SUMMARY
The function of the src-homology 3 (SH3) domain in class II myosins, a distinct β-barrel structure, remains unknown. Here we provide evidence, using electron cryomicroscopy, in conjunction with light scattering, fluorescence and kinetic analyses, that the SH3 domain facilitates the binding of the N-terminal extension of the essential light chain isoform (ELC-1) to actin. The 41-residue extension contains four conserved lysines followed by a repeating sequence of seven Pro/Ala residues. It is widely believed that the highly charged region interacts with actin, while the Pro/Ala-rich sequence forms a rigid tether that bridges the ~9 nm distance between the myosin lever arm and the thin filament. In order to localize the N-terminus of ELC in the actomyosin complex, an engineered Cys was reacted with undecagold-maleimide, and the labeled ELC was exchanged into myosin subfragment-1 (S1). Electron cryomicroscopy of S1-bound actin filaments, together with computer-based docking of the skeletal S1 crystal structure into 3D reconstructions, showed a well-defined peak for the gold cluster near the SH3 domain. Given that SH3 domains are known to bind proline-rich ligands, we suggest that the N-terminal extension of ELC interacts with actin and modulates myosin kinetics by binding to the SH3 domain during the ATPase cycle.
Keywords: electron cryomicroscopy, image reconstruction, SH3 domain, ELC, myosin II, motility, ATPase cycle, actin-binding protein
INTRODUCTION
A puzzling feature of the three-dimensional structure of chicken skeletal muscle myosin, first described in 1993, was the presence of a small six-stranded β barrel at the N-terminus of the heavy chain (K35 to M80), which appeared to be distinct from the rest of the structure1. The topology of this domain was noted as being similar to the Src-homology 3 (SH3) domain, originally identified in Src tyrosine kinases, but also observed in cytoskeletal proteins such as spectrin2. All class II myosins have this N-terminal domain, but it is absent in class I of the myosin superfamily3. The function of the SH3 domain in myosin remains unknown. Proximal to this structure is the essential light chain (ELC), which together with the regulatory light chain (RLC), stabilizes the C-terminal α-helical region of the heavy chain. The resulting >8 nm long “lever arm” is uniquely endowed to amplify small conformational changes at the nucleotide binding site into force and movement1.
The importance of the ELC for the structural integrity of the myosin motor has been well established: removal of the ELC from myosin results in a loss of movement of actin filaments4, and a reduction in isometric force, as determined by in vitro motility assays5. In striated slow/cardiac and fast muscle myosins, there are several isoforms of ELC (designated LC1 or A1), which include an N-terminal extension rich in lysine and proline residues. The prevailing view is that this extension interacts directly with the actin filament to modulate the kinetics of cross-bridge cycling6. How it modulates cycling is far less clear; does it do so by altering the actin filament structure, or by affecting the interdomain interactions in myosin, or by some combination of both? Another puzzling feature of the ELC is how the N-terminal peptide of ~40 amino acids bridges a distance of over 8 nm from the lever arm to the nearest actin monomer in the filament. Assuming the proline residues provide for a sufficiently rigid extended structure, what happens to this “antenna” when the myosin is dissociated from actin in the relaxed phase of the cycle?
The answers to questions of biological function ultimately benefit from structural observations. Unfortunately, there is no crystallographic structure of the N-terminus of the ELC. This region of the ELC is disordered, as is the shorter N-terminus of the RLC, in the available crystal structures of the myosin head1. In contrast to the strong protein-protein interactions between the C-terminal portion of the light chains and the heavy chain in the lever arm, the interactions of the N-terminal regions are more transitory, and probably have more of a regulatory role. To approach this problem, we have labeled the N-terminus of the ELC (A1) with a gold cluster, incorporated it into myosin subfragment-1 (S1), and determined the location of the gold cluster by electron cryomicroscopy (cryoEM) of actin filaments decorated with S1. By computer-based docking of the chicken skeletal S1 crystal structure into three-dimensional (3D) reconstructions of the decorated filaments, we have been able to show that the N-terminal region of the ELC is located near the SH3 domain in myosin. Given that SH3 domains recognize proline-rich ligands, and taken together with light scattering and spectroscopic evidence, we suggest that the proline-rich sequence in the ELC may modulate the kinetic properties of myosin by using the SH3 domain as a launching site for its interaction with actin, as well as by modulating interactions between the SH3 domain and neighboring elements in the myosin head.
RESULTS
Functional Properties of the ELC Isoform
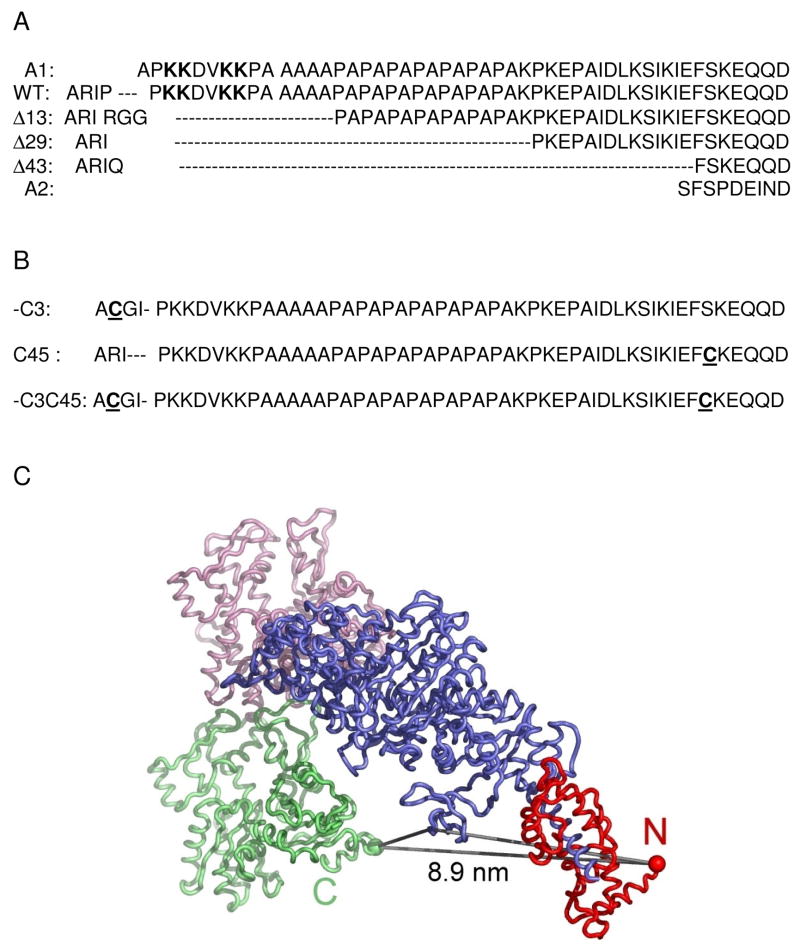
Data gathered over several decades for the fast skeletal muscle A1 (or LC1) have led to a consensus regarding certain unique features of this light chain isoform: a) myosin containing A1 has a 2-fold lower actin-activated maximum MgATPase activity (Vmax) and a 5 to 10-fold lower Michaelis constant (Km; i.e. higher affinity for actin) than myosin subfragment-1 (S1) containing the shorter A2 (or LC3) isoform (see Fig. 1A for partial sequences of the two native ELC isoforms and various deletion mutants of A1); b) the N-terminal sequence of A1 contains a cluster of lysine residues that bind to the C-terminus of actin; c) the Pro/Ala-rich region of A1 enables it to reach the actin-binding site. These conclusions were derived mainly from a comparison of the properties of S1(A1) versus S1(A2)7 and from peptides obtained by proteolysis of A18; 9. Here, we extend these studies through the use of genetically engineered chicken skeletal muscle light chains exchanged into chicken skeletal muscle S1 (for relevant earlier studies see review by Timson, 20036).
Figure 1.
(A) Sequences of wild-type and deletion mutants of ELC. The four lysines in the putative actin binding region are in bold. The sequences of the native isoforms, A1 and A2 are shown for comparison. Following the Asp (D) residue are ~ 140 residues common to both A1 and A2. (B) Mutants of A1 with one or two reactive cysteines underlined. (C) Schematic rendition of the crystal structures of chicken skeletal muscle S1 and actin docked on the reconstruction of the cryoEM actoS1 complex. S1 prepared by chymotryptic digestion of myosin does not contain the RLC. The upper actin (pink) and lower actin (green) are shown interacting with the S1 motor domain (blue). The calculated distance from residue 5 (the rest is disordered) of the ELC (red ball) to residue 365 in actin (green ball) is 8.9 nm. The SH3 (residue 70) distance to ELC is 6.6 nm and to actin (365) is 3.6 nm. (The choice of residues in SH3 is arbitrary, and only meant to give an approximate indication of distances).
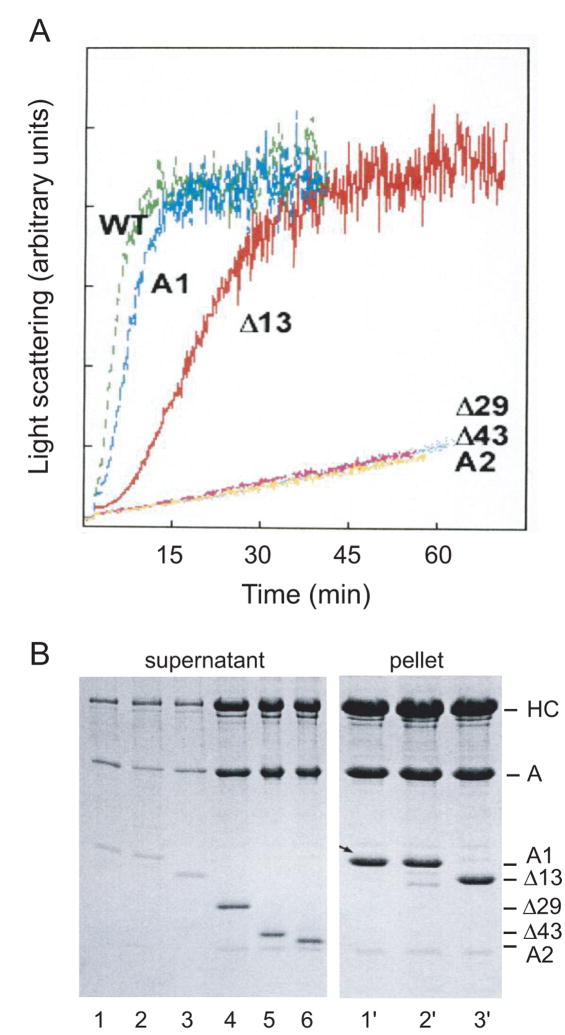
At low salt concentrations and in the absence of magnesium cations, actin is a stable monomer which does not contribute to light scattering. This property can be used to advantage to show how specific sequences in the N-terminus of A1 affect the ability of S1 to polymerize G-actin monomers into a filament, Fig. 2A. S1 with wild-type (WT) A1, similarly to S1 with the native A1 isoform, is able to rapidly initiate polymerization, unlike S1 with the shorter A2 isoform which binds to actin, but has little effect on G-actin polymerization in the course of an hour10. In fact, the extra Arg residue in the fusion peptide of WT has a slight enhancing effect. Deletion of the putative actin binding site (..K3K4DVK7K8..) reduces the rate of polymerization, but does not abolish it as long as charged Arg residues are introduced into the sequence (Fig. 2, Δ13). In the absence of these added arginines, however, the effect on G-actin is similar to that of A2 (data not shown). Irrespective of the N-terminal charge, deletion of the Pro/Ala sequence (Δ29, Δ43) results in a complete loss of actin polymerization, as evidenced by light scattering and SDS gel electrophoresis of samples taken from the light scattering experiments, Fig. 2A and B.
Figure 2.
(A) Time course of polymerization of G-actin by S1 reconstituted with ELC mutants (see Fig. 1). Light scattering was monitored at 325 nm in the ISS spectrofluorometer. S1 and G-actin, each at a concentration of 5μM, were in 5 mM Tris, 0.2 mM CaCl2, pH 8. S1 (WT) accelerated polymerization of G-actin even slightly more than native S1(A1), whereas deletion of the actin binding site, along with the ProAla region (Δ29), slowed polymerization to the rate observed for A2 or Δ43. The Δ13 mutant, which had two Arg residues at the N-terminus, did accelerate assembly of actin, although at a slower rate. Another mutant (Δ14), with only one Arg, had a very slight effect on assembly (data not shown; see similar results by cross-linking in Andreev et al., 199913). (B) Samples taken from the light scattering measurements after one hr were centrifuged, and the supernatants and pellets were analyzed by SDS-PAGE. As expected, S1 containing native A1, wild-type LC1, and Δ13 was found in the pellet (lanes 1′, 2′, and 3′), whereas S1 containing Δ29, Δ43, and A2 remained in the supernatant (lanes 4, 5, and 6). The amount of protein in the pellets for the S1 with A1 truncations that did not polymerize G-actin was very minor, and samples for lanes 4′– 6′ are omitted.
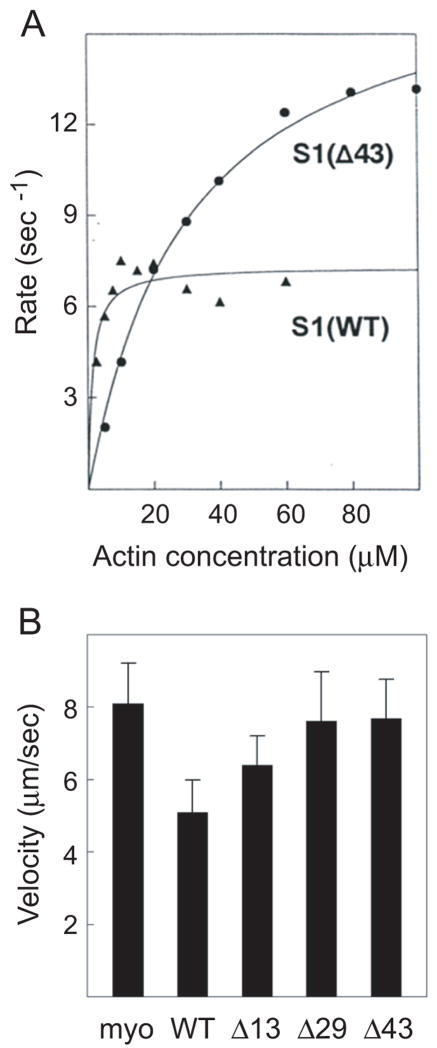
The extent of the interaction of these mutant light chains with actin is reflected in the enzymatic activity of the reconstituted S1s, Fig. 3A and Table 1. The absence of the N-terminus of A1, in particular the Pro/Ala region, raises the Vmax of the mutants to twice the levels found for WT, and the affinity for actin is greatly reduced, as evidenced by the larger Km, Table 1. The steady-state enzymatic activity can also be correlated with the velocity of actin filament movement, as measured by an in vitro motility assay using reconstituted myosins4. The absence of the Pro/Ala sequence (Δ29, Δ43) gives motilities that are faster than those of A1 or wild-type, and identical to A2, Fig. 3B and Table 1. The mutant lacking the N-terminal cluster of lysine residues (Δ13) slows the actin filament motility only slightly, but the parallel effect seen in the enhanced affinity for actin (Km) lends credence to the result. These studies support and extend earlier conclusions that the A1 light chain interacts with actin through weak electrostatic interactions11; 12; 13.
Figure 3.
(A) A representative sample of the actin-activated MgATPase activity for S1 reconstituted with wild-type and Δ43 light chain. The average value of four and seven independent determinations for WT and Δ43, respectively, are given in Table I. The rate in the absence of actin was subtracted from each value. The line is the best fit of the data to the Michaelis-Menten equation: for S1(WT), Vmax is 7.5 ± 0.4 s−1 and Km is 1.5 ± 0.6 μM; for S1(Δ43), Vmax is 15.6 ± 0.8 s−1 and Km is 25.5 ± 3.4 μM. (B) Actin filament velocities for myosin reconstituted with the stated expressed light chains. Values are given in Table I. The preparation entitled “myo” consisted of myosin reconstituted with equal amounts of the isolated native light chains A1 and A 2; its motility was the same as that of standard purified myosin4.
Table 1.
Summary of ATPase activity and motility of myosin reconstituted with various mutants of LC1
| Myosina (S1) | Vmax (s−1) | Km (μM) | n | Vactin (μm/s) |
|---|---|---|---|---|
| WT | 7.7 ± 1.3 | 1.7 ± 1.0 | 4 | 5.08 ± 0.89 |
| Δ13 | 13.3 ± 1.8 | 10.4 ± 4.7 | 8 | 6.39 ± 0.80 |
| Δ29 | 15.9 ± 6.2 | 21.7 ± 10.7 | 3 | 7.61 ± 1.35 |
| Δ43 | 15.1 ± 0.8 | 18.2 ± 3.0 | 7 | 7.68 ± 1.07 |
| A1+A2 | ---- | ---- | 8.09 ± 1.12 |
Myosin reconstituted with light chains was used only for the in vitro motility assays. The kinetic assays were all performed with S1(A2) exchanged with the various light chains cited here, as described in Methods. The mean and standard deviation for n independent determinations of ATPase activity are shown. The motility values (mean ± SD) represent two independent determinations of 30–40 filaments each.
Length of the N-terminal Extension by FRET
Although the evidence strongly favors an interaction between A1 and actin, the problem remains of how an extension of ~ 41 residues can bridge a distance of ~ 8 nm between the lever arm and the actin filament (Fig. 1C). This number of residues rich in prolines could easily form a long rigid structure with an average rise/residue of ~ 0.3 nm, but is there any evidence for such a structure? We have addressed this question by engineering a cysteine mutant (-C3/C45) that spans the entire N-terminal sequence of A1, Fig. 1B. The N-terminal Cys is in a tag (ACGI-) derived from the expression vector, followed by the native sequence, and C45 is created by a point mutation at Ser45 to Cys. The double Cys mutant was first labeled with a substoichiometric amount of the donor probe, 1,5-IAEDANS, fractionated by Mono Q ion-exchange chromatography to isolate the singly labeled dansylated species, and reacted with various acceptor chromophores on the second Cys, Table 2. Steady-state fluorescence resonance energy transfer (FRET) was used to measure the energy transfer distance (R) between the labeled cysteines. Although the efficiency of energy transfer (E) varied between 10 and 60%, depending on the acceptor used to quench donor fluorescence, the calculated distance between the cysteines was remarkably constant at 4.1 nm, Table 2. In the presence of a strong denaturant, such as 6M guanidine hydrochloride, the degree of donor quenching for the DAN/DAB pair was similar to that observed in a benign solvent (data not shown), suggesting that the N-terminal extension in A1 adopts a “random coil-like” conformation. A similar conformation was predicted by analysis of NMR spectra for an N-terminal peptide (residues 1–37) prepared from A19. Although an observed FRET value of ~ 4 nm is consistent with a stiff polymeric chain containing Pro/Ala repeats, it is still only half the length needed to span the distance between the lever arm and the actin filament, suggesting that additional interactions with the motor domain and/or actin are needed for the N-terminal extension to attain its full elongated structure.
Table 2.
Energy transfer parameters for the N-terminal extension of LC1
| Sample | Donor/Acceptor | E (%)a | R0 (Ǻ)b | R (Å)c |
|---|---|---|---|---|
| 1 | DAN/DDPM | 11 9.2 |
29 | 41 42 |
| 2 | DAN/DABM | 24 28 |
36 | 44 42 |
| 3 | DAN/IAF | 69 61 |
42 | 37 39 |
Efficiency of energy transfer between donor/acceptor probes on the cysteine residues in the -C3/C45 mutant (Figure 1).
Forster distance for which the efficiency of transfer is 50% (values taken from the literature).
Distance between the various donor/acceptor probes. DAN is the donor dansyl moiety of 1,5-IAEDANS; see chemical name for acceptor dyes under abbreviations.
Location of the N-terminal extension by cryoEM
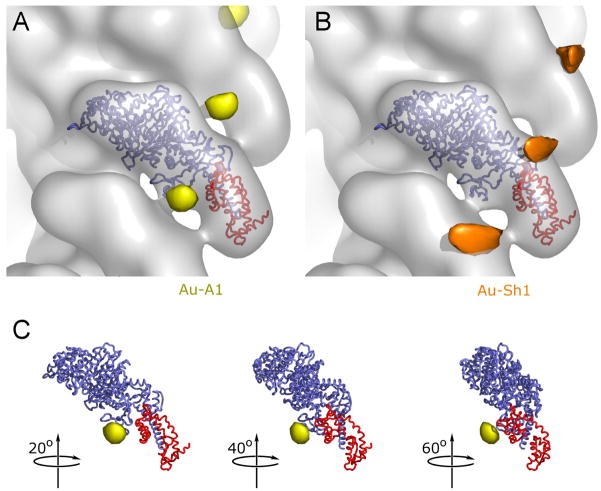
An earlier study using cryoEM and traditional helical reconstruction methods compared the three-dimensional (3D) maps generated by decorating actin with S1(A1) and S1(A2). The “difference map” showed an extra peak of density in the C-terminal region of actin, which was assigned to the extra N-terminal residues in A114. This finding was consistent with a biochemical study showing that the N-terminus could be cross-linked to acidic residues 361–364 of actin15. In order to improve the identification of the density peak seen in the difference maps, we created an ELC mutant containing a single Cys residue in a tag (-C3) at the N-terminus of A1, Fig. 1B. This Cys was reacted with undecagold and the labeled ELC was exchanged into S1(A2). After removal of excess ELC, actin was decorated with S1 for electron cryomicroscopy, and analyzed by conventional methods of helical image processing16. Unfortunately, not all attached S1 molecules contained gold, since the efficiency of labeling ELC was less than 100%, and therefore localization of the gold using difference mapping was ambiguous.
In order to resolve this problem, we adapted an alternative approach to standard helical reconstruction algorithms, the Iterative Helical Real Space Reconstruction (IHRSR) method17, and optimized it for use with actomyosin cryoEM reconstructions18. One of the main advantages of this method is the ability to sort mixed populations, which enabled us to significantly increase the signal for the gold cluster population. With the use of this technique, it was now possible to resolve the gold-labeled N-terminal sequence of A1 in the decorated filaments close to the SH3 domain, Fig. 4A (see Methods for description of image processing). Given the large size of the gold cluster and its surrounding organic shell (~ 2 nm), one can only expect the probe to lie in the vicinity of its intended target. Despite this uncertainty, the SH3 domain is clearly the nearest protein structure that can interact with the N-terminus of A1, Fig. 4C.
Figure 4.
Localization of the A1 extension by cryoEM. A reconstruction of actin decorated with S1 containing undecagold gold-labeled A1 (at –Cys3) is shown in transparent gray (pointed end up). The backbone of the docked crystal structure of chicken skeletal S1 is shown with the motor domain in blue and the ELC in red. The difference peak attributed to the A1 gold cluster label is shown in yellow (A and C); the difference peak attributed to the gold label on the reactive cysteine (SH1) of the heavy chain is shown in orange (B). This peak is located at the entrance of a narrow cleft between the converter/relay-loop and the rest of the myosin head, with the solvent exposed sulfhydryl group of SH1 as its bottom. This peak is approximately 10 times stronger than the peak in (A). Both contour levels were adjusted to optimize visibility.
Despite the use of the new IHRSR method, the statistical analysis of peaks in difference maps generated with unlabeled S1(A1) and S1(A2) remained inconclusive, owing to the relatively small difference in protein mass and the residual scaling errors resulting from differences in noise level and background. However, one of the three highest peaks in this difference map was between the SH3 domain and the location of the gold cluster label (data not shown). No peak was observed in the vicinity of the SH3 domain when difference maps of unlabeled S1(A1) versus unlabeled S1(A1), or S1(A2) versus S1(A2), were calculated.
As a control, we labeled the reactive sulfhydryl residue (SH1) in the myosin heavy chain with undecagold, and showed that its location was quite distinct from that of the light chain and consistent with labeling SH1, Fig. 4B. The peak in this control map is much larger, and almost one order of magnitude stronger than the SH3 peak. The most straightforward explanation for this difference is a higher degree of structural flexibility for the A1 label compared to the SH1 label. This is not surprising if in fact the labeled A1 residue is ~15 residues away from the proposed SH3/A1 interaction site (prolines), and the N-terminus must maintain a certain degree of mobility in order to reach actin when needed.
We were unable to reproduce the earlier electron microscope studies that suggested the N-terminal domain of A1 binds to the surface of F-actin14. Biochemical studies have indicated that cross-links between S1(A1) and F-actin can only occur when actin is present in large excess, presumably because A1 binds to the actin monomer below the one that provides the major binding surface for the S1 heavy chain12; 13. The nucleation of actin polymerization by S1(A1) shown in the light scattering results is consistent with this interpretation. Therefore, the interaction between A1 and actin would not be expected to be favored in a fully decorated filament. In muscle tissue, actin is in molar excess to myosin heads at all times, so that the N-terminal ELC extension can always bind to the actin below the corresponding myosin head.
DISCUSSION
We and others have firmly established that the N-terminus of the ELC isoform (A1) interacts transiently with actin filaments, and can thereby modify the cycling kinetics of the myosin motor in striated muscles. But the overriding question remains of how this actin interaction is coupled to the catalytic domain over a distance of > 8 nm? By means of improved cryoEM reconstructions using a hybrid approach that combines elements of single particle and helical reconstruction techniques17; 18, and a gold-labeled N-terminal residue in A1, we have shown here for the first time that this proline-rich sequence of A1 also has a binding site near the SH3 domain of myosin. Taken together with light scattering and spectroscopic evidence, crosslinking, and kinetic analysis, these data suggest that the communication pathway between the light chain/actin interface and the active site in the motor domain may be mediated by the SH3 domain, whose function in myosin has hitherto been unknown.
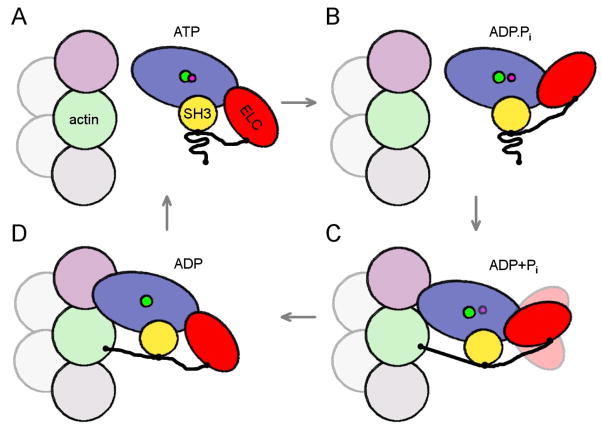
SH3 domains form small β-barrel structures that are found in many proteins, the best known of which are the Src tyrosine kinases, where the domain was first recognized. These adaptor domains bind polyproline-type helices and mediate interactions among different proteins, often in signal tranduction pathways. Interactions of this nature tend to be low affinity (μM), and reversible19, properties which would favor an interaction with the proline-rich sequence in the ELC. The SH3 domain in myosin is located near the N-terminus of the heavy chain, where it forms a six-stranded anti-parallel β-barrel (Lys35 to Met80) that is connected through a series of helices and β-strands to the active site. During the stage of the actomyosin cycle when S1 is detached from actin (the pre-power stroke), the N-terminus of the ELC may be predominantly bound to the SH3 domain (Figs. 5A,B). Only when S1 attaches to actin is the N-terminus of the ELC in a favorable position to bridge the interfilament distance between the lever arm and F-actin (Figs. 5C,D). These movements of the ELC could conceivably affect the interaction of SH3 with the nucleotide-binding region, and thereby modulate the ATPase activity.
Figure 5.
Schematic drawing of proposed events in the myosin II ATPase cycle. Motor domain is shown in blue, ELC in red with extension as thick black line, SH3 domain in yellow. The subunit corresponding to the upper actin in Fig 1C is shown in pink, the lower actin in Fig 1C in green. An additional actin subunit along the long-pitch helix is shown in gray and two subunits along the opposite long-pitch helix are shown in light-gray. The pointed end of the filament is at the top of the figure. (A, B) In the detached states, ATP, ADP.Pi, the proline-rich region of the ELC extension is bound to the SH3 domain, and does not interact with actin. (C) After initial weak binding to actin, the cross-bridge progresses to a stronger force-generating state (ADP+Pi), with the ELC extension still attached to the SH3 domain, but with the N-terminus of the extension now weakly bound to the lower actin. If the lower actin is occupied by another myosin molecule, as in our study, the ELC extension can not bind to it. (D) Strong binding to actin induces the release of Pi and the N-terminus of the ELC extension stabilizes the lever arm by binding to the C-terminus of the lower actin.
Apart from the structural evidence presented here, are there any solution studies that suggest an interaction between the N-terminus of ELC and the catalytic domain of myosin? We have previously measured the orientation and mobility of the N-terminus of LC1 by labeling the –C3 mutant (see Fig. 1B) with a rhodamine probe and exchanging it into S1. Fluorescence anisotropy measurements showed a large increase in anisotropy upon the addition of actin, and, more surprisingly, upon the addition of ATP20. The increase in anisotropy in the presence of actin is to be expected, but the increase in anisotropy with ATP (which completely dissociates the actoS1 complex) implied an interaction of the N-terminus with the myosin heavy chain that results in partial immobilization of the probe. We speculated at the time that the proline-rich extension of LC1 might be stabilized by weak interactions with the SH3 domain, but lacked any evidence for such an interaction. The 3D reconstructions presented here provide support for this interpretation of the spectroscopic data.
Can an Interaction Between the ELC and Actin Be Visualized?
Biochemical and biophysical solution studies provide the strongest evidence for an interaction between the N-terminus of the ELC isoform (A1) and the actin filament. In addition to the earlier NMR studies9 a more recent NMR study using synthesized peptides21 showed that the N-terminal four residues A1P2K3K4 constitute the major interaction with actin, with a more minor contribution from K7 and K8. Although a positive charge is a critical factor in the interaction, as shown here by the ability of Arg residues to compensate to some extent for the absence of the Lys-rich region (Δ13 mutant), the native sequence still has the strongest affinity for actin21. Several cross-linking studies have supported this interpretation of the NMR data13; 15. But the apparent visualization of this interaction by cryoEM in 199014 provided perhaps the most convincing evidence: the extra density at the actoS1 interface seen in a difference map between 3D reconstructions of S1(A1) and S1(A2) was ascribed to the N-terminal extension in A1. A later study of myosin subfragments, which included a comparison of negatively stained images of the same two S1 isoforms, also saw extra density at the actoS1 surface, although the presence of additional difference peaks elsewhere on actin was noted22. However, efforts here to reproduce the results of the original cryoEM study failed to identify any single, statistically significant peak at the actin interface. In order to increase the electron density of the N-terminus, we labeled mutagenized ELC with undecagold, as described above. Despite the presence of this electron dense probe, the difference maps still failed to yield a statistically significant peak on the surface of actin. This problem remained intractable until the introduction of a new approach, IHRSR, to the image analysis of helical filaments17. Unlike conventional helical reconstruction methods16, the new approach uses thousands of short segments (~15.5 asymmetric units) within filaments; which allows sorting segments into different populations such as those containing a gold cluster versus those that do not17; 18. By this single particle approach to helical structures, we were finally able to localize a statistically significant, well-defined peak in the difference map, but not on the actin surface, as expected, but instead near the SH3 domain. It will be of interest in future work to see if one can visualize a gold-labeled ELC (A1) on actin by using sparsely decorated actin filaments (i.e. a high molar ratio of actin to S1), which would favor an interaction between the N-terminal lysine sequence and unoccupied actin binding sites.
The Interaction Between ELC and F-Actin Slows the Velocity of Shortening
Even if structural studies can not unequivocally demonstrate an interaction between the ELC and F-actin filaments, physiological measurements on fibers, as well as in vitro motility studies, and kinetic analyses have all been consistent with this hypothesis. As early as 198823; 24, it was shown that unloaded shortening velocity of skinned skeletal muscle fibers depended on the ratio of the two ELC isoforms: fibers with a high content of LC3 (A2) shortened with a significantly greater velocity than those containing a high amount of LC1 (A1). Later studies corroborated these findings, and went on to show that the degree of shortening velocity depended on the myosin heavy chain composition (fast fiber types), as well as the ELC content25; 26. An in vitro motility assay, considered the solution analogue of unloaded fiber shortening, showed that skeletal muscle myosin containing only A1 moved actin filaments at a significantly slower velocity than myosin containing A24; 27. The in vitro motility measurements shown here for myosin reconsituted with recombinant ELC confirm and extend the earlier findings by showing that positive charges and a Pro/Ala sequence are essential prerequisites for slowing velocity. A simple explanation for these observations is that the ELC interaction with actin increases the lifetime of the attached state of the myosin heads (ton), and by delaying their dissociation, slows the shortening velocity (V=d/ton, where d is the step size). A related argument was used recently to explain the reduced isometric tension in fibers isolated from transgenic mice, which expressed an ELC with a truncated N-terminal extension28.
An Interaction Between the ELC and SH3 May Modulate the Steady-State ATPase Kinetics
It is well-established that the S1(A1) isoform has a lower actin-activated MgATPase activity (kcat) and a higher binding affinity (1/Km) for actin than the S1(A2) isoform7; 8. S1s reconstituted with recombinant ELC show that a charged N-terminus and a Pro/Ala-rich region are primarily responsible for these differences in ATPase activity (Table 1). Similar conclusions were reached earlier using expressed human atrial ELC constructs12; 21, but the data must be regarded as qualitative, since the maximum actin concentration for activation did not exceed 6 μM actin12; 21 which is far below the Km for many of the S1s. Reliable, individual kcat and Km values cannot be obtained from data measured at low substrate concentrations, but only their ratios, kcat/Km.
We find that the change in enzymatic activity is closely coupled to the change in in vitro motility. Although an ELC/actin interaction can explain a slower rate of ADP release (ton~1/kADP), and consequent reduced velocity, it is not readily apparent why Pi release, the rate-limiting step in ATPase activity for non-processive myosin motors, would be decreased by the ELC interaction. During activation, the A1 extension most likely binds to the actin monomer directly below the one that provides the major binding site for the S1 heavy chain: this was deduced from cross-linking experiments12; 13 as well as from the ability of S1(A1) but not S1(A2) to nucleate the rapid polymerization of G-actin10; 29. Here we show that the positive charge on the N-terminal extension is a critical factor in forming a nucleation oligomer in the polymerization process. It is possible that an electrostatic interaction between the ELC and the lower actin subunit could propagate structural changes to the adjacent upper actin subunit along the long pitch helix, and thereby affect the myosin active site. The actin filament is known to be a dynamic structure whose conformation is highly susceptible to actin binding proteins30; 31. However, the electron cryomicroscopic evidence showing that the ELC can bind to the SH3 domain suggests an alternative, more direct pathway to the active site through the motor domain.
This hypothesis has received support from a recent biochemical/genetic study32, aimed at understanding the function of the N-terminal region of the myosin heavy chain of Dictyostelium discoideum class II myosin. A construct that lacked the SH3 domain of myosin (residues 33–79) showed an ~8-fold reduction in in vitro motility, and a >2-fold loss in actin-activated ATPase activity. In addition, an 18-fold decrease in F-actin affinity in the absence of nucleotides, and a 12-fold increase in affinity for ADP in the actin-bound state were observed32. Although the construct was able to rescue function in myosin null cells in vivo32, these data demonstrate that removal of the SH3 domain affects the interaction with nucleotides and actin as well as communication within the motor domain.
Although no additional structural evidence exists that suggests a function for the SH3 domain in the myosin “head” region, recent NMR spectroscopy has revealed an interaction between the SH3 domain of Dictyostelium myosin VII and an adjacent proline-rich sequence in its “tail” region33. Involvement of the SH3 domain in regulating interactions in the tail region of Acanthamoeba myosin 1C has also been reported34; 35. These studies are all beginning to elucidate the different roles SH3 domains may play in the cellular function of members of the myosin superfamily.
Conclusions
Whether the effect of the ELC/actin interaction on the actomyosin ATPase cycle proceeds through conformational changes in actin or through the SH3 domain, or through some combination of both, the finding that the A1 isoform may bind to SH3 offers certain attractive features: the distance between the SH3 domain and actin is easier to bridge during activation than a distance of > 8 nm from the lever arm to actin (see Fig. 1C). The shorter pathway facilitates the diffusive search of a stiff “antenna-like” extension for an actin-binding site. Once the ELC extension is attached to actin, it may well serve to stabilize the interaction of the myosin head with actin by reducing the mobility of the lever arm. We also propose that by binding to the SH3 domain, the ELC may utilize this pathway to modulate ATPase activity: an upward rotation of the lever arm (Fig. 5B), leading to an increased distance between the SH3 domain and the ELC, could inhibit the swing sufficiently to reduce the rate of ATP hydrolysis36. Thus, multiple interactions of the ELC isoform may be instrumental in the execution of a power stroke by vertebrate muscle myosin.
MATERIALS AND METHODS
Protein Preparations
Chicken pectoralis muscle myosin was prepared as described in Margossian and Lowey (1982)37. Actin was prepared from chicken acetone powder38, and stored as F-actin at 4°C for up to 3 weeks. Skeletal muscle S1 was prepared by chymotryptic digestion of chicken muscle myosin, and fractionated into S1(A1) and S1(A2) isoforms as described in Waller et al. (1995)22.
Wild-type A1 was prepared from a chicken fast skeletal muscle cDNA clone provided by J. Robbins as described in Saraswat and Lowey (1998)39. The deletion mutants were prepared by site-directed mutagenesis or by PCR and cloned into the EcoRI site of the expression vector pT7-7. The Cys mutants had the endogenous Cys at position 178 changed to Ala (C178A). The pT7-7 vector was modified to encode Cys in the fusion peptide resulting in an N-terminal tag containing (ACGI) followed by the native sequence starting with Pro2 instead of Ala (see Fig. 1) The C45 mutant was created by changing Ser45 to Cys by site-directed mutagenesis. The double Cys mutant (-C3/C45) was prepared by cloning C45 into the Cys vector described above.
Protein expression and purification were done as described previously in Borejdo et al. (2001) and Wolff-Long et al. (1993)20; 40. The expressed and labeled ELC mutants were exchanged into S1(A2) in 4.7 M NH4Cl as described in Wagner and Weeds, 19777. Excess light chains and unexchanged S1 were removed by ion-exchange chromatography on a MonoQ column (FPLC) equilibrated in 25 mM imidazole, pH 7.0, 1 mM DTT, and eluted with a salt gradient of 100–200 mM NaCl. Myosin was reconstituted with expressed truncated ELC after removal of the endogenous light chains by gel filtration chromatography in 4.5 M NH4Cl as described in Waller et al. (1995)22.
Gold Labeling of Reactive Cysteines
Thiol-reactive undecagold cluster preparations (we thank D. Safer for his generous contribution of undecagold clusters in the early experiments) were prepared by reaction with N-methoxycarbonyl maleimide according to his procedure41. The cluster is composed of a dense, central 11-atom gold core (~ 0.8 nm in diameter) stabilized by a shell of organic groups (leading to a total diameter of ~ 2 nm) that is also necessary for solubility. Labeling of the -3Cys mutant was in 50 mM NaPi, pH 6.6, 10 mM NaCl, with a 4 to5- fold molar excess of undecagold maleimide at 4°C overnight. Free reducing agent was removed just prior to labeling. Unbound gold cluster was removed by DEAE-Sephacel chromatography. The extent of labeling was determined spectrophotometrically at 420 nm using an extinction coefficient of 47,000 M−1cm−1. Protein concentration was determined by the Bradford (Pierce) colorimetric assay. S1(A1) in 30 mM KCl, 20 mM Tris, pH 7.5, 3 mM NaN3 was reacted with a 2-fold molar excess of monomaleimido-undecagold (Nanoprobes, Inc.) for 3 hr on ice. Under these conditions the K+-ATPase was reduced relative to a control, indicating labeling of the reactive thiol, SH1.
Fluorophore Labeling of Cys mutants of the ELC
For the FRET experiments, labeling was essentially as described in Saraswat and Lowey (1998)39. Briefly, lyophilized light chains were incubated for 1 hr in 10 mM DTT, 8 M guanidine hydrochloride, PBS (10 mM NaPi, pH 7.2, 150 mM NaCl, 3 mM NaN3) in order to completely reduce the cysteines before dialysis against PBS and 50 μM DTT. The -3C mutant was labeled at RT with a 4-fold molar excess of 1,5-IAEDANS (Molecular Probes) over thiols for 3–4 hr. The reaction was stopped with 50 mM DTT, and the protein was dialyzed exhaustively against the appropriate buffer. The -C3/C45 mutant was reacted with a 3-fold molar excess of the dansyl donor for 10 min on ice. After dialysis into 25 mM imidazole, pH 7.0, 1 mM DTT, the protein was chromatographed on a 1 ml MonoQ ion-exchange column (Pharmacia, FPLC) equilibrated in the same buffer, and eluted with a 150–250 mM NaCl gradient. Urea-PAGE analysis of the fractions indicated that the major peak (–C3DAN/C45) could be separated from unlabeled and doubly reacted species, as well as from the other singly-labeled specie (–C3/C45DAN). The –C3DAN/C45 was subsequently labeled to completion with one of three acceptor probes: the non-fluorescent DABM and DDPM, or IAF by the procedure described above for the single cysteine mutant. The stoichiometry of labeling was determined using the following extinction coefficients: 6200 M−1cm−1 at 340nm (DAN), 2930 M−1cm−1 at 440 nm (DDP), 24,800 M−1cm−1 at 430 nm (DAB) and 45,000 M−1cm−1 at 495 nm (IAF).
Fluorescence Resonance Energy Transfer (FRET) Measurements
Steady-state fluorescence measurements were performed on an ISS PC1 photon counting spectrofluorometer. Corrected emission spectra of the donor (dansyl moiety) were recorded from 450 to 650 nm at an excitation wavelength of 340 nm with spectral band widths of 8 nm. The efficiency of energy transfer (E) was determined from the reduction in donor fluorescence (Fd) in the presence of acceptor (Fda) as calculated by E=1− (Fda/Fd). The degree of energy transfer depends upon the distance (R) between the donor and acceptor as given by R=(E−1 − 1)1/6 × Ro, where Ro, the Forster critical distance at which energy transfer is 50%, was determined as described in Saraswat and Lowey, 199839.
Actin-activated MgATPase measurements
The ATPase activity of S1 reconstituted with various deletion mutant LC1s was determined at 25°C in 5 mM KCl, 5 mM imidazole, pH 7.5, 1 mM DTT, 4 mM MgCl2, 2mM MgATP. Phosphate release was measured by the colorimetric method of White42. Actin concentrations ranged from 2.5 to 100 μM (details in Trybus, 200043). It is important that the actin concentrations extend to at least 4–5 times the Km in order to have a meaningful extrapolation to Vmax upon fitting the data to the Michaelis-Menten equation.
In vitro Motility Assays
The truncated ELC mutants were exchanged into myosin as described briefly above22. To remove rigor heads, samples were spun with equimolar concentrations of actin in the presence of MgATP prior to the assay. Myosin was perfused through the flow cell at ~50 μg/ml in 25 mM KCl, 25 mM imidazole, pH 7.5, 4 mM MgCl2, 1 mM EGTA, 10 mM DTT at 30°C. The final assay buffer contained 0.5 % methylcellulose, oxygen scavengers and 1 mM MgATP as described in Trybus, 200043. The velocities of 20–30 filaments for each of two independent experiments were averaged to obtain the mean ± standard deviation.
Electron microscopy
To prepare frozen-hydrated specimens for electron cryomicroscopy, F-actin (0.025–0.03 mg/ml) was applied to glow-discharged 400-mesh copper grids coated with holey carbon film in 20 mM NaCl, 5 mM NaPi, pH 7.0, 1 mM MgCl2, 2 mM NaN3. Following 1 min incubation in a humid chamber, the grids were rinsed twice with the respective myosin buffer without the myosin sample (10 mM NaCl, 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM DTT, 2 mM NaN3). Myosin sample, diluted to ~ 0.5 or 2 mg/ml was then applied to the grid for 30 s, and replaced by an additional drop of sample (30 s). The excess liquid was blotted, and the grids were plunged into liquid ethane cooled by liquid N2. Low-dose images were recorded using a CM12 electron microscope (FEI Electron Optics, the Netherlands) equipped with a LaB6 filament and a DH626 Gatan cryoholder (Gatan Inc., Pleasanton, CA), at a nominal magnification of 60,000 (at 120 keV), at ~1.5 μm defocus (electron dose ~10e−/Å−2). Micrographs were digitized with a SCAI scanner (Integraph, Phoenix, AZ) with a pixel size of 0.27 nm on the sample. One data set containing S1(A1) with gold-labeled A1, one set containing S1(A1) with gold-labeled SH1, one data set of S1(A2) and three data sets from different preparations of unlabeled S1(A1) were obtained.
Image analysis
Helical Reconstruction
The Brandeis Helical Imaging Package16 provided the alignment parameters for each filament that was introduced in the real space average. These include phase origin and particle tilt. These parameters were refined through minimization of the phase error in reciprocal space44. Using these alignment parameters, three-dimensional maps were computed separately for each individual filament in the data set. All reconstructions included 23 layer lines that were trimmed to 21 Å resolution. Since this is within the first node of the contrast transfer function, no phase correction was necessary. The abrupt edge in the data introduced by this procedure was smoothed to zero using a Gaussian fall-off. The layer line orders used were 2, −11, 4, −9, 6, −7, 8, −5, −3, −1, 14, 1, −12, 3, 5, −8, 7, −6, −4, −2, 13, 0 and the equator. The individual filament maps were aligned in real space, normalized, and averaged45. The real space averaging provides a single averaged value and its associated variance for each voxel in the volume.
Reconstruction by Iterative Helical Real Space Refinement
The iterative helical real space refinement (IHRSR) method17 is a hybrid approach that uses real-space, single-particle processing and imposition of helical symmetry in an iterative manner. Our implementation uses EMAN46 for the single-particle reconstruction portion and routines adapted from the CoAn suite47; 48 to determine and impose the helical symmetry. A box size of 80 × 80 pixels with a 5.4 Å pixel size was used. This corresponds to about 15.5 asymmetric units of the helix, a little over one actin crossover. An overlap of 60 pixels was chosen, allowing every asymmetric unit (a single actomyosin motif) to contribute to four different views of the helix. The data were arbitrarily divided into two subsets that were reconstructed independently. The variance for the IHRSR maps (necessary for calculating t-tests) was then estimated using the differences between the two subsets. Using a voxel-wise t-test procedure49, we established that there were no statistically significant differences (99.5% confidence level) in the structural features between maps of the same underlying data calculated by the two methods. However, the background was significantly reduced in the IHRSR reconstructions owing to the fact that a larger number of asymmetric units (~20000 versus ~5000) could be used.
Localization of the gold labels
Difference maps were calculated by subtracting the S1 maps from the gold-labeled maps giving a total of 4 difference maps per label (SH1 and A1 labels). Prior to subtracting, the maps were aligned to each other using the CoAn algorithm47. For each gold label, the difference maps were averaged and the voxel-wise variance was calculated which enabled us to test the significance of features in the (averaged) difference maps using a t-test at a confidence level of 99.5%. Difference maps between the helical reconstructions did not show any statistically significant peaks for the A1- labeled map, but did show a single significant peak for the SH1 label. For the IHRSR maps, there was one significant peak for each of the labels (SH1-labeled, and A1-labeled). For the SH1-labeled maps, the helical reconstruction difference peak corresponded to the highest peak in all IHRSR difference maps that were in the average. For the A1-labeled maps this was not the case, indicating disorder or partial decoration. Multi-reference refinement as described in Volkmann et al. (2005)18 was then applied to the A1- labeled data set using S1(A1) and S1(A1) with the A1-labeled peak from the averaged difference maps as references. Approximately 60% of the segments were sorted into the labeled class. The IHRSR reconstruction using that subset of the data was then used for difference mapping, which showed an enhanced peak in the same location, and now being, as in the case of the SH1-labeled map, the highest peak in each of the non-averaged difference maps.
S1(A1) - S1(A2) difference mapping
Difference maps between the various IHRSR S1(A1) and S1(A2) maps were constructed following the same procedure as used for the labeled maps, resulting in 3 difference maps that were averaged and used for variance calculations. No statistically significant peaks could be detected even after multi- reference refinement. However, one peak that becomes significant if the confidence level is dropped to 90% (p > 0.1) was ~ 7 Å closer to the SH3 domain than the center of the A1 gold-labeled peak, thus being consistent with the location of the A1 extension. This confidence level is clearly picking up a substantial amount of noise as exemplified by several peaks far away from the actomyosin density (data not shown). The SH3 peak is the third highest at this confidence level. There are no peaks detectable in the SH3 vicinity at the 90% confidence level if difference maps are calculated from unlabeled S1(A1) versus unlabeled S1(A1), or from S1(A2) versus S1(A2) (12 difference maps in total), providing further support that the peak in the S1(A1) – S1(A2) difference map represents the location of the N-terminal extension.
Abbreviations
- cryoEM
electron cryomicroscopy
- SH3
src-homology 3 domain
- SH1
reactive thiol in myosin heavy chain
- ELC
essential light chain
- LC1 or A1
the isoform of ELC containing the N-terminal extension of 41 residues
- A2
the smaller isoform of ELC that has eight unique residues at the N-terminus, but shares the remaining 140 residues with the A1
- S1
subfragment-1
- 1
5-IAEDANS, N-iodoacetyl-N′-(5-sulfo-1-napthyl)ethylenediamine
- DDPM
N-(4-dimethylamino-3,5-dinitrophenyl)maleimide
- DABM
(4-dimethylaminophenylazophenyl-4′ maleimide)
- IAF
5-(iodoacetamido) fluorescein
- DTT
dithiothreitol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–8. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A, Noble M, Pauptit R, Wierenga R, Saraste M. Crystal structure of a Src-homology 3 (SH3) domain. Nature. 1992;359:851–5. doi: 10.1038/359851a0. [DOI] [PubMed] [Google Scholar]
- 3.Cope MJ, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain: implications for structure and function. Structure. 1996;4:969–87. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 4.Lowey S, Waller GS, Trybus KM. Skeletal muscle myosin light chains are essential for physiological speeds of shortening. Nature. 1993;365:454–6. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]
- 5.VanBuren P, Waller GS, Harris DE, Trybus KM, Warshaw DM, Lowey S. The essential light chain is required for full force production by skeletal muscle myosin. Proc Natl Acad Sci U S A. 1994;91:12403–7. doi: 10.1073/pnas.91.26.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timson DJ. Fine tuning the myosin motor: the role of the essential light chain in striated muscle myosin. Biochimie. 2003;85:639–45. doi: 10.1016/s0300-9084(03)00131-7. [DOI] [PubMed] [Google Scholar]
- 7.Wagner PD, Weeds AG. Studies on the role of myosin alkali light chains. Recombination and hybridization of light chains and heavy chains in subfragment-1 preparations. J Mol Biol. 1977;109:455–70. doi: 10.1016/s0022-2836(77)80023-5. [DOI] [PubMed] [Google Scholar]
- 8.Hayashibara T, Miyanishi T. Binding of the amino-terminal region of myosin alkali 1 light chain to actin and its effect on actin-myosin interaction. Biochemistry. 1994;33:12821–7. doi: 10.1021/bi00209a013. [DOI] [PubMed] [Google Scholar]
- 9.Bhandari DG, Levine BA, Trayer IP, Yeadon ME. 1H-NMR study of mobility and conformational constraints within the proline-rich N-terminal of the LC1 alkali light chain of skeletal myosin. Correlation with similar segments in other protein systems. Eur J Biochem. 1986;160:349–56. doi: 10.1111/j.1432-1033.1986.tb09978.x. [DOI] [PubMed] [Google Scholar]
- 10.Chaussepied P, Kasprzak AA. Isolation and characterization of the G-actin-myosin head complex. Nature. 1989;342:950–3. doi: 10.1038/342950a0. [DOI] [PubMed] [Google Scholar]
- 11.Chalovich JM, Stein LA, Greene LE, Eisenberg E. Interaction of isozymes of myosin subfragment 1 with actin: effect of ionic strength and nucleotide. Biochemistry. 1984;23:4885–9. doi: 10.1021/bi00316a011. [DOI] [PubMed] [Google Scholar]
- 12.Timson DJ, Trayer HR, Trayer IP. The N-terminus of A1-type myosin essential light chains binds actin and modulates myosin motor function. Eur J Biochem. 1998;255:654–62. doi: 10.1046/j.1432-1327.1998.2550654.x. [DOI] [PubMed] [Google Scholar]
- 13.Andreev OA, Saraswat LD, Lowey S, Slaughter C, Borejdo J. Interaction of the N-terminus of chicken skeletal essential light chain 1 with F-actin. Biochemistry. 1999;38:2480–5. doi: 10.1021/bi981706x. [DOI] [PubMed] [Google Scholar]
- 14.Milligan RA, Whittaker M, Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–21. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 15.Sutoh K. Identification of myosin-binding sites on the actin sequence. Biochemistry. 1982;21:3654–61. doi: 10.1021/bi00258a020. [DOI] [PubMed] [Google Scholar]
- 16.Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: the Brandeis Helical Package. J Struct Biol. 1996;116:167–75. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 17.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–34. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 18.Volkmann N, Liu H, Hazelwood L, Krementsova EB, Lowey S, Trybus KM, Hanein D. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–92. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 20.Borejdo J, Ushakov DS, Moreland R, Akopova I, Reshetnyak Y, Saraswat LD, Kamm K, Lowey S. The power stroke causes changes in the orientation and mobility of the termini of essential light chain 1 of myosin. Biochemistry. 2001;40:3796–803. doi: 10.1021/bi002527u. [DOI] [PubMed] [Google Scholar]
- 21.Timson DJ, Trayer HR, Smith KJ, Trayer IP. Size and charge requirements for kinetic modulation and actin binding by alkali 1-type myosin essential light chains. J Biol Chem. 1999;274:18271–7. doi: 10.1074/jbc.274.26.18271. [DOI] [PubMed] [Google Scholar]
- 22.Waller GS, Ouyang G, Swafford J, Vibert P, Lowey S. A minimal motor domain from chicken skeletal muscle myosin. J Biol Chem. 1995;270:15348–52. doi: 10.1074/jbc.270.25.15348. [DOI] [PubMed] [Google Scholar]
- 23.Sweeney HL, Kushmerick MJ, Mabuchi K, Sreter FA, Gergely J. Myosin alkali light chain and heavy chain variations correlate with altered shortening velocity of isolated skeletal muscle fibers. J Biol Chem. 1988;263:9034–9. [PubMed] [Google Scholar]
- 24.Greaser ML, Moss RL, Reiser PJ. Variations in contractile properties of rabbit single muscle fibres in relation to troponin T isoforms and myosin light chains. J Physiol. 1988;406:85–98. doi: 10.1113/jphysiol.1988.sp017370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweeney HL. Function of the N terminus of the myosin essential light chain of vertebrate striated muscle. Biophys J. 1995;68:112S–118S. discussion 118S–119S. [PMC free article] [PubMed] [Google Scholar]
- 26.Bottinelli R, Betto R, Schiaffino S, Reggiani C. Unloaded shortening velocity and myosin heavy chain and alkali light chain isoform composition in rat skeletal muscle fibres. J Physiol. 1994;478 (Pt 2):341–9. doi: 10.1113/jphysiol.1994.sp020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowey S, Waller GS, Trybus KM. Function of skeletal muscle myosin heavy and light chain isoforms by an in vitro motility assay. J Biol Chem. 1993;268:20414–8. [PubMed] [Google Scholar]
- 28.Miller MS, Palmer BM, Ruch S, Martin LA, Farman GP, Wang Y, Robbins J, Irving TC, Maughan DW. The essential light chain N-terminal extension alters force and fiber kinetics in mouse cardiac muscle. J Biol Chem. 2005;280:34427–34. doi: 10.1074/jbc.M508430200. [DOI] [PubMed] [Google Scholar]
- 29.Chen T, Reisler E. Interactions of myosin subfragment 1 isozymes with G-actin. Biochemistry. 1991;30:4546–52. doi: 10.1021/bi00232a026. [DOI] [PubMed] [Google Scholar]
- 30.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanein D, Volkmann N, Goldsmith S, Michon AM, Lehman W, Craig R, DeRosier D, Almo S, Matsudaira P. An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat Struct Biol. 1998;5:787–92. doi: 10.1038/1828. [DOI] [PubMed] [Google Scholar]
- 32.Fujita-Becker S, Tsiavaliaris G, Ohkura R, Shimada T, Manstein DJ, Sutoh K. Functional characterization of the N-terminal region of myosin-2. J Biol Chem. 2006;281:36102–9. doi: 10.1074/jbc.M605171200. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Deloia MA, Kang Y, Litchke C, Zhang N, Titus MA, Walters KJ. The SH3 domain of a M7 interacts with its C-terminal proline-rich region. Protein Sci. 2007;16:189–96. doi: 10.1110/ps.062496807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa T, Cheng N, Liu X, Korn ED, Steven AC. Subdomain organization of the Acanthamoeba myosin IC tail from cryo-electron microscopy. Proc Natl Acad Sci U S A. 2004;101:12189–94. doi: 10.1073/pnas.0404835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang KJ, Mahmoodian F, Ferretti JA, Korn ED, Gruschus JM. Intramolecular interaction in the tail of Acanthamoeba myosin IC between the SH3 domain and a putative pleckstrin homology domain. Proc Natl Acad Sci U S A. 2007;104:784–9. doi: 10.1073/pnas.0610231104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malnasi-Csizmadia A, Toth J, Pearson DS, Hetenyi C, Nyitray L, Geeves MA, Bagshaw CR, Kovacs M. Selective perturbation of the myosin recovery stroke by point mutations at the base of the lever arm affects ATP hydrolysis and phosphate release. J Biol Chem. 2007 doi: 10.1074/jbc.M701447200. [DOI] [PubMed] [Google Scholar]
- 37.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 38.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–81. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 39.Saraswat LD, Lowey S. Subunit interactions within an expressed regulatory domain of chicken skeletal myosin. Location of the NH2 terminus of the regulatory light chain by fluorescence resonance energy transfer. J Biol Chem. 1998;273:17671–9. doi: 10.1074/jbc.273.28.17671. [DOI] [PubMed] [Google Scholar]
- 40.Wolff-Long VL, Saraswat LD, Lowey S. Cysteine mutants of light chain-2 form disulfide bonds in skeletal muscle myosin. J Biol Chem. 1993;268:23162–7. [PubMed] [Google Scholar]
- 41.Safer D, Bolinger L, Leigh JS., Jr Undecagold clusters for site-specific labeling of biological macromolecules: simplified preparation and model applications. J Inorg Biochem. 1986;26:77–91. doi: 10.1016/0162-0134(86)80001-0. [DOI] [PubMed] [Google Scholar]
- 42.White HD. Special instrumentation and techniques for kinetic studies of contractile systems. Methods Enzymol. 1982;85(Pt B):698–708. doi: 10.1016/0076-6879(82)85057-x. [DOI] [PubMed] [Google Scholar]
- 43.Trybus KM. Biochemical studies of myosin. Methods. 2000;22:327–35. doi: 10.1006/meth.2000.1085. [DOI] [PubMed] [Google Scholar]
- 44.DeRosier DJ, Moore PB. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J Mol Biol. 1970;52:355–69. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- 45.Hanein D, DeRosier D. A new algorithm to align three-dimensional maps of helical structures. Ultramicroscopy. 1999;76:233–8. doi: 10.1016/s0304-3991(98)00086-2. [DOI] [PubMed] [Google Scholar]
- 46.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 47.Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struct Biol. 1999;125:176–84. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- 48.Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–25. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
- 49.Trachtenberg S, DeRosier DJ. Three-dimensional structure of the frozen-hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]