Summary
To understand how a brain processes information, we must understand the structure of its neural circuits –especially circuit interconnection topologies and the cell and synapse molecular architectures that determine circuit signaling dynamics. Our information on these key aspects of neural circuit structure has remained incomplete and fragmentary, however, due to limitations of the best available imaging methods. Now, new transgenic tool mice and new image acquisition tools appear poised to permit very significant advances in our abilities to reconstruct circuit connection topologies and molecular architectures.
Introduction
The modern understanding of brain function grew from Ramon y Cajal’s beautiful and prescient india-ink reconstructions of neural circuit architectures (e.g., [1], Fig. 1). These drawing were based on observations using used two then-new imaging tools: Abbe’s apochromatic objective and Golgi’s silver impregnation stain. Ramon y Cajal’s drawings and insights were possible because the Golgi method could be titrated to stain a small fraction of cells intensely and completely while leaving the majority of adjacent cells unstained, allowing complete forms of the rare stained neurons to be visualized clearly by a well-corrected objective. Nearly every circuit reconstruction effort since has likewise relied upon sparse staining methods to overcome the difficulties of resolving the individual elements of very densely packed neural circuit elements. Thus, the best reconstructions of circuit connectivity available today still extrapolate from isolated observations of individual neurons and still provide only fragmentary and qualitative information about neural circuit architectures. Moreover, as our understanding of the vast molecular diversity of neurons and synapses has grown [2–5], it has become increasingly clear that reconstruction of neural circuits will require molecular information about cells and synapses much more detailed than any presently available.
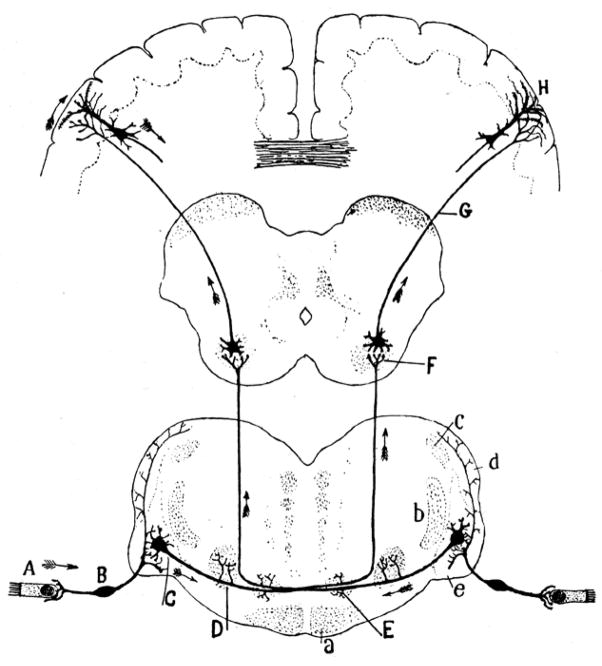
Fig. 1. Circuit reconstruction yesterday.
Drawings like this built the foundations of modern neuroscience, establishing the idea that brains process information and generate behavior as a result of the conduction of signals from cell to cell through anatomically defined circuits. Arrows in Ramon y Cajal’s india-ink reconstruction of the auditory pathway (Ref [1], fig. I-26) indicate information flow, from auditory hair cells (A) through the ventral cochlear nucleus (C) and the inferior colliculus (F) to cortical pyramidal cells (H), and then corticofugally to control behavior, via the axonal projections of cortical pyramidal cells. Like all subsequent reconstructions of brain circuitry, this early reconstruction is far from complete.
Today, rapid advances in molecular, physical and computational imaging tools are beginning to extend our sight far beyond what was possible with Ramon y Cajal’s apochromatic objective and Golgi stains and promising to extend our abilities to reconstruct far beyond those allowed by india-ink drawing. This commentary will provide an overview of some of these new imaging tools, focusing on (1) new genetic methods for neuroanatomical staining, (2) new physical methods for the high-resolution imaging of molecular architecture, (3) new strategies for high-throughput volume electron microscopy, and (4) new computational tools for the analysis of volume EM data. For brevity, this review will focus on a single target: the reconstruction of mammalian cerebral cortex. A summary section will consider the feasibility of a hypothetical project at the edge of today’s envelope for reconstruction technology.
Of Tool Mice and Men
The mouse cerebral cortex stands out today as a uniquely advantageous system for the study of cortical structure and function. The mouse offers a unique abundance of genetic information, transgenically labeled “tool mouse” lines, and genetic models for human disease. Meanwhile, the superficial location, relatively unfolded anatomy and small dimensions of the mouse cortex adapt it particularly well to physiological study by modern in vivo optical methods. These advantages are all the more valuable because of the strong similarities between mouse and human cerebral cortex.
A rapidly growing cornucopia of XFP tool mice is beginning to have an enormous impact on neuroscience. These transgenic mouse lines express genetically encoded fluorescent protein (XFP) markers in distinct subsets of neurons defined by intrinsic genetic control elements (e.g., [6–8]). In many cases, these subsets appear to correspond to classical morphologically and physiologically defined cell types. Sparseness of labeled subsets allows for Golgi-like optical resolution of individual neurons in many of these lines, but these genetic XFP labels offer enormous advantages over Golgi stains by allowing tagged cells to be imaged in live as well as fixed tissues and in being more predictable, repeatable and informative in their cell specificity. These advantages are being multiplied by cross breeding mouse lines carrying spectrally distinct XFP tags, to produce brain specimens exhibiting spectrally multiplexed labeling of distinct neural subsets [6]. Such multiplex tags can allow more complete (i.e., less sparse) labeling of individual circuits, because adjacent cells that otherwise would be too close for optical resolution may be resolved if they are distinct in color. The number of distinguishable tags may be extended beyond XFP spectral variants by the genetic encoding of additional, non-fluorescent epitope tags and reading those out in fixed specimens using antibodies [9] or other ligands [10,11].
The opportunities for parsing individual neurons from complex volume images are now being expanded still further by ingenious new strategies for patterning cell-specific tag expression. One spectacular and extremely promising new principle for driving highly multiplexed tags has been demonstrated by the recent introduction of “brainbow mouse” lines [12]. Neurons in brainbow mice express multiple tags drawn randomly from a palette of several XFPs, resulting in the cell-specific expression of a large number of combination colors as illustrated in figure 2. The large numbers (approximately 100 are distinguishable in lines described in reference 12) and random patterning of colors in such mice raises the exciting prospect of reconstructing complete circuits in animals where every neuron is labeled and yet resolvable by color from every one of its neighbors.
Fig. 2. A tool mouse of many colors. Cell-specific transgenic labels in a “brainbow” mouse.
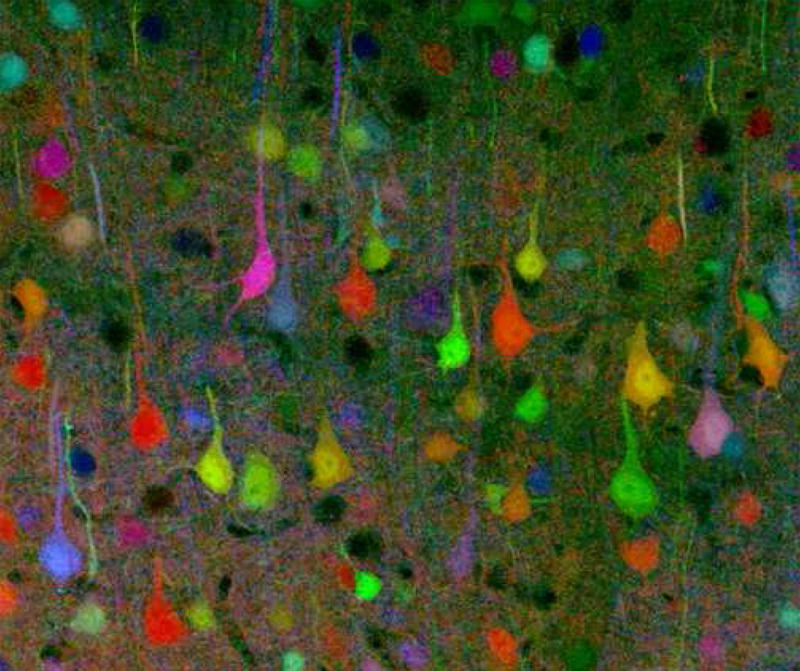
Reference 12 introduces a powerful new molecular genetic method for generating mice in which a large fraction of neurons express fluorescence tags drawn randomly from a large combinatorial color palette. (This image provided courtesy of Prof. Jeff Lichtman on behalf of all authors of Ref. 12). Such “color coded” brain cells promise new solutions to formerly intractable problems with resolving closely packed neurons and tracing their axons and dendrites reliably over long distances. It seems unlikely that neural information processing will ever be understood without solving such problems and reconstructing circuits in far more detail that presently possible.
Another important new approach to genetic targeting of cells-specific tags, known as Mosaic Analysis with Double Markers (MADM), allows (among other things) for sparse expression of a tag within a genetically defined subset of neurons at a density that can be adjusted by dosage of an otherwise silent drug pulse [13]. Yet another ingenious new circuit labeling principle has been demonstrated by showing the transmission of a genetically encoded fluorescent protein marker from one given cell to its direct synaptic partners and only to those cells [14]. Both of these approaches promise to be very useful in their own right and may also be useful in combination with other labeling strategies such as brainbow.
New High-Resolution Molecular Imaging Tools
A new approach to tissue immunofluorescence microcopy called “array tomography” provides a unique opportunity to image neural circuit molecular architecture at the level of individual synapses [15]. This new method overcomes past limitations of tissue immunofluorescence imaging by using a hydrophilic resin long known to permit efficient postembedding immunostaining and a new method for reliably and efficiently handling large numbers of serial ultrathin sections. A schematic of this technique and a rendering from a three-dimensional array tomographic image of cells and synapses in layer 5 of a mouse whisker barrel is shown in Fig. 3. Array tomography offers volumetric resolution much higher than that obtainable by confocal microscopy [16], eliminates depth-dependent variations in staining and imaging efficiency, and permits the multiplexing of large numbers of immunofluorescence channels via repeated cycles of antibody stripping and restaining. As noted below, array tomography also provides unique opportunities for conjugate immunofluorescence and electron microscopic imaging. As apparent in Fig. 3, the high volumetric resolution offered by array tomography allows for the clear optical resolution of individual synapses within cortical neuropil. Fig. 3 also demonstrates the extraordinary clarity with immunofluorescence array tomography can delineate fine cortical arborizations and dendritic spines. Automation of array tomographic image acquisition (see [15]) makes it straightforward to expand the area imaged on each section beyond a single optical field of view by tiling multiple, adjacent fields. Thus, array tomography appears ideally poised to exploit highly multiplexed tool mice and a growing arsenal of antibodies targeting key neural signaling molecules [17] to reconstruct cortical microcircuit connectivity and molecular architecture.
Fig. 3. Array Tomography. A new tool for conjugate molecular and ultrastructural imaging.
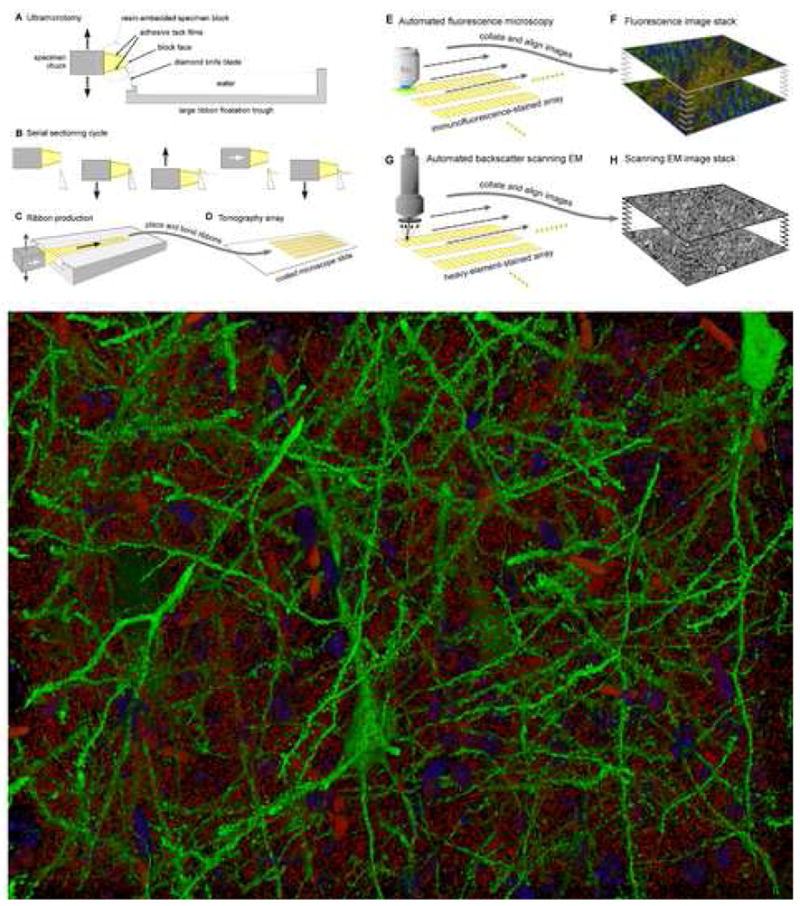
The reconstruction of neural circuits will benefit from complementing the molecular discrimination of immunofluorescence imaging with the structural precision of electron microscopy. A. - D. Tomography array production. An ultramicrotome (A) cuts a resin-embedded specimen into sections 50–200nm thick by motion (B) against a diamond knife blade. Adhesive block coatings cause serial sections to form continuous ribbons (C), which are transferred and bonded to a glass array slide (D). E., F. Immunofluorescence imaging. Array slides are immunostained and imaged using an automated fluorescence microscope (E). Resulting two-dimensional images are then aligned to form a three-dimensional image stack (F). Repeated cycles of immuno-staining, imaging and antibody elution allow multiplexing of very large numbers of immunofluorescence channels [15]. G., H. Electron-microscopy. After immunofluorescence imaging, arrays can be re-stained for imaging by SEM (G), providing unique opportunities to tap complementary strengths of immuno-fluorescence and electron microscopic volume imaging (H). I. Volume rendering of an array tomographic immunofluorescence image of a subset of layer 5 pyramidal cells (green) and putative synapses (anti-synapsin-I puncta, red) in a 180×140×30 um volume of mouse whisker barrel. Blue objects are DAPI stained nuclei of otherwise unstained cells. The larger red objects are erythrocytes within capillaries. The specimen is from a Line H Thy-1-YFP mouse [6]. See reference 15 for examples of conjugate immunofluorescence and SEM array tomography.
Complementing improvements in genetic and immunofluorescence staining techniques, several new light microscopy methods are bypassing nineteenth-century ideas about wave diffraction limits to resolution. Given that feature sizes much smaller than 200 nm are typical of mammalian neuropil, Abbe’s classic diffraction theory implies that the light microscope will fall far short of the needed resolution. Recently, however, imaging techniques based on non-linear optics have demonstrated sub-100 nm resolution with visible light wavelengths. Non-linear structured illumination fluorescence methods [18, 19] have been shown to provide lateral resolutions extending well below 100 nm, while methods based on fluorescence tag photoswitching [20–23] have been shown to offer the possibility of localizing switchable tags with precisions on the order of 10 nm. Though most of these new methods have been demonstrated so far only for two-dimensional imaging, they seem ideally suited to improve lateral resolution in three-dimensional imaging when used in conjunction with an automated serial sectioning method such as array tomography.
New Tools for Automated Electron Microscopy
Three serial sectioning EM techniques suitable for the analysis of three-dimensional neural circuits have been reviewed recently in these pages [24]. Of these three, the Serial Section Transmission Electron Microscopy (SSTEM) and Serial Section Electron Tomography (SSET) methods provide the very highest resolutions, while the Serial Block Face Scanning Electron Microscopy (SBFSEM) delivers slightly lower resolution but offers the two extremely important advantages of naturally excellent section-to-section image registration and full automation of operation. The SBFSEM imaging approach also lends itself particularly well to imaging specimen volumes that are large in lateral extent by using automated XY stage motion and image tiling techniques.
Additional new serial section EM methods introduced this year make use of the backscattered-electron scanning electron microscopy (BsSEM) imaging modality that was introduced to neural circuit analysis by Denk and Hortsmann [25]. One is based on the use of a Focused Ion Beam Scanning Electron Microscope (FIBSEM). As in SBFSEM, BsSEM is used to image the specimen block face rather than a thin section, but successive ultrathin sections are removed in the FIBSEM by an ablative focused ion beam instead of the diamond knife used in the SBFSEM. This method has produced extremely high-quality results from small circuit volumes (G Knott, Soc Neurosci Abstr 2007, 534.5), but it remains to be seen how it may be scaled to larger circuit volumes. The other new method is based on placing arrays of ultrathin sections on solid substrates, staining with heavy elements, and then imaging the sections themselves by BsSEM. Lichtman and colleagues (Soc Neurosci Abstr 2007, 534.11) have demonstrated excellent serial EM results from epoxy-embedded sections that were arrayed onto silicon wafer substrates using an automated tape-collecting lathe ultramicrotome [26]. Micheva and Smith [15] have meanwhile shown that good backscattered electron SEM images can be obtained also from the same acrylic sections used for immunofluorescence array tomography.
Segmentation and Reconstruction Tools
Several of the new high-resolution volume imaging methods discussed above have the potential to capture automatically the trillions of voxels of image data necessary to begin defining complete cortical circuit structures, but with such large data sets come enormous challenges of reliably abstracting biologically meaningful information about circuit connectivity and molecular architecture. Fortunately, the analysis of immunofluorescence data is simplified by the magic of antibody specificity. The antibodies themselves do the “heavy lifting” of discriminating neurons, arbors and synapses and the basis of distinctive antigens, making it relatively easy for known volume image analysis and visualization tools (e.g., [27, 28]; B Busse, Soc Neurosci Abs 2007, 534.1) to extract biologically meaningful information about these specific circuit elements. It is likely, however, that even with every “brainbow” and non-linear optical resolution-enhancement trick now known, the resolution obtainable by fluorescence imaging will not suffice to trace all arbors reliably from fluorescence data. It also seems certain that the resolution of electron microscopy will be necessary to measure many important details of circuit structure accurately (e.g., dendritic spine necks can be less than 50 nanometers wide, and spine signaling depends very strongly on spine width).
The abstraction of circuit information from EM data has proven difficult. Even before the introduction of high-throughput methods for the collection of volumetric EM images, the overwhelming “bottleneck” to the reconstruction of neural circuit features via serial EM was not the acquisition but instead the interpretation and segmentation steps [29]. EM images of cortical neuropil exhibit an enormous density of detail, but automated discrimination of neurobiologically meaningful objects, such as axons, dendrites, and synapses, and reliable automated tracking of long processes has yet to be demonstrated. Definitive results so far have been achieved only by manual tracing, performed by human hand and eye. Even with the help of the latest hardware and software for handling and tracing images (e.g., [30]), however, this processes is agonizingly slow - on the order of tens of person-hours per cubic micrometer. Considering that the newer automated serial EM approaches can acquire data at rates more like one cubic micron per second, and that even very small circuits extend through volumes of many millions of cubic micrometers, it is clear that progress in EM-based circuit analysis will depend heavily of the development of schemes for robust and efficient automated segmentation. Work is now under way to address this goal, including efforts to optimize EM staining specifically to ease segmentation (e.g., [31]; K Briggman, Soc Neurosci Abstr 2007, 534.8; J Buchanan, Soc Neurosci Abstr 2007, 534.4) as well as work on the segmentation algorithms themselves.
One promising new approach to EM segmentation uses “machine learning” algorithms, where a program automatically optimizes its own operation based on “training sets” pairing raw EM images and corresponding manual segmentation results [32]. After the assimilation of a sufficient quantity of sufficiently accurate training data, the learning algorithm should be able to automatically and reliably segment any new image data that is generally similar to that presented by the training sets. One potential limitation, however, lies in the difficulty of producing training sets of sufficient size and accuracy to train a sufficiently robustn and reliable learning algorithm.
The generation of conjugate, voxel-registered immunofluorescence (IF) and electron microscopic (EM) volume images (e.g., by array tomography) may help to solve EM segmentation problems, merging the molecular discrimination strengths of IF imaging with the high resolution strengths of EM. For instance, array tomographic IF image data could be used to pinpoint all synapses and to discriminate cell-specific axonal and dendritic tags in a tool mouse specimen and thus pass helpful “prior” information to an EM segmentation algorithm. One characteristic EM segmentation error that is very difficult to avoid and most devastating to the accurate abstraction of circuit topology analysis is a skip from one fine axon to the next when attempting to track densely packed axons over distance. Here, the availability low-resolution but cell-specific optical information (e.g., from a brainbow mouse) may prove crucial to the successful detection and avoidance skipping errors. Large and accurate EM segmentations training sets derived from conjugate IF/EM tool mouse data sets also might be used as large and highly accurate training sets to refine segmentation algorithms that might learn eventually to segment and interpret data sets comprising EM data alone. That is, such algorithms might lead eventually to an ability to reconstruct a circuit using a fully automated EM technique, such as SBFSEM, and to reconstruction of human cortical circuits, where tool-mouse genetic tricks are not available.
Tooling Up for a Whisker Barrel
Complete reconstruction of mammalian cortical circuit structure is an obvious and necessary goal for neuroscience, but one that still lies very far off. Were could we start today? How far could we go with presently available tools? To address such questions, the feasibility of one hypothetical, very ambitious project will be considered. That project would be to reconstruct one rodent “whisker barrel”, defining the morphologies of all neural arbors, all the sites of potential synaptic contact [33], and as many details of circuit molecular architecture as possible.
The whisker barrel is a patch of rodent sensorimotor cortex that processes information associated with one contralateral whisker (vibrissa), and is one element of a closely-packed somatotopic array of barrels, one per whisker [34, 35]. Each barrel occupies a columnar volume just under 0.5 mm in diameter and 1.2 mm tall, and includes approximately 15,000 neurons and 100 million synapses. Although a whisker barrel in isolation, bereft of its extrinsic cortical and subcortical connections, is not a functionally complete circuit, it is arguably the most widely recognized and agreed upon example of an anatomically distinct cortical circuit module. The barrel circumscribes a volume within which nearly all dendritic arbors are complete and where lateral connectivity within barrels is far denser than that between barrels [36]. The associated peripheral sensory-motor structure, a single vibrissa, is also exceptionally tractable to functional study and the barrel has been explored particularly well by a wide variety of sophisticated physiological methods (e.g., See references 37 and 38). Reconstruction of a single whisker barrel would provide therefore a bounty of new information about local circuit organization that is not now available for any cortical structure. Reconstructions of two or more whisker barrels would allow powerful new approaches to fundamental questions about circuit structure stereotypy and plasticity [39, 40].
Some practicalities of the hypothetical barrel reconstruction project are outlined in Table 1. Certain assumptions extrapolate (but just a little) from today’s demonstrated states of the relevant arts. It is assumed that transgenes would encode eight cell-marking epitope tags that would allow most or all nearby cells to be distinguished by brainbow-style immunofluorescence, as discussed above. It is assumed that an array comprising an entire whisker barrel cut into 50 nm sections could be fabricated reliably on 24 standard microscope slides, and that array tomographic immunofluorescence could discriminate 36 antibody channels, of which eight would be used to read the transgenic epitope tags and the remaining 28 would be used to read endogenous molecules useful for classifying and modeling neurons and synapses. It is also assumed that BsSEM imaging could read the ultrastructure of these 50 nm sections in enough detail to meaningfully measure spine necks and to generate useful learning algorithm training sets. Additional technical assumptions are stated in Table 1.
Table 1.
Imaging the whisker barrel local circuit. Estimates of requirements to acquire and analyze a data volumes of 0.6 × 0.6 × 1.2 um, contain one complete whisker barrel. The 1 × 1 × 5 mm block is certain to contain at least one complete whisker barrel, and parts of several more. Note that array fabrication and staining steps require only a very small fraction of the total process time, with the automated image acquisition process (steps 4 and 6), requiring the bulk of the time. Note also that the image acquisition rate can be scaled up linearly by using multiple automated microscopes in parallel on separate array slides.
| Task | Specifications | Time Requirement |
|---|---|---|
| 1. Specimen Preparation | Fix, Embed Mouse Whisker Barrel | 24 Hrs |
| Trim, Mount 1 × 1 × 1.5 mm Block | 1 Hr | |
| 2. Array Preparation | Cut 20,000 Ultrathin (50 nm) Sections | 56 Hrs |
| Mount Ribbons on 24 50×75mm Slides | 24 Hrs | |
| Map 24 Slides | 24 Hrs | |
| 3. Array Staining | Stain 24 Slides (for LM and EM cycles) | 96 Hrs |
| 4. Fluorescence Acquisition | 100 nm pixels (avg. exposure 250 msec/frame) 36 Immunofluorescence channels (9 cycles) 18 fields (2K×2K @ 63x) tile each section |
1620 Hrs (= 68 Days) (7 Days on 10 ‘Scopes) |
| 5. Fluorescence Analysis | 36,000 cycles/voxel on 1000 1GHz pipelines 36 Channels × 0.86 Teravoxels = 31 Terabytes |
8.6 Hrs |
| 6. BsSEM Acquisition | 12.5 nm pixels (1 MHz pixel acquisition rate) 288 fields (4K×4K) tile each section |
15,360 Hrs (= 640 Days) (64 Days on 10 ‘Scopes) |
| 7. BsSEM Analysis | (10,000 cycles/voxel on 1000 1GHz pipelines) 55 Teravoxels = 55 Terabytes |
154 Hrs |
Would this project be feasible today? The proposition of collecting, storing and analyzing a total of 86 terabytes of data would have been daunting until quite recently, but is no longer so. Computing equipment costing much less than $0.5M US would suffice easily for the data storage and image analysis tasks specified. The time requirements for image acquisition, especially for BsSEM phase, may sound formidable, but these steps are fully automated and the imaging throughput is scalable by adding more automated microscopes. The field emission gun SEM (FEG-SEM) required costs approximately $0.5M US each, so scaling up to 10 FEG-SEMs for a 10X increase in throughput would be a viable option. The major potential barrier to the success of this project may lie in the challenge of developing sufficiently robust EM segmentation algorithms, as discussed above. Given the talents, energy and good ideas being brought to bear on this challenge today, it is hard to believe that this final barrier will not yield.
Conclusion
Though the challenges of reconstructing cortical circuitry today are substantial, the potential payoffs are enormous. Given today’s universal agreement with the postulate of the neural circuit as the basis of the brain’s abilities to process information and generate behavior, it seems extremely unlikely that the brain will ever be understood without reconstructing circuit structure. A framework of complete and quantitative knowledge of circuit structure should also provide for deeper and more efficient physiological analysis of circuit function, and for deeper molecular exploration of potential complexity-management concepts such as “neuron type” [2–5], “synapse type” [41], “cortical modules” [42], and “network motifs” [43]. With the rapid growth in the power and availability of parallel computation (e.g., see [44]), accurate reconstruction of circuit connectivity and molecular architecture appears to be the last remaining obstacle to fulfilling the promise of computational circuit simulation as the next generation’s best tool for understanding the brain. With a little luck, the new reconstruction techniques reviewed may have an impact worthy of comparison with Ramon y Cajal’s drawings.
Acknowledgments
The author thanks JoAnn Buchanan, Winfried Denk, Kristen Harris, Jeff Lichtman, Kristina Micheva, Nancy O’Rourke and Sebastian Seung for many helpful discussions and comments on manuscripts, and the authors of reference 12 for providing the illustration used in Fig. 2. This work was supported in part by grant NS054252 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ramon y Cajal S. In: Histology of the Nervous System of Man and Vertebrates. Swanson N, Swanson L, editors. Oxford Press; 1995. Summarizes the work that first placed the concept of information processing by neural circuit structures on a firm anatomical foundation.** [Google Scholar]
- 2.McKay RD, Hockfield SJ. Monoclonal antibodies distinguish antigenically discrete neuronal types in the vertebrate central nervous system. Proc Natl Acad Sci U S A. 1982;79:6747–6751. doi: 10.1073/pnas.79.21.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson SB, Hempel C, Sugino K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006 Oct;16(5):571–6. doi: 10.1016/j.conb.2006.08.006. Epub 2006 Sep 7. [DOI] [PubMed] [Google Scholar]
- 4.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;6:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 5.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 6.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. This landmark work introduced strategies that have proven very versatile and powerful for making mice with color-coded neuronal subsets.** [DOI] [PubMed] [Google Scholar]
- 7.Hatten ME, Heintz N. Large-scale genomic approaches to brain development and circuitry. Annu Rev Neurosci. 2005;28:89–108. doi: 10.1146/annurev.neuro.26.041002.131436. [DOI] [PubMed] [Google Scholar]
- 8.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monro S, Pelham HRB. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp70. EMBO J. 1984;3:3087–3093. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin BA, Adams SR, Jones J, Tsien RY. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- 11.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 12.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis R, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. Introduces a powerful new approach to circumventing optical resolution limits that have limited optical imaging of complete neural circuits.** [DOI] [PubMed] [Google Scholar]
- 13.Luo L. Fly MARCM and mouse MADM: Genetic methods of labeling and manipulating single neurons. Brain Res Rev. 2007 doi: 10.1016/j.brainresrev.2007.01.012. [Epub ahead of print]. A very promising new set of schemes for labeling and manipulating circuits.** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. A new strategy for labeling cells and their monsynaptically partners only.** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Micheva KD, Smith SJ. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conchello JA, Lichtman JW. Optical sectioning microscopy. Nat Methods. 2005 Dec;2(12):920–31. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J Neurosci. 2006;26:8017–8020. doi: 10.1523/JNEUROSCI.2728-06.2006. Reviews important recent progress in validating antibody specificity.* [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. * [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. * [DOI] [PubMed] [Google Scholar]
- 21.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–53. doi: 10.1126/science.1146598. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. * (References 18–23 introduce new methods for fluorescence imaging at resolutions far higher than formerly respected diffraction limits. Most of these methods have been demonstrated mainly for two-dimensional imaging, but may be very useful to enhance fluorescence lateral resolution in three-dimensional imaging if used in conjunction with array tomography mechanical sectioning procedures). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. Reviews the nascent field of high-throughput electron microscopy.* [DOI] [PubMed] [Google Scholar]
- 25.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;11:e329. doi: 10.1371/journal.pbio.0020329. Introduces the first fully automated method for the acquisition of volumetric data with resolution and accuracy adequate to allow tracing of neural circuit elements, and inspires a new field of high-throughput electron microscopic neuroanatomy.** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayworth KJ, Kasthuri N, Schalek R, Lichtman JW. Automating the collection of ultrathin serial sections for large volume TEM reconstructions. Microscopy and Microanalysis. 2006;12:86–87. Describes exciting an new microtome and an automated section collection scheme that are likely to be very useful for producing very-large-scale tomography arrays.* [Google Scholar]
- 27.Levoy M. Display of surfaces from volume data. IEEE Computer Graphics and Applications. 1988;8:29–37. [Google Scholar]
- 28.Cai H, Xu X, Lu J, Lichtman JW, Yung SP, Wong ST. Repulsive force based snake model to segment and track neuronal axons in 3D microscopy image stacks. Neuroimage. 2006 Oct 1;32(4):1608–20. doi: 10.1016/j.neuroimage.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 29.Fiala JC, Harris K. Computer-based alignment and reconstruction of serial sections. Microscopy Anal USA Edition. 2002;52:5–7. [Google Scholar]
- 30.Fiala JC. Reconstruct: a free editor for serial section microscopy. Journal of Microscopy. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 31.Toni N, Teng EM, Bushong EA, Aimone JB, Zhao C, Consiglio A, van Praag H, Martone ME, Ellisman MH, Gage FH. Synapse formation on neurons born in the adult hippocampus. Nat Neurosci. 2007;10:727–34. doi: 10.1038/nn1908. [DOI] [PubMed] [Google Scholar]
- 32.Jain V, Murray JF, Roth F, Turaga S, Zhigulin V, Briggman KL, Helmstaedter MN, Denk W, Seung HS. Supervised learning of image restoration with convolutional networks. IEEE International Conference on Computer Vision (ICCV); 2007. [Google Scholar]
- 33.Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–94. doi: 10.1016/j.tins.2005.05.006. Introduces a very powerful way to make the most of incomplete information about circuit topologies.* [DOI] [PubMed] [Google Scholar]
- 34.Woolsey TA, Dierker ML, Wann DF. Mouse SmI cortex: qualitative and quantitative classification of golgi-impregnated barrel neurons. Proc Natl Acad Sci U S A. 1975;72:2165–2169. doi: 10.1073/pnas.72.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinfeld D, Ahissar E, Diamond ME. Active sensation: insights from the rodent vibrissa sensorimotor system. Curr Opin Neurobiol. 2006;16:435–444. doi: 10.1016/j.conb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Petersen CC, Sakmann B. The excitatory neuronal network of rat layer 4 barrel cortex. J Neurosci. 2000;20:7579–86. doi: 10.1523/JNEUROSCI.20-20-07579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brecht M, Roth A, Sakmann B. Dynamic receptive fields of reconstructed pyramidal cells in layers 3 and 2 of rat somatosensory barrel cortex. J Physiol. 2003;553:243–265. doi: 10.1113/jphysiol.2003.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato TR, Gray NW, Mainen ZF, Svoboda K. The functional microarchitecture of the mouse barrel cortex. PLoS Biol. 2007;5:e189. doi: 10.1371/journal.pbio.0050189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micheva KD, Beaulieu C. Development and plasticity of the inhibitory neocortical circuitry with an emphasis on the rodent barrel field cortex: a review. Can J Physiol Pharmacol. 1997;75:470–8. [PubMed] [Google Scholar]
- 40.Knott GW, Quairiaux C, Genoud C, Welker E. Formation of dendritic spines with GABAergic synapses induced by whisker stimulation in adult mice. Neuron. 2002;34:265–73. doi: 10.1016/s0896-6273(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 41.Grant SG. Toward a molecular catalogue of synapses. Brain Res Rev. 2007;55:445–9. doi: 10.1016/j.brainresrev.2007.05.003. Clearly articulates the importance of finding ways to measure and classify individual synapses in their native brain tissue contexts.* [DOI] [PubMed] [Google Scholar]
- 42.Swindale NV. Is the cerebral cortex modular? Trends Neurosci. 1990;13:487–92. doi: 10.1016/0166-2236(90)90082-l. [DOI] [PubMed] [Google Scholar]
- 43.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 44.Migliore M, Cannia C, Lytton WW, Markram H, Hines ML. Parallel network simulations with NEURON. J Comput Neurosci. 2006;21:119–129. doi: 10.1007/s10827-006-7949-5. A strong argument that the computational power for circuit simulation will be ready as soon as the structural and molecular data for the model is available. [DOI] [PMC free article] [PubMed] [Google Scholar]