Abstract
Hypermutation and class switch recombination of immunoglobulin genes are antigen-activated mechanisms triggered by AID, a cytidine deaminase. AID deaminates cytidine residues in the DNA of the variable and the switch regions of the immunoglobulin locus. The resulting uracil induces error-prone DNA synthesis in the case of hypermutation or DNA breaks that activate non-homologous recombination in the case of class-switch recombination. In vitro studies have demonstrated that AID deaminates single-stranded but not double-stranded substrates unless AID is in a complex with RPA and the substrate is actively undergoing transcription. However, it is not clear whether AID deaminates its substrates primarily as a monomer or as a higher order oligomer. To examine the oligomerization state of AID alone and in the presence of single stranded DNA substrates of various structures, including loops embedded in double-stranded DNA, we used atomic force microscopy (AFM) to visualize AID protein alone or in complex with DNA. Surprisingly, AFM results indicate that most AID molecules exist as a monomer and that it binds single-stranded DNA substrates as a monomer at concentrations where efficient deamination of single-stranded DNA substrates occur. The rate of deamination, under conditions of excess and limiting protein, also imply that AID can deaminate single-stranded substrates as a monomer. These results imply that non-phosphorylated AID is catalytically active as a monomer on single stranded DNA in vitro, including single-stranded DNA found in loops similar to those transiently formed in the immunoglobulin switch regions during transcription.
1. Introduction
B cells undergo three mechanisms of somatic gene alteration that profoundly impact the recognition and binding of foreign antigen by antibodies. V(D)J recombination is a developmentally programmed mechanism that generates the diverse pre-immune antibody repertoire. Class switch recombination (CSR) and somatic immunoglobulin (Ig) hypermutation (SHM) are antigen-activated mechanisms that lead to the formation of isotype-switched, high affinity antibodies characteristic of the secondary immune response [1–7]. Both CSR and SHM as well as Ig gene conversion in other species such as rabbit and chicken require the cytidine deaminase, AID [8–12]. AID deaminates cytidines, generating uracils in the DNA encoding the variable and the switch regions of the immunoglobulin locus [13,14]. In CSR, the uracil is removed from the DNA of the involved switch regions by uracil DNA glycosylase (UNG), generating abasic sites [14]. The requirement for at least part of the non-homologous end-joining double-strand break DNA repair machinery in CSR suggests that these abasic sites lead to breaks that trigger the switch reaction [15–17]. Indeed, AID-dependent DNA breaks have been extensively documented for the switch regions [17–22]. In SHM, the resolution of the AID-generated uracil in the DNA is less clear. It appears that hypermutation at G:C base pairs that results from deamination of cytidine residues in the DNA of Ig V regions can originate from replication over the uracil leading to G:C transitions. Alternatively, it can originate from excision of uracil by UNG presumably via a base excision repair mechanism leading to both transitions and transversions at G:C base pairs [14]. Mutations at A:T base pairs, while entirely dependent on AID (AID deficient mice lack mutations from both A:T and G:C base pairs), are likely an indirect result of AID-mediated deamination involving the mismatch DNA repair proteins and translesion synthesis DNA polymerases (TLS) [23–36; for a review see 37]. The requirement for mismatch repair proteins and TLS DNA polymerases for the A:T phase of hypermutation suggests that the G:U mismatch generated by AID initiates a mismatch repair synthesis patch that utilizes the error-prone TLS polymerases instead of the high fidelity DNA polymerases [37]. This model is still speculative since the resolution of the AID-generated uracil in the DNA of Ig V regions leading to mutations at A:T base pairs remains unclear. Finally, it is possible that in addition to the DNA encoding Ig V regions, AID may also deaminate an RNA encoding molecules critical to SHM and CSR [38], although an RNA target has not been identified.
AID deaminates single-stranded but not double-stranded DNA in vitro [39–42] except for double-stranded DNA that is actively undergoing transcription [42–45], particularly if AID is phosphorylated by PKA and in a complex with RPA. AID preferentially deaminates microsequences associated with SHM hotspots (WRCY) [39, 46–49], providing a rationale for the existence of SHM hotspots that are predominantly found in the complementarity determining regions (CDR’s). Data from “pull-down” assays using tagged-AID proteins [50] and from structural modeling based on homologous cytidine deaminases [51] suggest that AID may function as a dimer to deaminate DNA. However, it remains unclear whether or not AID is catalytically active as a monomer. Herein, we examine AID deamination of single-stranded DNA alone or embedded within double-stranded DNA in the form of loops. We determined the oligomeric state of the protein by direct visualization using atomic force microscopy (AFM) [52]. Surprisingly, the AFM experiments demonstrate that the dominant AID species is a monomer and that dimerization is not promoted by the presence of single-stranded DNA. These AFM data taken together with a kinetic analysis of deamination suggest that a monomer of AID is sufficient to catalyze the deamination of single-stranded DNA in vitro.
2. Methods and Materials
2.1 Preparation of oligonucleotide substrates
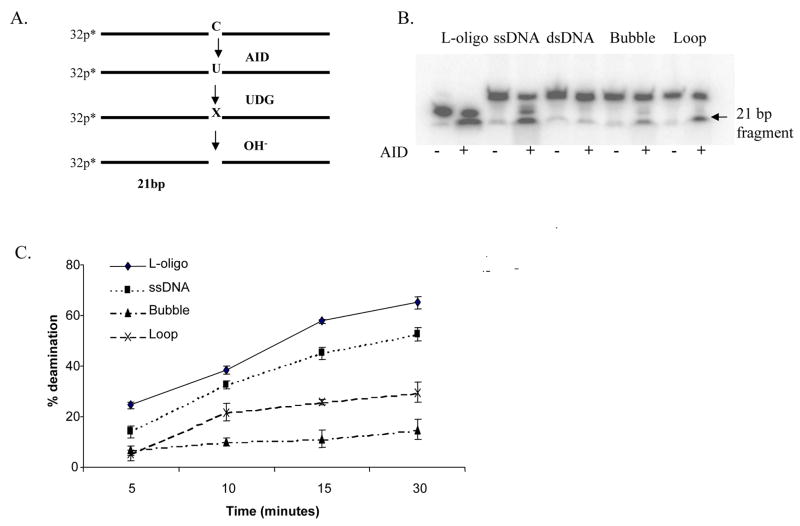
The oligonucleotides used in the deamination assays and the predicted structures of the double-stranded oligonucleotides are depicted in figure 1. The “L-oligo” was designed based on the primer used by Yu and colleagues [46] while the single-stranded DNA oligonucleotide (AID1 primer) was based on the CDR2 sequence of the human Ig heavy chain VH3-30-3. Oligonucleotides (100 μM) were 5′-end labeled with γ-32P ATP (3000 Ci/mmol, Amersham Biosciences) using Optikinase (United States Biochemical Corp.) according to the manufacturer’s instructions. The reaction was terminated by the addition of 20 mM EDTA, and the enzyme was heat denatured by incubation for 10 minutes at 65 °C. After labeling, the free nucleotides were removed by gel filtration chromatography (Micro Bio-spin-6, Bio-Rad). For the generation of double-stranded substrates, the labeled oligonucleotide was annealed with the complimentary strand [(AID1+AID2 = double strand), (AID1+AID8 = Bubble), and (AID1+AID9 = Loop) see Fig. 1] using 1.2 molar excess of cold oligonucleotides in the presence of 100 mM KCl, heated at 65°C for 5 minutes and slowly cooled to room temperature. The duplexes were analyzed on a native 10% polyacrylamide gel.
Figure 1.
Sequence of substrates used for deamination assays and predicted structures for the duplexed oligonucleotides.
2.2 Deamination assays
Typically, a reaction mixture (10 μl) containing 2–100 nM radioisotope-labeled oligonucleotide substrate, 5nM-2000nM purified human AID protein (Enzymax, University of Kentucky), 100ng of RNase A (Qiagen) and 1 unit of uracil DNA glycosylase (Invitrogen) was incubated for 15 min at 37 °C in a buffer containing: 25 mM Tris-Cl, pH 8.0, 50 mM NaCl, and 5 mM EDTA (see scheme in Fig. 2a). C’terminus (histidine)6-tagged AID protein was purified from E. coli by Enzymax (The University of Kentucky, Advanced Science and Technology Commercialization Center) by affinity column (HiTrap Chelating HP ™ followed by HiTrap Heparin HP ™, GE Healthcare) using an elution buffer consisting of 50 mM Tris (pH of 7.5), 5 mM β-mercaptoethanol, 10% glycerol and 500 Mm NaCl. Also, a mutant form of human AID wherein the second cysteine in the active site (position 90) was mutated to alanine was used as a control and it was purified under identical conditions as the wild type AID, by Enzymax. We confirmed the purity of the AID protein preparation using a Coomassie stained gel with SimplyBlue SafeStain (Invitrogen, Carlsbad, USA) per manufacturer’s instructions, and in deamination assays, the wild type protein but not the mutant form, deaminated a single-stranded DNA substrate (Supplemental figure). To examine deamination at limiting and excess AID protein concentrations, deamination reactions were carried out at final AID protein concentrations ranging from 5 nM to 2 μM and oligo concentrations ranging from 2 nM to 100 nM. For a subset of samples, RNAse A (400 ng) (Qiagen) was pre-incubated with AID for 30 minutes prior to addition of the oligo. The reaction was stopped by adding 1 μl of 2 M NaOH and heated for 5 min at 95°C. Ten μl of formamide was added and samples were heated at 95°C for 5 min and loaded on 10% denaturing polyacrylamide gels containing 7 M urea. Gels were run in 1X Tris borate-EDTA (TBE) buffer under constant voltage at ambient temperature. Gels were dried and visualized by autoradiography using the phosphor-imager Typhoon 9400 (Amersham Biosciences) and the bands quantified by ImageQuant software (Molecular Dynamics). Deamination rates were determined by fitting the percentage of deamination with time to first order exponentials. All deamination reactions were repeated at least two times.
Figure 2.
AID deaminates single-stranded DNA in various configurations. A) Schematic representation of deamination reaction and expected product following incubation of end-labeled oligonucleotide with AID and UNG. B) AID deaminates most oligonucleotides where single-stranded DNA (ssDNA) is exposed but fails to deaminate double-stranded DNA (dsDNA), C) Quantitation of autoradiographs after deamination reactions. The substrate devoid of secondary structure (L-oligo) deaminated best but there was substantial deamination of loop structures over time. All reactions contained 2 nM oligonucleotide and 500 nM AID protein and were repeated at least three times.
2.2 Atomic Force Microscopy
AID protein was diluted to final concentrations of 5, 10, and 20 nM in imaging buffer (20 mM HEPES, pH 7.3/10 mM MgOAc/100mM NaOAc). In a subset of experiments AID was incubated at 500 nM and then quickly diluted to 10 nM for deposition. Deposition of AID at concentrations higher than 20 nM resulted in too many proteins on the surface, thereby preventing a statistical analysis. For depositions in the presence of DNA, AID was incubated with various concentrations of DNA oligonucleotides (5 nM-50 nM) for 30 minutes at room temperature prior to deposition. After incubation, 10 μl of AID in the presence or absence of DNA was deposited onto freshly cleaved mica (Spruce Pine Mica Company, Spruce Pine, NC). The sample was rinsed immediately with nanopure water, excess water was wicked from the surface using filter paper, and the surface was then dried using a stream of nitrogen. AFM images were captured in air using either a Nanoscope III or IIIa (Digital Instruments, Santa Barbara, CA) microscope in tapping mode. Pointprobe Plus tapping mode silicon probes (Molecular Imaging, Tempe, AZ) with resonance frequencies of approximately 170 kHz were used for imaging. Images were collected at a speed of 2–3 Hz with an image size of 1 μm × 1 μm at 512 × 512 pixel resolution. Each experiment was repeated at least twice. Volume analysis was done as previously described [52]. The program Kaleidagraph (Synergy Software, Reading PA) was used to generate statistical plots.
3. Results
3.1 AID deaminates DNA loops and bubbles embedded in double-stranded DNA
To examine the deamination activity of AID on various single-stranded DNA substrates including substrates containing loops or bubbles, and to determine substrates for AFM analysis, we designed oligonucleotides containing the structures depicted in Fig. 1. Deamination reactions were done for the various substrates following the scheme depicted in Fig. 2a. AID deaminated all substrates tested, including the loop and bubble substrates, but not double-stranded DNA (Figs. 2b and c). The best substrate for AID is the L-oligo, modeled after the oligonucleotide designed by Yu and colleagues [46], with over 50% deamination after 15 minutes, followed by the single-stranded oligonucleotide modeled after V regions of human IgV heavy chains (over 40% deaminated product at 15 minutes) (Fig. 2c). Even at high protein concentrations, in the single-stranded oligonucleotide that contains multiple dC’s throughout its length, the majority of product is deaminated at or near the site resembling AID-mediated immunoglobulin somatic hypermutation hotspots (AGCA) (Figs. 1 and 2b), as seen previously by us and other groups [39, 46–49]. Although the substrate containing a single-stranded 8-nucleotide loop is not deaminated as rapidly as the single-stranded oligonucleotides, its deamination is significantly faster than that of the substrate containing a 10-nucleotide bubble. For example, after a 15 minute incubation with AID, greater than 20 % of the loop substrate was deaminated while only 11% of the bubble substrate was deaminated (Fig. 2c). Deamination of single-stranded hairpin loops was consistently better than that of double-stranded loops, regardless of the size of the loops, at all concentrations of protein and DNA tested (data not shown). This lower rate of deamination of bubble substrates probably results from formation of non-canonical base pairs in the loop, thereby disrupting the accessibility of the cytidine to AID [53]. Taken together, these results indicate that AID preferentially deaminates cytosines that are maximally exposed. This preference for single-stranded vs. double-stranded loops might reflect AID-deamination of exposed single-stranded DNA in hairpin loops that might be formed due to the palindromic sequences often associated with CSR regions [54] or single-stranded DNA regions formed during transcription.
3.2 AFM reveals AID exists predominantly as a monomer, even in the presence of substrate
AFM, which yields topographic images of molecules deposited on a surface, has proven to be a powerful tool for determining the oligomerization state of proteins as well as protein-protein association constants [52, 55–62]. Specifically, we have demonstrated that the volume measured by AFM exhibits a linear dependence on the molecular weight of proteins with V = 1.2(MW) − 14.7, where V is the volume as measured by AFM and MW is the molecular weight of the protein [52, 61]. Using this relationship, the oligomerization states for many different proteins and protein-protein complexes have been determined in the presence and absence of DNA [52, 55–58, 61–62]. Accordingly, we have used this method to determine the oligomerization state of AID in the presence and absence of DNA substrates (either the L-oligo or the single-stranded loop DNA substrate).
Figure 3a shows representative AFM images of AID in the absence of DNA. To determine the oligomerization state of AID, the volume of each of the peaks (AID proteins) in the AFM images was determined from a large number of images (3028 peak volumes). Figure 3c shows the distribution of volumes in the absence and presence of DNA. In the absence of DNA, the distribution shows a single peak at ~15 nm3. This volume is consistent with the predicted volume of 15 nm3 for a monomer of His-tagged AID (AID monomer MW = 24.8kDa). Notably, only a few percent of the peaks have volumes corresponding to the predicted volume of a dimer of AID (45 nm3). Increasing the protein concentration from 5 nM to 20 nM or incubating at 500nM and immediately diluting to 10 nM before deposition (see methods) did not increase the population of species with larger volumes (data not shown). These results indicate that AID exists predominantly as a monomer at these concentrations and suggests that if a dimer forms it has a relatively weak association constant [52].
Figure 3.
Atomic Force Microscopy analysis of the oligomeric state of AID. A) 2 and 3-dimensional view of AID protein alone deposited at a final concentration of 10 nM. B) Representative gel image of the rate of deamination by AID when present at 20 nM concentration acting on 10 nM of L-oligo. The range of deaminated product at 30 minutes from 4 experiments for this ratio of AID protein to oligonucleotide was 32%-49%. C) Volume distributions of AID in the absence of DNA (black bars) and of RNaseA-treated AID in the presence of the L-oligo DNA substrate (grey bars). These results are representative of 5, 10, and 20 nM AID alone (black bars) and of 5, 10, and 20 nM AID in the presence of 10–50 nM of either the L-oligo and the loop substrate oligonucleotide (grey bars). Similar results were obtained when the protein was pre-incubated at 500nM with or without substrate, immediately diluted, and deposited at 10nM concentrations. In all cases, the major peaks in the distributions are consistent with the expected volume of an AID monomer (The predicted volumes (see text) of the monomer: (15 nm3) and dimer (45 nm3) are shown on the x-axis.
To determine if DNA induces dimerization, AID was incubated with 10nM to 50nM of the L-oligo or the loop substrate for 30 minutes before deposition. The concentrations of AID protein and oligonucleotide used in these AFM experiments are within the range of concentrations where significant deamination of substrate by AID after 30 minutes of incubation is observed (for example see deamination at 20 nM AID with 10 nM oligo, Fig. 3b). We used 5, 10, and 20 nM of AID protein as well as an incubation concentration of 500nM, immediately followed by deposition at 10nM (see above). In addition, because addition of RNase A is necessary for deamination, it is possible that bound RNA, known to inhibit the deamination reaction [39], might interfere with dimerization and/or DNA binding. To test this possibility, we pre-incubated AID with RNase A for 30 minutes before adding DNA (L-oligo), and imaged as usual. To assure that AID was active under these conditions, we tested the deamination activity of AID when it was preincubated with RNase A for 30 minutes versus adding AID at the start of the deamination assay. As seen previously, RNase A was required for deamination to occur, however, pre-incubation with RNase A did not change the rate of deamination (Fig. 4). The concentration of AID was 20-fold higher than that of RNase A so that RNase A would only be a minor population (5 %) of the protein in the AFM images. We analyzed between 800 and 4000 protein data points for each AID and oligo concentrations. Figure 3c shows the distribution of volumes (810 peak volumes) for AID that was treated with RNaseA and incubated with the L-oligo. This distribution is indistinguishable from that of AID that was not treated with RNaseA and incubated with the L-oligo or with the loop substrate, or from any of the protein/oligo concentration combinations (data not shown). The major peak in the distribution is found at ~18 nm3. This peak, while still corresponding to monomeric AID, is shifted to a slightly higher volume compared to AID in the absence of oligonucleotide (Figure 3c). This increase in volume in the presence of oligonucleotide is consistently seen for both AID treated and untreated with RNaseA, and it likely represents oligonucleotide being bound to monomeric AID. These results suggest that binding of AID to DNA does not induce dimerization of AID, because there is no significant increase in the fraction of peaks with volumes corresponding to the dimer (~45 nm3) in the presence of DNA substrate.
Figure 4.
RNAse A is required for deamination, but pre-incubation does not alter the rate of deamination. 10 nM of L-oligo was incubated with 500 nM of AID protein without any RNaseA (triangles) or with 400 ng RNaseA added either with the oligo (diamonds) or 30 minutes before the additon of oligo (squares).
3.3 AID can deaminate single-stranded DNA as a monomer
To determine if AID can function as a monomer, we measured the rates of deamination of the L-oligo substrate (100 nM) as a function of AID concentration (5 nM to 500nM) (Fig. 5). For 5 nM and 20 nM AID, where our AFM data indicate that AID is primarily monomeric, with 100 nM oligonucleotide, the initial rates (determined by linear fits to the data for timepoints ≤ 20 minutes) are 0.15 min−1 and 0.73 min−1, respectively (Fig. 5a). This increase in rate (4.8-fold) is within error of the expected increase for the 4-fold increase in enzyme concentration [63]. Furthermore, increasing the AID concentration to 100 nM increases the rate to 1.27 min−1, but increasing it further to 500 nM results in only a small increase in deamination rate (1.34 min−1) (Fig. 5a). These latter results indicate that the DNA is nearly saturated at 100 nM AID, which is a one-to-one ratio of AID monomer to DNA. If the observed activity were due solely to AID dimers, we would expect to see at least a 2-fold increase in rate going from 100 nM to 500 nM AID. These results strongly suggest that AID can function as a monomer to deaminate cytosine in single-stranded DNA.
Figure 5.
Concentration dependence of deamination rates indicates that most of the catalysis on single-stranded DNA oligonucleotide is from the monomer. A) Rate of deamination of 100 nM L-oligo in the presence of 5 nM (diamonds), 20 nM (triangles), 100 nM (circles), and 500 nM (squares) AID. The straight lines are linear fits to data points for times ≤ 20 minutes. The initial rates, which were determined from the linear fits, are 0.15, 0.73, 1.27, and 1.34 min−1 for 5 nM, 20 nM, 100 nM, and 500 nM AID, respectively. B) Rate of deamination of 10 nM L-oligo in the presence of 20 nM (squares) and 500 nM (circles) AID. The curves are first order exponential fits to the data. The error bars represent the standard error from 3 or more experiments.
To further verify that this observed activity is due to the monomer and not to an increase in the population of dimers, we examined the rates of deamination of 10 nM L-oligo with 2-fold (20 nM, where less than 20% of the population has volumes that could be attributed to the dimer in the presence of oligo) and 50-fold (500 nM) excess AID. If a small fraction of AID dimers were responsible for the observed activity, we would expect to see an increase in deamination rate with increasing AID concentration from 20 nM to 500 nM, because at 20 nM AID, the dimer concentration (20% of the population corresponds to ~ 3 nM AID dimer) would be below the DNA concentration. Consequently, if the dimer were responsible for the observed activity, we should see an increase in deamination activity upon increasing the AID concentration from 20 to 500 nM, as we observed for the experiments with the DNA in excess (Fig. 5a). Inspection of Figure 5b reveals that the rate of deamination is the same for both of these concentrations. These results indicate that at 10 nM L-oligo, a two-fold excess of AID is sufficient to bind all of the L-oligo and catalyze deamination. This result is consistent with the measured binding affinities of AID for oligonucleotide substrates, which are in the sub to low nanomolar range [64]. These kinetic data together with the AFM data strongly support the conclusion that AID is functional as a monomer on single-stranded DNA in vitro.
4. Discussion
We found that AID is catalytically active as a monomer on single-stranded DNA in vitro, even when the single-stranded DNA region is embedded in double-stranded DNA in the form of loops. We did not observe a significant population of volumes consistent with AID dimers by AFM for any of the protein/oligo concentrations, including concentrations where well over 50% of the oligo is deaminated (Figure 3c, data not shown). Pre-incubation of AID at concentrations up to 500nM also failed to alter the percentage of volumes corresponding to the dimer. In addition, the dependence of the deamination rates on AID concentration is consistent with AID functioning as a monomer for both the experiments in which the AID concentration is lower than the DNA concentration (Fig. 5a) and for those in which the AID monomer concentration is higher than the DNA concentration (Fig. 5b). While these results demonstrate that AID is active as a monomer on single-stranded DNA in vitro, they do not preclude the possibility that AID may also function as a dimer in vivo (or in vitro), where it may be post-translationally modified via phosphorylation [45] (see below).
Comparison of the rates of deamination on single-stranded DNA loops within double-stranded DNA with deamination rates on substrates devoid of any secondary structure such as the L-oligo indicates that even short loops of single-stranded DNA can be deaminated by AID relatively well, albeit with less efficiency than DNA sequences in which the cytidine is expected to be completely exposed, such as the L-oligo (Figs. 1 and 2). These results imply that even at low concentrations, as found in the nucleus of cells expressing AID, monomeric AID could catalyze the deamination of cytidine to uracil as long as it can find a suitable substrate in the form of exposed single-stranded DNA patches or loops. In support of this idea, recent data suggests that AID can bind and deaminate single-stranded DNA in a variety of configurations such as displaced single-stranded DNA loops or patches [64–67] and even in negatively supercoiled double-stranded DNA [68] formed during transcription. These results might explain why AID can access and deaminate DNA in non-natural substrates such as in DNA of E. coli cells, non-lymphoid cells, and non-Ig genes that are actively transcribed [14, 69–71], and wherein B-cell specific post-translational modifications of AID do not occur. If for example, dimerization is optimally promoted when AID is specifically phosphorylated in activated B cells, these results imply that non-phosphorylated monomeric AID in other cells could potentiate inappropriate deamination of dC in non-immunoglubulin gene targets. It will be interesting to determine whether monomeric AID contributes to non-targeted AID-mediated deamination in B cell lymphomas or when expressed in non-B cells. If so, the oligomeric status of AID may contribute yet another layer of regulation similar to transcriptional regulation and intracellular localization that keep cells expressing AID from experiencing its mutagenic activity in non-intended targets [72–74]. In addition it is likely that, in B cells, targeting co-factors sharpen AID’s substrate specificity uniquely to the Ig V and CSR regions [75], although these putative co-factors have yet to be identified.
Our results do not imply that the dimer does not form or is not functional. In the AFM images, a few percent of the molecules have volumes that correspond to the dimer and other higher order oligomeric states, although increasing the concentration from 5 nM to 20 nM or following incubation at 500nM, does not result in any detectable changes in the population of molecules with higher volumes. However, It is clear from our studies that in vitro, i) the monomer is the predominant species in the absence of substrate (at the concentrations examined), ii) the monomer is catalytically active on single-stranded DNA, and iii) binding to single-stranded DNA substrate does not induce any significant dimerization. A recent study that examined the binding of GST-tagged and His-tagged AID supports our conclusion that AID can function as a monomer [64]. Specifically, in band-shift assays of AID with a DNA substrate containing a single-stranded bubble, the GST-tagged-AID, which is forced to be a dimer by GST dimerization [76], causes dramatically greater shifts of the DNA than does the His-tagged-AID, consistent with His-tagged-AID binding to DNA as a monomer.
A recent report by Wang and colleagues, suggests that AID dimerization is required for optimal class switch recombination in vivo; however, these studies examined co-immunoprecipitation of AID with two different tags from cell extracts, and therefore, the apparent dimerization in these studies could be dimerization mediated by other proteins and not a direct interaction of two AID molecules [77]. In addition, whether or not this “dimerization” is also required for immunoglobulin hypermutation where deamination of dC’s throughout the length of the V region is evident was not tested. Several other reasons may account for the differences between our results and those reported in the previous study [77]. Namely, we have found that in its most proximal activity, as a deaminase, monomeric AID can deaminate dC’s in vitro, whereas class switch recombination in vivo, may require a higher order oligomeric state of AID to help align specific dC’s in switch regions or for other functionalities. Also, it is possible that AID acts both as a monomer and as a dimer in vivo, perhaps differentially for hypermutation and class switch recombination; a hypermutation phenotype was not reported in that study for the dimer mutants. While both of these processes require AID, they also likely require different co-factors, occur independently of each other, and target different regions of the heavy chain Ig locus. Finally, it is conceivable that phosphorylation or other post-translational modifications of AID in activated B cells may promote dimerization, while the non-modified form of AID acts as a monomer, with less efficiency. In fact, ectopically-expressed AID is not phosphorylated and yet it can deaminate dC in non-lymphoid cells, but at a lower frequency (14, 44, 69–71).
The observation that AID exhibits catalytic activity as a monomer in vitro is in contrast to other deaminases studied to date, including the RNA editing enzymes such as Apobec-1, and the adenosine deaminases [78–81]. Interestingly, these deaminases require dimerization not only for their enzymatic activity but also for specificity of the substrate. For example, homodimers comprising a wild type monomer and a mutant monomer of ADAR1 or ADRAR2 lose only half the ability to deaminate a non-specific dsRNA substrate, in a sequence-independent manner but lose nearly 70% site-selective activity on natural substrates, such as the RNA encoding serotonin 2C subtype receptor [81]. Consequently, it is possible that while monomeric AID is catalytically active, a homodimer may confer some kind of substrate specificity wherein interaction between the monomers helps align a specific cytosine residue for deamination. This idea is interesting because deamination of cytosine in an RNA target would require exquisite targeting not only to a particular RNA species, but a particular cytosine in its sequence. Such a level of specificity may require dimerization as seen with RNA editing enzymes such as Apobec-1 [78]. However, an RNA target for AID has not been identified although indirect evidence supports the possibility [reviewed in 38]. Alternatively, an AID homodimer, while not required for deamination, may help the simultaneous deamination of cytosine on both DNA strands, as previously suggested [49], an idea supported by our findings predicting that each monomer of AID in a putative dimer can catalyze deamination of single-stranded DNA independently. Additional support for this notion comes from theoretical analysis based on a comparison of yeast RNA-editing deaminase to AID suggesting that for the homodimer, both monomers are able to catalyze deamination of two cytosines within the same nucleic acid strand or in opposite strands [51]. In addition, a recent structure of the AID homolog APOBEC2 shows that while APOBEC2 forms a tetramer, which is a dimer of dimers, the putative active site is not near either the dimer or tetramer interface and each of the monomers appears to have a fully intact active site [82]. Finally, it is important to note that although it is likely that AID can form a dimer, there is no direct evidence for dimerization in vitro or in vivo [other than the small population of dimers observed in this study (Fig. 3)], only inference from homology models and co-immunoprecipitation of cell extracts [51, 77]. It will be interesting to determine whether AID catalyzes deamination as a monomer and/or dimer in vivo, and whether the oligomeric state of AID contributes a previously unappreciated layer of functional plasticity by modifying substrate specificity differentially for SHM or CSR, or for DNA vs RNA substrates depending on whether a specific cytosine needs to be deaminated (as seen in RNA deamination) versus several cytosines in a DNA substrate with some preference (i.e. hotspots) but without requiring deamination of a specific residue.
Supplementary Material
Acknowledgments
We are grateful to David Schatz for providing the AID-DN vector, to Hong Wang for technical advice, and to Jan Drake for helpful discussions. We also thank William Copeland and Thomas Darden for critical reading of the manuscript. This work was supported by the American Cancer Society (DAE) and NIH grant R01 GM54136 (DAE) and by the Intramural Research Program of the NIH, and NIEHS (MD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eisen HN, Siskind GW. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 2.Klinman NR, Rockey JH, Frauenberger G, Karush F. Equine anti-hapten antibody 3. The comparative properties of {gamma}G- and {gamma}A-antibodies. J Immunol. 1966;96:587–595. [PubMed] [Google Scholar]
- 3.Klinman NR. The mechanism of antigenic stimulation of primary and secondary clonal precursor cells. J Exp Med. 1972;136:241–260. doi: 10.1084/jem.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the {lambda} light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 5.Crews S, Griffin J, Huang CK, Hood L. A single VH gene segment encodes the immune response to phosphorylcholine: somatic mutation is correlated with the class of the antibody. Cell. 1981;25:59–66. doi: 10.1016/0092-8674(81)90231-2. [DOI] [PubMed] [Google Scholar]
- 6.Clarke SH, Huppi K, Ruezinski D, Staudt L, Gerhard W, Weigert M. Inter- and intraclonal diversity in the antibody response to influenza hemagglutinin. J Exp Med. 1985;161:687–704. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121–1129. doi: 10.1016/0092-8674(91)90289-b. [DOI] [PubMed] [Google Scholar]
- 8.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 9.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 10.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 11.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 12.Yang G, Obiakor H, Sinha RK, Newman BA, Hood BL, Conrads TP, Veenstra TD, Mage RG. Activation-induced deaminase cloning, localization, and protein extraction from young VH-mutant rabbit. Proc Natl Acad Sci USA. 2005;102:17083–17088. doi: 10.1073/pnas.0501338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 14.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 15.Manis JP, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt FW. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J Exp Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin XF, Besmer E, Kenter A, Rajewsky K, Nussenzweig MC. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalan N, Selz F, Imai K, Revy P, Fischer A, Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 19.Rush JS, Fugmann SD, Schatz DG. DNA double strand breaks are the predominant DNA lesions targeted to S mu in Ig class switch recombination. Int Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- 20.Arudchandran A, Bernstein RM, Max EE. Single-stranded DNA breaks adjacent to cytosines occur during Ig gene class switch recombination. J Immunol. 2004;173:3223–3229. doi: 10.4049/jimmunol.173.5.3223. [DOI] [PubMed] [Google Scholar]
- 21.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, Manis JP, Chen J, Carpenter PB, Alt FW. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs H, Fukita Y, van der Horst GT, de Boer J, Weeda G, Essers J, de Wind N, Engelward BP, Samson L, Verbeek S, de Murcia JM, de Murcia G, te Riele H, Rajewsky K. Hypermutation of immunoglobulin genes in memory B cells of DNA repair-deficient mice. J Exp Med. 1998;187:1735–1743. doi: 10.1084/jem.187.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 26.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 29.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 30.Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 31.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 32.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 43.Besmer E, Market E, Papavasiliou FN. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol Cell Biol. 2006;26:4378–4385. doi: 10.1128/MCB.02375-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 45.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 46.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 47.Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–3384. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- 48.Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 49.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 50.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 51.Xie K, Sowden MP, Dance GS, Torelli AT, Smith HC, Wedekind JE. The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proc Natl Acad Sci USA. 2004;101:8114–8119. doi: 10.1073/pnas.0400493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ratcliff GC, Erie DA. A novel single-molecule study to determine protein-protein association constants. J Am Chem Soc. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- 53.Chou SH, Chin KH, Wang AH. Unusual DNA duplex and hairpin motifs. Nuc Acids Res. 2003;31:2461–2474. doi: 10.1093/nar/gkg367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tashiro J, Kinoshita K, Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- 55.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J Biol Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 56.Tessmer I, Moore T, Lloyd RG, Wilson A, Erie DA, Allen S, Tendler SJ. AFM studies on the role of the protein RdgC in bacterial DNA recombination. J Mol Biol. 2005;350:254–262. doi: 10.1016/j.jmb.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Sass L, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, Redinbo MR. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′-5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry. 2002;41:2901–2912. doi: 10.1021/bi0157161. [DOI] [PubMed] [Google Scholar]
- 59.Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell. 2005;17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Verhoeven EE, Wyman C, Moolenaar GF, Goosen N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. Embo J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, Wang H, Erie DA. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003;29:175–187. doi: 10.1016/s1046-2023(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- 63.Segal IH. Enzyme Kinetics. John Wiley and Sons; New York: 1975. [Google Scholar]
- 64.Larijani M, Petrov AP, Kolenchenko O, Berru M, Krylov SN, Martin A. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol Cell Biol. 2007;27:20–30.H.M. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 66.Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 70.Martin A, Scharff MD. Somatic hypermutation of the AID transgene in B and non-B cells. Proc Natl Acad Sci USA. 2002;99:12304–12308. doi: 10.1073/pnas.192442899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito S, Nagaoka H, Shinkura R, Begum N, Muramats M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 74.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 76.Dirr H, Reinemer P, Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur J Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, Shinkura R, Muramatsu M, Nagaoka H, Kinoshita K, Honjo T. Identification of a specific domain required for dimerization of activation-induced cytidine deaminase. J Biol Chem. 2006;281:19115–19123. doi: 10.1074/jbc.M601645200. [DOI] [PubMed] [Google Scholar]
- 78.Lau PP, Zhu HJ, Baldini A, Charnsangavej C, Chan L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallo A, Keegan LP, Ring GM, O’Connell MA. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 2003;22:3421–3430. doi: 10.1093/emboj/cdg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J Biol Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 81.Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–170102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 82.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.