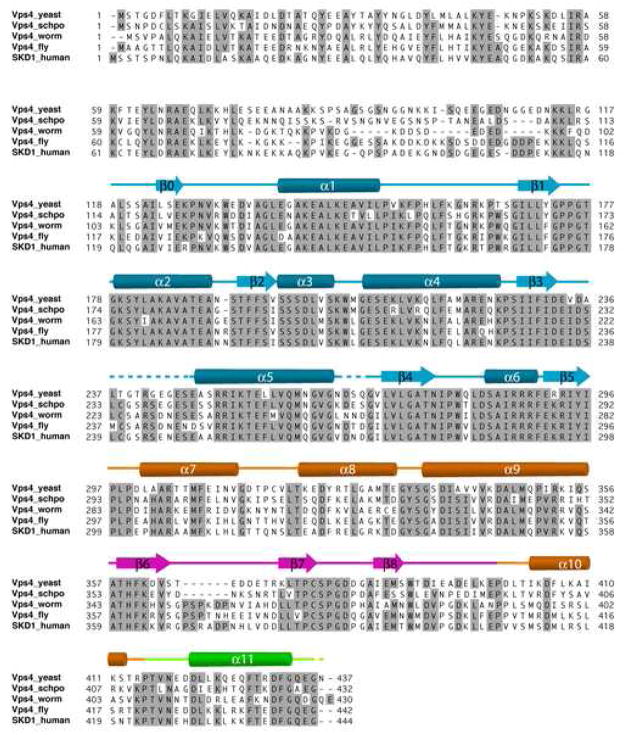
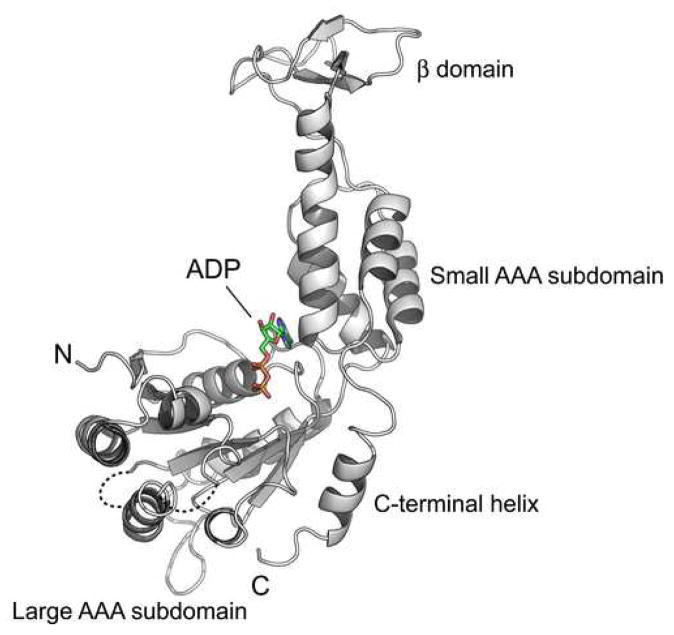
The structure of yeast Vps4 contains a single canonical AAA-domain fold20,21 that can be further divided into a large subdomain and a small subdomain (Figure 1A). The large subdomain has an α/β fold that includes α-helices α1-α6 and β-strands β0-β5. Strands β1-β5 form a parallel β-sheet in the order of β2-β3-β4-β1-β5 with the additional strand β0 running anti-parallel next to strand β2. Six helices are distributed on both sides of the sheet with α1-α2 on one side and α3-α6 on the other. The small subdomain has an all-α fold that includes four α-helices α7-α10. Sequence alignment of Vps4 homologs reveals two unique structural features not seen in other AAA-family proteins, a stretch of 42 to 48 residues inserted between α9 and α10 in the small subdomain (colored pink) and a stretch of extra 23 residues at the C-terminus of the protein (colored green) (Figure 1B). In yeast Vps4, the small subdomain insertion sequence folds into an independent anti-parallel β-strand-containing domain (β6-β8) that has few interactions with the rest of the protein. This domain has been shown to interact with Vps4 regulator Vta118. The C-terminal sequence is structured as a four-turn α-helix (α11) that packs itself onto the large subdomain at the same side of the parallel β-sheet as helices α3-α6. Deletion of the C-terminal helix resulted in an insoluble and unstable mutant protein suggesting that the helix is an integral part of the large subdomain structure (data not shown). The function of the helix is not known but it is thought to participate in subunit interaction upon Vps4 oligomerization5. Vps4 belongs to the ‘meiotic’ clade of the AAA protein family that also includes katanin, spastin and fidgetin22. Like Vps4, these proteins all contain a single AAA domain in their sequences. While the β-domain insertion appears to be a unique feature in Vps4 proteins only, the conserved C-terminal helix has been observed in these other members of the clade suggesting a common mechanism for protein oligomerization23.
Figure 1. Crystal structure of yeast Vps4 and sequence alignment of Vps4 proteins.
(A) Stereo view of the monomeric yeast Vps4 structure in a ribbon representation. The large AAA ATPase subdomain is colored blue; the small AAA ATPase subdomain is colored orange; the β domain is colored pink; and the C-terminal helix is colored green. Structurally disordered regions are represented as dash lines. α-Helices are labeled as α1-α11 and β-strands β0-β8. The N- and C-termini of the structure are also indicated. (B) Sequence alignments of Vps4 proteins from S. cerevisiae, S. pombe, C. elegans, D. melanogaster and H. sapiens. Secondary structural elements identified in the crystal structure of yeast Vps4 are displayed above its sequence. α-Helices shown as cylinders, β-strands arrows and loops as lines. They are labeled and colored using a same scheme as in A. Structurally disordered regions are shown as dash lines. Conserved residues are shaded with gray boxes.
Comparison to human VPS4B structure
The human genome contains two Vps4 genes, VPS4A and VPS4B24. Of the two, VPS4B has an overall 60% sequence identity to that of yeast Vps4 and is able to complement the MVB sorting defect in vps4-null yeast cells24. Comparison between the structures of yeast Vps4 and human VPS4B (in its nucleotide-free form) showed that the overall similarity between the two protein structures is high18. The root mean square difference between the two structures based on 285 Cα positions is 2.2 Å. Each of the eleven α-helices and nine β-strands in yeast Vps4 corresponds closely to one of the secondary structural elements in VPS4B, except for the short two-turn helix α3 where the sequence in VPS4B adopts a more extended loop conformation (Figure 1B). In addition, VPS4B has a slightly larger β insertion domain where a six-residue insertion was seen in the loop that connects strands β6 and β7. The high degree of structural similarity between the two proteins supports the genetic complementation results that the structure and function of Vps4 is well preserved during evolution.
The quaternary structure of Vps4 in its nucleotide-free form
The most apparent structural change of Vps4 during its ATPase reaction cycle is the change in its quaternary structure. Based on results from gel filtration and chemical cross-linking analyses and the fact that Vps4 is a AAA-protein, it has been suggested that the protein inter-converts between a dimer in the absence of nucleotide and a dodecamer in the presence of ATP10,18. If the ground state of Vps4 in the ATPase reaction cycle is indeed a dimer, the dimer structure should exist in both the nucleotide-free and the ADP-bound Vps4 crystals. Examination of crystal packing within the two crystal forms showed a “crystallographic dimer” structure (Supplementary Figure 2). However, we reasoned that it could not represent a structure of a molecular dimer for the following reasons. Each “subunit” in this pair buries 3% of its total solvent-accessible surface and residues at the interface are not conserved. More importantly, mutagenesis of interface residues (M330D/L407D) resulted in a mutant protein whose gel filtration profile did not change appreciably as compared with the wild-type protein (data not shown).
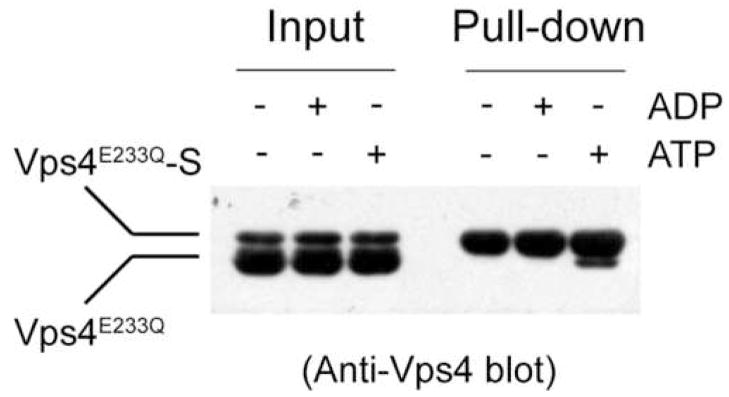
Since the mechanism and regulation of Vps4 ATPase cycle depend on its ground state structure, we asked whether the predominant species of Vps4 in the absence of nucleotide binding is a dimer. To address this, we co-expressed and co-purified Vps4E233Q with C-terminal S-tagged Vps4E233Q from bacteria E. coli. If Vps4 forms a stable dimer, mixing the co-purified proteins with S-protein agarose should lead to retention of both S-tagged and non-tagged proteins due to interaction between the two proteins. The result showed that only S-tagged Vps4 is retained on the beads suggesting that protein-protein interaction between Vps4 in the absence of nucleotide is weak (Figure 2). As a positive control, we also preformed the same experiment in the presence of ATP. Both proteins were retained on the beads indicating protein-protein interaction in the presence of ATP (Figure 2). In addition, we co-expressed N-terminal His-tagged Vps4 together with wild-type Vps4 and performed pull-down experiments using Ni2+-NTA resin. Again, only the His-tagged Vps4 is retained on the beads (data not shown). These results strongly argue that the predominant species of Vps4 in the ground state of its ATPase reaction cycle is a monomer.
Figure 2. Yeast Vps4 exists as a monomer in the absence of ATP.
Untagged Vps4 and C-terminal S-tagged Vps4 were co-expressed and co-purified from E. coli. Protein samples were applied to S-protein agarose under different nucleotide conditions as indicated. After extensive wash, bound proteins were separated on SDS-PAGE and detected by western blotting using anti-Vps4 antibody.
The quaternary structure of Vps4 in its ADP-bound form
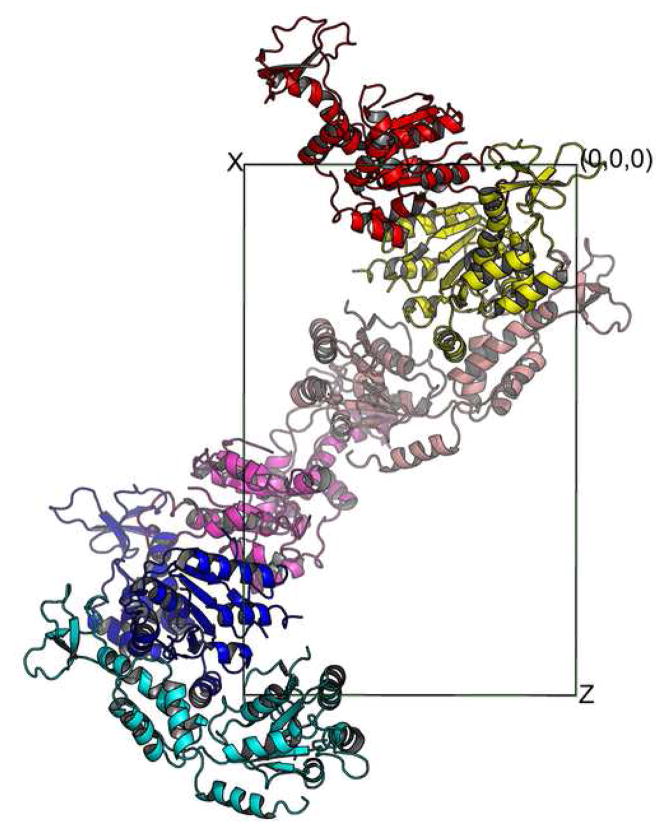
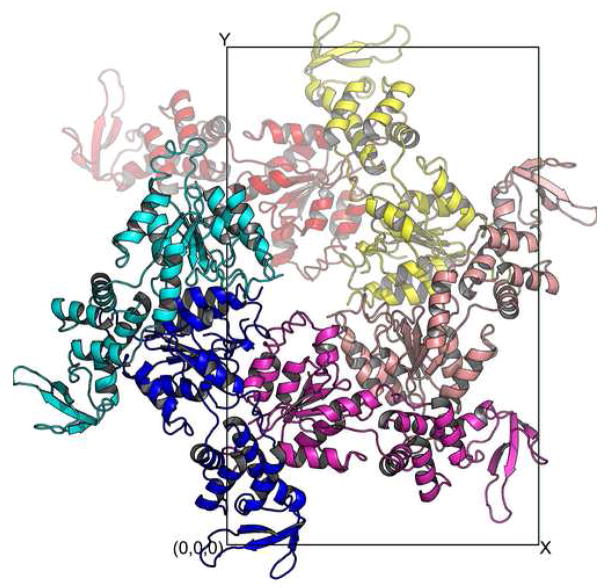
Analysis of protein-protein interaction within the lattice of the ADP-bound Vps4 crystal shows that the protein aggregates into a form that suggests the molecular symmetry for the oligomerized species of Vps4. There are three molecules in the asymmetric unit of the ADP-bound Vps4 crystal. Together with other molecules related by crystallographic symmetry, they form a left-handed helix with a pseudo six-fold screw axis along the z-axis. Each molecule is related to its neighbor by a 60° rotation and 1/6 of unit cell translation along the z axis (Figure 3A). Therefore, if viewed along the z-axis, the projection of these molecules forms a hexameric ring (Figure 3B).
Figure 3. Crystal packing within the lattice of the ADP-bound Vps4 structure.
Six Vps4 molecules along the z-axis in the P212121 space group are shown either down the z-axis (A) or y-axis (B). These six molecules form a pseudo-left-handed helix with a 6-fold screw symmetry.
Although AAA-ATPase with a helical, open assembly has been described25, overwhelming majority of them assemble into ring-shaped assemblies25. However, it is not uncommon that ring-shaped AAA-ATPases crystallize in a helical packing arrangement similar to what is seen in Vps4. For example, bacterial molecular chaperone ClpB crystallizes into a similar helix but forms a ring structure in solution in the presence of ATP as determined by cryo-EM imaging26. Therefore, the molecular arrangement within Vps4-ADP crystal provides strong evidence that supports a hexameric/dodecameric model previously proposed based on structural similarity of Vps4 to the D1 domain of AAA-protein p9718.
ADP binding
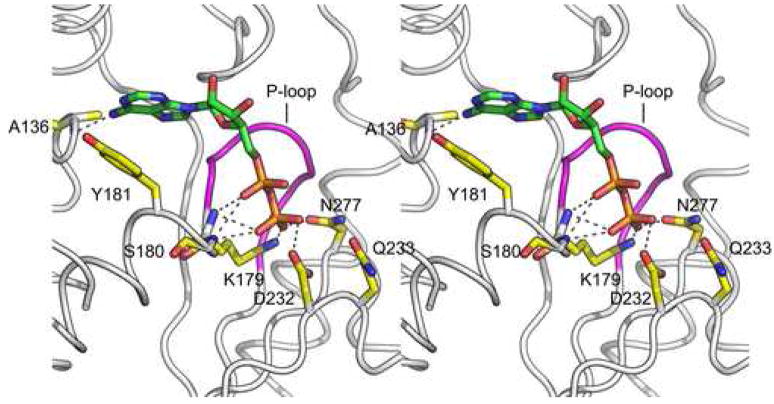
Consistent with what have been observed in other AAA-protein systems20, ADP is bound in a shallow cleft located between the large and small AAA subdomains (Figure 4A). It adopts a rather extended conformation with the ribose ring arching over the P-loop, a structurally conserved motif with an invariant GPPGTG sequence between strand β1 and helix α2. The diphosphate sits in a pocket formed by C-terminal residues of the parallel β-strands β1, β3 and β4 as well as residues from helix α1. Specifically, the oxygen atoms of the β-phosphate form electro-static and/or hydrogen bond interactions with the side chains of highly conserved Lys179, Asp232 and Asn277 (Figure 4B). Lys179 is a Walker A motif (GxxxxGKT, x = any residue) residue and a K179A mutation has been shown to result in a loss in nucleotide binding10. Asp232 is a Walker B motif (hhhhDExx, h = hydrophobic residue) residue and its neighbor Glu233 is catalytically important10. In the ADP-bound structure, Glu233 has been mutated to glutamine. The corresponding residue is about 4.3Å away from the β-phosphate oxygen and is poised to catalyze the cleavage of the β-γ phosphate bond. In addition to aforementioned side chain interactions, the β-phosphate is also engaged in hydrogen bond interactions with backbone amide nitrogen atoms of Lys179 and Ser180.
Figure 4. ADP binding causes conformational changes in Vps4.
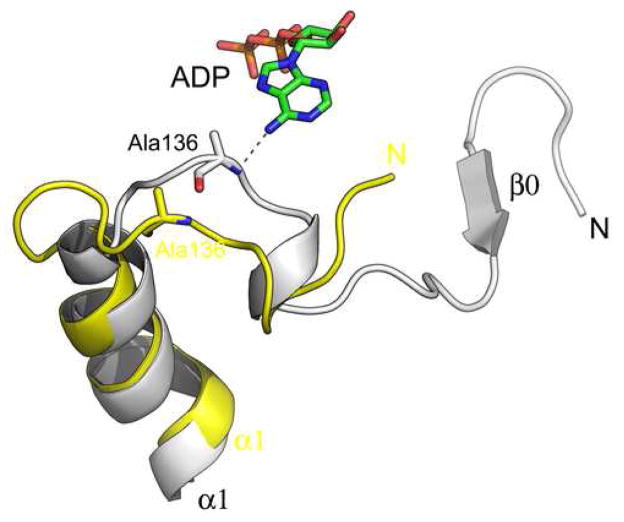
(A) Ribbon representation of the yeast Vps4 structure in complex with ADP. The structure is shown in the same orientation as in Figure 1A. ADP molecule bound in the cleft between the large AAA and small subdomains is shown as a stick model and colored based on its atomic property. (B) Stereo view of the nucleotide-binding pocket of Vps4 showing interactions between the protein and the ADP molecule. The P-loop is highlighted in magenta. Potential hydrogen bond interactions are shown as dash lines. (C) Conformational changes in yeast Vps4 as induced by nucleotide binding. The nucleotide-free structure is colored yellow and the ADP-bound structure is white. Residues 119–128, which forms the β0 strand in the ADP-bound structure are disordered in the nucleotide-free structure therefore not present in the model.
Interactions between the rest of ADP and Vps4 are dominated by van der Waals forces with only a few specific hydrogen bonds. The adenine-binding pocket is largely hydrophobic and is formed by residues from the loop preceding helix α1, helix α2 and small subdomain helix α7 and the loop between α8 and α9 (Figure 4B). Ring stacking interaction is observed between the purine ring and the side chain of the conserved Tyr181, together with a hydrogen bond between the 6′-amine nitrogen and the backbone amide nitrogen of Ala136. Interaction between the adenine and Ala136 appears to be important in communicating the presence of nucleotide in the binding pocket to the N-terminus of Vps4 through which the protein binds substrates including the ESCRT-III subunits. In the absence of ADP, helix α1 bends near its N-terminus and pulls the preceding loop away from the adenine-binding pocket (Figure 4C). As a result, residues 119–129 that form an anti-parallel β-strand (β0) in the ADP-bound structure become disordered in the nucleotide-free structure.
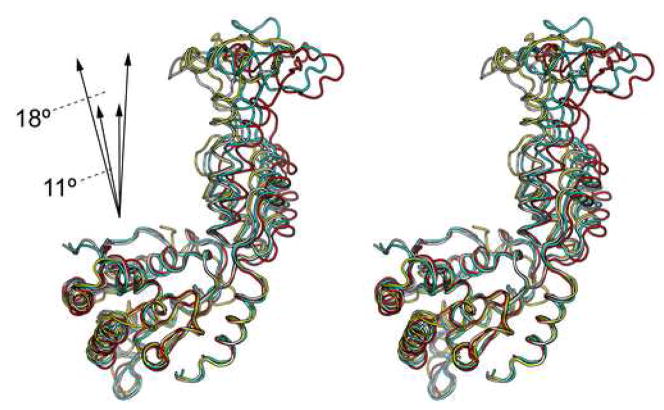
Structural flexibility within Vps4
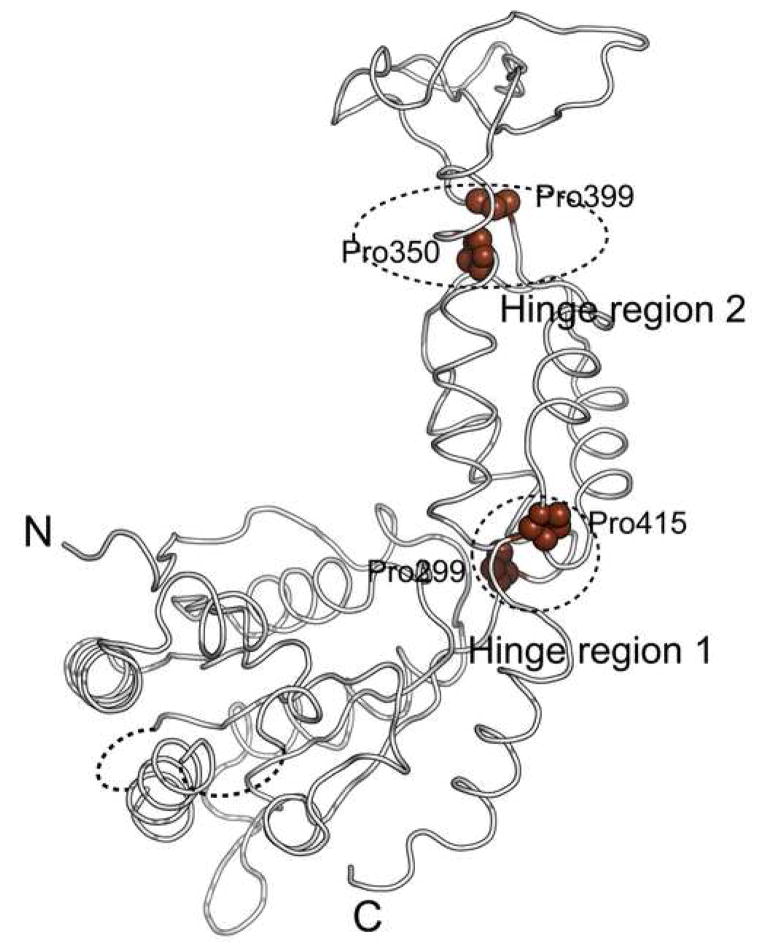
Comparison of the four independently determined Vps4 structures in our study shows that there is significant structural flexibility within the molecule (Figure 5A). The flexibility is displayed as en bloc domain movements as there is little conformational change within the individual domains of Vps4. Two general hinge regions can be identified in the structure. One is located between the large AAA sub-domain and the small AAA sub-domain, consisting of residues Pro299 and Pro415. The other is located between the small AAA sub-domain and the β-domain, consisting of residues Pro350 and Pro399 (Figure 5B). These four proline residues are highly conserved among Vps4 homologs, suggesting that the observed structural flexibility is a general feature inherent to the function of Vps4. The en bloc motion involves an 11° swing between the large and small AAA subdomains and an 18° swing between the small AAA subdomain and the β domain around axes defined by respective hinge residues.
Figure 5. Structural flexibility within Vps4.
(A) Superimposition of the four independently determined Vps4 structures in the current study. The three molecules (ADP-bound) in the asymmetric unit of the P212121 space group are colored white, cyan, and red, respectively. The one molecule (nucleotide-free) in the asymmetric unit of P6522 space group is colored yellow. The magnitude of the en bloc motions between the large AAA subdomain and small AAA subdomain and between the small AAA subdomain and the β domain is illustrated schematically. (B) Two potential hinge regions within Vps4 structure. The conserved proline residues in the hinge regions are shown as spheres and highlighted in brown.
The four Vps4 structures display a progressive opening of the nucleotide-binding site. As a consequence, interactions between the protein and ADP in particular those involving the ribose and adenine rings vary considerably. Although clear electron-density is observed for the di-phosphate in all three ADP-bound molecules, density for the rest of the nucleotide is poor in the two more open structures. This observation suggests that structural flexibility of Vps4 is important for the nucleotide-binding induced conformational change within the molecule.
Structural basis of ATP-dependent ESCRT-III binding
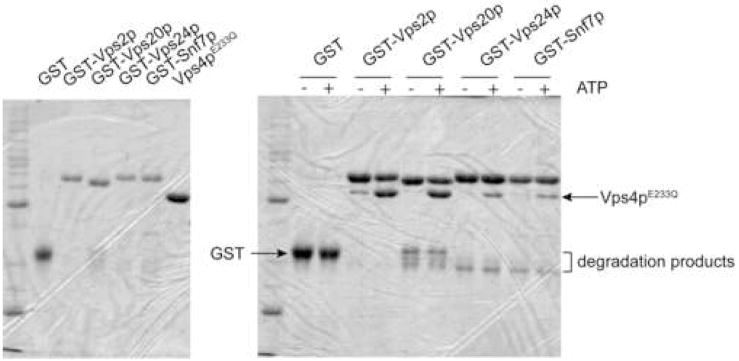
The N-terminal domain of Vps4, whose structure is missing in our current model, has been shown to be responsible for ESCRT-III binding19. To determine whether the interactions are ATP dependent, GST-tagged ESCRT-III proteins (Vps2, Vps20, Vps24 and Snf7) were expressed and purified from E. coli and their interactions with Vps4E233Q were examined using a GST pull-down assay in the absence and presence of ATP. As shown in Figure 6A, it is evident that these interactions either require ATP or are strongly enhanced by ATP.
Figure 6. Nucleotide binding induces conformational change within the N-terminal region of Vps4.
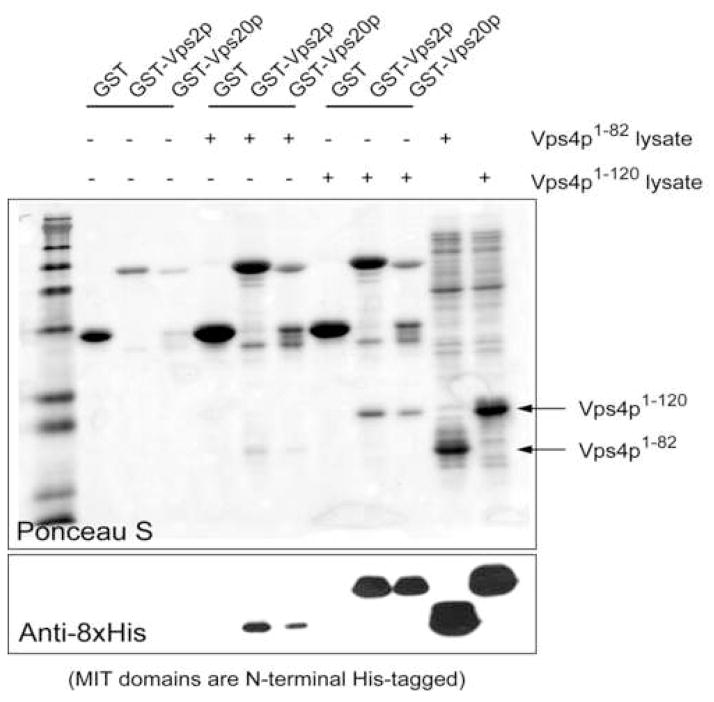
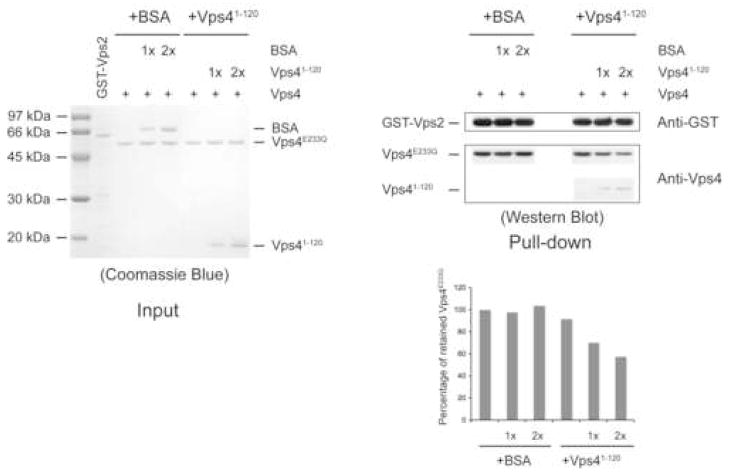
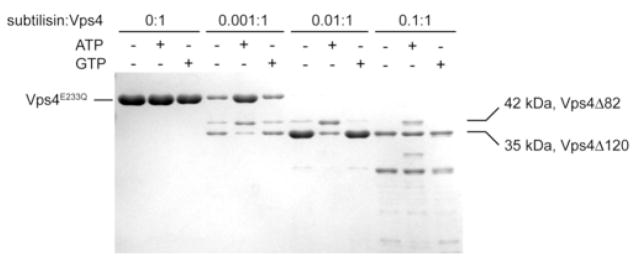
(A) The ESCRT-III subunits interact with Vps4 in the presence of ATP. ESCRT-III subunits Vps2, Vps20, Vps24 and Snaf7 were expressed as GST-tagged fusion proteins and bound to glutathione-agarose beads (left panel). Purified Vps4E233Q was loaded onto the ESCRT-III subunit-bound matrix either in the absence (−) or presence (+) of ATP. Proteins retained on the matrix after extensive washes were separated on 12% SDS-PAGE gel and stained with Coomassie Blue (right panel). (B) The N-terminal domain of Vps4 interacts with the ESCRT-III subunits. Vps2 and Vps20 were expressed as GST-tagged fusion proteins and bound to glutathione-agarose beads. Cell lysate containing His8-Vps41–82 or His8-Vps41–120 was loaded onto GST-Vps2 or GST-Vps20 bound matrix. Proteins retained on the matrix after extensive washes were separated on the SDS-PAGE gel and detected by either Ponceau S staining (top panel) or anti-His antibody (bottom panel). (C) Vps41–120 competes with full-length Vps4 for binding to the ESCRT-III subunit Vps2. GST-Vps2 was bound to glutathione-agarose beads. Purified Vps4E233Q was loaded onto Vps2-bound matrix in the presence of ATP and increasing amounts of BSA or Vps41–120. Proteins retained on the matrix were separated on SDS-PAGE gel and detected by western blotting with anit-Vps4 antibody. The amount of Vps4E233Q was quantified by program ImageJ and shown in a bar diagram (the amount in the first lane was set as 100%). (D) Vps4 undergoes conformational change at the linker region upon ATP binding. Vps4E233Q was incubated with increasing amounts of subtilisin at 4°C for 30 min with different nucleotides. Digestion products were separated on 15% SDS-PAGE, followed by Coomassie staining.
There are two possible explanations for the ATP dependency of these interactions. The N-terminal domain might not be accessible to the ESCRT-III protein in the absence of ATP. Alternatively, stable association may require the ESCRT-III protein to simultaneously bind to more than one Vps4 subunit; a condition only met in the ATP-induced Vps4 oligomeric structure. To distinguish between these two possibilities, we expressed and purified two Vps4 N-terminal fragments, Vps41–82 and Vps41–120. Both protein fragments were able to bind to the ESCRT-III subunit Vps2 and Vps20 though the interaction with Vps41–120 appears to be stronger (Figure 6B). Purified Vps41–120 also effectively competes with full-length Vps4E233Q for binding to Vps2 as shown by decreased amounts of full-length Vps4E233Q retained by GST-Vps2 when Vps41–120 was included in the binding assay (Figure 6C). These results showed that an isolated, monomeric N-terminal domain is sufficient for binding to the ESCRT-III protein. Rather than the valency requirement, it is the ATP-binding induced conformational change in Vps4 structure that leads to the increased accessibility of the N-terminal domain. Similar ATP-binding induced conformational change in the substrate-binding domain has been observed in other AAA protein systems such as p9727,28.
To probe ATP-binding induced conformational change within Vps4 structure, we subjected Vps4 to mild protease treatment in the absence and presence of ATP. Purified proteins were incubated with increasing amounts of subtilisin and the reaction was stopped after 30 minutes on ice. As can be seen in Figure 6D, in the absence of ATP, Vps4 was quickly converted into a fragment of 35 kDa that corresponds to residues 121 to the C-terminus (Vps4Δ120). In contrast, a stable fragment of 42 kDa accumulated before the appearance of the 35 kDa fragment in the presence of ATP. The new fragment contains residues 83 to the C-terminus (Vps4pΔ82). Interestingly, this region corresponds to the linker that connects the ESCRT-III binding domain and the ATP-binding AAA domain. This effect was specific for ATP as inclusion of GTP in the digestion mixture did not lead to the accumulation of Vps4pΔ82 (Figure 6D). As expected, ATP had no effect on the subtilisin digestion pattern of trigger factor, a molecular chaperone that does not bind to ATP29 (Supplementary Figure 3). Together, these results demonstrated that the linker undergoes conformational change in response to ATP binding and this is likely the structural basis for the ATP-dependent ESCRT-III binding to the full-length Vps4.
Discussion
Vps4 catalyzes the dissociation of the ESCRTs from the endosomal membrane in the MVB pathway. This function is critical to a number of cell biology processes including cell surface growth hormone receptor down-regulation and budding of retroviral particles from host cells as dysfunction of Vps4 will lead to the blockage of these processes. Among proteins that have been implicated in the proper function of the MVB pathway, Vps4 is one of the most conserved proteins with a high level of sequence identity between yeast and human proteins. Comparison of yeast and human Vps4 crystal structures confirms the structural conservation suggesting that all Vps4 proteins likely function through a common mechanism.
As a member of the AAA protein family, Vps4 derives its ability to dissociate and remodel protein complexes from ATP binding and hydrolysis driven conformational changes within its structure. While most AAA proteins contain two AAA domains with one defective in ATP hydrolysis thus enabling the formation of a stable protein assembly, Vps4 has only one AAA domain23. As ATP is hydrolyzed, the assembly is weakened resulting in cyclic quaternary structural change. Based on results from gel filtration and cross-linking experiments, it has been proposed that Vps4 cycles between a dimer structure and a structure containing 10–12 subunits10. Molecular packing within the Vps4-ADP crystal favors a model of Vps4 high-order oligomer with a six-fold symmetry. Therefore, it is likely that the high-order oligomer contains 12 subunits arranged as two hexameric rings as observed in most AAA proteins. Our structural and biochemical analysis, however, does not support that Vps4 oligomer dissociates into a stable dimmer upon ATP hydrolysis and subsequent release of the nucleotide. Instead, it suggested that the ground state structure of Vps4 ATPase reaction cycle is predominantly a monomer. Our results, however, does not rule out weak protein-protein interaction between Vps4 subunits in the absence of nucleotide. In deed, analytical ultracentrifugation analysis suggests that the solution behavior of Vps4 is complicated and cannot be fitted into a simple one species model (data not shown).
While ADP binding to Vps4 is not sufficient to support a stable oligomeric structure as would ATP, insights regarding conformational change of Vps4 induced by ATP binding can be gained from the Vps4-ADP structure. It is likely that ATP binding results in similar en bloc domain movements within the Vps4 structure, albeit larger in magnitude, which favors protein-protein interactions between Vps4 subunits leading to protein oligomerization. The reason that ADP is unable to support a closed ring structure may be due to the lack of additional inter-subunit interactions stabilized by the presence of the γ-phosphate19. Interestingly, nucleotide binding also causes en bloc movement of the β domain relative to the core AAA domain (Figure 5A). Given that the β domain is responsible for binding to Vta1 in the presence of ATP, this observation suggests that ATP binding to Vps4 not only causes local conformational changes at the lower portion of the molecule but its effect is also propagated to the distant part of Vps4 resulting conformational changes that support its interaction with Vta1 as well.
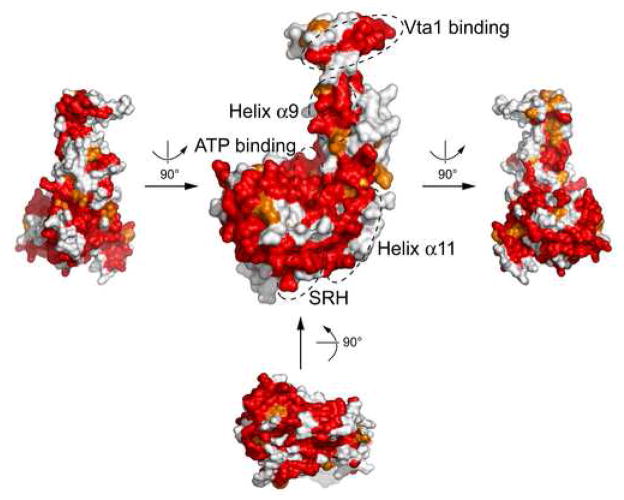
Examination of yeast Vps4 structure reveals several highly conserved surface patches. They are mostly located on the large AAA subdomain but the small AAA subdomain and the β domain each contains one prominent patch (Figure 7). A significant fraction of the conserved surfaces on the large subdomain are involved in binding nucleotide as has been shown in the ADP-bound Vps4 structure. The function for the rest of the conserved surfaces is less clear but probably involved in ATP-dependent oligomerization. Similarly, the conserved patch on the small subdomain formed by residues of helix α9 is probably also involved in protein oligomerization. The conserved patch on the β domain formed by loop residues between strands β7 and β8 is likely involved in Vta1 binding as mutation of Ser377 and Asp380 of Vps4 in the region abolishes its ability to bind to its regulator18.
Figure 7. Conserved surface patches on Vps4.
Surface representation of yeast Vps4 is shown in several different orientations as indicated. They are colored based on the conservation of underlying residues as presented in Figure 1B: identical residues red and homologous ones orange. Surface patches known to be involved in ATP binding and Vta1 binding are indicated. In addition, those formed by residues from helix α9 and α11 as well as SRH (Second Region of Homology, a region likely involved in inter-subunit interactions) are also highlighted.
In summary, our structural and biochemical analyses of yeast Vps4 structure suggest that Vps4 likely functions in the MVB pathway via a highly conserved mechanism supported by similar protein-protein interactions through its ATPase reaction cycle. Our study provides the first structural view illustrating how nucleotide binding might induce conformational changes within Vps4 that leads to oligomerization and binding to its substrate ESCRT-III. In addition, inherent hinged domain movements within the Vps4 structure may also play a role in supporting nucleotide binding. In contrast to previous models, our characterization of Vps4 now supports a model where the ground state structure of Vps4 ATPase reaction cycle is predominantly a monomer and the activated state structure is a dodecamer. While a high resolution structure of Vps4 oligomer will ultimately be required to define the ATP driven structural changes within Vps4, our study provides a platform where these changes can now be probed experimentally by mutagenesis and other methods.
METHODS
Cloning, expression and purification
DNA fragment encoding Vps4 was amplified from the Saccharomyces cerevisiae genomic DNA and cloned into the BamHI/XhoI sites of a modified pET21a vector that contains an N-terminal His8-tag and a TEV protease cleavage site. E. coli strain Rosetta(DE3) harboring the His8-TEV-Vps4 expression plasmid was grown at 37°C in LB medium to an OD600 of 0.6–0.8 before induced with 0.4 mM IPTG at 25°C for 4 hours. The cells were harvested by centrifugation and frozen at −80°C.
All protein purification steps were performed at 4°C. Frozen cell pastes were resuspended in 100 ml buffer A (50 mM Tris, pH 8.0, 300 mM NaCl, 1 mM MgCl2) supplemented with 10 μg/ml PMSF and disrupted by sonication. Supernatant from centrifugation was loaded on a Ni2+-NTA affinity column. After the column was washed extensively with buffer A, His8-TEV-Vps4 was eluted with a 0–250 mM imidazole gradient in buffer A. Fractions containing Vps4 were pooled, incubated with TEV protease (100:1), and dialyzed against 50 mM Tris, pH 8.0, 50 mM NaCl, 1 mM MgCl2 overnight. A second Ni2+-NTA step removed the tag and TEV protease. Vps4-containing fractions was then placed into buffer C (25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM MgCl2) and further purified using an anion exchange Resource-Q column (Amersham Pharmacia) with a 0.1–0.5 M NaCl gradient. For MAD phasing, selenomethionyl protein were expressed in E. coli B834(DE3) using a minimal medium where methionines were replaced with selenomethionines30. Derivative proteins were purified in a similar way as native proteins.
Crystallization and data collection
For crystallization, purified protein was stored in a final buffer containing 20 mM Tris, pH 7.5, 400 mM NaCl, 5 mM MgCl2 and concentrated to 25 mg/ml. The nucleotide-free crystal form was obtained by the sitting drop method at 20°C. Proteins were mixed in a 1:1 ratio with a reservoir solution containing 1.0 M sodium acetate, 0.2–0.25 M cadmium sulfate, and 0.1 M PIPES, pH 6.5 in a final volume of 4 μl and equilibrated against the reservoir solution. Crystals grew to full size in several days and were transferred into 1.05 M sodium acetate, 0.25 M cadmium sulfate, 0.1 M HEPES, pH 7.0, and 35% sucrose and equilibrated against the same solution for several hours before flash-frozen under liquid nitrogen. The adjustment of buffer, pH and the soaking step improved the diffraction quality of the crystal. Selenomethionyl crystals were prepared in a similar manner.
The ADP crystal form was also grown by the sitting drop method at 20°C. Proteins in the presence of 5mM ADP were mixed in a 1:1 ratio with a reservoir solution of 1.3–1.4 M Na/K-phosphate, pH 5.8, 10% glycerol, 5% glucose in a final volume of 4 μl and equilibrated against the reservoir solution. Crystals grew to full size in a week and were often clustered. Single crystals used for data collection were obtained by seeding. Crystals were transferred into 1.6 M Na/K-phosphate, pH 5.8, 1 mM ADP, and 35% sucrose and flash-frozen in liquid nitrogen before data collection. All diffraction data were collected at Advanced Photon Source beam line 23-ID. Diffraction data were integrated and scaled by using the program HKL2000 (HKL Research).
Structure determination and refinement
The nucleotide-free structure was solved initially at 3.2 Å resolution using the MAD method31. Four of the seven expected selenium sites were found with SnB32. Two additional sites were identified using anomalous difference Fourier method as implemented in SHARP33. Parameters for these six sites were refined and MAD phases were calculated and improved by solvent flattening with SHARP. The resulting experimental map was interpretable to allow the building of a polyalanine model in O34. Cycles of rebuilding and refinement were carried out using the program O and CNS35, while phases were extended to 2.9 Å resolution using a native data set. Initial refinement was carried out with torsion angle dynamics simulated annealing using the maximum likelihood target function with the experimental phases as a prior phase distribution (MLHL). A randomly selected 5% of the total reflections were used for cross-validation. Later rounds of refinement were performed with REFMAC5 in the CCP4 suite with TLS parameters incorporated36. The final structure contains residues 129–206, 210–239, 246–264, 268–365, and 371–433. The refined model has 86.2% of the residues with the most favored backbone conformations and only one residue with disallowed conformation on the Ramachandran plot.
The ADP structure was determined by the molecular replacement method with the program PHASER37 using the partially refine nucleotide-free structure as a search model. The β-domain and most of the loop regions were removed during the molecular replacement process. The model was refined using CNS with intersperse manual rebuilding with O. Three Vps4 molecules were found in the asymmetric unit and they were initially refined with strict non-crystallographic symmetry (NCS) constraint. However, as the refinement progressed, it was clear that they differ from each other quite significantly therefore only weak NCS restraint was applied during the later stage of the refinement. The final structure contains residues 119–237, 249–262, and 270–435 for molecule A and B, and 123–237, 249–262, and 270–432 for molecule C. A complete ADP molecule was built in molecule A but only a phosphate group was built in molecule B and C due to poor electron density quality for the rest of the ADP molecule.
In vitro binding experiments
For the S-tag pull-down experiments, untagged Vps4E233Q and C-terminal Stagged Vps4 E233Q were co-expressed using pETDuet-1 vector (Novagen) and co-purified by anino-exchange chromatography followed by gel filtration chromatography. Protein samples were applied to S-protein agarose (Novagen) in the absence or presence of 1 mM adenosine nucleotides. After extensive wash, bound proteins were separated by SDS-PAGE and detected by Western blotting with anti-Vps4 antibody (Santa Cruz, sc-21821).
For GST pull-down experiments, DNAs encoding yeast Vps2, Vps20, Vps24, and Snf7 were amplified from the S. cerevisiae genomic DNA and cloned into a modified pET41a vector (Novagen). GST tagged proteins were expressed in E. coli BL21 (DE3) cells, induced with 0.4 mM IPTG at 16°C overnight. These cells were resuspended in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) supplemented with 1 mM DTT and 0.1% Triton X-100 and lysed by mild sonication. 20 ml glutathione-agarose (Sigma) pre-equilibrated with PBS/DTT buffer was incubated with soluble cell lysates at 4°C for 40 minutes. The beads were subsequently washed with the above buffer and incubated with purified Vps4E233Q proteins or cell lysate containing His8-tagged Vps4 N-terminal domains at 4°C for 40 minutes. ATP was added to a final concentration of 1 mM where indicated. Bound proteins were analyzed by SDS-PAGE.
Limited proteolysis
Subtilisin was used in the assay with protease to protein ratios in the following order: 0/1, 0.001/1, 0.01/1, and 0.1/1. For a typical reaction, 1 mg/ml Vps4E233Q protein was incubated with subtilisin in the reaction buffer (20 mM HEPES, pH 7.5, 200 mM NaCl, 10% glycerol). Reactions were allowed to proceed for 30 minutes on ice before terminated by addition of protease inhibitor PMSF. Samples were boiled in SDS-loading buffer, subjected to SDS-PAGE and visualized by Coomassie staining.
Protein Data Bank accession number
Atomic coordinates and structure factors for yeast Vps4 have been deposited in the RCSB Protein Data Bank with accession code 2QP9 for the nucleotide-free form and 2QPA for the ADP-bound form.
Supplementary Material
Acknowledgments
We thank the staffs of the Center for Structural Biology at the University of Michigan for maintaining the X-ray facility and the staffs at the APS GM/CA-CAT beamline in the Argonne National Laboratory for access and help with data collection; X. Chen for technical help; A. Ludlam for helps at early stage of this project; R. Matthews and D. Katzmann for critically reading the manuscript. This work was supported in part by NIH grants to Z.X. (R01-DK65980).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 2.Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 3.Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 4.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 5.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 6.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 7.Hurley JH, Emr SD. The ESCRT Complexes: Structure and Mechanism of a Membrane-Trafficking Network. Annu Rev Biophys Biomol Struct. 2006 doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 9.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. Embo J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. Embo J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 12.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 15.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 16.Sachse M, Strous GJ, Klumperman J. ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J Cell Sci. 2004;117:1699–1708. doi: 10.1242/jcs.00998. [DOI] [PubMed] [Google Scholar]
- 17.Fujita H, Yamanaka M, Imamura K, Tanaka Y, Nara A, Yoshimori T, Yokota S, Himeno M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 18.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. Embo J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A. 2005;102:13813–12818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 21.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 22.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheuring S, Rohricht RA, Schoning-Burkhardt B, Beyer A, Muller S, Abts HF, Kohrer K. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol. 2001;312:469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 25.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 27.DeLaBarre B, Brunger AT. Nucleotide Dependent Motion and Mechanism of Action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 28.Davies JM, Tsuruta H, May AP, Weis WI. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure (Camb) 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Ludlam AV, Moore BA, Xu Z. The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae. Proc Natl Acad Sci U S A. 2004;101:13436–13441. doi: 10.1073/pnas.0405868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z, Knafels JD, Yoshino K. Crystal structure of the bacterial protein export chaperone secB. Nat Struct Biol. 2000;7:1172–1177. doi: 10.1038/82040. [DOI] [PubMed] [Google Scholar]
- 31.Hendrickson WA, Ogata CM. Phase Determination from Multiwavelength Anomalous Diffraction Measurements. Methods in Enzymology. 1997;276:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 32.Weeks CM, Miller R. The design and implementation of SnB v2.0. J Appl Cryst. 1999;32:120–124. [Google Scholar]
- 33.Fortelle EdlBG. Maximum-Likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Methods in Enzymology. 1997;276 doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 34.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.