Summary
The suboptimal DNA repair capacity is a risk factor for cancer that may be modulated by dietary nutrient intake, and the serine hydroxymethyltransferase (SHMT) participates in folate metabolism and synthesis of purine and pyrimidine needed for DNA repair. Therefore, we tested our hypothesis that genetic variants of the cytosolic SHMT (SHMT1) gene are associated with lung cancer risk. In a hospital-based case-control study of 1032 non-Hispanic white lung cancer patients and 1145 matched cancer-free controls, we genotyped five common SHMT1 polymorphisms either in the promoter, exons, or 3′-untranslated regions. Although the genotype and allele frequency distribution of each SNP did not differ between cases and controls statistically significantly in the single-locus analysis, the rs638416 polymorphism in the promoter alone and the combined putative risk variant genotypes containing rs643333C, rs638416G, rs1979277T, rs3738G, and rs1979276C were associated with altered risk. Those carrying the combined 3+ risk variant genotypes had an increased risk of lung cancer (adjusted OR = 1.65, 95% CI = 1.05–2.57, compared with those having 0–1 risk genotypes; and OR = 1.21, 95% CI = 1.01–1.45, compared with those having 0–2 risk genotypes). The risk was more pronounced among older individuals (>61 years) or those having a low total folate intake or a high methionine intake. No evidence of interactions between the putative SHMT risk variant genotypes and the selected variables was found. These results suggest that SHMT1 variants may play a role in the etiology of lung cancer, and our findings need to be verified in larger prospective studies.
Keywords: DNA repair, genetic susceptibility, lung cancer, serine hydroxymethyltransferase, tetrahydrofolate metabolism
1. Introduction
Lung cancer is the leading cause of cancer-related death for men and women in the world, and there were estimated 1.35 million new cases worldwide in 2002 [1]. Tobacco consumption has been well documented as the primary risk factor for lung cancer [2], and smoking cessation has been proven to be the most important and cost-effective management to date. However, fewer than 20% of lifetime smokers develop lung cancer, suggesting that other factors may modulate individual risk associated with exposure to tobacco carcinogens. Therefore, both inter-individual genetic variation and one’s dietary habits may contribute to susceptibility to lung cancer [3, 4].
DNA repair is a critical mechanism to protect human genomic integrity against DNA damage generated by environmental factors, including tobacco carcinogens [5]. Studies have shown that suboptimal DNA repair capacity was a risk factor for cancer [6] and that low dietary folate intake was associated with suboptimal DNA repair capacity [7]. The methionine is an essential amino acid and its derivative S-adenosyl methionine (SAM) serves as a methyl donor, involving in the tetrahydrofolate synthesis by the methionine synthase and vitamin B12 in humans. Meanwhile, the serine hydroxymethyltransferase (SHMT) and its coenzyme, vitamin B6, catalyze the reversible conversion of serine and tetrahydrofolate to glycine and 5,10-methylene tetrahydrofolate that serves as a provider of one-carbon units during the synthesis of pyrimidine and purine[8]. Deficiencies in such nutrients could lead to DNA damage including single- and double-strand breaks, or oxidative lesions, or both [9]. Therefore, it appears that there is a possible link among dietary nutrient intake, tetrahydrofolate metabolism, DNA repair, and one’s susceptibility to lung cancer.
Human SHMT1 gene is located at chromosome 17p11.2, encoding SHMT1, a cytosolic isoform of SHMT[10, 11]. A recent case-control analysis reported that genetic variants of SHMT1 were associated with risk of squamous cell carcinoma of the head and neck in non-Hispanic whites [12]. To date, no report has investigated the role of SHMT1 variants in the development of lung cancer, even though both reduced DNA repair capacity and low intake of dietary folate were associated with lung cancer risk [13]. We hypothesized that SHMT1 variants are associated with lung cancer risk that may be also modified by dietary nutrient intake. Therefore, we genotyped five common, potentially functional single nucleotide polymorphisms (SNPs) in SHMT1 and tested this hypothesis in an ongoing hospital-based case–control study of lung cancer.
2. Materials and methods
2.1. Study population
The recruitment of lung cancer patients and frequency-matched cancer-free controls has been previously described [3, 14]. Briefly, the patients were recruited consecutively between September 1995 and December 2003, without any restrictions on age, sex, cancer stage or histology, from an ongoing molecular epidemiologic study of lung cancer conducted in the Department of Epidemiology, The University of Texas M. D. Anderson Cancer Center in Houston, Texas. The control subjects were selected from a pool of cancer-free subjects recruited through the largest multi-specialty physician practice, the Kelsey Seybold Foundation, with multiple clinics throughout the Houston metropolitan area. The controls were frequency matched to the cases on age (±5 years), sex, ethnicity, and smoking status. The exclusion criteria included previous treatments (by radiotherapy or chemotherapy or both), previous cancer, and recent (in last 6 months) blood transfusions. After the informed consent was obtained, each subject was scheduled for an interview, and the information about demographic and the selected variables was collected by a structured questionnaire administered and maintained by interviewers. The study protocol was approved by the institutional review boards of M. D. Anderson Cancer Center and the Kelsey Seybold Foundation.
2.2. Dietary Analysis
We used a modified version of the National Cancer Institute’s Health Habits and History Questionnaire to collect the dietary data [15, 16], including a food-frequency list, an open-ended food section, and other dietary behavior questions pertaining to use of supplements, restaurant dining, and food preparation methods. The food frequency instrument assessed diet in cases the year prior to diagnosis and in the controls the year prior to enrollment in the study. Data entry was performed using DietSys (version 4.01) and DietSYS+Plus (DietSYS+Plus Analysis Software, Version 5.9 Block Dietary Data Systems, Berkeley CA, 1999) programs. Dietary analysis was conducted using DietSYS+Plus (Version 5.9). The source of total folate values was Standard Release 14 [17]. Recipe adjustments for moisture changes and nutrient losses due to cooking were also made. Dietary total folate intake was adjusted by daily calorie intake and expressed as dietary total folate in μg/1,000 kcal/day. There were 932 lung cancer patients and 1073 controls whose dietary information was complete and used in the final analysis.
2.3. Genotyping
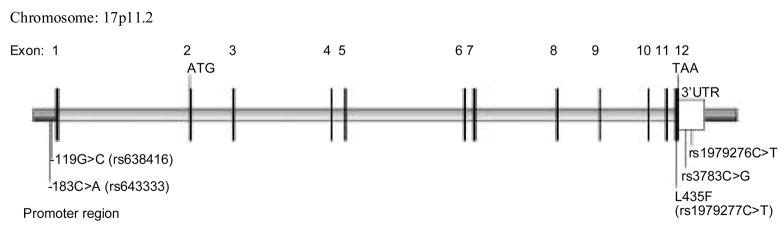
The SNP rs1979277 (34761C>T) is located in exon 12 (codon 435 in isoforms 1) or exon 13 (codon 474 in isoforms 2) of SHMT1 mRNA transcripts, whereas the SNPs rs3783 (34840C>G) and rs1979276 (34859C>T) are located in the 3′-untranslated region. We used the previously published methods to genotype these three SNPs [12]. We also identified additional 20 SNPs by using bioinformatics analysis in the dbSNP database of National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Snp), of which two SNPs were found to be located in the SHMT1 promoter region as predicted by the online software TSSW (http://www.softberry.com/berry.phtml?topic=tssw&group=programs&subgroup=promoter). The locations of these two promoter SNPs are -183 nt (rs643333) and -119 nt (rs638416) from the putative transcription starting site of SHMT1mRNA transcripts, as shown in Figure 1. The allele frequencies in Caucasian populations were reported as ~0.65 for the rs643333C allele and ~0.69 for the rs638416 G allele in the dbSNP database. We determined their genotypes by using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. Briefly, genomic DNA fragments harboring the rs643333C>A (168 bp), the rs638416C>G (215 bp), the rs1979277C>T (108 bp), and both rs3783C>G and rs1979276C>T (203 bp) were amplified with the primer sets and conditions shown in Table 1. The Hha I, Ava I, Ear I, EcoN I and Dde I restriction enzymes (New England Biolabs, Beverly, Massachusetts, USA) were used to distinguish the nucleotides at the polymorphic site by generating RFLP patterns in the presence of rs643333C, or rs638416G, or rs1979277C, or rs3783C, or rs1979276C. More than 10% of the DNA samples were randomly selected for genotype confirmation and the results were 100% in concordance.
Figure 1.
Genomic structure and locations of five functional SNPs in the SHMT1 gene
Table 1.
The primers information of five SNPs used in this study
| SNP locationa | db SNP | PCR primers (5′ → 3′) | Annealing temp. | Restriction Enzyme | Note |
|---|---|---|---|---|---|
| Promoter region: 32515G>Tb | rs643333 | Forward: TGGACGCACATTTGTCCTACTT |
62°C | Hha I | Newly designed |
| Reverse: CAGGGACCTGCAGAACTGACGCc | |||||
| Promoter region: 32451G>C | rs638416 | Forward: TGAGGAAGGCCCTGTGTAGT |
62°C | Ava I | Newly designed |
| Reverse: ATCAGAGAGCGCAGCCAAG | |||||
| Exon 12: 34761C>T(Codon L435F) | rs1979277 | Forward: CTGGCAGGGGATAAGTACCA |
64°C | Ear I | Reference (12) |
| Reverse: CCCGCTCCTTTAGAAGTCAG | |||||
| 3′-UTR: 34840C>G | rs3783 | Forward: TGGCAGGGGATAAGTACCAG |
56°C | EcoN I | Reference (12) |
| 3′-UTR: 34859C>T | rs1979276 | Reverse: GTCAACAGTTCCCCTTTGGA |
Dde I | Reference (12) |
2.4. Statistical analysis
We used χ2 test to evaluate differences in the frequency distributions of selected categorical variables, including demographic variables, smoking status, alcohol use, and frequencies of the SHMT1 genotypes or alleles, between the cases and controls. Participants who had smoked less than 100 cigarettes in their lifetime were categorized as never smokers, and all others were categorized as ever smokers. For ever smokers, those who had quit smoking more than 1 year previously were considered former smokers, and the others (recent quitters) were included with current smokers. All subjects were also regrouped as never, light, and heavy smokers based on the pack-years they smoked (0, ≤35.4, and >35.4 years, respectively). Participants who had drunk alcoholic beverages at least once a week for one year or more were categorized as ever drinkers, and the rest were defined as never drinkers. The Student’s t tests were performed to distinguish differences in mean values of age, pack-years smoked, and dietary intake of total folate, vitamin B6, vitamin B12, and methionine between the cases and controls. The mean values for all continuous variables in controls were used as the cut-off to dichotomize the subjects into two groups for further statistical analysis. Those smokers with the number of pack-years smoked more than the median number in controls were defined as heavy smokers. Univariate and multivariate unconditional logistic regression analyses were used to calculate odds ratios (ORs) and their 95% confidence intervals (CIs) for assessing risk of lung cancer. The putative risk genotypes and alleles were determined, if their frequencies were higher in the lung cancer patients than in the controls [18]. Because there was no prior information about the effects of these SNPs on lung cancer risk, we used Akaike’s information criterion (AIC) to select the best genetic-effect model for each SNP. In this process, AIC combines a measure of the lack of fit of a particular model with a penalty for the number of parameters in the model, and a model with smaller AIC values was preferably selected [19]. In the dominant model, the common homozygous genotype in the controls was defined as the reference group; the rare homozygous and heterozygous genotypes were variant genotypes, and their effects were individually estimated by comparison with the reference. Alternatively, the rare homozygotes and the heterozygotes were combined to form the reference group and compared with that of the common homozygote. In the recessive model, only the rare homozygotes were defined as the variant genotype; the other two genotypes were combined as the reference. Considering possible effects of other covariates, we calculated the AIC in unconditional logistic regression model with adjustment for age, sex, smoking and alcohol usage. To take into account the overall effect of these five SNPs, we generated a categorical variable from a combination of five variant genotypes based on the dichotomized genotypes of each SNP. We also used the SAS/Genetics software to detect the linkage disequilibrium (LD) between any pair of SNPs and generated the haplotype based on the observed genotypes [20]. The haplotype data were further analyzed with these variables and stratified by age, sex, smoking status, number of pack-years smoked, alcohol drinking status, and dietary intake of total folate, vitamin B6, vitamin B12, and methionine. Potential multiplicative and additive interactions were also evaluated by performing the logistic regression analysis [21]. All tests were two-sided and P value of <0.05 was defined as the significance level. The statistical analyses were performed by using the Statistical Analysis System software (Version 9.1; SAS Institute, Cary, NC).
3. Results
3.1. Characteristics of the study population
We included 1032 lung cancer patients and 1145 controls in this analysis whose demographic variables were similar to those described in previous reports [22–24]; because of the frequency matching design, these covariates were only used for the adjustment in this analysis. However, both smoking status and the number of pack-years smoked still showed differences between the case and controls (P = 0.015 and P < 0.001, respectively), further suggesting that additional adjustment is needed. The dietary intake for total folate was lower in the lung cancer patients (mean value of 208.0 ± 69.0) compared with the cancer-free controls (220.7 ± 86.2), and this difference was also statistically significant (P < 0.001; Student t-tests). We also detected statistically significant difference in dietary intake of vitamin B6 (P < 0.004; Student t-tests) but not in vitamin B12 and methionine (P = 0.528 and P = 0.791; Student t-tests) between the case and controls. In addition, there were more alcohol users among the case than among the controls (P < 0.001). Therefore, to remove the residual impact of these variables on the main effects of SNPs, we included all these variables in the adjustment in the unconditional multivariate logistic regression analysis.
3.2. Frequency distributions of SHMT1 genotypes and alleles and their associations with lung cancer risk
The genotype and allele frequency distributions and the associations between each of the five SHMT1 functional SNPs and lung cancer risk are presented in Table 2. Although frequency distributions of all genotypes in the controls did not deviate statistically significantly from those expected under the Hardy-Weinberg equilibrium (P > 0.05 for all SNPs), there were no statistically significant differences in the frequency distributions of these SHMT1 genotypes and alleles between cases and controls, except for the promoter rs638416G>C, the only SNP that was associated with statistically significantly increased risk of lung cancer in the multivariate logistic regression analysis with adjustment for all covariates (adjusted OR = 1.30 and 95% CI = 1.00–1.70 for G allele carriers). The allele frequencies of the promoter SNPs (rs643333C and rs638416G), the exon 12 codon 435 SNP (rs1979277T), and the 3′UTR SNPs (rs3783G and rs1979276C) were slightly increased in the cases than in the controls and thus considered as the risk variant alleles in further analyses. We found that the effects of the promoter SNPs (rs643333C and rs638416G) best fit the dominant genetic model, as indicated by the AIC values (2711.37 and 2707.58, respectively) being the smallest among all possible assumed models. Similarly, we also found that the effects of the exon 12 codon 435 SNP (rs1979277T), and the SNPs (rs3783G and rs1979276C) in the 3′UTR best fit the recessive genetic models, with the smallest AIC value (2710.56, 2710.90 and 2710.69, respectively) among all possible models.
Table 2.
SHMT1 genotype and allele frequencies of lung cancer cases and cancer-free controls and their association with risk of lung cancer
| Genotype | Number of subjects (%) |
P value b | Crude OR (95% CI) | Adjusted OR (95% CI) c | |
|---|---|---|---|---|---|
| Patients | Controls a | ||||
| Total | 1032 (100.0) | 1145 (100.0) | |||
| Promoter region | |||||
| rs643333 | |||||
| AA | 104 (10.1) | 117 (10.2) | 0.836 | 1.00 | 1.00 |
| CA | 476 (46.1) | 541 (47.3) | 0.99 (0.74–1.33) | 0.97 (0.71–1.32) | |
| CC | 452 (43.8) | 487 (42.5) | 1.04 (0.78–1.40) | 0.98 (0.72–1.34) | |
| C allele | 0.669 | 0.662 | 0.624 | ||
| CA+CC | 928 (89.9) | 1028 (89.8) | 0.914 | 1.02 (0.77–1.34) | 0.97 (0.72–1.31) |
| rs638416 | |||||
| CC | 127 (12.3) | 160 (14.0) | 0.381 | 1.00 | 1.00 |
| CG | 505 (48.9) | 532 (46.4) | 1.20 (0.92–1.56) | 1.35 (1.02–1.79) | |
| GG | 400 (38.8) | 453 (39.6) | 1.11 (0.85–1.46) | 1.24 (0.93–1.65) | |
| G allele | 0.632 | 0.628 | 0.768 | ||
| CG+GG | 905 (87.7) | 985 (86.0) | 0.251 | 1.16 (0.90–1.49) | 1.30 (1.00–1.70) |
| Exon 12 (L435F) | |||||
| rs1979277 | |||||
| CC (LL) | 459 (44.5) | 495 (43.2) | 0.535 | 1.00 | 1.00 |
| CT (LF) | 431 (41.7) | 504 (44.0) | 0.92 (0.77–1.11) | 0.95 (0.78–1.15) | |
| TT (FF) | 142 (13.8) | 146 (12.8) | 1.05 (0.81–1.37) | 1.10 (0.83–1.46) | |
| T allele | 0.654 | 0.652 | 0.935 | ||
| CC+CT | 890 (86.2) | 999 (87.3) | 0.488 | 1.00 | 1.00 |
| TT | 142 (13.8) | 146 (13.8) | 1.09 (0.85–1.40) | 1.13 (0.87–1.47) | |
| 3′-UTR | |||||
| rs3783 | |||||
| CC | 457 (44.2) | 500 (43.7) | 0.805 | 1.00 | 1.00 |
| CG | 464 (45.0) | 529 (46.2) | 0.96 (0.80–1.15) | 0.98 (0.81–1.18) | |
| GG | 111 (10.8) | 116 (10.1) | 1.05 (0.78–1.40) | 1.10 (0.81–1.50) | |
| G allele | 0.332 | 0.332 | 0.997 | ||
| CC+CG | 921 (89.2) | 1029 (89.9) | 0.634 | 1.00 | 1.00 |
| GG | 111 (10.8) | 116 (10.1) | 1.07 (0.81–1.41) | 1.11 (0.83–1.49) | |
| rs1979276 | |||||
| TT | 110 (10.7) | 131 (11.4) | 0.461 | 1.00 | 1.00 |
| CT | 457 (44.3) | 528 (46.1) | 1.03 (0.78–1.37) | 1.04 (0.77–1.41) | |
| CC | 465 (45.0) | 486 (42.5) | 1.14 (0.86–1.51) | 1.12 (0.83–1.51) | |
| C allele | 0.672 | 0.655 | 0.237 | ||
| TT+CT | 567 (54.9) | 659 (57.5) | 0.220 | 1.00 | 1.00 |
| CC | 465 (45.0) | 486 (42.5) | 1.11 (0.94–1.32) | 1.08 (0.90–1.30) | |
The observed genotype frequency distributions in the controls were in agreement with Hardy-Weinberg equilibrium (p2 + 2pq + q2 = 1) (χ2 = 3.484, P = 0.062 for rs643333; χ2 = 0.036, P = 0.849 for rs638416; χ2 = 0.995, P = 0.318 for rs1979277; χ2 = 1.936, P = 0.164 for rs3783; and χ2 = 0.474, P = 0.491 for rs1979276).
Two-sided χ2 test for the difference in the genotype or allele distributions between the cases and the controls.
Due to missing data, 932 cases and 1073 controls were analyzed with adjustment for age, sex, smoking status, square root of pack-years smoked, alcohol drinking status, and dietary intake of total folate, vitamin B6, vitamin B12, and methionine in logistic regression models.
As shown in Table 3, there was incomplete LD between the promoter SNPs (rs643333 and rs638416) and the other three SNPs; but the LD between the exon 12 codon 435 SNP (rs1979277) and either the 3′UTR rs3783 or rs1979276 SNPs was relatively higher (data not shown). In further haplotype analysis, we identified 29 haplotypes from the five SHMT1 SNPs but only 3 haplotype alleles (i.e., AGTGT, CGCCC, and CCCCC) were common (allele frequency ≥ 5%), representing 85% of the chromosomes in the 1145 controls. However, the frequency distribution of haplotype alleles between the cases and controls did not differ statistically significantly. When we used the most frequent haplotype CCCCC in controls as the reference, we found that other haplotypes were not associated with any altered lung cancer risk (data not shown). Therefore, we then focused on the combined effect of all five loci by generating a categorical variable, the putative SHMT1 risk variant genotypes, from among all individual dichotomized genotypes.
Table 3.
The linkage disequilibrium analysis for 5 SNPs of SHMT gene in pair-wise pattern (D′ and r2)a
| D′ | rs643333 | rs638416 | rs1979277 | rs3783 | rs1979276 |
|---|---|---|---|---|---|
| r2 | |||||
| rs643333 | 0.7160 | 0.7769 | 0.7939 | 0.7600 | |
| rs638416 | 0.1554 | 0.6216 | 0.6682 | 0.6117 | |
| rs1979277 | 0.5794 | 0.1220 | 0.9608 | 0.9147 | |
| rs3783 | 0.6132 | 0.1316 | 0.8623 | 0.9297 | |
| rs1979276 | 0.5610 | 0.1168 | 0.8270 | 0.8169 |
D′ values were illustrated in the part of right up, and r2 values were illustrated in the part of left down.
3.3. Associations of the putative SHMT risk variant genotypes with lung cancer risk
The frequency distributions and the associations of the number of variant genotypes with lung cancer risk are presented in Table 4. Although the overall frequency distribution of the combined putative risk variant genotypes was not statistically significant between cases and controls, individuals who carried the combined genotypes having 3+ risk variant genotypes had an increased risk of lung cancer (adjusted OR = 1.65 and 95% CI = 1.05–2.57, compared with those having 0–1 risk genotypes and OR = 1.21 and 95% CI = 1.01–1.45, compared with those having 0–2 risk genotypes). In addition, the trend of the ORs for 0–1, 2, and 3+ risk genotypes was statistically significant (P = 0.013). The stratified associations between the dichotomized number of risk genotypes (0–2 vs 3+) and lung cancer risk are presented in Table 5. The risk was statistically significantly increased for older individuals with age older than 61 (P = 0.045) but borderline significant for women, heavy smokers, and never drinkers. Furthermore, a statistically significantly increased risk was observed for those who had total folate dietary intake less than 220.7 μg/day/1000kcal (adjusted OR = 1.39, 95% CI = 1.09–1.78, P = 0.009) and those who had higher methionine intake (adjusted OR = 1.32, 95% CI = 1.01–1.71, P = 0.040), whereas this risk was only borderline significant for those who had high intake of vitamin B6 (P = 0.057) or lower vitamin B12 intake (P = 0.075). However, we did not find any evidence of interactions between the putative SHMT risk variant genotypes and the selected variables in either multiplicative or additive models (data not shown).
Table 4.
Distributions of the number of putative SHMT1 risk variant genotypes in the combined genotypes of lung cancer cases and cancer-free controls and the association with risk of lung cancer
| Putative SHMT1 risk variants genotype a | Number of subjects (%) |
Pb | Crude OR (95% CI) | Adjusted OR (95% CI)c | |
|---|---|---|---|---|---|
| Patients | Controls | ||||
| Total | 1032 (100.0) | 1145 (100.0) | |||
| 0 | 1 (0.1) | 1 (0.1) | 0.269 | ||
| 1 | 37 (3.6) | 62 (5.4) | |||
| 2 | 498 (48.3) | 573 (50.0) | |||
| 3 | 468 (45.4) | 484 (42.3) | |||
| 4 | 26 (2.5) | 24 (2.1) | |||
| 5 | 2 (0.2) | 1 (0.1) | |||
| 0–1 | 38 (3.7) | 63 (5.5) | 0.056 | 1.00 | 1.00 |
| 2 | 498 (48.3) | 573 (50.0) | 1.44 (0.95–2.19) | 1.39 (0.89–2.18) | |
| 3+ | 496 (48.1) | 509 (44.5) | 1.62 (1.06–2.46) | 1.65 (1.05–2.57) | |
| Ptrend = 0.029 | Ptrend = 0.013 | ||||
| 0–2 | 536 (51.9) | 636 (55.5) | 0.092 | 1.00 | 1.00 |
| 3+ | 496 (48.1) | 509 (44.5) | 1.16 (0.98–1.37) | 1.21 (1.01–1.45) | |
Constructed from the combination of dichotomized genotypes as shown in Table 2.
Two-sided χ2 test for the difference in the genotype or allele distributions between the cases and the controls.
Due to missing data, 932 cases and 1073 controls were analyzed with adjustment for age, sex, smoking status, square root of pack-years smoked, alcohol drinking status, and dietary intake of total folate, vitamin B6, vitamin B12, and methionine in logistic regression models.
Table 5.
Frequency distribution and adjusted ORs and 95% CIs for the putative SHMT1 risk variants genotype stratified by selected variables in the lung cancer patients and controls
| Variables | Number of patients (%) |
Number of controls (%) |
Adjusted OR (95% CI) a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | 0–2 | 3+ | Total | 0–2 | 3+ | 0–2 | 3+ | ||
| Total | 932 | 485 (52.0) | 447 (48.0) | 1073 | 599 (55.8) | 474 (44.2) | 1.00 | 1.21 (1.01–1.45) | |
| Age | ≤61 | 452 | 236 (52.2) | 216 (47.8) | 520 | 291 (56.0) | 229 (44.0) | 1.00 | 1.17 (0.91–1.52) |
| >61 | 480 | 249 (51.9) | 231 (48.1) | 553 | 308 (55.7) | 245 (44.3) | 1.00 | 1.30 (1.01–1.67) | |
| Sex | Female | 436 | 217 (49.8) | 219 (50.2) | 553 | 308 (55.7) | 245 (44.3) | 1.00 | 1.27 (0.98–1.64) |
| Male | 496 | 268 (54.0) | 228 (46.0) | 520 | 291 (56.0) | 229 (44.0) | 1.00 | 1.19 (0.92–1.53) | |
| Smoking status | Never | 156 | 78 (50.0) | 78 (50.0) | 182 | 105 (57.7) | 77 (42.3) | 1.00 | 1.35 (0.87–2.10) |
| Former | 397 | 205 (51.6) | 192 (48.4) | 515 | 283 (54.9) | 232 (45.1) | 1.00 | 1.23 (0.94–1.61) | |
| Current | 379 | 202 (53.3) | 177 (46.7) | 376 | 211 (56.1) | 165 (43.9) | 1.00 | 1.18 (0.87–1.60) | |
| Pack-years smoked | Never | 156 | 78 (50.0) | 78 (50.0) | 182 | 105 (57.7) | 77 (42.3) | 1.00 | 1.35 (0.87–2.10) |
| Light | 199 | 100 (50.2) | 99 (49.7) | 353 | 187 (53.0) | 166 (47.0) | 1.00 | 1.16 (0.81–1.66) | |
| Heavy | 577 | 307 (53.2) | 270 (46.8) | 538 | 307 (57.1) | 231 (42.9) | 1.00 | 1.25 (0.98–1.60) | |
| Alcohol use | Never | 333 | 170 (51.0) | 163 (49.0) | 326 | 186 (57.1) | 140 (42.9) | 1.00 | 1.32 (0.97–1.81) |
| Former | 306 | 167 (54.6) | 139 (45.4) | 267 | 159 (59.5) | 108 (40.5) | 1.00 | 1.25 (0.89–1.75) | |
| Current | 293 | 148 (50.5) | 145 (49.5) | 480 | 254 (52.9) | 226 (47.1) | 1.00 | 1.16 (0.86–1.57) | |
| Total folate intake | High | 388 | 210 (54.1) | 178 (45.9) | 539 | 290 (53.8) | 249 (46.2) | 1.00 | 1.04 (0.79–1.36) |
| Low | 544 | 275 (50.5) | 269 (49.5) | 534 | 309 (57.9) | 225 (42.1) | 1.00 | 1.39 (1.09–1.78) | |
| Vitamin B6 intake | High | 420 | 218 (51.9) | 202 (48.1) | 529 | 297 (56.1) | 232 (43.9) | 1.00 | 1.30 (0.99–1.69) |
| Low | 512 | 267 (52.1) | 245 (47.9) | 544 | 302 (55.5) | 242 (44.5) | 1.00 | 1.17 (0.91–1.49) | |
| Vitamin B12 intake | High | 479 | 260 (54.3) | 219 (45.7) | 534 | 304 (56.9) | 230 (43.1) | 1.00 | 1.20 (0.92–1.55) |
| Low | 453 | 225 (49.7) | 228 (50.3) | 539 | 295 (54.7) | 244 (45.3) | 1.00 | 1.26 (0.98–1.63) | |
| Methionine intake | High | 451 | 236 (52.3) | 215 (47.7) | 538 | 311 (57.8) | 227 (42.2) | 1.00 | 1.32 (1.01–1.71) |
| Low | 481 | 249 (51.8) | 232 (48.2) | 535 | 288 (53.8) | 247 (46.2) | 1.00 | 1.12 (0.87–1.44) | |
Adjusted for age, sex, smoking status, square root of pack-years smoked, alcohol drinking status, and dietary intake of total folate, vitamin B6, vitamin B12, and methionine in logistic regression models.
4. Discussion
To the best of our knowledge, this is the first large case-control study to date to show that the rs638416G>C polymorphism in the promoter alone and the combination of SHMT1 genetic variants was associated with risk of lung cancer in non-Hispanic whites. Furthermore, the finding that an increased risk was more pronounced among older subjects and those who had low total folate intake or high methionine intake may be a chance finding because of smaller observations in the stratified analyses. Nevertheless, our results appear to support our hypothesis that the SHMT1 genetic variants are associated with risk of lung cancer and that this risk may be modulated by dietary nutrient intake.
Although it is likely that stratification analysis may lead to some spurious findings, our results appear to have some biological plausibility. SHMT1 involves in the metabolism of one-carbon units critical for the synthesis of pyrimidine and purine. Therefore, it may have a role in the maintenance of genomic DNA integrity through effective DNA repair. One study showed that a deletion of chromosome 17p11.2 harboring SHMT1 was associated with the Smith-Magenis syndrome whose phenotype included midface hypoplasia and growth retardation [25]. Further, the mothers, to whom a child of the SHMT1 codon 435 CC homozygote with neural tube defects was born, had elevated homocysteine levels and reduced folate levels [26]. There were several reports on the association between the SHMT1 codon 435 SNP and risk of cancer [27–29], but the results were not conclusive, although this SHMT1 SNP causes an amino acid change from leucine (L) to phenylalanine (F). Therefore, it was still not clear what role the SHMT1 genetic variants may play in the etiology of cancer, because only one SNP was included in these published studies.
The SNPs altering the conserved amino acids, i.e., non-synonymous SNPs, are more likely to be associated with cancer susceptibility and thus often recommended for their inclusion in association studies [30–33]. However, other types of SNPs may also have an impact on gene functions at the transcription level through modulating its mRNA amount, such as those SNPs that are located in the promoter and 3′-untranslated region, i.e., the regulatory SNPs. However, such regulatory SNPs are not often included in the current molecular epidemiological studies, because there is no information or a consensus standard for their selection for association studies.
The relatively large sample size is a strength of this study, which made the combined and subsequent stratification analyses possible and meaningful. Another strength of our study is the approach used to select SNPs. We identified two new promoter SNPs of SHMT1 and their frequency information in the NCBI dbSNP database and confirmed their locations in the SHMT1 gene, i.e., the putative promoter region, by using bioinformatics analysis. We believe that the set of these two SNPs together with three previously reported SNPs [12] may collectively have some impact on the functions of SHMT1 at the transcription level. Indeed, we found a statistically significant association between the genotypes having 3+ SHMT1 variants and risk of lung cancer in this study population. This finding is also consistent with previous findings of SHMT1 variants in squamous cell carcinoma of the head and neck and adult acute lymphocytic leukemia, although fewer SHMT1 SNPs were included in these studies [12, 27]. Our data presented here support the notion that a single polymorphism may only contribute to a modest effect, if any, but the use of combined variant genotypes are more likely to identify an association. Therefore, it appears that it is more efficient and effective to include as many functional SNPs in one gene as possible in such association studies [34]. Such an approach should be of special value for evaluating the roles of those genes with low penetration in one’s genetic susceptibility to cancer.
5. Conclusion
We observed an increased lung cancer risk associated with the rs638416G>C polymorphism in the promoter and the combined genotypes of three or more SHMT1 putative risk variant genotypes containing the alleles of the promoter SNPs rs643333C and rs638416G, the exon 12 codon 435 SNP rs1979277T, or the SNPs rs3783G and rs1979276C in the 3′-untranslated region, in a large, non-Hispanic white population. The increased risk was also more pronounced among older individuals or those with a low intake of total folate or a high intake of methionine, but these results need to be verified in larger studies. Overall, our data support the hypothesis that the SHMT1 genetic variants are associated with susceptibility to lung cancer. Further prospective studies are needed to validate this finding.
Acknowledgments
We thank Susan Honn for assistance in recruiting the subjects; Li-E Wang and Zhensheng Liu for their technical support; Jianzhong He, Kejin Xu and Yinyan Li for their laboratory assistance, Patricia Pillow and Ladia Hernandez for nutrition data analysis, and Joanne Sider for manuscript preparation. The present study was supported by National Institutes of Health grants CA55769 (M.R. Spitz), ES11740 (Q. Wei), and CA16672 (The University of Texas M.D. Anderson Cancer Center).
Abbreviations
- AIC
Akaike information criterion
- CI
confidence interval
- LD
linkage disequilibrium
- OR
odds ratio
- PCR-RFLP
the polymerase chain reaction-restriction fragment length polymorphism
- SHMT
serine hydroxymethyltransferase
- SNP
single nucleotide polymorphisms
Footnotes
Conflict of interest statement
None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Shields PG. Molecular epidemiology of smoking and lung cancer. Oncogene. 2002;21(45):6870–6. doi: 10.1038/sj.onc.1205832. [DOI] [PubMed] [Google Scholar]
- 3.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12(8):689–98. [PubMed] [Google Scholar]
- 4.Glade MJ. Food, nutrition, and the prevention of cancer: a global perspective. American Institute for Cancer Research/World Cancer Research Fund, American Institute for Cancer Research, 1997 Nutrition. 1999;15(6):523–6. doi: 10.1016/s0899-9007(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 5.Wei Q, Spitz MR. The role of DNA repair capacity in susceptibility to lung cancer: a review. Cancer Metastasis Rev. 1997;16(3–4):295–307. doi: 10.1023/a:1005852211430. [DOI] [PubMed] [Google Scholar]
- 6.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42(2):65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 7.Wei Q, Shen H, Wang LE, Duphorne CM, Pillow PC, Guo Z, et al. Association between low dietary folate intake and suboptimal cellular DNA repair capacity. Cancer Epidemiol Biomarkers Prev. 2003;12(10):963–9. [PubMed] [Google Scholar]
- 8.Fowler B. The folate cycle and disease in humans. Kidney Int Suppl. 2001;78:S221–9. doi: 10.1046/j.1523-1755.2001.59780221.x. [DOI] [PubMed] [Google Scholar]
- 9.Ames BN. DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutat Res. 2001;475(1–2):7–20. doi: 10.1016/s0027-5107(01)00070-7. [DOI] [PubMed] [Google Scholar]
- 10.Shane B. Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam Horm. 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- 11.Girgis S, Nasrallah IM, Suh JR, Oppenheim E, Zanetti KA, Mastri MG, et al. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene. 1998;210(2):315–24. doi: 10.1016/s0378-1119(98)00085-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Shi Q, Sturgis EM, Spitz MR, Wei Q. Polymorphisms and haplotypes of serine hydroxymethyltransferase and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Pharmacogenet Genomics. 2005;15(8):557–64. doi: 10.1097/01.fpc.0000170915.19522.b2. [DOI] [PubMed] [Google Scholar]
- 13.Wei Q, Cheng L, Hong WK, Spitz MR. Reduced DNA repair capacity in lung cancer patients. Cancer Res. 1996;56(18):4103–7. [PubMed] [Google Scholar]
- 14.Shen H, Wei Q, Pillow PC, Amos CI, Hong WK, Spitz MR. Dietary folate intake and lung cancer risk in former smokers: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2003;12(10):980–6. [PubMed] [Google Scholar]
- 15.Block G, Hartman AM, Dresser CM, Carroll MD, Gannon J, Gardner L. A data-based approach to diet questionnaire design and testing. Am J Epidemiol. 1986;124(3):453–69. doi: 10.1093/oxfordjournals.aje.a114416. [DOI] [PubMed] [Google Scholar]
- 16.Block G, Coyle LM, Hartman AM, Scoppa SM. Revision of dietary analysis software for the Health Habits and History Questionnaire. Am J Epidemiol. 1994;139(12):1190–6. doi: 10.1093/oxfordjournals.aje.a116965. [DOI] [PubMed] [Google Scholar]
- 17.U.S, Department of Agriculture, ARS. USDA Nutrient Database for Standard Reference, Release 14. Nutrient Data Laboratory Home Page. http://www.nal.usda.gov/fnic/foodcomp/2001.
- 18.Neumann AS, Lyons HJ, Shen H, Liu Z, Shi Q, Sturgis EM, et al. Methylenetetrahydrofolate reductase polymorphisms and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Int J Cancer. 2005;115(1):131–6. doi: 10.1002/ijc.20888. [DOI] [PubMed] [Google Scholar]
- 19.Akaike H. A new look at statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 20.Lu J, Wei Q, Bondy ML, Li D, Brewster A, Shete S, et al. Polymorphisms and haplotypes of the NBS1 gene are associated with risk of sporadic breast cancer in non-Hispanic white women <=55 years. Carcinogenesis. 2006;27(11):2209–16. doi: 10.1093/carcin/bgl077. [DOI] [PubMed] [Google Scholar]
- 21.Wang LE, Xiong P, Strom SS, Goldberg LH, Lee JE, Ross MI, et al. In vitro sensitivity to ultraviolet B light and skin cancer risk: a case-control analysis. J Natl Cancer Inst. 2005;97(24):1822–31. doi: 10.1093/jnci/dji429. [DOI] [PubMed] [Google Scholar]
- 22.Shi Q, Zhang Z, Li G, Pillow PC, Hernandez LM, Spitz MR, et al. Polymorphisms of methionine synthase and methionine synthase reductase and risk of lung cancer: a case-control analysis. Pharmacogenet Genomics. 2005;15(8):547–55. doi: 10.1097/01.fpc.0000170916.96650.70. [DOI] [PubMed] [Google Scholar]
- 23.Shi Q, Zhang Z, Li G, Pillow PC, Hernandez LM, Spitz MR, et al. Sex differences in risk of lung cancer associated with methylene-tetrahydrofolate reductase polymorphisms. Cancer Epidemiol Biomarkers Prev. 2005;14(6):1477–84. doi: 10.1158/1055-9965.EPI-04-0905. [DOI] [PubMed] [Google Scholar]
- 24.Shi Q, Zhang Z, Neumann AS, Li G, Spitz MR, Wei Q. Case-control analysis of thymidylate synthase polymorphisms and risk of lung cancer. Carcinogenesis. 2005;26(3):649–56. doi: 10.1093/carcin/bgh351. [DOI] [PubMed] [Google Scholar]
- 25.Smith AC, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, et al. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet. 1986;24(3):393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- 26.Heil SG, Van der Put NM, Waas ET, den Heijer M, Trijbels FJ, Blom HJ. Is mutated serine hydroxymethyltransferase (SHMT) involved in the etiology of neural tube defects? Mol Genet Metab. 2001;73(2):164–72. doi: 10.1006/mgme.2001.3175. [DOI] [PubMed] [Google Scholar]
- 27.Skibola CF, Smith MT, Hubbard A, Shane B, Roberts AC, Law GR, et al. Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood. 2002;99(10):3786–91. doi: 10.1182/blood.v99.10.3786. [DOI] [PubMed] [Google Scholar]
- 28.Hishida A, Matsuo K, Hamajima N, Ito H, Ogura M, Kagami Y, et al. Associations between polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and susceptibility to malignant lymphoma. Haematologica. 2003;88(2):159–66. [PubMed] [Google Scholar]
- 29.Chen J, Kyte C, Valcin M, Chan W, Wetmur JG, Selhub J, et al. Polymorphisms in the one-carbon metabolic pathway, plasma folate levels and colorectal cancer in a prospective study. Int J Cancer. 2004;110(4):617–20. doi: 10.1002/ijc.20148. [DOI] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Spitz MR, Amos CI, Lin J, Schabath MB, Wu X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004;64(6):2251–7. doi: 10.1158/0008-5472.can-03-2800. [DOI] [PubMed] [Google Scholar]
- 32.Bhatti P, Church DM, Rutter JL, Struewing JP, Sigurdson AJ. Candidate single nucleotide polymorphism selection using publicly available tools: a guide for epidemiologists. Am J Epidemiol. 2006;164(8):794–804. doi: 10.1093/aje/kwj269. [DOI] [PubMed] [Google Scholar]
- 33.Nakken S, Alseth I, Rognes T. Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes. Neuroscience. 2006 doi: 10.1016/j.neuroscience.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Liu H, Zhang Z, Spitz MR, Wei Q. Association of genetic variants of O6-methylguanine-DNA methyltransferase with risk of lung cancer in non-Hispanic Whites. Cancer Epidemiol Biomarkers Prev. 2006;15(12):2364–9. doi: 10.1158/1055-9965.EPI-06-0437. [DOI] [PubMed] [Google Scholar]