Abstract
Human anti-V3 monoclonal antibodies (mAbs) generated from HIV-1 infected individuals display diversity in the range of their cross-neutralization that may be related to their immunogenetic background. The study of the immunoglobulin (Ig) variable region gene usage of heavy chains have shown a preferential usage of the VH5-51 gene segment which was detected in 35% of 51 human anti-V3 mAbs. In contrast, human mAbs against other envelope regions of HIV-1 (anti-Env), including the CD4-binding domain, the CD4-induced epitope, and gp41 preferentially used the VH1-69 gene segment, and none of them used the VH5-51 gene. Furthermore, the usage of the VH4 family by anti-V3 mAbs was restricted to only one gene segment, VH4-59, while the VH3 gene family was used at a significantly lower frequency by all of the analyzed anti-HIV-1 mAbs. Multivariate analysis showed that usage of VH gene segments was significantly different between anti-V3 and anti-Env mAbs, and compared to antibodies from healthy subjects. In addition, the anti-V3 mAbs preferentially used the JH3 and D2-15 gene segments. The preferential usage of selected Ig gene segments and the characteristic pattern of Ig gene usage by anti-V3 mAbs can be related to the conserved structure of the V3 region.
Keywords: Immunoglobulin gene usage, human monoclonal antibodies, anti-HIV-1 antibodies
1. Introduction
Immunoglobulin (Ig) gene usage is not random. B cell epitopes select for and stimulate B cells carrying surface-bound Ig that provide the best fit. This process implies a structure/fuction relationship between epitope and the Ig gene-encoded antibody (Ab) produced by the B cell which it stimulates. Examples of preferential Ig gene usage include the differential VH gene usage by human Abs specific for different pathogens. Thus, Abs against the capsular polysaccharide of Hemophilus influenzae type b primarily use the VH3-23 gene (Lucas et al., 2003), Abs against Streptococcus pneumoniae preferentially use the VH3 gene family (Sun et al., 1999), and the gene segment VH1-46 was dominant for Abs against rotavirus (Weitkamp et al., 2003).
The antigen combining site of the Ab is encoded by genes generated by the combinatorial rearrangement of five gene segments, including the variable (VH), diversity (D) and joining (JH) segments for the heavy chain, and the variable (VL) and joining (VJ) segments for the light chain (Cook and Tomlinson, 1995). The VH gene segment encodes a leader peptide and the largest part of the variable (V) fragment of an Ab, containing 96 to 101 amino acids. This part includes two complementarity determining regions 1 and 2 (CDR 1 and 2) which interact with antigen, and three framework regions (FR) which help adapt CDRs for binding. The CDR3 of the heavy chain is a component of the region created by the joining of the C-terminal end of VH to the D and JH segments plus palindromic (P) and non-templated (N) nucleotides; CDR3 length of human antibodies is, on average, 14 amino acids (Tiller et al., 2007).
The VH gene segments are divided into seven gene families, VH1-VH7, each being at least 80% homologous at the nucleotide sequence level. In healthy individuals, the percentage of VH gene family usage is generally dependent upon the number of gene segments in each family. For example, the VH3 gene family contains 21 functional gene segments and is the most frequently used, whereas the VH5 family has only two genes and is only used by a low percentage of Abs (http://imgt.cines.fr).
Studies of human anti-HIV-1 monoclonal Abs (mAbs) and VH gene usage show a reduced representation of the VH3 gene family in the repertoire of various anti-gp120 and anti-gp41 mAbs (David et al., 1995a; Wisnewski et al., 1996). The decreased usage of the VH3 family genes may be related to the activity of gp120 of HIV-1 as a superantigen which causes a depletion of B cells expressing the VH3 gene-encoded surface Ig (Berberian et al., 1993). Among a number of human mAbs against HIV-1, only one group of mAbs, those specific for the CD4-induced epitope (CD4i), has been analyzed for VH gene usage (Huang et al., 2004). This study showed that 12 human mAbs and Fabs specific for the CD4i epitope selectively use the VH1 gene family (Huang et al., 2004).
The human anti-V3 mAbs generated from HIV-1 infected individuals are able to cross-react with different viruses and neutralize primary isolates from various HIV-1 subtypes (Gorny et al., 1997; Gorny et al., 2002; Gorny et al., 2006). Using several animal models, passive administration of these Abs has also been shown to protect against HIV-1 infection (Andrus et al., 1998; Emini et al., 1992). Based on these data, we hypothesize that anti-V3 Abs induced by a vaccine in healthy individuals may play an important role in protecting against HIV-1 infection. Therefore, a number of human anti-V3 mAbs were produced in our laboratory from the cells of HIV-1 infected individuals in order to study the mechanism of neutralization and to characterize the V3 region of the virus envelope (Gorny et al., 1998; Gorny et al., 1997; Gorny et al., 2002; Gorny et al., 2006; Gorny et al., 1991; Gorny et al., 1993). These anti-V3 mAbs exhibit a broad range of activity; they can be type-specific and neutralize a few viruses belonging to one subtype, or the mAbs can broadly neutralize viruses from different HIV-1 subtypes. Ig gene usage has been examined only in few human anti-V3 mAbs (Andris et al., 1991; Ditzel et al., 1997; Lewis et al., 1995; Liu et al., 2003; van der Donk et al., 1994), and the role of different VH gene segments in V3 mAb function remains unclear. Because of the potential importance of inducing such Abs with a prophylactic vaccine, we hypothesized that understanding the immunogenetics of V3 Abs could have implications for vaccine design. Therefore, using a large panel of human anti-V3 mAbs, we undertook this study and have found preferential use of the rarely employed VH5-51 gene segment, significantly decreased usage of VH3, and restriction in the use of the VH4 gene family to only the VH4-59 gene segment. The analysis of JH and D gene families for the V region showed preferential usage of JH3 and D2-15. Overall, human anti-V3 mAbs display a pattern of Ig gene usage with biased usage of several gene segments, which differs significantly from other anti-HIV-1 human mAbs and normal antibodies.
2. Materials and methods
2.1. Human anti-V3 mAbs
A panel of 51 human anti-V3 mAbs was analyzed for the VH, D and JH gene segment usage. Forty-eight mAbs were developed in our laboratory and three mAbs, MN215, F425 and Fab DO142, were developed in other laboratories (Ditzel et al., 1997; Liu et al., 2003; van der Donk et al., 1994). The sequences of the latter are available at GeneBank. Thirty-five anti-V3 mAbs produced in this laboratory were previously described (Gorny et al., 1997; Gorny et al., 2002; Gorny et al., 2006; Gorny et al., 1991; Gorny et al., 1993), and 13 mAbs are described in this study for the first time (Table 1).
Table 1.
New human mAbs produced from subjects infected with non-clade B and clade B HIV-1
| mAb | Specificity | Isotype | Subtype of infecting virus | Country of origin |
|---|---|---|---|---|
| 3694 | V3 | IgG1λ | H | Cameroon |
| 3697 | V3 | IgG1λ | G | Cameroon |
| 3869 | V3 | IgG1λ | nd | Cameroon |
| 3881 | V3 | IgG1κ | CRF02_AG | Cameroon |
| 4085 | V3 | IgG1λ | nd | Cameroon |
| 3791 | V3 | IgG1κ | C | India |
| 3792 | V3 | IgG1λ | C | India |
| 3904 | V3 | IgG1κ | nd | India |
| 3906 | V3 | IgG1λ | nd | India |
| 4022 | V3 | IgG1λ | C | India |
| 4025 | V3 | IgG1λ | nd | India |
| 4121 | V3 | IgG1λ | nd | India |
| 3402 | V3 | IgG1κ | B | USA |
nd – not determined
Thirty-one mAbs were generated from individuals living primarily in the New York City (NYC) area who were presumably infected with clade B viruses. Twenty mAbs were produced from donors infected with non-clade B viruses (Table 2) while living in Cameroon or India with the exception of two mAbs, 1324E and 2182, whose donors were infected in Thailand and Ivory Coast (Gorny et al., 1998; Gorny et al., 2002). Twelve of the non-clade B mAbs were newly developed and their characteristics are shown in Table 1. This study has been reviewed and approved by the New York University School of Medicine Institutional Review Board.
Table 2.
Immunoglobulin gene usage for variable heavy chain of anti-V3 mAbs
| mAb | Cells | IGHV | IGHD | D-RF | IGHJ | Reference |
|---|---|---|---|---|---|---|
| 419-D | Hybridoma | 5-51*01 | 6-19 | 3 | 4 | (Gorny et al., 1993) |
| 782-D | “ | 5-51*01 | 5-24 | 1 | 4 | (Gorny et al., 1997) |
| 838-D | “ | 5-51*01 | 2-2 | 2 | 4 | (Gorny et al., 1997) |
| 908-D | “ | 5-51*01 | 1-14 | 2 | 4 | (Gorny et al., 1997) |
| 2456c | “ | 5-51*01 | 3-22 | 2 | 3 | (Gorny et al., 2002) |
| 2557a | “ | 5-51*01 | 3-22 | 2 | 3 | (Gorny et al., 2004) |
| 3906a | “ | 5-51*01 | 3-22 | 3 | 3 | This study |
| 4022a | “ | 5-51*01 | 2-8 | 2 | 5 | This study |
| 4025a | “ | 5-51*01 | 3-3 | 1 | 6 | This study |
| 257-2D | EBV | 5-51*03 | 3-22 | 2 | 4 | (Andris et al., 1991) |
| 1006-15D | Hybridoma | 5-51*03 | 3-22 | 2 | 3 | (Gorny et al., 1997) |
| 2219b | “ | 5-51*03 | 4-17 | 2 | 3 | (Gorny et al., 2002) |
| 2483b | “ | 5-51*03 | 3-10 | 2 | 3 | (Gorny et al., 2004) |
| 2558a | “ | 5-51*03 | 1-26 | 3 | 4 | (Gorny et al., 2004) |
| 3019a | “ | 5-51*03 | 2-21 | 2 | 3 | (Gorny et al., 2006) |
| 3694a | “ | 5-51*03 | 3-9 | 2 | 1 | This study |
| 4085a | “ | 5-51*03 | 2-21 | 2 | 4 | This study |
| 3792a | “ | 5-51*03 | 3-22 | 2 | 4 | This study |
| 447-52D | “ | 3-15*07 | 3-10 | 3 | 6 | (Lewis et al., 1995) |
| MN215 | EBV | 3-21*01 | 2-21 | 3 | 4 | (van der Donk et al., 1994) |
| 2601a | Hybridoma | 3-30*02 | 2-2 | 2 | 4 | (Gorny et al., 2006) |
| 504-D | “ | 3-30*03 | 3-16 | 2 | 3 | (Gorny et al., 1993) |
| 1324Ea | “ | 3-30*03 | 2-15 | 3 | 5 | (Gorny et al., 1998) |
| 3904a | “ | 3-30*03 | 3-16 | 3 | 4 | This study |
| 412-D | “ | 3-33*01 | 3-10 | 1 | 6 | (Gorny et al., 1993) |
| 2424 | “ | 3-53*01 | 5-12 | 3 | 6 | (Gorny et al., 2004) |
| 418-D | “ | 3-53*03 | 3-22 | 2 | 6 | (Gorny et al., 1993) |
| F425 | “ | 3-64*01 | 3-22 | 2 | 3 | (Liu et al., 2003) |
| 537-D | “ | 3-7*01 | 2-2 | 2 | 6 | (Gorny et al., 1993) |
| 3402 | “ | 3-7*02 | 3-22 | 2 | 6 | This study |
| 3791a | “ | 1-18*01 | 4-17 | 2 | 4 | This study |
| 4121a | “ | 1-18*01 | 1-1 | 2 | 4 | This study |
| 391/95-D | “ | 1-24*01 | 3-3 | 1 | 4 | (Gorny et al., 1993) |
| 311-11D | “ | 1-3*01 | 3-10 | 2 | 4 | (Gorny et al., 1993) |
| 1334 | “ | 1-3*01 | 3-9 | 2 | 4 | (Gorny et al., 2000) |
| 1027-15D | “ | 1-f*01 | 3-3 | 2 | 5 | (Gorny et al., 1997) |
| 2191 | “ | 1-f*01 | 3-3 | 2 | 5 | (Gorny et al., 2002) |
| 3224a | “ | 1-f*01 | 6-19 | 2 | 6 | (Gorny et al., 2006) |
| 3697a | “ | 1-f*01 | 5-24 | 3 | 4 | This study |
| 3869a | “ | 1-f*01 | 2-15 | 2 | 4 | This study |
| DO142 | Phage | 1-f*01 | 2-15 | 2 | 4 | (Ditzel et al., 1997) |
| 268-11D | EBV | 4-59*01 | 5-12 | 3 | 3 | (Andris et al., 1991) |
| 386-D | Hybridoma | 4-59*01 | 2-8 | 3 | 4 | (Gorny et al., 1993) |
| 453-D | “ | 4-59*01 | 2-15 | 3 | 3 | (Gorny et al., 1993) |
| 1108 | “ | 4-59*01 | 2-15 | 3 | 3 | (Zolla-Pazner et al., 1999) |
| 2442 | “ | 4-59*01 | 6-13 | 1 | 4 | (Gorny et al., 2002) |
| 3074a | “ | 4-59*01 | 3-22 | 2 | 3 | (Gorny et al., 2006) |
| 3881a | “ | 4-59*01 | 2-2 | 3 | 3 | This study |
| 2182a | “ | 4-59*04 | 2-15 | 3 | 3 | (Gorny et al., 2002) |
| 2412c | “ | 2-5*04 | 3-22 | 2 | 4 | (Gorny et al., 2002) |
| 694/98-D | “ | 2-5*04 | 5-12 | 3 | 4 | (Gorny et al., 1992) |
mAbs derived from non-clade B HIV-1 infected individuals living or being infected abroad.
These mAbs were generated from blood samples derived from one patient at different time. D-RF, D gene reading frame.
2.2. Human anti-gp120 and gp41 mAbs
The Ig gene usage was also analyzed for 44 human mAbs specific for epitopes in the envelope proteins of HIV-1 other than V3; these included 11 anti-CD4bd mAbs, 12 anti-CD4i mAbs and 21 anti-gp41 mAbs (Table 3). Thirty-four nucleotide sequences for the Ig variable fragment of the heavy chain were obtained from published data or GenBank (Andris et al., 1991; Bagley et al., 1994; Barbas et al., 1993; Cavacini et al., 1998; David et al., 1995b; de Haard et al., 1998; Felgenhauer et al., 1990; Huang et al., 2004; Kunert et al., 1998; Kunert et al., 2004; Marasco et al., 1992; Moran et al., 1993; Thali et al., 1993; van der Donk et al., 1994; Zhang et al., 2004; Zwick et al., 2001) while 10 human anti-gp41 mAbs, 50–69 (Gorny et al., 1989), 126-6, 167-7, 181-D, 240-D and 246-D (Xu et al., 1991), 1281, 1342, 1367 and 1379 (Gorny et al., 2000), were produced and sequenced in our laboratory (Table 3).
Table 3.
Immunoglobulin gene usage for variable heavy chains of human anti-Env mAbs
| mAb | Cells | IGHV | IGHD | D-RF | IGHJ | Reference |
|---|---|---|---|---|---|---|
| Anti- CD4bd mAbs | ||||||
| b12a | Phage | 1-3*01 | 2-21 | 2 | 6 | (Barbas et al., 1993) |
| GP44 | EBV | 1-46*01 | 3-16 | 3 | 4 | (van der Donk et al 1994) |
| GP68 | “ | 1-69*01 | 3-16 | 2 | 4 | (van der Donk et al 1994) |
| S1-1 | Hybridoma | 1-69*09 | 2-2 | 3 | 4 | (Moran et al., 1993) |
| 21h | EBV | 3-11*03 | 4-17 | 3 | 5 | (Bagley et al., 1994) |
| b3a | “ | 3-15*06 | 3-9 | 2 | 6 | (Barbas et al., 1993) |
| b13a | “ | 3-30*04 | 3-9 | 2 | 4 | (Barbas et al., 1993) |
| m14 | “ | 4-30-4*01 | 3-10 | 1 | 4 | (Zhang et al., 2004) |
| 15e | “ | 4-39*02 | 2-8 | 1 | 6 | (Bagley et al., 1994) |
| F105 | Hybridoma | 4-59*01 | 4-17 | 2 | 5 | (Marasco et al., 1992) |
| GP13 | EBV | 5-a*03 | 3-9 | 2 | 6 | (van der Donk et al 1994) |
| Anti-CD4i mAbs | ||||||
| 411gb | EBV | 1-24*01 | 1-26 | 1 | 4 | (Huang et al., 2004) |
| X5 | Phage | 1-69*01 | 3-22 | 2 | 4 | (Huang et al., 2004) |
| E51c | EBV | 1-69*01 | 2-2 | 3 | 6 | (Huang et al., 2004) |
| 412dc | “ | 1-69*01 | 4-4 | 2 | 2 | (Huang et al., 2004) |
| 47e3 | “ | 1-69*01 | 4-17 | 2 | 6 | (Huang et al., 2004) |
| 23eb | “ | 1-69*06 | 3-3 | 2 | 6 | (Huang et al., 2004) |
| 17b6 | “ | 1-69 | nd | nd | 1 | (Thali et al., 1992) |
| Sb1d,f | Phage | 1-69 | nd | nd | 6 | (Huang et al., 2004) |
| C12d,f | “ | 1-69 | nd | nd | 4 | (Huang et al., 2004) |
| m16f | “ | 1-69 | nd | nd | 6 | (Huang et al., 2004) |
| 16c | “ | 1-f*01 | 5-12 | 3 | 4 | (Huang et al., 2004) |
| 48d | EBV | 1-f*01 | 1-26 | 2 | 3 | (Thali et al., 1992) |
| Anti-gp41 mAbs | ||||||
| DaB2B3 | EBV | 1-46*01 | 3-16 | 3 | 3 | (David et al., 1995b) |
| Ab 31 | Phage | 1-46*01 | 2-2 | 1 | 3 | (de Haard et al., 1998) |
| 246-D+ | Hybridoma | 1-69*01 | 6-19 | 2 | 4 | (Xu et al., 1991) |
| 181-D+ | “ | 1-69*01 | 3-16 | 1 | 6 | (Xu et al., 1991) |
| 50-69D+ | “ | 1-69*01 | 2-21 | 2 | 4 | (Gorny et al., 1989) |
| No.86 | “ | 1-69*06 | 3-9 | 2 | 4 | (Moran et al., 1993) |
| 1281+ | “ | 1-69*06 | 3-9 | 1 | 5 | (Gorny et al., 2000) |
| 126-7+,e,h | “ | 1-69*06 | 2-2 | 3 | 4 | (Xu et al., 1991) |
| 167-7+ | “ | 1-69*09 | 3-10 | 3 | 4 | (Xu et al., 1991) |
| 240-D+ | “ | 1-69*10 | 3-9 | 2 | 5 | (Xu et al., 1991) |
| 4E10f | “ | 1-69 | 3-16 | nd | 1 | (Kunert et al., 2004) |
| F240 | “ | 3-11*01 | 3-22 | 2 | 5 | (Cavacini et al., 1998) |
| 1379+ | “ | 3-11*03 | 4-17 | 1 | 4 | (Gorny et al., 2000) |
| NG3B7 | EBV | 3-23*04 | 1-26 | 3 | 3 | (David et al., 1995b) |
| 1342+ | Hybridoma | 3-23*04 | 3-10 | 3 | 4 | (Gorny et al., 2000) |
| 3D6 | “ | 3-9*01 | 3-22 | 2 | 3 | (Felgenhauer et al. 1990) |
| 120-16e | EBV | 4-30-2*01 | 5-12 | 3 | 2 | (Andris et al., 1991) |
| 1367+ | Hybridoma | 4-30-4*01 | 2-2 | 2 | 6 | (Gorny et al., 2000) |
| Z13 | Phage | 4-59*03 | 3-16 | 3 | 6 | (Zwick et al., 2001) |
| 98-6e | EBV | 4-61*02 | 4-23 | 3 | 4 | (Andris et al., 1991) |
| 2F5g | Hybridoma | 2-5*10 | nd | nd | 6 | (Kunert et al., 1998) |
These mAbs were derived from five HIV-infected individuals;
only VH nucleotide sequence available;
nucleotide sequence is not available;
mAb 126-7 was previously known as 126-6;
the mAbs sequenced in our laboratory for this study; nd – not defined. D-RF, D gene reading frame.
2.3. Control data
Tiller et al. (Tiller et al., 2007) used single-cell PCR to analyze the sequence of antibodies in IgG memory B cells from three healthy donors. These data were used as normal controls for our studies.
2.4. Heterohybridoma cell lines
Thirteen heterohybridoma cell lines secreting human anti-V3 mAbs (Table 1) were produced in our laboratory for this study using a previously described technique (Gorny, 1994; Gorny et al., 1991). Briefly, peripheral blood mononuclear cells (PBMC) derived from HIV-1-infected individuals were transformed with Epstein-Barr virus (EBV), and reactive cells producing specific anti-V3 Abs were subsequently fused with heteromyeloma cells SHM-D33 (Teng et al., 1983). The resulting hybridomas were cloned by limiting dilution to monoclonality. PBMCs derived from non-clade B infected individuals were screened using a mixture of three V3-fusion proteins (V3-FP) containing the V3 sequence of clade A primary virus 92UG037.8 (V3A-FP), clade B primary isolate JR-CSF (V3B-FP) and consensus clade C (V3C-FP) (Kayman et al., 1994; Krachmarov et al., 2005). In the case of the subject infected with a clade B virus (Table 1), the cells were selected using the neutralization assay, as previously described (Gorny et al., 2005).
2.5. RT-PCR amplification of the Ig variable region and sequence analysis
Hybridoma cell lines producing human mAbs were analyzed by sequencing the variable fragment of the heavy chains. The mRNA was isolated from hybridoma cell lines using Micro-FastTrack 2.0 Kit (Invitrogen, CA). First strand cDNA was synthesized using oligodT(12–18) and Superscript II RNase H Reverse Transcriptase (Invitrogen, CA). Amplification of the variable fragment was performed by PCR using different gene family-specific sets of primers and cDNA as template. Five forward primers, VH 1/5 [5′CAG GTG CAG CTG GTG CAG TCT GG ′3], VH2 [5′CAG GTC AAC TTA AGG GAG TCT GG′3], VH3 [5′GAG GTG CAG CTG GTG GAG TCT GG′3], VH4 [5′CAG GTG CAG CTG CAG GAG TCG GG′3] and VH6 [5′CAG GTA CAG CTG CAG CAG TCA GG′3] were located at 5′ end of V genes. Reverse primer [5′CTTGGTGGARGCTGARGAGACGGTGACC′3] was located at 3′ end of JH segment and up to 12 nucleotides at the 5′ of the constant region of IgG (Marks et al., 1991). In order to confirm the sequence of the V region, the PCR amplification of V genes was repeated twice using the same forward primers, but with two reverse primers designed in our lab: [AbHC1:5′AGATGTAGGTCTGGGTGCCCAAGCTGC′3] and [AbHC2: 5′ GGAGATCATGAAGGTGTCCTTGGGTT′3] located in the constant region C1 and C2, respectively. PCR amplification was performed using cycling program of 2 min at 94°C, 35 cycles of 60 s at 94°C, 60 s at 56°C, and 90 s at 72°C, followed 8 min at 72°C. Ethidium bromide-stained 0.8% agarose gels were used to visualize the PCR products. The bands of the appropriate size were excised and cleaned with GeneElute Minus EtBr Spin Column (Sigma). PCR products were sequenced in both directions using the primers applied for amplification. All sequencing reactions were performed by a sequencer, ABI Prism model 377, at New York University Core Facility. The sequence data were analyzed using Pregap4, BioEdit software and the International ImMunoGene Tics (IMGT) information system (http://imgt.cines.fr). The sequences determined in our laboratory have been submitted to GenBank (accession numbers reserved: EU794398-EU794443).
2.6. Sequencing the Env HIV-1 infecting the donor of mAb
Extraction of total RNA was performed on 100 μl of plasma as described by Boom et al. (Boom et al., 1990). A single-tube reverse transcriptase (Access RT-PCR system, Promega, Madison, WI) was then performed to amplify the C2-V5 region of the Env of HIV-1 (Zhong et al., 2003). The amplified products were sequenced and phylogenetically analyzed with reference subtype sequences to determine the virus subtypes as previously described (Delwart et al., 1993; Zhong et al., 2002).
2.7. Statistical analysis
Fisher’s exact test was used to compare frequency of Ig gene usage. Multivariate analysis of the VH gene segment usage was performed using Chi-Squared tests.
3. Results
3.1. Generation of new human anti-V3 mAbs
Thirteen new human anti-V3 mAbs were developed from 12 subjects infected with non-clade B HIV-1 isolates and living in Cameroon and India, and one was developed from an individual infected with clade B HIV-1 living in the NYC area (Table 1). All of the newly generated mAbs belong to the IgG1 subclass of Abs, with nine of the mAbs using lambda light chains, and the remaining four using kappa light chains. Sequencing of the C2-V5 region of the gp120 HIV-1 was done to determine the infecting virus subtype for the donors of seven of the mAbs (Table 1).
3.2. VH gene family usage by anti-V3 mAbs
Forty-five out of 51 tested human anti-V3 mAbs were sequenced in our laboratory using mRNA from human heterohybridoma cell lines producing anti-V3 mAbs; the remaining six sequences of anti-V3 human mAbs were obtained from published data (Table 2). All amplified V regions were productively rearranged sequences without stop codons and were within the frame junction as determined using the IMGT information system (http://imgt.cines.fr). The sequences determined in our laboratory have been deposited in the GenBank database (accession numbers reserved: EU794398-EU794443).
The frequencies of Ig gene usage by human anti-HIV-1 mAbs were compared to the normal repertoire of Ig genes calculated from published data (Tiller et al., 2007). The Abs from 183 single IgG memory B cells derived from three healthy individuals were analyzed and the use of each gene family is as follows: VH1-15.8%, VH2-0%, VH3-54.8%, VH4-23.5%, VH5-6%, VH6-0% and VH7-0.7% (Tiller et al., 2007). These results are comparable to other single B cell analyses (Brezinschek et al., 1995) and therefore, are used as normal controls for our studies because human mAbs are generated from memory B cells (Traggiai et al., 2004) (unpublished data).
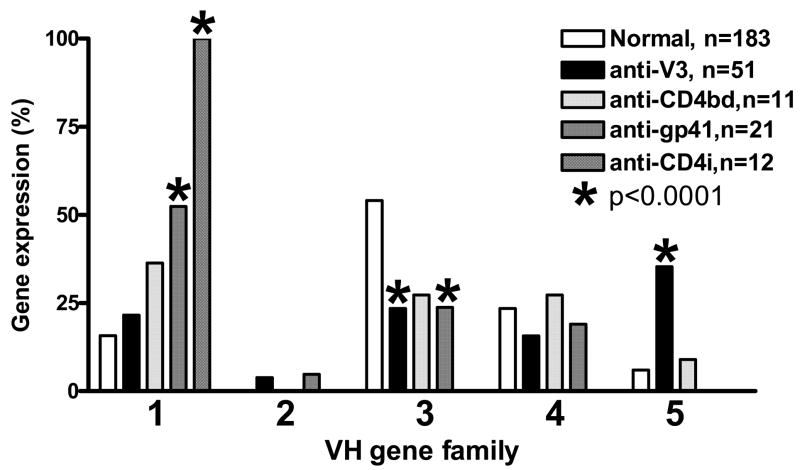
The VH genes from five families, VH1 to VH5, were used by the anti-V3 mAbs while neither the VH6 nor the VH7 gene family was detected. The VH5-51 gene segment was the only member of the VH5 gene family that was represented and was preferentially used by 18 out of 51 (35%) anti-V3 mAbs (Fig.1 and Table 2). Use of the VH5-51 gene segment is significantly increased in anti-V3 mAbs as compared to control Abs from healthy individuals, which use the VH5-51 gene segment in only 6% of the Ig sequences (Tiller et al., 2007) (p<0.0001) (Fig.1). The VH5-51 gene is one of the two gene segments belonging to the VH5 gene family, however, the second gene segment, VH5-a, is rarely used by normal human Abs (Tiller et al., 2007).
Fig.1.
The VH gene family usage by human mAbs against V3, CD4bd, CD4i and gp41 compared to control antibodies from healthy individuals. Anti-V3 mAbs preferentially use the VH5 gene family (p<0.0001), represented exclusively by VH5-51 gene segment, while the VH3 gene family use was significantly reduced (p<0.0001). Anti-CD4i mAbs exclusively used the VH1 gene family (p<0.0001) while the anti-gp41 mAbs used the VH1 gene family significantly more (p<0.0001) and used the VH3 gene family significantly less (p<0.0001).
The IMGT system, which was used for Ig gene usage analysis, also identified alleles for each gene segment. Alleles represent the sequence polymorphism at the nucleotide level compared to the reference sequence designated *01 (http://imgt.cines.fr). VH5-51 has five alleles, *01 to *05, which differ from each other by one nucleotide. These alleles were used in a restricted manner by the anti-V3 mAbs: the VH5-51 gene segments were equally aligned with only two alleles, *01 and *03 (Table 2).
Twenty of the fifty-one anti-V3 mAbs were generated from individuals infected outside of the United States with non-clade B HIV-1 strains (Table 2). The VH5-51 gene segment was preferentially used by both clade B and non-B derived mAbs; forty-five percent (9/20) of non-clade B derived anti-V3 mAbs and twenty-nine percent (9/31) of the V3 mAbs from clade B-infected individuals used the VH5-51 gene.
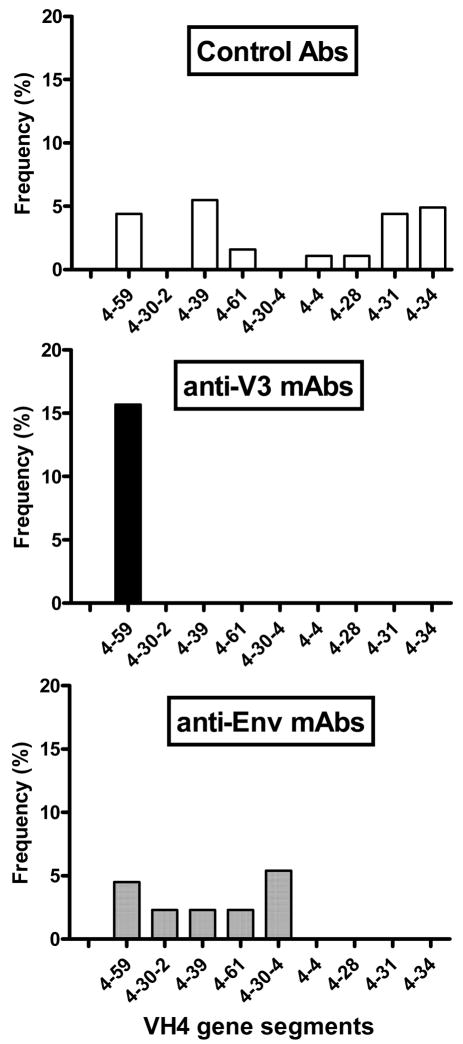
A restricted usage of one gene segment was observed in the VH4 gene family used by anti-V3 mAbs (Table 2). The VH4 gene family contains nine gene segments but only one of these genes, VH4-59, was used by anti-V3 mAbs, in contrast to seven and five VH4 genes used by the normal Abs and anti-Env mAbs, respectively. Thus, in contrast to anti-V3 mAbs, mAbs against the CD4bd, the CD4i, and gp41 use several VH4 gene segments (Fig. 2). In terms of frequency, the VH4-59 gene segment was used by 8 of 51 (16%) V3 mAbs (p=0.1) as compared to 8 of 183 (4%) control Abs and 2 of 44 (4%) anti-Env mAbs (Fig.2).
Fig. 2.
VH4 gene segment use. Anti-V3 mAbs used only one VH4 gene segment, VH4-59, out of nine possible VH4 gene segments. This is in contrast to seven and five VH4 gene segments used by control Abs and anti-Env mAbs, respectively.
The VH3 is the largest gene family, containing 21 gene segments, and it is most frequently used by Abs in uninfected individuals (Brezinschek et al., 1995; Tiller et al., 2007). Our analyses show that the VH3 gene family was used by 12 anti-V3 mAbs (23%), a frequency that is significantly lower (p<0.0001) than in control Abs (54%) (Fig 1). This analysis of VH3 gene usage corroborated previous observations that the VH3 gene family is less frequently used by anti-HIV-1 antibodies, possibly due to gp120-induced depletion of the B cells producing Abs encoded by the VH3 gene family (Berberian et al., 1993; David et al., 1995a; David et al., 1995b).
The two other VH gene families, VH1 and VH2, were used by anti-V3 mAbs with slightly increased frequencies compared to controls (Fig.1).
Overall, the anti-V3 mAbs displayed varying levels of VH gene family usage. The anti-V3 mAbs preferentially used the VH5 gene family, but were represented only by the VH5-51 gene segment and restricted to two of the five alleles. The VH4 gene family was used less frequently by anti-V3 mAbs, and was restricted to only one gene segment, VH4-59, in contrast to control Abs and anti-Env mAbs which used several VH4 gene segments. The VH3 family genes were used significantly less frequently by the anti-V3 mAbs than in control Abs.
3.3. VH gene family usage by human mAbs to the CD4 binding domain (anti-CD4bd), the CD4-induced epitope (anti-CD4i) and gp41 (collectively, anti-Env mAbs)
Preferential VH5-51 gene usage by the anti-V3 mAbs may be a compensatory mechanism for the decreased usage of the VH3 gene family in all anti-HIV-1 Abs. If this is true, it would be expected that other anti-HIV-1 mAbs would also use the VH5 gene family at a higher frequency than the normal repertoire. To test this hypothesis, we analyzed the sequences of 44 human mAbs specific for various regions of the HIV-1 envelope (Env) including 11 mAbs with specificity for the CD4bd, 12 mAbs against the CD4i epitope, and 21 mAbs against gp41. The Ig gene sequences of 34 mAbs were obtained from published data while 10 mAbs against gp41 were produced and sequenced in our laboratory (Table 3).
In contrast to the anti-V3 mAbs, the non-V3 anti-Env mAbs only used the VH5 gene family 2% of the time (1/44), and instead preferentially used the VH1 gene family. Anti-CD4i mAbs exclusively used the VH1 gene family (100%) and 36% and 52% of anti-CD4bd and anti-gp41 mAbs used the VH1 gene family, respectively, compared to 16% for normal Abs (Fig.1). The VH3 gene family usage by the anti-Env mAbs was reduced in a manner that was similar to the anti-V3 mAbs. The percentages of the VH2, VH4 and VH5 gene family usage were all within an average range of the normal repertoire.
3.4. Pattern of VH gene segment usage by anti-V3 mAbs and anti-Env mAbs
The usage of the VH gene segments was different for anti-V3, anti-Env mAbs and normal antibodies. One remarkable characteristic was the usage of different dominant gene segments. Thirty-five percent of the anti-V3 mAbs used the VH5-51 gene segment, which was significantly increased compared to anti-Env mAbs and control Abs (p<0.0001) while 45% of the anti-Env mAbs preferentially used the VH1-69 gene segment (p<0.0001) (Fig.3).
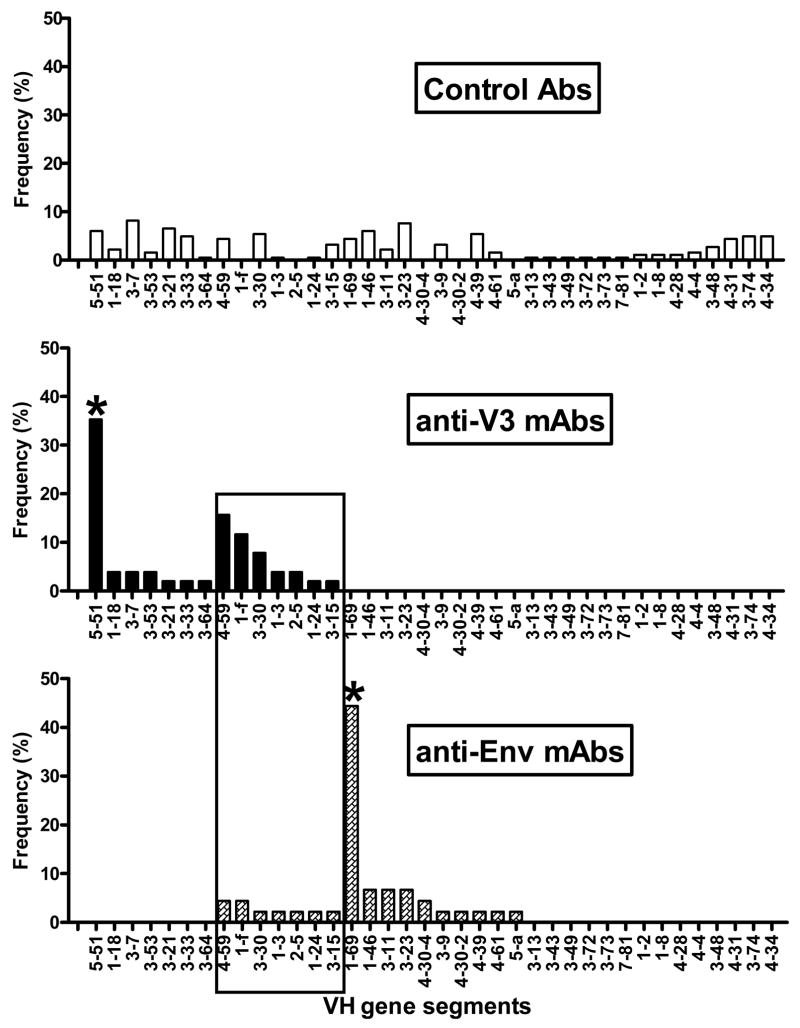
Fig.3.
Individual VH gene segment use by human anti-V3, anti-Env mAbs and control antibodies. The panel of anti-Env mAbs includes mAbs against CD4bd, CD4i and gp41. The profile of VH gene segment usage by anti-V3 mAbs is very different from anti-Env mAbs as several genes, 5-51, 1-18, 3-7, 3-53, 3-21, 3-33 and 3-64, were used by anti-V3 mAbs only, while the other genes, 1-69, 1-46, 3-11, 3-23, 4-30-4, 3-9, 4-30-2, 4-39, 4-61 and 5-a were used exclusively by anti-Env mAbs. Seven VH gene segments, which were used by both types of mAbs, are highlighted in the box. VH5-51 was used by 18 of 51 (35%) of anti-V3 mAbs, while VH1-69 gene segment was utilized by 20 of 44 (45%) of anti-Env mAbs; both gene segments were significantly (*) increased compared to control antibodies (p<0.0001). Multivariate analysis determined highly significant difference in the VH gene usage between anti-V3 and anti-Env mAbs and compared to normal antibodies (p<0.0001).
Furthermore, seven VH gene segments were used exclusively by anti-V3 mAbs and ten VH genes were used only by anti-Env mAbs while seven genes were used by both types of mAbs, however, with a higher frequency of usage by anti-V3 mAbs; these data are highlighted in the box in Fig. 3.
The distribution of VH gene segments used by anti-V3 and anti-Env mAbs was significantly different (p<0.0001) when analyzed by multivariate comparisons using Chi-Squared tests. Both panels of mAbs also differed significantly from normal Ab gene segment usage (p<0.0001). These results suggest that there are separate profiles of VH gene segments used by anti-V3 mAbs and anti-Env mAbs and they both differ compared to the repertoire of VH genes used by control Abs (Fig. 3).
3.5. JH use by anti-V3 and anti-Env mAbs
The variable fragment of the heavy chain contains the CDR H3 which is formed by recombination of the D gene segment to a JH gene followed by the rearrangement of VH to the D-JH complex. There are six JH gene segments, JH1 to JH6, which encode part of the CDR H3 of the variable fragment.
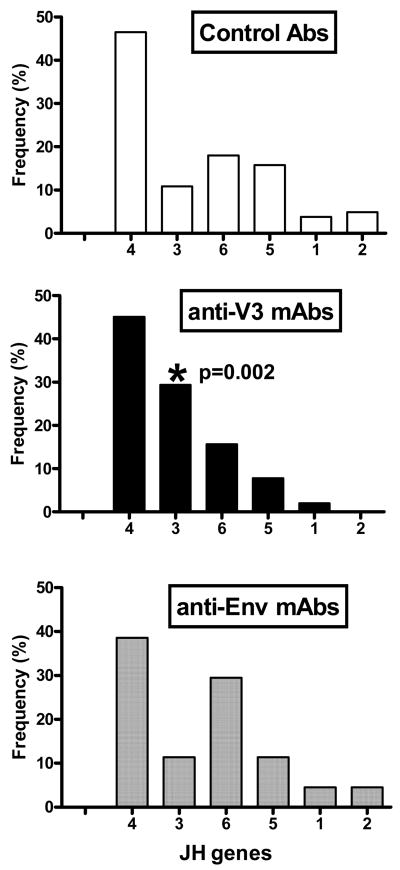
There was one significant difference in JH gene usage by anti-V3 mAbs: 29% of mAbs preferentially used the JH3 gene compared to 11% of anti-Env mAbs and control Abs (p=0.002) (Fig. 4). The use by anti-Env mAbs of the JH genes was not significantly different from that of control Abs (Fig. 4).
Fig.4.
JH gene segment use by anti-V3, anti-Env and control antibodies. The anti-V3 mAbs utilized the JH3 gene segment significantly more frequently (p=0.002) than other antibodies, while anti-Env mAbs had a tendency to use the JH6 gene segment with an increased frequency.
3.6. Diversity (D) gene segment use by mAbs
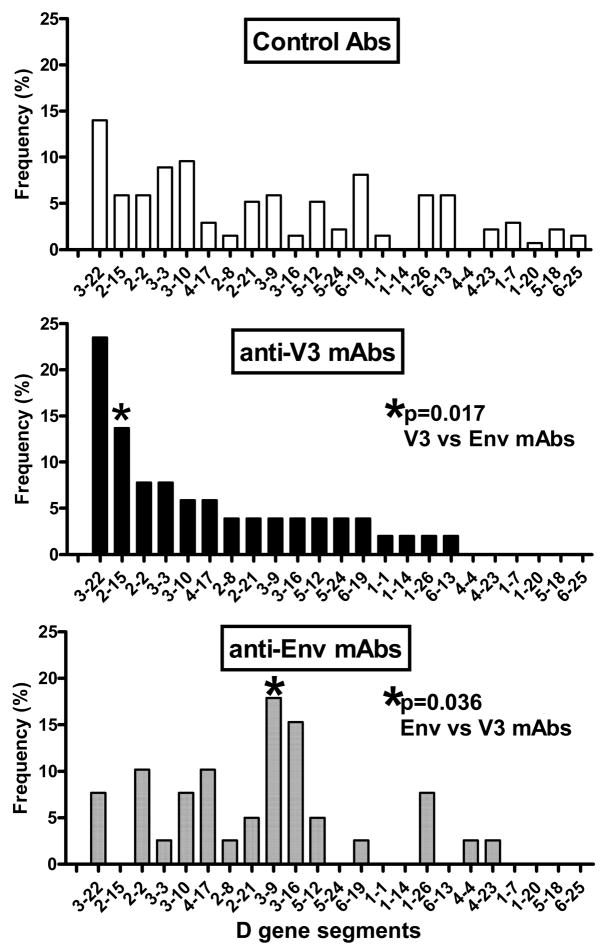
There are 27 D gene segments according to the IMGT system (http://imgt.cines.fr) which contribute to the formation of the middle section of the CDR H3 loop. Due to exonuclease activity during recombination with the JH and subsequently with the VH gene segments, the D gene is shortened and its identity with germline genes may not always be defined. We analyzed individual D gene segments and D gene reading frame use in the V3 and Env mAbs (Table 2 and 3) compared to control Abs. Twenty-one individual D gene segments were used by the control Abs while 17 and 14 gene segments were used by anti-V3 and anti-Env mAbs, respectively (Fig. 5). There was no significant difference in the frequency of D gene usage between control Abs and anti-HIV-1 mAbs, however, such differences were noted between the two panels of mAbs. The D2-15 gene was used significantly more frequently (14%) by anti-V3 mAbs compared to 0% by anti-Env mAbs (p=0.017). The anti-Env mAbs used the D3-9 gene segment with significantly increased frequency (18%) compared to 4% of anti-V3 mAbs (p=0.036) (Fig. 5).
Fig.5.
D gene segment use by V3 and Env mAbs compared to control antibodies. The D genes were used by anti-V3 and anti-Env mAbs in a comparable frequency to control antibodies. The profile of D gene segments used by anti-V3 mAbs was, however, different then by anti-Env mAbs; the gene D2-15 was used significantly more frequently (p=0.017) by the anti-V3 mAbs and the gene D3-9 was used more often by anti-Env mAbs (p=0.036).
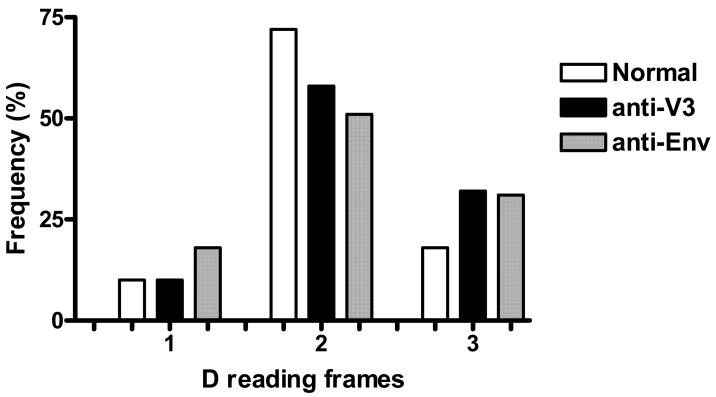
The D genes can be read in all three reading frames and this may be an inherent solution to imperfect joining during recombination of VH, D and JH genes. Reading a D gene in three reading frames increases the chances of proper recognition of CDR H3 by the antigen. Distribution of D-reading frames was not significantly different between the two panels of mAbs and normal Abs (Fig. 6).
Fig.6.
The D reading frames used by human V3 and Env mAbs and control antibodies. The D-reading frame 3 was overused and D-reading frame 2 was underused by anti-V3 and anti-Env mAbs as compared to control antibodies.
4. Discussion
The analysis of the Ig genes encoding the variable fragment of the heavy chain of human mAbs against epitopes in the envelope glycoproteins of HIV-1 revealed a pattern of the VH gene segment usage in anti-V3 mAbs that is completely different from anti-Env mAbs (including anti-CD4bd, anti-CD4i and anti-gp41 mAbs) and control Abs which represent the repertoire of VH genes used by healthy individuals. Normal Abs displayed a broadly distributed usage of VH gene segments with usage frequencies <~10%, while anti-V3 mAbs preferentially used the VH5-51 gene segment, a segment which was not used by any of the non-V3 anti-Env mAbs. In contrast, almost half of the anti-Env mAbs preferentially used the VH1-69 gene segment which was not used by any of the anti-V3 mAbs (Fig. 3). Additionally, the pattern of other VH gene segment usage differed for the two panels of mAbs derived from HIV-1-infected individuals (Figs. 3). These results suggest that specific epitopes located in the various regions of the HIV-1 envelope glycoproteins select for Abs using particular VH gene segments, and that, in particular, the epitopes in the crown of V3 select a unique set of VH gene segments, preferentially using different gene segments than those used by anti-Env mAbs and control Abs.
The immunologic literature suggests preferential VH gene usage by Abs of particular specificities indicating that germline genes, the genomic VH precursors, code for specific protein sequences which dictate the best fit of the Ig receptor on naïve B cells. Somatic hypermutation further improves affinity during Ab maturation, but the initial binding activity is provided by the germline gene-encoded sequences.
Dominant VH gene usage has been observed in Abs to several pathogens. For example, Abs against the capsular polysaccharide of Hemophilus influenzae type b primarily use the VH3-23 gene (Lucas et al., 2003), Abs against Streptococcus pneumoniae preferentially use genes from the VH3 family (Sun et al., 1999), Abs against rotavirus primarily use the VH1-46 gene segment (Weitkamp et al., 2003), monoclonal cold agglutinins with I/i antigen specificity exclusively use the VH4-34 gene segment (Silberstein et al., 1996), and monoclonal rheumatoid factors use the VH1-69 gene segment (De Re et al., 2002). The two human mAbs derived from dominant clones in the pneumococci-specific memory B cell repertoire are of particular note in that they are both coded for by the VH3-48 gene segment and associated with high affinity and opsonic activities when compared to mAbs with the same specificity but different VH gene usage (Baxendale and Goldblatt, 2006). The data from anti-HIV-1 mAbs provide further evidence for selective gene usage by epitopes of human pathogens. Moreover, the finding that V3 mAbs preferentially use the VH5-51 gene suggests that this gene segment has been selected to optimally accommodate the conserved structural motifs that characterize this region of the HIV-1 envelope (Cardozo et al., 2007; Sharon et al., 2003).
Thirty-five percent (18/51) of anti-V3 mAbs contained VH segments coded by the VH5-51 gene. All but two of these 18 VH5-51-derived mAbs were isolated from separate individuals infected with various subtypes of HIV-1; this indicates that the V3 region of the infecting viruses from approximately one-third of donors had a related conformation which selected B cells using this gene segment. Crystallographic analysis of mAb 2219 complexed with three V3 peptides confirmed that the V3 conformation is important for Ab binding since all three peptides had different amino acid sequences but displayed the same shape when bound to the mAb (Stanfield et al., 2006). The peptide adapts to the Ab combining site upon binding, apparently reflecting the template of the V3 region on the primary virus that originally bound to and activated the naïve B cell.
Except for the analysis of the anti-CD4i mAbs, previous studies have been limited to a general analysis of anti-HIV-1 mAbs and showed a substantial reduction in the VH3 and enrichment for VH1 and VH4 gene family usage compared to the normal repertoire (David et al., 1995b; Wisnewski et al., 1996). It was suggested that the decreased usage of the VH3 family genes might be the result of depletion of B cells expressing the VH3-encoded Ig receptor due to the binding of gp120 to VH3 Ig-bearing B cells given the capacity of gp120 to act as a superantigen for these cells (Berberian et al., 1993). It is improbable that depletion of VH3 B cells by this method would lead to a preferential use of the VH5-51 gene to encode anti-V3 Abs or to preferential use of the VH1 gene family by other anti-HIV-1 Abs. It is more probable that the V3 epitope preferentially selects naïve B cells expressing the Ig receptor coded by the VH5-51 gene segment, thereby resulting in reduced usage of VH3 by anti-V3Abs. Similarly, since the CD4bd epitope, the CD4i epitope and gp41 primarily select the VH1 family gene, this may further reduce the usage of the VH3 gene family for anti-HIV-1 Abs. The possibility of the compensated reduction of VH3 gene usage is supported by a recent study which showed that the increased number of VH4 plasma cells in cerebrospinal fluid in multiple sclerosis was accompanied by a significant reduction in cells expressing VH3 genes (Owens et al., 2007).
In summary, the anti-V3 mAbs display a characteristic and non-random pattern of variable fragment heavy chain Ig gene usage in comparison to anti-Env mAbs and to control antibodies. There was biased usage of VH, D and JH genes by anti-V3 mAbs including preferential usage of the rarely employed VH5-51 gene segment. Furthermore, multivariate analysis showed that usage of VH gene segments was significantly different between anti-V3 and anti-Env mAbs and compared to normal antibodies. As the V3 region is structurally conserved and distinct in conformation from other Env regions, the data suggest that the V3 region selects for naïve B cells bearing surface Ig molecules characterized by the use of particular Ig genes that form Abs with an optimal fit for the epitopes at the crown of the V3 loop.
Acknowledgments
The study was supported in part by NIH grants AI077451, HL59725 and AI36085, by the Immunology Core of the NYU Center for AIDS Research (NIH grant AI27742), by research funds from Department of Veterans Affairs and by the Gates Foundation. We are grateful to Dr. Christopher A. Anyangwe for help in providing the blood samples from Cameroon and to Drs. Kalpana Luthra and Suman Laal for help in procurement of the blood samples from India. We would also like to thank Alok Kumar Choudhary for isolation the virus and Dr. Jennifer Fuller for help in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andris JS, Johnson S, Zolla-Pazner S, Capra JD. Molecular characterization of five human anti-HIV-1 antibody heavy chains reveals extensive somatic mutation typical of an antigen-driven immune response. Proc Natl Acad Sci U S A. 1991;88:7783–7787. doi: 10.1073/pnas.88.17.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus L, Prince AM, Bernal I, McCormack P, Lee DH, Gorny MK, Zolla-Pazner S. Passive immunization with a human immunodeficiency virus type-1 neutralizing monoclonal antibody in Hu-PBL-SCID mice: Isolation of a neutralization escape variant. J Infect Dis. 1998;177:889–897. doi: 10.1086/515251. [DOI] [PubMed] [Google Scholar]
- Bagley J, Dillon PJ, Rosen C, Robinson J, Sodroski J, Marasco WA. Structural characterization of broadly neutralizing human monoclonal antibodies against the CD4 binding site of HIV-1 gp120. Mol Immunol. 1994;31:1149–1160. doi: 10.1016/0161-5890(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- Baxendale HE, Goldblatt D. Correlation of molecular characteristics, isotype, and in vitro functional activity of human antipneumococcal monoclonal antibodies. Infect Immun. 2006;74:1025–1031. doi: 10.1128/IAI.74.2.1025-1031.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberian L, Goodglick L, Kipps TJ, Braun J. Immunoglobulin VH3 gene products: natural ligands for HIV gp120. Science. 1993;261:1588–1591. doi: 10.1126/science.7690497. [DOI] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural Basis of Co-Receptor Selectivity by the HIV-1 V3 Loop. AIDS Res Hum Retroviruses. 2007;23:415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, Posner MR. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res Hum Retroviruses. 1998;14:1271–1280. doi: 10.1089/aid.1998.14.1271. [DOI] [PubMed] [Google Scholar]
- Cook GP, Tomlinson IM. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237–242. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- David D, Demaison C, Bani L, Zouali M, Theze J. Selective variations in vivo of VH3 and VH1 gene family expression in peripheral B cell IgM, IgD and IgG during HIV infection. Eur J Immunol. 1995a;25:1524–1528. doi: 10.1002/eji.1830250608. [DOI] [PubMed] [Google Scholar]
- David D, Goossens D, Desgranges C, Theze J, Zouali M. Molecular characterization of human monoclonal antibodies specific for several HIV proteins: analysis of the VH3 family expression. Immunol Lett. 1995b;47:107–112. doi: 10.1016/0165-2478(95)00078-j. [DOI] [PubMed] [Google Scholar]
- de Haard JJ, Kazemier B, Oudshoorn P, Boender P, van Gemen B, Koolen MJ, van der Groen G, Hoogenboom HR, Arends JW. Selection of human anti-human immunodeficiency virus type 1 envelope single-chain antibodies from a peripheral blood cell-based phage repertoire. J Gen Virol. 1998;79:2883–2894. doi: 10.1099/0022-1317-79-12-2883. [DOI] [PubMed] [Google Scholar]
- De Re V, De Vita S, Gasparotto D, Marzotto A, Carbone A, Ferraccioli G, Boiocchi M. Salivary gland B cell lymphoproliferative disorders in Sjogren’s syndrome present a restricted use of antigen receptor gene segments similar to those used by hepatitis C virus-associated non-Hodgkins’s lymphomas. Eur J Immunol. 2002;32:903–910. doi: 10.1002/1521-4141(200203)32:3<903::AID-IMMU903>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rubsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: Analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- Ditzel HJ, Parren PW, Binley JM, Sodroski J, Moore JP, Barbas CF, 3rd, Burton DR. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- Emini EA, Schleif WA, Nunberg JH, Conley AJ, Eda Y, Tokiyoshi S, Putney SD, Matsushita S, Cobb KE, Jett CM, Eichberg JW, Murthy KK. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- Felgenhauer M, Kohl J, Ruker F. Nucleotide sequences of the cDNAs encoding the V-regions of H- and L-chains of a human monoclonal antibody specific to HIV-1-gp41. Nucleic Acids Res. 1990;18:4927. doi: 10.1093/nar/18.16.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK. Production of human monoclonal antibodies via fusion of Epstein-Barr virus-transformed lymphocytes with heteromyeloma. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Vol. 2. Academic Press; 1994. pp. 276–281. [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse HIV-1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to HIV. Proc Natl Acad Sci. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Mascola JR, Israel ZR, VanCott TC, Williams C, Balfe P, Hioe C, Brodine S, Burda S, Zolla-Pazner S. A human monoclonal antibody specific for the V3 loop of HIV type 1 clade E cross-reacts with other HIV type 1 clades. AIDS Res Hum Retroviruses. 1998;14:213–221. doi: 10.1089/aid.1998.14.213. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov CP, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 Loop Is Accessible on the Surface of Most Human Immunodeficiency Virus Type 1 Primary Isolates and Serves as a Neutralization Epitope. J Virol. 2004;78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. Effects of oligomerization on the epitopes of the Human Immunodeficiency Virus Type 1 envelope glycoproteins. Virology. 2000;267:220–228. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov CP, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize HIV-1 primary isolates from various clades. J Virol. 2002;76:9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Koning FA, Nadas A, Anyangwe C, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of HIV-1. J Virol. 2006;80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Gianakakos V, Karwowska S, Williams C, Sheppard HW, Hanson CV, Zolla-Pazner S. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the HIV-1 envelope glycoprotein. Proc Natl Acad Sci. 1991;88:3238–3242. doi: 10.1073/pnas.88.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol. 1994;68:400–410. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79:780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert R, Ruker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: Identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res Hum Retroviruses. 1998;14:1115–1128. doi: 10.1089/aid.1998.14.1115. [DOI] [PubMed] [Google Scholar]
- Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses. 2004;20:755–762. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Hollis GF, Mark GE, 3rd, Tung JS, Ludmerer SW. Use of a novel mutagenesis strategy, optimized residue substitution, to decrease the off-rate of an anti-gp120 antibody. Mol Immunol. 1995;32:1065–1072. doi: 10.1016/0161-5890(95)00079-8. [DOI] [PubMed] [Google Scholar]
- Liu F, Bergami PL, Duval M, Kuhrt D, Posner M, Cavacini L. Expression and functional activity of isotype and subclass switched human monoclonal antibody reactive with the base of the V3 loop of HIV-1 gp120. AIDS Res Hum Retroviruses. 2003;19:597–607. doi: 10.1089/088922203322230969. [DOI] [PubMed] [Google Scholar]
- Lucas AH, McLean GR, Reason DC, O’Connor AP, Felton MC, Moulton KD. Molecular ontogeny of the human antibody repertoire to the Haemophilus influenzae type B polysaccharide: expression of canonical variable regions and their variants in vaccinated infants. Clin Immunol. 2003;108:119–127. doi: 10.1016/s1521-6616(03)00094-9. [DOI] [PubMed] [Google Scholar]
- Marasco WA, Bagley J, Zani C, Posner M, Cavacini L, Haseltine WA, Sodroski J. Characterization of the cDNA of a broadly reactive neutralizing human anti-gp120 monoclonal antibody. J Clin Invest. 1992;90:1467–1478. doi: 10.1172/JCI116014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks JD, Tristem M, Karpas A, Winter G. Oligonucleotide primers for polymerase chain reaction amplification of human immunoglobulin variable genes and design of family-specific oligonucleotide probes. Eur J Immunol. 1991;21:985–991. doi: 10.1002/eji.1830210419. [DOI] [PubMed] [Google Scholar]
- Moran MJ, Andris JS, Matsumato Y, Capra JD, Hersh EM. Variable region genes of anti-HIV human monoclonal antibodies: non-restricted use of the V gene repertoire and extensive somatic mutation. Mol Immunol. 1993;30:1543–1551. doi: 10.1016/0161-5890(93)90462-k. [DOI] [PubMed] [Google Scholar]
- Owens GP, Winges KM, Ritchie AM, Edwards S, Burgoon MP, Lehnhoff L, Nielsen K, Corboy J, Gilden DH, Bennett JL. VH4 gene segments dominate the intrathecal humoral immune response in multiple sclerosis. J Immunol. 2007;179:6343–6351. doi: 10.4049/jimmunol.179.9.6343. [DOI] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic B hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure (Camb) 2003;11:225–236. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Silberstein LE, George A, Durdik JM, Kipps TJ. The V4-34 encoded anti-i autoantibodies recognize a large subset of human and mouse B-cells. Blood Cells Mol Dis. 1996;22:126–138. doi: 10.1006/bcmd.1996.0020. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of HIV-1 neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Park MK, Kim J, Diamond B, Solomon A, Nahm MH. Repertoire of human antibodies against the polysaccharide capsule of Streptococcus pneumoniae serotype 6B. Infect Immun. 1999;67:1172–1179. doi: 10.1128/iai.67.3.1172-1179.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng NN, Lam KS, Calvo Riera F, Kaplan HS. Construction and testing of mouse-human heteromyelomas for human monoclonal antibody production. Proc Natl Acad Sci. 1983;80:7308–7312. doi: 10.1073/pnas.80.23.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Ho DD, Robinson J, Tilley S, Pinter A, Sodroski J. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 1992;66:5635–5641. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved HIV-type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Donk EM, Schutten M, Osterhaus AD, van der Heijden RW. Molecular characterization of variable heavy and light chain regions of five HIV type 1-specific human monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:1639–1649. doi: 10.1089/aid.1994.10.1639. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Bures E, Williams JV, LaFleur B, Greenberg HB, Crowe JE., Jr Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J Immunol. 2003;171:4680–4688. doi: 10.4049/jimmunol.171.9.4680. [DOI] [PubMed] [Google Scholar]
- Wisnewski A, Cavacini L, Posner M. Human antibody variable region gene usage in HIV-1 infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;11:31–38. doi: 10.1097/00042560-199601010-00004. [DOI] [PubMed] [Google Scholar]
- Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Shu Y, Sidorov I, Dimitrov DS. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res. 2004;61:161–164. doi: 10.1016/j.antiviral.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Zhong P, Burda S, Konings F, Urbanski M, Ma L, Zekeng L, Ewane L, Agyingi L, Agwara M, Saa, Afane ZE, Kinge T, Zolla-Pazner S, Nyambi P. Genetic and biologic properties of HIV type 1 isolates prevalent in villagers of the Cameroon equatorial rain forests and grass fields: Further evidence of broad HIV-1 genetic diversity. AIDS Res Hum Rertroviruses. 2003;19:1167–1178. doi: 10.1089/088922203771881284. [DOI] [PubMed] [Google Scholar]
- Zhong P, Burda S, Urbanski M, Kenfack H, Tongo M, Heyndrickx L, Nanfack A, Shang J, Agyingi L, Zolla-Pazner S, Zekeng L, Nyambi P. HIV Type 1 Group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J Acquir Immune Defic Syndr. 2002;31:495–505. doi: 10.1097/00126334-200212150-00007. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, Gorny MK, Nyambi PN, VanCott TC, Nadas A. Immunotyping of human immunodeficiency virus type 1 (HIV): an approach to immunologic classification of HIV. J Virol. 1999;73:4042–4051. doi: 10.1128/jvi.73.5.4042-4051.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]