Abstract
Kinetoplast maxicircle DNA sequence organization was investigated in Leishmania donovani, strain 1S LdBob. Gene arrangement in the coding (conserved) region of the maxicircle is collinear with that of most trypanosomatids, with individual genes showing 80-90% nucleotide identity to Leishmania tarentolae, strain UC. The notable exception was an integration of a full-size minicircle sequence in the ND1 gene coding region found in L. donovani. Editing patterns of the mitochondrial mRNAs investigated also followed L. tarentolae UC patterns, including productive editing of the components of respiratory complexes III, IV and V, and ribosomal protein S12 (RPS12), as well as the lack of productive editing in five out of six pan-edited cryptogenes (ND3, ND8, ND9, G3, G4) found in these species. Several guide RNAs for the editing events were localized in minicircles and maxicircles in the locations that are conserved between the species. Mitochondrial activity, including rates of oxygen consumption, the presence and the levels of respiratory complexes and their individual subunits and the steady-state levels of several mitochondrial-encoded mRNAs were essentially the same in axenically grown amastigotes and in promastigotes of L. donovani. However, some modulation of mitochondrial activity between these developmental stages was suggested by the finding of an amastigote-specific component in Complex IV, a down-regulation of mitochondrial RNA-binding proteins (MRP) [define] and MRP-associated protein (MRP-AP) in amastigotes, and by variations in the levels of RPS12, ND3, ND9, G3 and G4 pre-edited transcripts.
Keywords: Amastigote, Kinetoplast DNA, Leishmania donovani, Mitochondrion, RNA editing
1. Introduction
The life cycle of dixenous Trypanosoma and Leishmania spp. includes two types of hosts (vertebrate and invertebrate) which represent drastically different types of environments. Adaptations that evolved in Trypanosoma brucei to meet the challenges for survival and propagation in its hosts include well documented reversible changes in mitochondrial metabolism, and are accompanied by far less understood changes in mitochondrial gene activity, including editing of mRNA (recently reviewed by Hannaert et al., 2003; Lukeš et al., 2005; Stuart et al., 2005; Bringaud et al., 2006; Fenn and Matthews, 2007). Thus, while insect stage trypanosomes have a fully functional oxidative phosphorylation system, in bloodstream trypanosomes the cytochrome c oxidase and bc1 complexes are no longer present and the function of ATP synthase functions to hydrolyze ATP in order to maintain a transmembrane potential (Clarkson et al., 1989; Bienen et al., 1991; Schnaufer et al., 2005; Vertommen et al., 2008). On the contrary, relatively little is known about changes in mitochondrial activity that might accompany differentiation of Leishmania during the insect (promastigote) and mammalian (intracellular amastigotes) stages of the life cycle. An early report indicated that promastigotes and lesion-derived amastigotes in Leishmania mexicana had similar oxygen uptake rates and sensitivity to inhibitors of the respiratory enzymes, indicating that oxidative phosphorylation was active in both developmental stages (Hart et al., 1981). The current view is generally that the metabolic changes between the stages of Leishmania are much less pronounced compared with T. brucei (Opperdoes and Coombs, 2007). Although a number of genes with stage-specific expression patterns were identified earlier (Bahr et al., 1993; Joshi et al., 1993; Charest and Matlashewski,1994; Kar et al., 2000; Nugent et al., 2004; Walker et al., 2006), the recent genome-wide transcriptome and proteome analyses revealed that only ∼3.5% of genes demonstrate a stage-specific expression pattern (Holzer et al., 2006; Leifso et al., 2007; Morales et al., 2008).
There is still some controversy with respect to the presence and function of NADH dehydrogenase (Complex I) in the respiratory chain at any stage in these organisms (Santhamma and Bhadurri, 1995; Bermúdez et al., 1997; Opperdoes and Michels, 2008). The investigated nuclear and mitochondrial genomes of several Leishmania species encode subunits of this complex (Simpson et al., 1998; Hertz-Fowler et al., 2004; Peacock et al., 2007), although expression of these genes has not yet been verified. In spite of the presence of these genes, the complex could not be visualized by Blue Native gel analysis of mitochondrial lysate from promastigotes of Leishmania amazonensis and Leishmania tarentolae (Maslov et al., 2002). Moreover, Complex I is not detected in L. tarentolae promastigotes by in gel activity staining or in vitro NADH-ubiquinone oxidoreductase activity measurements (A. Horváth, personal communication). Finally, a disruption of productive editing of several genes due to the loss of minicircle-encoded guide RNAs did not affect the viability of L. tarentolae cells in culture (Thiemann et al., 1994). These results suggests that, if this complex is indeed present, it is a small amount and/or is dispensable for proliferation of promastigotes, at least in culture. This also leaves open the possibility that the complex might be required in amastigotes or in some other life cycle stages, such as metacyclic promastigotes.
The mitochondrial genetic system, including RNA editing, has been we characterized only in one species of Leishmania, L. tarentolae, a parasite of geckos. Unfortunately, this organism has not been propagated as amastigotes, and that renders it unsuitable for investigation of mitochondrial adaptations during the life cycle. However, axenic cultivation of amastigotes of several human pathogenic species is possible by exposing the organisms to acidic pH and increased temperature (Bates, 1993). One such strain is a clonal derivative (LdBob) of the Leishmania donovani strain 1S (MHOM/SD/62/1S-Cl2D) (Joshi et al., 1993; Goyard et al., 2003). This species represents an important agent of human visceral leishmaniasis in the Old World. In addition, a large collection of minicircle sequences from this and closely related species (Leishmania infantum, Leishmania chagasi) species is already available in public databases and can be used to search for guide RNAs once the sequences of homologous pre-edited and edited maxicircle mRNA sequences are determined. The work presented below describes the initial characterization of the mitochondrial genome and RNA editing of L. donovani 1S LdBob, as well as an investigation of the possibility of mitochondrial gene regulation during the life cycle of these parasites.
2. Materials and methods
2. 1. Leishmania cultures and isolation of mitochondria
Promastigotes of L. donovani 1S clonal line LdBob were grown at 26 °C in M199 medium, while amastigotes of the same strain were cultivated at 37 °C with 5% CO2 in the ‘amastigote’ medium as described previously (Goyard et al., 2003). Promastigotes of L. tarentolae UC strain were cultivated in brain heart infusion medium supplemented with 10 μg/m hemin (Simpson and Braly, 1970). Mitochondria from all types of cells were isolated by hypotonic lysis followed by Renografin density gradient centrifugation (Braly et al., 1974). Typically 1-2 L cultures with cell densities of 20-40 × 106 cell m-1 were used to obtain ∼0.5 g of isolated mitochondria (wet weight).
2.2. Measurement of respiration rate
Oxygen uptake by L. donovani cells was measured with a biological oxygen monitor, YSI 5300, equipped with the YSI 5331 oxygen probe. The rate (expressed as μmol of O2 consumed per cell per min) was calculated by assuming the oxygen content of air-saturated Ringers solution of 0.227 mM at 28 °C or 0.2 mM at 37 °C (according to the YSI 5300 manual). KCN was used at 1 mM, and salicylhydroxamic acid (SHAM) at 0.1 mM. The cell concentration was 20–30 × 106 ml-1 for promastigotes and 2-6 × 106 ml-1 for amastigotes.
2.3. Protein electrophoretic and immunochemical procedures
Samples were analyzed by single-dimension Tris-glycine SDS-polyacrylamide gels (Laemmli, 1970) and two-dimensional Blue Native/Tris-tricine SDS-polyacrylamide gels (Schagger et al., 1994). Gel loading was normalized by using 70 × 106 cells or 50 μg of mitochondrial proteins, as appropriate. The resolved polypeptides were transferred onto nitrocellulose membranes by semidry blotting, as described previously (Horváth et al., 2000). After electrophoresis, the gels were stained either with Coomassie Brilliant Blue R250 (Sigma) or SYPRO Ruby (Molecular Probes). For autoradiography, dried gels were exposed to low energy screens and analyzed by using the PhosphorImager (Molecular Dynamics). Antibodies used were described previously: mouse antibody against L. tarentolae Rieske iron-sulfur protein of cytochrome bc1 (Neboháčová et al., 2004), the rabbit antibodies against L. tarentolae subunit IV of cytochrome c oxidase (Maslov et al., 2002), L. tarentolae p18 protein (subunit b of mitochondrial ATP synthase) (Bringaud et al., 1995), L. tarentolae MRP1/2 complex (formerly named Ltp26/28 complex) (Aphasizhev et al., 2003). Rabbit polyclonal antibodies against Leishmania major adenylate kinase 2 (AK2) were provided by D. Nierlich (University of California - Los Angeles, Los Angeles, USA). Mouse antibody against the amastigote-specific protein A2 family (Zhang et al., 1996) was provided by S. Beverley (Washington University, Missouri, USA). Western blots were processed by using the SuperSignal West Pico chemiluminescent system (Pierce).
2.4. DNA isolation, cloning and sequence analysis procedures
Kinetoplast DNA was isolated by sedimentation through a CsCI cushion as described previously (Simpson and Berliner, 1974). The maxicircle component was purified after release from kinetoplast DNA networks by digestion with EcoRI followed by equilibrium centrifugation in CsCI-Hoechst 33258 gradients (Simpson, 1979). In the initial stage of the project, the maxicircle DNA was randomly sheared to fragments of average size of 3 kb using a GeneMachine HydroShear, a computer-controlled repetitive syringe-driven device with an occlusion of about 10 μm (Thorstenson et al., 1998). The sheared fragments were cloned into the pCR2.1 vector (Invitrogen) using standard procedures. Subsequently, oligonucleotide primers were designed based on the sequence conservation between L. donovani and L. infantum and used to amplify several missing regions of the maxicircle. Gaps between the contigs were joined using direct sequencing of the respective PCR products. The final sequence was assembled using the ContigExpress program of Vector NTI (Invitrogen). Both strands of the maxicircle DNA were sequenced. The sequences were analyzed using Vector NTI programs. Guide RNA genes were searched for using the UWGCG program BESTFIT as previously described (Simpson et al., 1994; Maslov and Simpson, 2007).
2.5. RNA isolation and RT- PCR
Total cell RNA was isolated by Trizol extraction (Invitrogen) following the manufacturer's protocol. The same protocol was used for isolation of RNA from purified mitochondria. The isolated RNA was additionally treated with RNAse-free DNAse I (La Roche). cDNA was synthesized and amplified using SuperScript™ III One-Step reverse transcriptase (RT)-PCR System (Invitrogen). The amplification conditions included cDNA synthesis at 45 °C for 45 min, initial denaturation at 94 °C for 2 min, followed by 40 cycles composed of incubations at 94 °C for 15 s, 45 °C for 30 s and 65 °C for 1 min 30 s, concluding with incubation at 65 °C for 5 min. The oligonucleotides used for the amplification of L. donovani mitochondrial mRNA are listed in Supplementary Tabe S1. The amplified DNA was resolved in agarose gels, appropriate size products were extracted from electrophoretic gels using a QIAquick gel extraction kit (Qiagen), and cloned in a pT7Blue plasmid vector (EMD Bioscience). The recombinant plasmids were purified using a PureLink™ Quick Plasmid Miniprep kit (Invitrogen). DNA was sequenced by the UC Riverside Institute for Integrative Genome Biology Core Instrumentation facility (Riverside, California, USA) with an Applied Biosystems 3730xl DNA Sequencer.
2.6. Quantitative RT-PCR
The oligonucleotide primers were designed using Integrated DNA Technologies' Primer Quest online design tool (http://biotools.idtdna.com/Scitools/Applications/Primerquest/). Sequences of primers can be found in Supplementary Tabe S1. Four micrograms of total RNA were reverse transcribed by using random hexamers and SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen) according the manufacturer's protocol. Quantitative RT-PCR was performed by using an ABI Prism 7900HT sequence detection system (Applied Biosystems) and SYBR Green detection (QuantiTect SYBR Green PCR Kit, Qiagen) in triplicate according to the manufacturer's protocols. The PCR was performed as a 15-min step at 94 °C and 50 cycles of 94 °C for 15s and 57 °C for 1 min, ending with a slow heating step from 55 °C to 95 °C to generate the melting curve data. cDNA levels in each sample were calculated using standard curves generated from serial fivefold dilutions of pooled RNA. Each sample of cDNA was analyzed separately and normalized to endogenous controls, such as the L. donovani 24Sa ribosomal RNA gene (designated Ld24S) and the L. infantum 40S subunit ribosomal protein S12 gene (designated Li40Srp). The data are presented as a scattergram with the mean and the standard errors indicated on the graph. The ANOVA t-test was performed using Statview (version 5.0) (Abacus Concepts, Berkeley, CA) to compare differences among groups in gene expression. The differences were considered statistically significant at P < 0.05. The final data are expressed as an average relative expression of amastigotes versus promastigotes.
3. Results
3.1. Mitochondrial gene organization
The determined sequence of the maxicircle kinetoplast DNA of the strain L. donovani 1S LdBob covers almost the entire coding region of the molecule. The gene order with the 12S rRNA gene on one side and the ND5 gene on the other is collinear with that found in other trypanosomatids (Simpson et al., 1998). Gene sequences in L. donovani were, as expected, highly similar to L. tarentolae, with the nucleotide identity levels averaging close to 90%. Sequence conservation also included small pre-edited regions of 5′-edited and internally edited mitochondrial cryptogenes, such as ND7, COIII, Cyb, COII and MURF2, found in the same positions in both species. The high genomic sequence conservation extended to extensively edited pan-edited cryptogenes identifiable as intergenic G-rich regions: G1 (=ND8), G2 (=ND9), G3, G4, G5 (=ND3) and G6 (=RPS12), as well as the extensively edited 5′-region of A6, which were ∼80-90% identical, with RPS12 and A6 being the most conserved.
The only notable exception from the overall highly conserved architecture of the determined sequence was a full-length minicircle insertion (positions 7,913-8,697 of GenBank™ entry FJ416603) found in the 3′ region of the ND1 gene (see Supplementary Information). The insertion is 99.1 % identical to one of the known minicircle sequences from L. infantum (GenBank™ L19877) (Tripp et al., 1993). The putative integration sites on both molecules do not have any significant sequence similarity, leaving no obvious clue as to what might have triggered this recombination event. Such inserts have not been observed in other trypanosomatids studied to date. Moreover, PCR amplification and size analysis of the respective maxicircles region from L. donovani strains KA-Jeddah, THAK35 (LG10), GILANI (LG12), and L. infantum strains LPN114 (LG2), PM1 (LG3), IMT260 (LG6) (Lukes et al., 2007), as well as a partial DNA sequencing of the amplicons from the first two of these, showed that the minicircle insert is absent in these strains (see Supplementary Information). Therefore, this insert represents a peculiar feature of the strain 1S LdBob.
The integrated minicircle has all typical features including Conserved Sequence Blocks (CSB) 1-3, a discernable bend sequence region and a guide RNA gene that is properly positioned with respect to the CSB-3 region (Ray, 1989; Simpson, 1997). By showing a match with the edited sequences of L. tarentolae, this guide RNA might represent gND9-XIV. However, this could not be verified, because the cognate edited sequence is not available (see below).
The insertion interrupts the open reading frame (ORF) of the ND1 gene at the position 51 amino acids upstream of the conserved termination codon. Instead, the reading frame continues from the upstream coding sequence into the minicircle sequence for 92 amino acid residues and then terminates at a TAA codon. It is not known whether the resulting polypeptide product is functional. An ND1 gene product has not yet been detected in any trypanosomatid species. It also remains to be investigated how this insertion affects the processing of the ND1 mRNA.
3.2. Editing of mRNA
RNA editing patterns of several maxicircle transcripts have been investigated in promastigotes by RT-PCR amplification, cloning and sequencing using the experimental approach summarized previously (Simpson et al., 1996; Maslov and Simpson, 2007). In brief, the putative pre-edited regions were delineated by sequence comparison with maxicircle DNA from L. tarentolae. Oligonucleotide primers flanking pre-edited regions were used to obtain cDNA clones representing a mixture of pre-edited and various edited molecules. Edited sequences were aligned according to the known 3′-to-5′ progression of editing, and an edited sequence consensus was derived whenever possible. When translation of an edited consensus yielded a conserved amino acid sequence, that consensus was regarded as representing a mature (fully) edited mRNA pattern.
The fact that the editing leads to a noticeable net size increase of the mRNA allows identification of a sub-population of the PCR products enriched in edited cDNA molecules (Fig. 1). Edited RPS12, COIII, Cyb, ND7 and A6 cDNA molecules (Fig. 1A and B, arrows) were present in a relatively large amount among the amplified products, and the respective consensus edited patterns could be easily derived from the cloned sequences. Because the edited patterns and the respective derived amino acid products in each case were highly conserved with those in L. tarentolae, these patterns most likely represented functional mature edited mRNAs (Fig. 2A-E). A non-canonical intiation codon AUU (Ile) is present in the edited RPS12 mRNA of L. donovani instead of a canonical AUG (Met) observed in L. tarentolae (Fig. 2A). Putative non-canonical initiation codons were observed previously (Shaw et al., 1988; Landweber and Gilbert 1993; Maslov et al., 1999), although experimental proof of their utilization is still required. Nonetheless, it is interesting to note that a conserved in-frame AUU is also observed in the L. tarentolae RPS12 mRNA sequence upstream of the AUG codon (Fig. 2A).
Fig. 1.
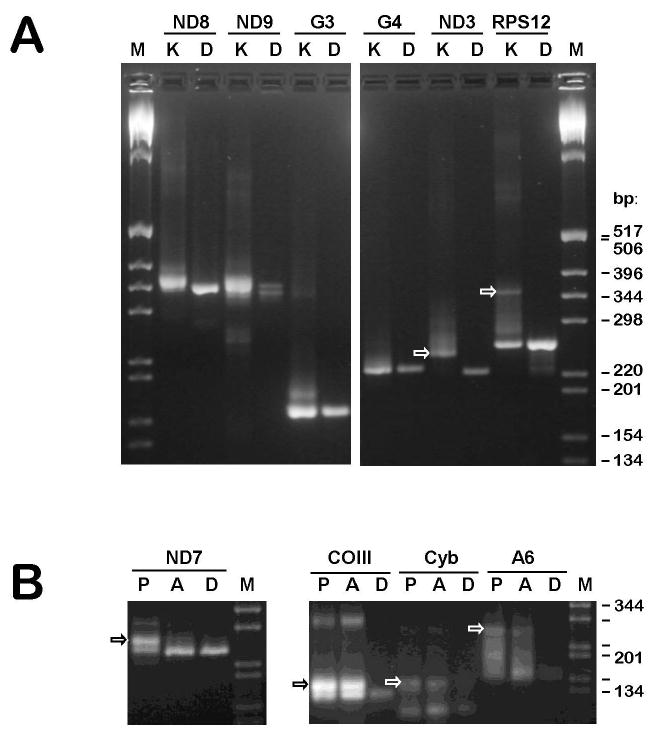
Reverse transcriptase-PCR analysis of RNA editing of several mitochondrial transcripts in promastigotes and axenic amastigotes of Leishmania donovani. (A) Amplification of mRNA derived from pan-edited cryptogenes ND8 (G1), ND9 (G2), G3, G4, ND3 (G5) and RPS12 (G6) using RNA isolated from purified kinetoplast-mitochondria of promastigotes, K. Control amplifications of the respective gene regions, D, were used to identify pre-edited cDNA products. Arrows indicate bands with the sizes expected for fully edited products that were used for cloning and sequencing. A 1 kb DNA ladder (Invitrogen) was used as a size marker, M. The amplified products were resolved in 3% NuSieve agarose gels. (B) Amplification of mRNA derived from 5′-edited and internally edited mRNA encoded by the genes ND7, COIII, Cyb and A6 using total cell RNA isolated from promastigotes (P) and axenically grown amastigotes (A).
Fig. 2.
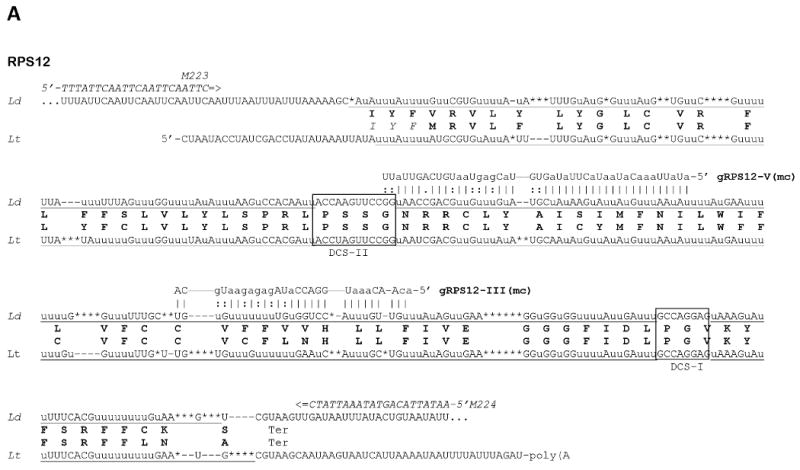
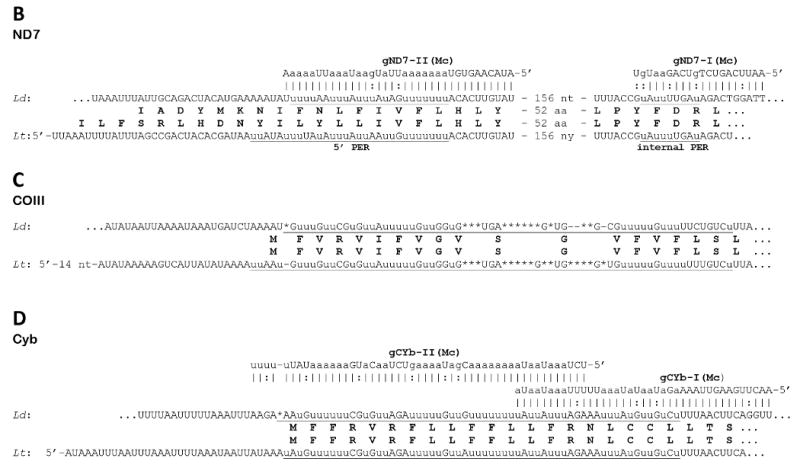

Sequences of edited mitochondrial mRNA in Leishmania donovani (Ld) and their comparison with respective transcripts of Leishmania tarentolae (Lt). Genomically encoded nucleotides are shown in upper case, inserted uridylates with lower case u, and deleted residues are indicated with asterisks. Complete editing sequence patterns are underlined. Gaps (shown with dashes) were introduced for the purpose of aligning the sequences. The predicted amino conserved acid sequences from both species are shown in boldface. The identified guide RNA molecules (mc - minicircle encoded, Mc - maxicircle encoded) are shown above the edited sequences of L. donovani with canonic bp shown with vertical lines (|) and G-U bp with colons and A-C mismatches with dots. The 5′ and 3′ ends of the L. donovani transcripts are not mapped. (A) The entire edited region of the pan-edited mRNA of mitochondrial ribosomal protein S12 (RPS12). Domain connection sequences (DCS) I and II are boxed. Notice that the translation sequence of L. tarentolae includes three additional conserved residues (italicized) which precede the proposed N-terminal methionine. (B) 5′-edited region (5′-ER) and internal edited region of the NADH dehydrogenase subunit 7 (ND7) mRNA. (C) 5′-edited region of the cytochrome c oxidase subunit III (COIII) mRNA. (D) 5′-edited region of the apocytochrome b (Cyb) mRNA. (E) A mature editing pattern of the first two out of six expected editing blocks of the 5′-edited mRNA of ATP synthase subunit 6 (A6). (F) Mis-editing of a partially edited transcript of the pan-edited ND3 mRNA. The transcript is correctly edited with cognate maxicircle-encoded gND3 block I guide RNA (gND3-l) and mis-edited further upstream by gND8 block XIV guide RNA (gND8-XIV) that is encoded by the minicircle insert discovered in the maxicircle of L. donovani 1S LdBob.
In the cases where a fully edited cDNA amplicon could be obtained by standard RT-PCR, this amplicon was observed with both promastigote and amastigote total cell RNA, although some difference in the relative amount was noticed for ND7 and, to a lesser extent, A6 (Fig. 1B). In the case of fully edited RPS12, efficient amplification of the full-length transcript (Fig. 1A) was only possible using mitochondrial mRNA.
Several putative guide RNA genes were identified using these edited mRNA patterns. Genes for both guide RNAs required for Cyb mRNA editing (gCyb-I and gCyb-II) and one gene for ND7 mRNA editing (gND7-I) were located in identical maxicircle locations as in L. tarentolae (Blum et al., 1990) and C. fasciculata (Van der Spek et al., 1991). The second guide RNA gene for ND7 editing (gND7-II) is expected to localize in a conserved position upstream of the 12S gene outside of the sequenced region. This was confirmed by investigating the respective locus of the maxicircle sequence from a closely related species, L. infantum (S. Beverley, personal communication). For each mRNA, the editing pattern observed is highly similar to that of L. tarentolae in (Fig. 2B and D).
A maxicircle-encoded putative COII guide RNA gene is identifiable at the 3′ end of the gene (not shown), as in other species, assuming that the edited COII pattern is as in L. tarentolae. Similarly, at least one of the two maxicircle-encoded MURF2 guide RNA genes (gMURF2-II), based on comparison with the edited L. tarentolae MURF2 mRNA sequence (not shown), is found in a conserved position.
We searched for minicircle-encoded guide RNAs using the edited mRNA sequences from L. donovani determined in this paper and minicircle DNA sequences from L. donovani and the closely related spp., L. infantum and L. chagasi, which are available from GenBank™. The identified genes included block I (GenBankTM entry AJ010082) and block II (X98347) guide RNAs for A6 mRNA editing (Fig. 2E), as well as guide RNA block III (AJ010079) and block V (multiple entries including AF167712-AF167717) of RPS12 mRNA (Fig. 2A). In each case a single guide RNA gene is located in a respective minicircle molecule downstream of the CSB-3 sequence (5′-GGGGTTGGTGA) at a distance of ∼0.45 kb on the opposite strand, in an arrangement that is similar to that in minicircles of L. tarentolae (Simpson, 1997). It is interesting to note that the extent of guide RNA overlaps and positions of block boundaries closely parallel those observed in L. tarentolae (Maslov and Simpson, 1992).
To date, only pre-edited and minimally edited transcripts were found for the pan-edited cryptogenes, ND8, ND9, G3 and G4, in cultured promastigotes and amastigotes of L. donovani. Upon completion of editing, these transcripts are nearly double the size of pre-edited transcripts in the L. tarentolae LEM 125 strain (Thiemann et al., 1994; Gao et al., 2001) and the same size increase is expected for L. donovani. However, no prominent cDNA band with such a significant size increase was found among the RT-PCR products even when purified mitochondrial RNA was used as template (Fig. 1A). The partially edited sequences obtained contained a variety of patterns that could not be reduced to a consensus (not shown). An interesting exception from this was G5. It can be seen in the gel representing the respective cDNA products that most of those (Fig. 1A, arrow) are noticeably larger than the size of the respective pre-edited molecule. Cloning and sequencing showed that these transcripts contained an identical short editing pattern at the very 3′ end, apparently representing a single block of editing (Fig. 2F). The respective maxicircle-encoded guide RNA gene (gND3-I) was found in the conserved position upstream of the 12S rRNA gene. Various editing patterns were seen further upstream of the correctly edited block I, and one of these patterns was attributable to mis-editing by the putative gND9-XIV guide RNA encoded by the aforementioned minicircle insert. An attempt was made to extend the determined correctly edited sequence further upstream by using a 3′ PCR primer annealing within the edited block I mRNA sequence. However, the clones obtained were still pre-edited or mis-edited (data not shown), indicating the lack of productive editing upstream of block I.
It is still unknown whether the apparent lack of productive editing of the ND8, ND9, G3 and G4 was due to technical problems, such as an inappropriate choice of the primer sequences, or to a low amount of fully-edited products. However, it is likely that this, together with the ND3 case, represents another example of the situation first observed with the old laboratory strain L. tarentolae UC (Thiemann et al., 1994), whence the lack of productive editing was due to the loss of respective minicircle-encoded guide RNAs.
3.3. Mitochondrial respiratory complexes in promastigotes and amastigotes
Oxygen uptake by in vitro grown promastigotes and amastigotes was investigated with the aim to verify the presence of an active electron transport chain in mitochondria of these cell types. No substantial difference in uptake was detected, with both forms consuming approximately 0.250-0.300 nmol O2 min-1 per 106 cells (Table 1). Only a relatively small decrease in uptake (∼16%) was observed in both forms in the presence of 0.1 mM SHAM, in line with the absence of a functional alternate oxidase in Leishmania (Opperdoes and Coombs, 2007). On the contrary, the respiration was completely (in amastigotes) or 95% (in promastigotes) inhibited by 1 mM KCN which, in accordance with earlier observations (Hart et al., 1981; Santhamma and Bhaduri, 1995; Bermudez et al., 1997) indicated an exclusive role of cytochrome c oxidase in oxygen consumption.
Table 1. Respiration of Leishmania donovani amastigote and promastigote cells.
The oxygen consumption is expressed as nanomole per min by 106 cells.
| Cells | No inhibitor | 1 mM KCN | 0.1 mM SHAM |
|---|---|---|---|
| amastigotes | 0.241 ±0.063 | Not detectable | 0.144 |
| promastigotes | 0.263 ±0.018 | 0.008 ±0.007 | 0.239 |
SHAM, salicylhydroxamic acid
The presence of respiratory complexes in isolated mitochondria from both types of cells was directly verified by two-dimensional Blue Native gel system (Schägger et al., 1994) (Fig. 3). The complexes observed were identified by their relative migration order in the native dimension and by characteristic subunit band patterns in the denaturing dimension, with both of these features being highly reminiscent of those observed in L. tarentolae and L. mexicana amazonensis (Maslov et al., 1999; Horváth et al., 2000; Maslov et al., 2002). The same set of complexes including cytochrome bc1 (Complex III), cytochrome c oxidase (Complex IV) and ATP synthase (Complex V, apparently oligomerizing) was detected in promastigotes and amastigotes. A noticeable difference between these cells was observed for the nuclear encoded subunit trCOVI of cytochrome c oxidase. This subunit represents the third largest visible band in the gel because the three mitochondrial encoded subunits COI, COII and COIII, the largest in the complex, are virtually undetectable by staining (Horváth et al., 2000). Thus, the largest visible band is actually subunit IV (trCOIV) (Maslov et al., 2002). In amastigotes, the subunit VI band is reduced in amount relative to the other subunits of the complex. Also, a different, slightly faster migrating band appears close to the original band. It still remains to be investigated whether the new band represents a completely different polypeptide or is due to a proteolytic cleavage of the original polypeptide. Similarly, the appearance of an amastigote-specific band in cytochrome c oxidase has been observed in L. mexicana amazonensis (Dmitri A. Maslov, unpublished observations).
Fig. 3.
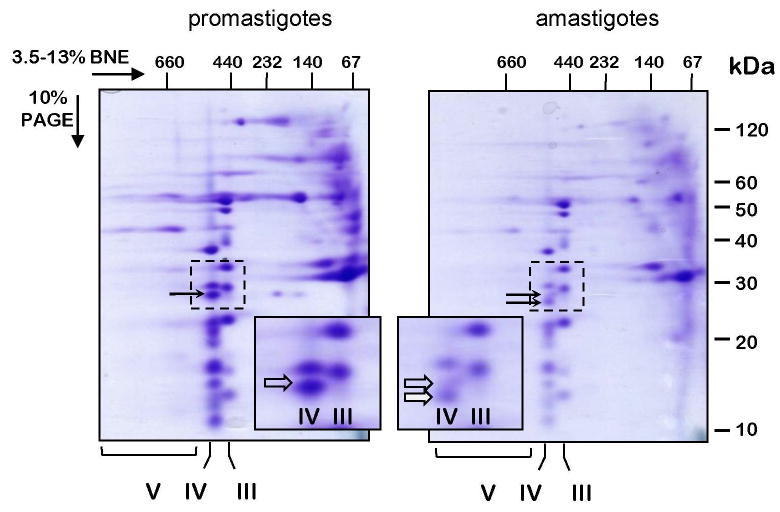
Separation of Leishmania donovani respiratory complexes in native gels. Mitochondria were isolated from promastigote and amastigote cells, lysed with 1% dodecyl maltoside and fractionated in 3.5-13% gradient Blue Native/10% Tris-tricine-SDS two-dimensional polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue R250. Positions of the respiratory Complex V (ATP synthase), Complex IV (cytochrome c oxidase) and Complex III (cytochrome bc1) are shown below the gel. The inset panels represent the gel areas (bordered with a dotted line) containing an amastigote-specific band of Complex IV (filled arrow). The promastigote-specific subunit trCOVI, which is highly reduced in amastigotes, is shown with a transparent arrow. The native gel dimension was calibrated with an HMW Native Marker Kit (GE Healthcare) and the denaturing dimension with BenchMark™ Pre-stained Protein Ladder (Invitrogen). The size markers are shown above and to the right of the gel panels.
In order to estimate relative amounts of the respiratory complexes in amastigotes and promastigotes, normalized loads of proteins in whole cell and mitochondrial lysates were resolved by gel electrophoresis and probed with polyclonal antibodies against L. tarentolae nuclear encoded subunits trCOIV (cytochrome c oxidase), Rieske (cytochrome bc1), subunit b (p18) (ATP synthase) and the intermembrane space marker AK2 (Fig. 4). Expression of the amastigote-specific A2 protein family (Zhang et al., 1996) was used as a differentiation control (Fig. 4C). No significant difference in Western blot signal intensity was observed for the mitochondrial proteins tested, suggesting an approximately equal content of these components and, therefore, of the respective complexes in both types of cells.
Fig. 4.
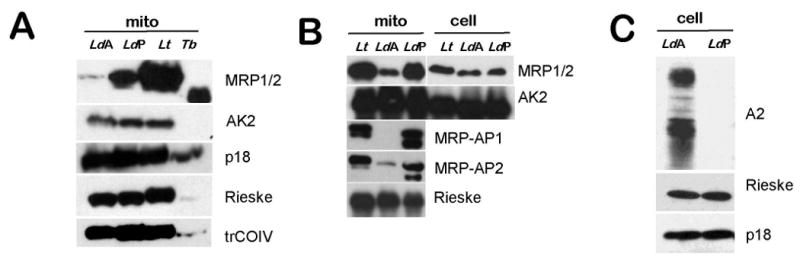
Protein levels of mitochondrial proteins in different trypanosomatids. (A) Mitochondrial extracts from Leishmania donovani amastigote (LdA), L. donovani promastigote (LdP), Leishmania tarentolae (Lt) and Trypanosoma brucei (Tb) cells were analyzed by Western blot. The antisera used are shown to the right of each panel. (B) Mitochondrial extracts and whole cell lysates were analyzed by Western blotting with antibodies against Mitochondrial RNA-binding proteins 1 and 2 (MRP1/2) and adenylate kinase 2 (AK2). In addition, mitochondrial extract were probed for MRP-AP1, MRP-AP2 and Rieske proteins. (C) Differentiation of L. donovani was verified by probing cell lysates for an amastigote-specific marker A2. Notice that this protein is represented by multiple bands (Charest and Matlashewski, 1994). Rieske and p18 proteins were probed as indicated. Total cell lysate loading was normalized by using the same number of cells (70 × 106) per lane and loading of mitochondrial lysates was normalized by using 50 μg protein per lane.
3.4. RNA editing and mRNA levels in promastigotes and amastigotes
The results described above have demonstrated that mitochondria remain active in amastigotes as they are in promastigotes. To investigate whether some modulation of mitochondrial gene expression still might take place during the development of these organisms, we compared the levels of several mitochondrial transcripts by quantitative RT-PCR (qRT-PCR). Since nuclear genome expression in trypanosomatids is largely constitutive (Holzer et al., 2006; Leifso et al., 2007; Morales et al., 2008), we used two nuclear genes encoding components of the cytoplasmic ribosomes, 24Sa ribosomal RNA of L. donovani (GenBankTM entry L19408) and ribosomal protein S12 of L. infantum (GeneDB entry LinJ13_V3.0460). The results, presented in Fig. 5A and B, were essentially the same with both normalization procedures. Analysis of the β-tubulin gene (LinJ33.0830) used as a control showed a reduced level in amastigotes, as expected (Rainey et al., 1991; Mojtahedi et al., 2008). The levels of immature (pre-edited) mRNAs encoded by six pan-edited cryptogenes were found to display various patterns. One of these transcripts (ND8) remained unchanged, three transcripts (ND9, G4 and ND3) were up-regulated in promastigotes, and two transcripts (G3 and RPS12) were up-regulated in amastigotes. Remarkably, unlike the pre-edited mRNAs, the levels of the edited RPS12 transcripts were approximately equal in both stages. Edited sequences for the other pan-edited mRNAs are not available so their levels could not be investigated. The levels of the 12S and 9S mitochondrial ribosomal RNAs also remained unchanged, as were levels of the unedited (‘never-edited’) COI mRNA. Thus, in the four functional transcripts tested (RPS12 edited, COI, 12S and 9S), the levels were approximately constant. The levels of edited ND7 and A6 transcripts which might differ in promastigotes and amastigotes (Fig. 1B) were not investigated.
Fig. 5.
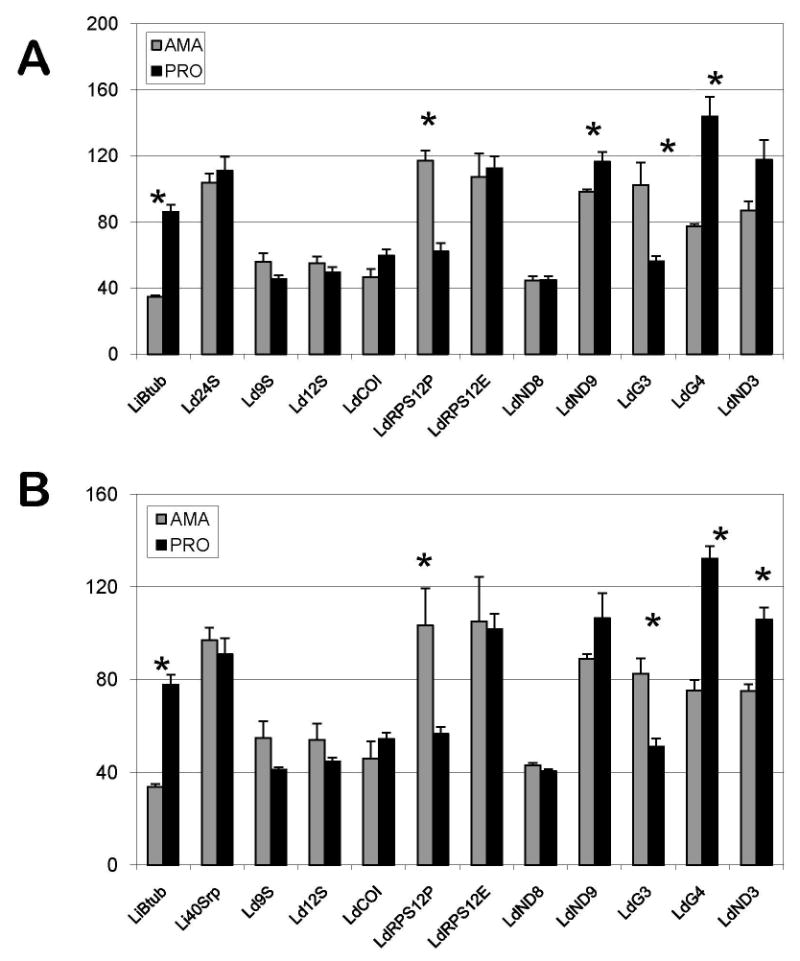
Quantitative reverse transcriptase-PCR analysis of nuclear and mitochondrial transcripts in amastigotes and promastigotes of Leishmania donovani. The columns represent averages of three replicates normalized to mRNA of Leishmania infantum cytoplasmic 40S subunit ribosomal protein S12 (A), designated Li40Srp, and L. donovani 24Sa ribosomal RNA (B), designated Ld24S. Other designations for L. donovani transcripts are: Ld9S and Ld12S - mitochondrial ribosomal 9S and 12S RNA, respectively; LdRPS12P and LdRPS12E - pre-edited and edited mRNA, respectively, of mitochondrial ribosomal protein S12; LdCOI - cytochrome c oxidase subunit I mRNA; LdND3, LdND8 and LdND9 -subunits 3, 8 and 9 of NADH dehydrogenase mRNA; LdG3 and LdG4 - pan-edited cryptogenes G3 and G4 mRNA. LiBtub - L. infantum (β-tubulin mRNA. The vertical axis shows the relative abundance of the respective mRNAs in amastigotes (grey) and promastigotes (black) with a standard error as shown. The asterisks positioned above some columns (β-tubulin, pre-edited mitochondrial RPS12, ND9, G3, G4 and ND3 transcripts) indicate that the observed differences in relative amount statistically significant (with P < 0.05, derived from Student's test).
This constant level of the edited RPS12 mRNA together with the relative decrease in the ratio of pre-edited to edited RPS12 mRNA in promastigotes, along with the differences in levels of other pre-edited mRNAs, suggest a differential stability of pre-edited transcripts in these cells. The MRP1/2 complex has been implicated in the stability of several mRNA species in T. brucei (Zíková et al., 2006, 2008). To this end, we compared the levels of MRP1/2 and MRP Associated Proteins 1 and 2 (AP1 and AP2) proteins in promastigotes and amastigotes by Western blot analysis. There was no noticeable difference in the levels of the MRP1/2 proteins in total cell lysate (Fig. 4B), however the levels in isolated mitochondria differed significantly, with a highly reduced amount of this complex in amastigotes (Fig. 4A, B). The mitochondrial levels of AP1 and AP2 proteins were also highly reduced in amastigotes compared with promastigotes (Fig. 4B). The levels in total cell lysates could not be investigated.
4. Discussion
In this work we investigated RNA editing and mitochondrial gene expression in L. donovani. To our knowledge, this represents the only medically important Leishmania species for which such analyses have been performed to date. We also employed axenically grown amastigotes of the strain 1S LdBob to investigate the possibility of mitochondrial gene regulation during the life cycle of these cells. Our main finding was that, although there are some differences between these two life cycle stages, the investigated aspects of mitochondrial activity are largely constitutive. Thus, both forms possess cyanide-sensitive respiration that is due to the constitutive presence of cytochrome c oxidase (Complex IV), as well as the respiratory Complexes III and V. The levels of several nuclear encoded subunits tested (trCOIV of cytochrome c oxidase, Rieske subunit of cytochrome bc1, subunit b (p18) of ATP synthase) also do not show noticeable changes in amastigotes compared with promastigotes. Although we have not directly investigated levels of the mitochondrial-encoded subunits of these complexes, we have obtained indirect evidence that their expression is also likely constitutive. The evidence represents the qRT-PCR data demonstrating that the components of mitochondrial ribosomes (12S and 9S rRNA, and edited mRNA for ribosomal protein S12), which are necessary for the synthesis of mitochondria-encoded polypeptides, are constitutively expressed, as is the mRNA for mitochondrial subunit COI of cytochrome c oxidase.
The functionality of the mitochondrial respiratory chain in promastigotes and amastigotes of L. donovani is further corroborated by the presence of fully edited transcripts for the subunits of these complexes: edited COIII, Cyb and A6 are present in both stages, although we have not compared their relative levels.
The finding of an amastigote-specific band among the nuclear subunits of cytochrome c oxidase indicates that, in addition to its constitutive functionality, some modulation of its activity might be taking place in amastigotes.
By the contrast with the subunits of Complexes III, IV and V, the expression of some mitochondrial-encoded products, especially the putative subunits of NADH dehydrogenase (Complex I), has not been convincingly demonstrated in both promastigotes and amastigotes. In general, the existence of this complex in trypanosomatids is controversial and its importance unclear (Hernandez and Turrens, 1998; Allemann and Schneider, 2000; Opperdoes and Michels, 2008). It has been proposed that, when present, the trypanosomatid complex does not perform proton-pumping across the inner membrane but is only involved in regeneration of mitochondrial NAD+ (Opperdoes and Michels, 2008). An enzymatically active NADH dehydrogenase complex has been detected in T. brucei (Fang et al., 2001) and Phytomonas serpens (Čermáková et al., 2007; González-Halphen and Maslov, 2004), organisms that either temporarily (bloodstream trypanosomes) (Vickerman, 1994) or permanently (Phytomonas spp.) (Nawathean and Maslov, 2000) lack a cytochrome-mediated electron transport chain. In these organisms, Complex I might be required only at specific stages of the natural life cycle, as proposed during the transitional metabolism in short stumpy trypanosomes (Bienen et al., 1991). At other stages or in culture this complex might be fully dispensable.
The repression of cytochrome-containing complexes or the existence of viable natural mutants similar to dyskinetoplastic trypanosomes are not known for Leishmania (Hannaert et al., 2003; Opperdoes and Coombs, 2007), and the role of Complex I is even less clear. Earlier inhibition studies indicated that this complex is absent in L. donovani (Santhamma and Bhadurri, 1995), albeit present in L. mexicana (Bermúdez et al., 1997). Yet, the nuclear-encoded homologs of Complex I subunits are present in the three Leishmania genomes (L. major, L. infantum, L. braziliensis) investigated (Peacock et al., 2007). This evolutionary conservation strongly indicates that this complex is functional at least during some stages of the Leishmania life cycle.
It must be mentioned that the Leishmania natural life cycle includes a succession of three replicating stages (amastigotes, procyclic promastigotes, leptomonad promastigotes) and two non-replicating stages (nectomonad and metacyclic promastigotes) (Sacks, 2001; Gossage et al., 2003). To date, only the first two of these stages in L. donovani are amenable to biochemical analysis by availability in culture (Goyard et al., 2003; Debrabant et al., 2004). The Blue Native gel results presented in this paper suggest that Complex I, if present in cultured promastigotes and amastigotes, must be in present in a relatively low amount compared with respiratory complexes III, IV and V. Moreover, expression of some of its subunits seems to be impaired in the 1S LDBob strain of L. donovani: a minicircle insert interrupts the reading frame of ND1, and no productively edited mRNAs could be found for the ND3, ND8 and ND9 subunits. Taken together, these results indicate that Complex I in Leishmania is fully dispensable in culture. This suggests that the lack of any selective pressure to preserve the integrity of editing cascades for the pan-edited cryptogenes, ND3, ND8, ND9, and also possibly G3 and G4, encoding components of Complex I, is then responsible for the absence of the respective fully edited mRNAs in the 1S LdBob strain of L. donovani. This case may be analogous to the disruption of productive editing for the same set of cryptogenes that was observed in L. tarentolae UC (Thiemann et al., 1994) and for the ND8 cryptogene in P. serpens 1G (Maslov et al., 1998). It was shown that at least in L. tarentolae UC, these defects were caused by the loss of minicircle-encoded guide RNAs that resulted from an intrinsic randomness of minicircle segregation in the absence of a selective pressure (Simpson et al., 2000). Characteristically, more recently isolated strains of L. tarentolae and P. serpens did show productive editing of the respective mRNAs (Nawathean and Maslov, 2000; Gao et al., 2001) suggesting that the random loss of minicircle classes has not yet proceeded as far as it did in the older laboratory strains. It is remarkable that the ND7 mRNA is still productively edited with the aid of two cognate maxicircle-encoded guide RNAs. Moreover, the first block of editing in the ND3 mRNA is fully completed by a cognate maxicircle-encoded guide RNA. These results show that the potential of the enzymatic machinery to process mitochondrial-encoded Complex I mRNA is intact, and when guide RNAs are still present (because they are encoded by a maxicircle) there is productive editing. Therefore, in cases when no Complex I mRNA editing is found it must be due to the lack of the respective guide RNAs, as in L. tarentolae UC. To verify whether minicircle loss did occur in L. donovani strain 1S LdBob, the minicircle content of this strain needs to be investigated and compared with that of a recently isolated strain, as well as the extent of RNA editing in the latter.
Acknowledgments
The strain L. donovani 1S LdBob and the amastigote-specific monoclonal antibodies were kindly provided by S. Beverley. We also thank J. Lukes for other strains of L. donovani and L. infantum, C. Brunk for the permission to use the DNA shearing device, J. Lusis for help with qPCR and A. Horváth for discussions. This work was supported by the National Institutes of Health grant AI070927 to D.A.M.
Footnotes
Note: Nucleotide sequence data reported in this work are available in the GenBank™ database under the accession number FJ416603.
Note: Supplementary information associated with this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allemann N, Schneider A. ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Mol Biochem Parasitol. 2000;111:87–94. doi: 10.1016/s0166-6851(00)00303-0. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr V, Stierhof YD, Ilg T, Demar M, Quinten M, Overath P. Expression of lipophosphoglycan, high-molecular weigth phosphoglycan and glycoprotein 63 in promastigotes and amastigotes of Leishmania mexicana. Mol Biochem Parasitol. 1993;58:107–122. doi: 10.1016/0166-6851(93)90095-f. [DOI] [PubMed] [Google Scholar]
- Bates PA. Axenic culture of Leishmania amastigotes. Parasitol Today. 1993;9:143–146. doi: 10.1016/0169-4758(93)90181-e. [DOI] [PubMed] [Google Scholar]
- Bermúdez R, Dagger F, D'Aquino JA, Benaim G, Dawidowicz K. Characterization of mitochondrial electron-transfer in Leishmania mexicana. Mol Biochem Parasitol. 1997;90:43–54. doi: 10.1016/s0166-6851(97)00131-x. [DOI] [PubMed] [Google Scholar]
- Bienen EJ, Saric M, Pollakis G, Grady RW, Clarkson AB., Jr Mitochondrial development in Trypanosoma brucei brucei transitional bloodstream forms. Mol Biochem Parasitol. 1991;45:185–192. doi: 10.1016/0166-6851(91)90085-k. [DOI] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Braly P, Simpson L, Kretzer F. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J Protozool. 1974;21:782–790. doi: 10.1111/j.1550-7408.1974.tb03752.x. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Peris M, Zen KH, Simpson L. Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae. Mol Biochem Parasitol. 1995;71:65–79. doi: 10.1016/0166-6851(95)00023-t. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Čermáková P, Verner Z, Man P, Lukeš J, Horváth A. Characterization of the NADH:ubiquinone oxidoreductase (complex I) in the trypanosomatid Phytomonas serpens (Kinetoplastida) FEBS J. 2007;274:3150–3158. doi: 10.1111/j.1742-4658.2007.05847.x. [DOI] [PubMed] [Google Scholar]
- Charest H, Matlashewski G. Developmental gene expression in Leishmania donovani: differential cloning and analysis of an amastigote-stage-specific gene. Mol Cell Biol. 1994;14:2975–2984. doi: 10.1128/mcb.14.5.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson AB, Bienen EJ, Pollakis G, Grady RW. Respiration of bloodstream forms of the parasite Trypanosoma brucei brucei is dependent on a plant-like alternative oxidase. J Biol Chem. 1989;264:17770–17776. [PubMed] [Google Scholar]
- Debrabant A, Joshi MB, Pimenta PF, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. Int J Parasitol. 2004;34:205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Fang J, Wang YD, Beattie DS. Isolation and characterization of complex I, rotenone-sensitive NADH:ubiquinone oxidoreductase, from the procyclic forms of Trypanosoma brucei. Eur J Biochem. 2001;268:3075–3082. doi: 10.1046/j.1432-1327.2001.02205.x. [DOI] [PubMed] [Google Scholar]
- Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GG, Kapushoc ST, Simpson AM, Thiemann OH, Simpson L. Guide RNAs of the recently isolated LEM125 strain of Leishmania tarentolae: An unexpected complexity. RNA. 2001;7:1335–1347. doi: 10.1017/s1355838201018076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Halphen D, Maslov DA. NADH-ubiquinone oxidoreductase activity in the kinetoplasts of the plant trypanosomatid Phytomonas serpens. Parasitol Res. 2004;92:341–346. doi: 10.1007/s00436-003-1058-4. [DOI] [PubMed] [Google Scholar]
- Gossage SM, Rogers ME, Bates PA. Two separate growth phases during the development of Leishmania in sand flies: implications for understanding the life cycle. Int J Parasitol. 2003;33:1027–1034. doi: 10.1016/s0020-7519(03)00142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyard S, Segawa H, Gordon J, Showalter M, Duncan R, Turco SJ, Beverley SM. An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol. 2003;130:31–42. doi: 10.1016/s0166-6851(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Hannaert V, Bringaud F, Opperdoes FR, Michels PA. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis. 2003;2:11. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart DT, Vickerman K, Coombs GH. Respiration of Leishmania mexicana amastigotes and promastigotes. Mol Biochem Parasitol. 1981;4:39–51. doi: 10.1016/0166-6851(81)90027-x. [DOI] [PubMed] [Google Scholar]
- Hernandez FR, Turrens JF. Rotenone at high concentrations inhibits NADH-fumarate reductase and the mitochondrial respiratory chain of Trypanosoma brucei and T-cruzi. Mol Biochem Parasitol. 1998;93:135–137. doi: 10.1016/s0166-6851(98)00015-2. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, Parkhill J, Ivens AC, Rajandream MA, Barrell B. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucl Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Horváth A, Kingan TG, Maslov DA. Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. J Biol Chem. 2000;275:17160–17165. doi: 10.1074/jbc.M907246199. [DOI] [PubMed] [Google Scholar]
- Joshi M, Dwyer DM, Nakhasi HL. Cloning and characterization of differentially expressed genes from in vitro-grown ‘amastigotes’ of Leishmania donovani. Mol Biochem Parasitol. 1993;58:345–354. doi: 10.1016/0166-6851(93)90057-5. [DOI] [PubMed] [Google Scholar]
- Kar S, Soong L, Colmenares M, Goldsmith-Pestana K, Mahon-Pratt D. The immunologically protective P-4 antigen of Leishmania amastigotes. A developmentally regulated single strand-specific nuclease associated with the endoplasmic reticulum. J Biol Chem. 2000;275:37789–37797. doi: 10.1074/jbc.M002149200. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landweber LF, Gilbert W. RNA editing as a source of genetic variation. Nature. 1993;363:179–182. doi: 10.1038/363179a0. [DOI] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KW, Jirků M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci USA. 2007;104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, Hollar L, Haghighat P, Nawathean P. Demonstration of mRNA editing and localization of guide RNA genes in kinetoplast-mitochondria of the plant trypanosomatid Phytomonas serpens. Mol Biochem Parasitol. 1998;93:225–236. doi: 10.1016/s0166-6851(98)00028-0. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Nawathean P, Scheel J. Partial kinetoplast-mitochondrial gene organization and expression in the respiratory deficient plant trypanosomatid Phytomonas serpens. Mol Biochem Parasitol. 1999;99:207–221. doi: 10.1016/s0166-6851(99)00028-6. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Simpson L. Strategies of kinetoplastid cryptogene discovery and analysis. In: Gott JM, editor. RNA Editing, Methods in Enzymology. Vol. 424. Academic Press; San Diego: 2007. pp. 127–139. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Zíková A, Kyselová I, Lukeš J. A putative novel nuclear-encoded subunit of the cytochrome c oxidase complex in trypanosomatids. Mol Biochem Parasitol. 2002;125:113–125. doi: 10.1016/s0166-6851(02)00235-9. [DOI] [PubMed] [Google Scholar]
- Mojtahedi Z, Clos J, Kamali-Sarvestani E. Leishmania major: Identification of developmentally regulated proteins in procyclic and metacyclic promastigotes. Exp Parasitol. 2008;119:422–429. doi: 10.1016/j.exppara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Morales MA, Watanabe R, Laurent C, Lenormand P, Rousselle JC, Namane A, Spath GF. Phosphoproteomic analysis of Leishmania donovani pro- and amastigote stages. Proteomics. 2008;8:350–363. doi: 10.1002/pmic.200700697. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Maslov DA. The absence of genes for cytochrome c oxidase and reductase subunits in maxicircle kinetoplast DNA of the respiration-deficient plant trypanosomatid Phytomonas serpens. Curr Genet. 2000;38:95–103. doi: 10.1007/s002940000135. [DOI] [PubMed] [Google Scholar]
- Neboháčová M, Maslov DA, Falick AM, Simpson L. The effect of RNA interference down-regulation of RNA editing 3′-terminal uridylyl transferase (TUTase) 1 on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J Biol Chem. 2004;279:7819–7825. doi: 10.1074/jbc.M311360200. [DOI] [PubMed] [Google Scholar]
- Nugent PG, Karsani SA, Wait R, Tempero J, Smith DF. Proteomic analysis of Leishmania mexicana differentiation. Mol Biochem Parasitol. 2004;136:51–62. doi: 10.1016/j.molbiopara.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Coombs GH. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 2007;23:149–158. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Michels PA. Complex I of Trypanosomatidae: does it exist? Trends Parasitol. 2008 doi: 10.1016/j.pt.2008.03.013. in press. [DOI] [PubMed] [Google Scholar]
- Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, Kerhornou A, Ivens A, Fraser A, Rajandream MA, Carver T, Norbertczak H, Chillingworth T, Hance Z, Jagels K, Moule S, Ormond D, Rutter S, Squares R, Whitehead S, Rabbinowitsch E, Arrowsmith C, White B, Thurston S, Bringaud F, Baldauf SL, Faulconbridge A, Jeffares D, Depledge DP, Oyola SO, Hilley JD, Brito LO, Tosi LR, Barrell B, Cruz AK, Mottram JC, Smith DF, Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PM, Spithill TW, McMahon-Pratt D, Pan AA. Biochemical and molecular characterization of Leishmania pifanoi amastigotes in continuous axenic culture. Mol Biochem Parasitol. 1991;49:111–118. doi: 10.1016/0166-6851(91)90134-r. [DOI] [PubMed] [Google Scholar]
- Ray D. Conserved sequence blocks in kinetoplast DNA minicircles from diverse species of trypanosomes. Mol Cell Biol. 1989;9:1365–1367. doi: 10.1128/mcb.9.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DL. Leishmania-sand fly interactions controlling species-specific vector competence. Cell Microbiol. 2001;3:189–196. doi: 10.1046/j.1462-5822.2001.00115.x. [DOI] [PubMed] [Google Scholar]
- Santhamma KR, Bhaduri A. Characterization of the respiratory chain of Leishmania donovani promastigotes. Mol Biochem Parasitol. 1995;75:43–53. doi: 10.1016/0166-6851(95)02510-3. [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker CD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Feagin JE, Stuart K, Simpson L. Editing of mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- Simpson L. Isolation of maxicircle component of kinetoplast DNA from hemoflagellate protozoa. Proc Natl Acad Sci USA. 1979;76:1585–1588. doi: 10.1073/pnas.76.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L. The genomic organization of guide RNA genes in kinetoplastid protozoa: Several conundrums and their solutions. Mol Biochem Parasitol. 1997;86:133–141. doi: 10.1016/s0166-6851(97)00037-6. [DOI] [PubMed] [Google Scholar]
- Simpson L, Berliner J. Isolation of the kinetoplast DNA of Leishmania tarentolae in the form of a network. J Protozool. 1974;21:382–393. doi: 10.1111/j.1550-7408.1974.tb03675.x. [DOI] [PubMed] [Google Scholar]
- Simpson L, Braly P. Synchronization of Leishmania tarentolae by hydroxyurea. J Protozool. 1970;17:511–517. doi: 10.1111/j.1550-7408.1970.tb04719.x. [DOI] [PubMed] [Google Scholar]
- Simpson L, Frech GC, Maslov DA. RNA Editing in Trypanosomatid Mitochondria. In: Attardi GM, Chomyn A, editors. Methods in Enzymology. Vol. 264. Academic Press; San Diego: 1996. pp. 99–121. [DOI] [PubMed] [Google Scholar]
- Simpson L, Simpson AM, Blum B. RNA editing in mitochondria. In: Higgins SJ, Hames BD, editors. RNA Processing. IRL Press; Oxford: 1994. pp. 69–105. [Google Scholar]
- Simpson L, Thiemann OH, Savill NJ, Alfonzo JD, Maslov DA. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci USA. 2000;97:6986–6993. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Wang SH, Thiemann OH, Alfonzo JD, Maslov DA, Avila HA. U-insertion/deletion Edited Sequence Database. Nucl Acids Res. 1998;26:170–176. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Thiemann OH, Maslov DA, Simpson L. Disruption of RNA editing in Leishmania tarentolae by the loss of minicircle-encoded guide RNA genes. EMBO J. 1994;13:5689–5700. doi: 10.1002/j.1460-2075.1994.tb06907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstenson YR, Hunicke-Smith SP, Oefner PJ, Davis RW. An automated hydrodynamic process for controlled, unbiased DNA shearing. Genome Res. 1998;8:848–855. doi: 10.1101/gr.8.8.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp CA, Myler PJ, Stuart KD. Nucleotide sequence of a minicircle from Leishmania infantum. Mol Biochem Parasitol. 1993;62:319–320. doi: 10.1016/0166-6851(93)90122-e. [DOI] [PubMed] [Google Scholar]
- Van der Spek H, Arts GJ, Zwaal RR, Van den Burg J, Sloof P, Benne R. Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J. 1991;10:1217–1224. doi: 10.1002/j.1460-2075.1991.tb08063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertommen D, Van RJ, Szikora JP, Rider MH, Michels PA, Opperdoes FR. Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol Biochem Parasitol. 2008;158:189–201. doi: 10.1016/j.molbiopara.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Vickerman K. The evolutionary expansion of the trypanosomatid flagellates. Int J Parasitol. 1994;24:1317–1331. doi: 10.1016/0020-7519(94)90198-8. [DOI] [PubMed] [Google Scholar]
- Walker J, Vasquez JJ, Gomez MA, Drummelsmith J, Burchmore R, Girard I, Ouellette M. Identification of developmentally-regulated proteins in Leishmania panamensis by proteome profiling of promastigotes and axenic amastigotes. Mol Biochem Parasitol. 2006;147:64–73. doi: 10.1016/j.molbiopara.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Zhang WW, Charest H, Ghedin E, Matlashewski G. Identification and overexpression of the A2 amastigote-specific protein in Leishmania donovani. Mol Biochem Parasitol. 1996;78:79–90. doi: 10.1016/s0166-6851(96)02612-6. [DOI] [PubMed] [Google Scholar]
- Ziková A, Horáková E, Jirků M, Dunajčíková P, Lukeš J. The effect of down-regulation of mitochondrial RNA-binding proteins MRP1 and MRP2 on respiratory complexes in procyclic Trypanosoma brucei. Mol Biochem Parasitol. 2006;149:65–73. doi: 10.1016/j.molbiopara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zíková A, Kopečná J, Schumacher MA, Stuart K, Trantírek L, Lukeš J. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int J Parasitol. 2008;38:901–912. doi: 10.1016/j.ijpara.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]


