Abstract
To elucidate the role of the histone tails in chromatin compaction and in higher-order folding of chromatin under physiological conditions, we extend a mesoscale model of chromatin [Arya, Zhang, and Schlick, Biophys. J. 91, 133 (2006); Arya and Schlick, Proc. Natl. Acad. Sci. USA 103, 16236 (2006)] to account for divalent cations (Mg2+) and linker histones. Configurations of 24-nucleosome oligonucleosomes in different salt environments and in the presence and absence of linker histones are sampled by a mixture of local and global Monte Carlo methods. Analyses of the resulting ensembles reveals a dynamic synergism between the histone tails, linker histones, and physiological ions in forming compact higher-order structures of chromatin. In the presence of monovalent salt alone, oligonucleosomes remain relatively unfolded and the histone tails do not mediate many internucleosomal interactions. Upon the addition of linker histones and divalent cations, the oligonucleosomes undergo a significant compaction triggered by: a dramatic increase in the internucleosomal interactions mediated by the histone tails; formation of a rigid linker DNA “stem” around the linker histones' C-terminal domains; and reducion in the electrostatic repulsion between linker DNAs via sharp bending in some linker DNAs caused by the divalent cations. Among all histone tails, the H4 tails mediate the most internucleosomal interactions, consistent with experimental observations, followed by the H3, H2A, and H2B tails in decreasing order. Apart from mediating internucleosomal interactions, the H3 tails also contribute to chromatin compaction by attaching to the entering and exiting linker DNA to screen electrotatic repulsion among the linker DNAs. This tendency of the H3 tails to attach to linker DNA, however, decreases significantly upon the addition of linker histones due to competition effects. The H2A and H2B tails do not mediate significant internucleosomal interactions but are important for mediating fiber/fiber intractions, especially in relatively unfolded chromatin in monovalent salt environments.
Keywords: chromatin, histone tails, nucleosome, DNA, linker histone, Monte Carlo
1 Introduction
All eukaryotic organisms store their genome (DNA) inside cell nuclei in the form of chromatin. Chromatin is the array of nucleosomes—cylindrical units composed of DNA wrapped around histone proteins—separated by protein-free DNA (linker DNA) that folds in a zigzag or solenoidal-like arrangement to yield a ~30-nm thick fiber. The tightly-packed and organized structure of the chromatin fiber in eukaryotes serves two essential cellular functions: it condenses meters-long genomic DNA by several orders of magnitude to enable its packaging into micrometer-sized cell nucleus, and it regulates the template-directed transcription of genes through local unfolding of chromatin.1,2 The fully-compact state of the 30-nm chromatin fiber at physiological conditions is stabilized by a complex interplay of each constituent histone tails as well as the linker histone and cellular counterions (monovalent and divalent).
The histone tails are positively-charged unstructured termini of the core histones (H3, H4, H2A, and H2B) that project outwards from the nucleosome core. The histone tails play a critical role in chromatin compaction by mediating internucleosomal interactions, i.e., attaching to the wound DNA or acidic histone domains of neighboring nucleosomes to bring them into closer proximity. Without these tail-mediated attractive interactions, the repulsive electrostatic interactions between the negatively-charged DNA (wound and linker DNA) would dominate and trigger the unfolding of chromatin. Indeed, sedimentation coefficient measurements on in vitro reconstituted chromatin with intact and missing histone tail regions reveal a dramatic unfolding of chromatin upon the removal of histone tails.3-5 It is also likely that the histone tails mediate interactions across distant portions of the fiber to promote higher-order folding through fiber inter-digitation. The histone tails play an equally important role in the regulation of gene transcription through chemical modification of their residues. These modifications alter the compaction state of chromatin to either promote gene transcription through partial unfolding of chromatin or inhibit transcription by stabilizing tightly packed chromatin. Such epigenetic factors have been linked to human disorders, including cancer.
The linker histones (H1 and H5) are highly positively-charged proteins consisting of a structured globular domain, a highly charged unstructured C-terminal domain, and a short N-terminal domain.6Experiments suggest that the globular domain of the linker histone (LH) binds to the nucleosome core at or near the dyad axis between the entering/exiting linker DNAs,7-9 while the C-terminal domain extends outwards and triggers formation of linker DNA “stems”10,11 (i.e., first 20 bp of the entering/exiting linker DNAs juxtapose with the C-terminal of the linker histone). By constraining the path of the linker DNAs, these stems could cause a reduction in the entry/exit angle of the linker DNAs at the nucleosome core.10,12,13 It is not, however, clear how this linker DNA stem promotes the global condensation of chromatin.
Physiological salt conditions, containing monovalent (K+, Na+) and divalent cations (Mg2+) at roughly 150 mM and 1 mM concentrations, respectively, also help compact chromatin by screening electrostatic repulsion between the linker DNAs.14 It is well known that Mg2+ at physiological concentrations causes dramatic compaction of chromatin15,16 that cannot be achieved with elevated levels of monovalent cations. This suggests that Mg2+ ions play roles other than merely providing an increased level of screening to linker DNA electrostatic repulsion. Recent studies on DNA bending in the presence of multivalent ions17,18 lead us to speculate that the magnesium ions help reduce the persistence length of DNA to promote bending of linker DNAs, but how this could lead to better packing of nucleosomes within chromatin remains known.
Thus, the detailed mechanisms by which the histone tails, linker histone, and physiological salt consort to compact chromatin are not well understood. Furthermore, the contribution of each histone tail and that of the linker histone and physiological salt in chromatin folding is also unclear. Many experimental studies have attempted to dissect the role of histone tails in chromatin compaction over the past decade. Most have examined in vitro reconstituted chromatin to assess the impact of tail removal on chromatin folding through sedimentation coeficient analyses.3,4,19 All studies point towards the greater importance of the H3 and H4 tails in chromatin compaction compared to the H2A and H2B tails. Furthermore, sophisticated chemical crosslinking approaches have attempted to quantify the number of inter- and intranucleosomal interactions mediated by the histone tails within chromatin.5,20,21 Recent studies have shown that the H4 tails, and especially the H4K16 residue, play a critical role in compacting chromatin by mediating interactions with an acidic patch on the nucleosome surface.22,23 However, because experimental approaches cannot resolve the positional distribution of each histone tail as well as their energetic interactions within chromatin, the detailed structural changes in chromatin accompanying each histone tail modification are not understood.
Computational approaches have the potential to dissect histone tail interactions and dynamics at molecular lengths and time scales. However, conventional approaches based on atomistic simulations like molecular dynamics cannot probe the dynamics of systems as large as a nucleosome, let alone a short chromatin fiber composed of tens of nucleosomes. This argues for more specialized coarse-grained approaches that attempt to significantly reduce the total number of degrees of freedom by grouping clusters of atoms into “interaction” sites involved with other sites through “effective” energetic interactions. Though atomistic details of chromatin are lost, essential physio-chemical interactions—such as electrostatics, excluded volume interactions, solvation, and conformational flexibility—can be captured through careful modeling. The many innovative macroscopic and mesoscale models of chromatin developed thus far have captured the mechanics of DNA and nucleosomes but out of necessity have either neglected to model the histone tails or approximated them as rigid bodies.14,24-29
Recently, we have developed a mesoscale model of chromatin that captures the essential physics of chromatin—such as its electrostatics, DNA and nucleosome mechanics, structural irregularity, and histone tail flexibility—and which averages over other effects—protein/DNA sequence effects, hydrogen-bonding, atomistic fluctuations, and solvation effects.30-32 The model makes chromatin fibers as large as 48-nucleosomes amenable to long-time, large-scale simulations by Brownian dynamics (BD) and Monte Carlo (MC) methods and allows generation of a representative ensemble of thermodynamically feasible structures of chromatin. Calibrations to experimental sedimentation and diffusion coefficients of oligonucleosomes over a broad range of monovalent salt concentrations30,31 have indicated the reasonableness of the model; analyses of the histone tails have shed insights into the positional fluctuations and roles of each histone tail in compacting chromatin to a moderately folded, irregular zigzag morphology.31 The model, however, did not account for the binding of linker histones and the effect of divalent cations like Mg2+ on chromatin architecture.
Here we systematically investigate the role of histone tails in chromatin compaction under different salt conditions, as well as a function of the presence and absence of linker histones, to dissect the role of the tails in chromatin compaction. Specifically, we seek answers to the following questions: How frequently does each histone tail attach to neighboring nucleosomes, parent nucleosomes, and linker DNA in physiological conditions? What is the individual role of each component? Which regions of the nucleosome mediate the most number of interactions with the histone tails? What is the positional distribution of the histone tails, and how is it affected by the binding of linker histones?
We investigate these questions here by extending the mesoscale model of Arya and Schlick30,31 to incorporate the effects of divalent cations and linker histones and to modify the DNA entry/exit geometry to better reproduce the internucleosomal interaction patterns and linker DNA trajectories obtained experimentally under monovalent salt conditions. We then analyze configurational ensembles generated from Monte Carlo simulations of oligonucleosomes with one linker histone molecule per nucleosome, 1 mM divalent salt, and 150 mM monovalent salt, and compare their structural features and the tails' positional ditributions and interactions to those obtained for linker-histone inclusive but divalent cation-deficient oligonucleosomes, as well as to both linker histone and divalent ion deficient oligonucleosomes. An elaborate mechanism of how the histone tails combine with the linker histones and physiological salt to promote chromatin compaction emerges. The biological implications of our results are also discussed.
2 Computational Methods
Our first-generation macroscale models of chromatin developed in 200126,33 captured the essential monovalent-salt dependent mechanics of chromatin and the thermal fluctuations of the nucleosome and linker DNA but did not account for the irregular surface of the nucleosome and also neglected the effect of histone tails and linker histones. The next generation mesoscale models developed in 2005-0714,30,31,34 treated both the irregular surface of the nucleosome and the role of histone tails in mediating internucleosomal interactions, and reproduced most experimental data very well. The latest mesoscale model of chromatin developed and used in the present study further improves upon these models by: (a) treating linker histones bound to nucleosomes; (b) treating divalent salt/cations (Mg2+); and (c) improving the handling of the nucleosome-DNA geometry and mechanics. Details of these modeling developments are described below, followed by a description of the Monte Carlo methodology used generate equilibrium ensembles of oligonucleosome configurations. We also summarize for completeness the various tests conducted against experimental measurements.
2.1 Mesoscale Oligonucleosome Model
Modeling of oligonucleosome components
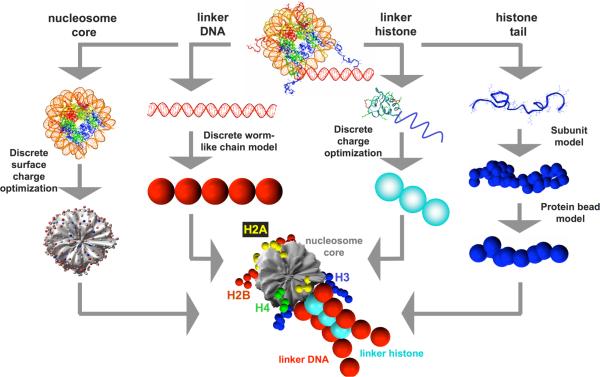
The nucleosome core, linker DNA, histone tails, linker histone, and the physiological environment (salt + solvent) are treated using separate mesoscale modeling strategies, which are all integrated to yield the final mesoscale model of the oligonucleosome.
Nucleosome-core modeling
The nucleosome core, defined as the histone octamer and wound DNA minus the N-termini of all four histones and short C-termini of H2A, is treated as a charged, rigid body (Fig. 1). Using the irregular discrete surface charge optimization (DiSCO) algorithm,33,34 300 “pseudo” charges are uniformly distributed across the surface of the nucleosome core to mimic the surrounding electrostatic potential and electric field. The charge magnitudes are optimized at each salt concentration so that their electric field obtained via a Debye-Hückel formulation approximates closely the salt-dependent electric field of the atomistic nucleosome core at distances > 5Å away from the surface of the core. This is achieved through minimization of an objective function that represents this error, with the optimization performed numerically using the truncated-Newton (TNPACK) routine35,36 integrated within the DiSCO software. The electric field landscape of the atomistic nucleosome is computed using the nonlinear Poisson-Boltzmann equation solver QNIFFT 1.237-39 where the atomic radii are assigned using the default extended atomic radii based loosely on Mike Connolly's Molecular Surface program,40 and the charges are assigned using the AMBER 1995 force field.41 In general, we obtain excellent agreement between the Debye-Hückel electric fields and the electric field obtained by solving the complete Poisson-Boltzmann equation (>90% accuracy) as analyzed in detail elsewhere.26 In addition, each nucleosome charge is also assigned an effective excluded volume modeled using a Lennard-Jones potential to account for the excluded volume of the nucleosome core. Overall, the above treatment results in a significant reduction in the number of degrees of freedom (from ~75,000 corresponding to the ~25,000 atoms in the nucleosome core to six degrees of freedom of a rigid body) and significantly reduces the number of charge-charge interaction computations (from ~25,000 atomic partial/full charges in the atomistic nucleosome to 300 charges in the mesoscale nucleosome).
Figure 1.
Mesoscale modeling of linker-histone bound oligonucleosomes. The nucleosome core is modeled as an irregular-shaped rigid body with 300 pseudo charges on it surface. The linker DNA is treated using the discrete version of the worm-like chain model. The linker histone is treated as charged beads connected rigidly to the nucleosome and the histone tails are treated using the protein bead model. The bottom figure at center shows the final integrated mesoscale model.
Linker DNA modeling
The linker DNA connecting adjacent nucleosome cores is treated using the discrete elastic wormlike chain model of Allison et al.,42 i.e., as a chain of charged beads where each bead represents a 3-nm (~10 bp) of relaxed DNA (Fig. 1). Each bead is assigned a salt concentration-dependent negative charge (see Table 1) to mimic the electrostatic potential of linear DNA using the procedure of Stigter43 and an excluded volume using the Lennard-Jones potential to prevent overlap with other components of chromatin. Each linker bead-chain is also assigned an internal force field comprising of stretching, bending, and twisting potential energy terms, as used for macroscopic models of supercoiled DNA. Again, this mesoscopic modeling approach to the DNA leads to significant reduction in the degrees of freedom of DNA (from ~800 atoms per DNA twist to 1 bead per twist).
Table 1.
Parameter values for the linker-histone inclusive mesoscale oligonucleosome model.
| Parameter | Description | Value |
|---|---|---|
| l0 | Equilibrium DNA segment length | 3.0 nm |
| Lp | Persistence length of DNA | 50 nm |
| h | Stretching constant of DNA | 100 kBT/ |
| g | Bending constant of DNA | LpkBT/l0 |
| s | Torsional rigidity constant of DNA | 3.0 × 10-12 erg.nm |
| θ0 | Angular separation between linker segments at core | 108° |
| 2w0 | Width of wound DNA supercoil | 3.6 nm |
| r0 | Radius of wound DNA supercoil | 4.8 nm |
| htc | Stretching constant for tail bead attached to core | h |
| σtt | Excluded volume distance (EVD) for tail/tail interactions | 1.8 nm |
| σtc | EVD for tail/core interactions | 1.8 nm |
| σcc | EVD for core/core interactions | 1.2 nm |
| σtl | EVD for tail/linker interactions | 2.7 nm |
| σcl | EVD for core/linker interactions | 2.4 nm |
| σgLHc | EVD for globular linker histone bead/core interactions | 2.4 nm |
| σgLHl | EVD for globular linker histone bead/linker interactions | 3.6 nm |
| σcLHc | EVD for C-terminal linker histone bead/core interactions | 2.2 nm |
| σcLHl | EVD for C-terminal linker histone bead/linker interactions | 3.4 nm |
| kev | Excluded volume interaction energy parameter | 0.001 kBT |
| kevt | Tail/tail excluded volume interaction energy parameter | 0.1kBT |
| ql | Charge on linker DNA bead at 0.15 M monovalent salt | -24.1e |
| ql | Charge on linker DNA bead at 0.01 M monovalent salt | -7.5e |
| qgLH | Charge on globular linker histone bead at 0.15 M monovalent salt | 12.4e |
| qcLH | Charge on C-term linker histone bead at 0.15 M monovalent salt | 29.9e |
| ε | Dielectric constant of solvent | 80 |
Histone tail modeling
The ten histone tails, which include eight N-termini of H2A, H2B, H3, and H4 and two C-termini of H2A, are treated using a protein bead-chain model. In this model, we represent five residues using a single bead located at the Cβ atom of the central amino acid (Fig. 1). Thus, 50 tail beads per nucleosome are used to model ~250 histone tail residues (~4000 tail atoms) associated with each nucleosome. Each bead is assigned a charge equal to the sum of the charges on the five amino acids it represents multiplied by a scaling factor ftc close to unity that accounts for salt-dependence in the “effective” charge. See Table 1 and text of Arya et al.30 for the charge values assigned to each tail bead and the magnitude of ftc as a function of monovalent salt concentration, respectively. Each tail bead is also assigned an excluded volume treated via the Lennard-Jones potential. Finally, each tail unit is assigned a customized intramolecular force field comprising of bond stretching, and bond-angle bending terms. The parameters for this force field, i.e., equilibrium bond lengths and bond angles, and the related force constants, are optimized to reproduce the configurational properties of the atomistic histone tails. See Tables 3 and 4 of Arya et al.30 for the values of intramolecular force field parameters.
Linker histone modeling
The linker histone is modeled based on the structure of rat H1d linker histone predicted by Bharath et al.44,45 via fold recognition and molecular modeling. H1d consists of an N-terminal region of 33 residues, globular-shaped central region of 76 residues, and highly charged C-termini of 110 residues. In our model, we represent the globular domain by a single charged bead and the C-terminal domain by two charged beads. The three beads are positioned in a straight line and separated by a distance of 2.6 nm (see Fig. 1). Each bead is assigned an optimized charge at its center such the resulting Debye-Hückel electric field of the model reproduces the electric field of the atomistic linker histone at distances >5Å from the linker histone surface, obtained by solving the non-linear Poisson-Boltzmann equation using QNIFFT 1.2.37-39 In addition, each bead is assigned a Lennard-Jones potential to account for the excluded volume of the two domains. We neglect the short, relatively uncharged, N-terminal region. The linker histones beads interact with all chromatin components except their parent nucleosomes through excluded volume interactions (Lennard-Jones potential) and electrostatic interactions (Debye-Hückel potential), as detailed below. The charge values and excluded volume parameters for the linker histone beads are provided in Table 1.
Monovalent and divalent salt modeling
The solvent surrounding oligonucleosomes is treated as a continuum with a dielectric constant of 80. The screening effect of monovalent salt on electrostatic interactions within an oligonucleosome is treated using the Debye-Hückel formulation (i.e., the Debye-Hückel potential given in Eq. 7 below, where the magnitude of κ depends strongly on the salt concentration).
The effect of divalent salt (e.g., Mg2) on chromatin calls for a different treatment. The analytical estimate of the electrostatic screening length of 150 mM NaCl and 1 mM MgCl2 using the Debye-Hückel theory (κ = 1.31 nm-1), is only nominally larger than that obtained for 150 mM monovalent salt alone (1.27 nm-1) and not sufficient to account for the extensive chromatin compaction obtained with the addition of Mg2+. This has also been verified through simulations of our mesoscale model where κ−1 has been set to this value. Such dramatic condensation likely arises due to underestimation of the condensed magnesium ion concentration at the DNA surface due to the Debye-Hückel formulation (leading to substantial underestimation of salt-screening) as well as the charge-correlation effects described in detail below. To account for this divalent-ion induced effects as well as the concomittant increase in the flexibility of DNA (decrease in persistence length) with divalent cations,17,18 we resort to a first-order approximation. First, we further reduce the repulsion among linker DNA by setting an inverse Debye length of κ = 2.5 nm-1 based on the argument that, at the fully condensed state of chromatin, the linker DNAs are almost touching each other (which can also be deduced directly from the nucleosome packing ratios of 10-11 nucleosome/11nm exhibited by chromatin in the presence of linker histone and Mg2+. Note that this modified value κ is only employed when computing linker/linker interactions. The original value of κ = 1.27 nm-1 is still used for the remaining electrostatic interactions. Second, we reduce the persistence length of the linker DNAs from 50 to 30 nm according to several studies.17,18 With this phenomenological model, we expect to capture the essence of Mg2+ ion induced chromatin compaction. A more specialized modeling of Mg2+ effects that includes charge correlation effects is not compatible with the mesoscale chromatin model and would be far more computationally intensive. This limitation along with others related to proper treatments of specific interactions and solvation energies are described in more detail below.
Physico-chemical interactions not treated in the mesoscale model
The mesoscale model described here neglects the effect of three physico-chemical interactions: charge-charge correlation, specific protein-protein interactions (hydrogen bonding), and solvation (desolvation) energies, on chromatin compaction.
Charge-charge correlations arises from the ability of mobile counterions within the diffuse layer surrounding a charged surface to dynamically orient themselves in an arangement that minimizes their mutual repulsion and/or maximizes favorable interactions with an external charged surfaces.46,47 Such charge correlations generally become significant in systems with closely-spaced and highly charged surfaces and multivalent counterions (valency> 2), leading to the remarkable attraction observed between like-charged particles in multivalent ions. Mean field theories such as the Poisson-Boltzmann/Debye-Hückel formulations fail to account for such correlation effects despite their excellent handling of salt-induced electrostatic screening. Hence, our mesoscale model based on the Debye-Hückel formulation does not account for charge-correlation effects that are likely to be important for chromatin at physiological conditions (highly charged macromolecule in the presence of Mg2+ (multivalent) ions. However, an exact description of these effects would entail an explicit treatment of Mg2+ ions, and detailed calculations (via Monte Carlo sampling) to determine their positions within chromatin for each Monte Carlo sampled chromatin configuration, a task that would be prohibitively expensive. Though related simulations were performed by Nordenskiold and coworkers48,49 for two DNA-based systems (nucleosomes models in the presence of divalent ions and double-stranded DNA in the presence of linear polyamines), the systems studied were too small and/or highly simplified compared to our oligonucleosome system. Implicit treatment of charge-correlation effects, on the other hand, may not be as computationally expensive but will lack in accuracy and will be difficult to implement for such a large macromolecule.50 To this end, we have resorted to the above-mentioned phenomenological approach that accounts for Mg2+ ioncondensation and charge-charge correlation that lead to linker DNAs almost touching each other.
Specific protein-protein interactions such as hydrogen bonding are also not included in our mesoscale model. Such interactions have to be accounted either at the atomistic level and or through high-resolution mesoscale approaches.51,52 Most of these protein-protein interactions across nucleosomes are expected to be relatively weak compared with the dominating electrostatic interactions; however, some specific interactions such as those between the H4 tails and the acidic patch on nucleosomes may be quite strong.53 As discussed below, our neglect of this specific interaction likely prevents our oligonucleosomes from achieving complete compaction in the presence of linker histones and magnesium ions.
Finally, our model also neglects desolvation effects that are also assumed to be secondary with respect to strong electrostatic interactions within chromatin. Again, a proper treatment of solvation effects in our mesoscale model is impractical as it either requires a molecular-scale treatment of the solvent and protein/DNA, or at minimum, a residue-level description of the proteins and DNA.54 The simplest approaches will also involve fairly detailed calculations at each Monte Carlo step involving surface area determination for computing desolvation energies for nonpolar contributions and Poisson-Boltzmann calculations for the electrostatic contributions.55
Integration of oligonucleosome components
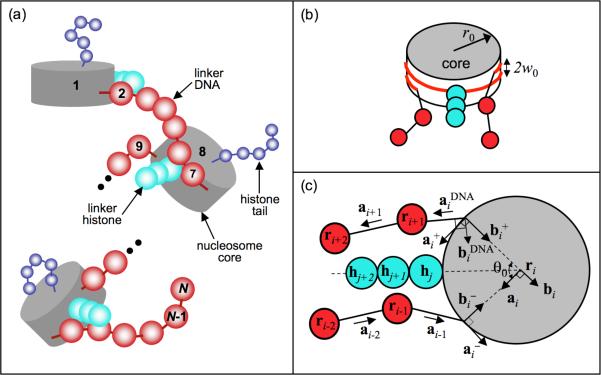
The mesoscale models for the nucleosome cores, linker DNA, histone tails, and linker histone are integrated to yield the repeating oligonucleosome motif shown in Fig. 1. The histone tails bead-chains are attached to the nucleosome core via stiff harmonic springs at locations consistent with the nucleosome crystal structure. The three beads corresponding to the globular and C-terminal domains of the linker histone are placed on the dyad axis of each nucleosome at distances r = 6.2, 8.8, and 11.4 nm from the nucleosome center, as suggested by Sheng et al.56 and Bharath et al.45We consider that the linker histones beads, similar to the nucleosome pseudo charges, are rigidly attached to their parent nucleosomes. Each nucleosome core other than the first nucleosome core of the nucleosomal array is attached to two linker DNAs, termed the “entering” and “exiting” linker DNA to yield the oligonucleosome chain in Fig. 2. The two points on the nucleosome at which the entering and exiting linker beads are attached enclose an angle θ0 about the center of the nucleosome core, and are separated by a distance of 2w0 normal to the plane of the nucleosome core, consistent with the geometry of the nucleosome crystal structure of Luger et al.53
Figure 2.
Mesoscale oligonucleosome model showing the: (a) assembly of oligonucleosome motifs into a chain; (b) the entering and exiting linker DNA geometry at the nucleosome core; and (c) the linker-DNA/nucleosome mechanics in terms of individual coordinate systems for the linker DNA beads and the nucleosome core.
Hence, if the oligonucleosome contains NC nucleosomes and each linker is z nm long, the number of beads in one linker DNA is (z/3) - 1 and the total number of linker DNA beads is ND = (z/3)NC. The total number of linker histone beads is NL = 3NC and the total number histone tails and histone tail beads is NT = 10NC and NTb = 50NC, respectively. The nucleosome and linker DNA beads are numbered/indexed in the direction of the oligonucleosome chain starting from i = 1 for the first nucleosome core to i = NC + ND(= N) the last linker DNA bead. IC and IL denote the subset of indices that represent the nucleosomes and linker DNA beads, respectively. The nucleosome center of mass positions and linker DNA bead positions are denoted by vectors ri, and the center of mass positions of the linker histone beads and histone tail beads are denoted by vectors hi and ti, respectively.
Refinement in linker DNA attachment positions
Our earlier oligonucleosome model30,31 was based on 1.75 turns of the nucleosome-wound DNA; consequently, θ0 was set to 90°. Though the model correctly reproduces many experimental observations of chromatin in monovalent salt conditions (e.g., irregular zigzag morphology, salt-dependent folding/unfolding, histone tail dynamics, diffusion coefficients, internucleosomal interaction energies), it predicts dominant interactions between nucleosome i and nucleosomes i± 3 and i± 4 at monovalent salt conditions, while recent experiments by Grigoryev and coworkers57 indicate dominant interactions between nucleosomes i and i±2. The model also does not yield the correct mixture of open and crossed linker DNA conformations seen experimentally by Toth and coworkers.58 We have improved this model based on recent crystal data that suggest a reduced wrapping of 1.7 turns of wound DNA,59 or a corresponding value of θ0 = 108° for the linker DNA entry/exit trajectory orientation. These parameters produce a mixture of crossed and uncrossed linker DNA geometries at high monovalent salt (20% open and 80% crossed) and an internucleosomal interaction pattern dominated by (i±2) interactions, in agreement with the experimental findings of Toth et al.58 We have verified that this parameter adjustment did not affect other properties like sedimentation coefficients and internucleosomal interaction energies, as reported previously.30,31
Linker-DNA/nucleosome mechanics
Each linker DNA bead and nucleosome is allowed to twist about the DNA axis with a twisting energy penalty. This is implemented by assigning local coordinate systems to all linker DNA beads and the nucleosome core, as shown in Fig. 2. The coordinate system for component i comprises of three orthonormal unit vectors {ai, bi, ci}, where i = 1, 2, … , N. If i is a nucleosome core (i ∈ IC, ai and bi lie in the plane of the nucleosome core, where ai points along the tangent at the attachment site of the exiting linker DNA and bi points in the direction normal to this tangent and inwards toward the nucleosome center. The vector ci lies normal to the nucleosome plane: ci = ai × bi. For the linker DNA, the vector ai points from ri toward ri+1 when i + 1 is also a linker DNA bead. When i + 1 is a nucleosome core, the vector ai points from ri in the direction of the linker bead's attachment point. When i + 1 is a nucleosome core and i + 1 is a linker DNA bead, we have to define another coordinate system . Here points along the exiting linker DNA, i.e., toward ri+1 from its point of attachment at the nucleosome core i.
Two additional coordinate systems are required to describe the trajectory of the wrapped DNA on the nucleosome cores at the point where it diverges from the core to form the two linker DNA, as given by and . The former represents the local tangent on the nucleosome core at the point of attachment of the entering linker DNA, while the latter represents the tangent corresponding to the exiting linker DNA. Note that with this formalism, = {ai, bi, ci}. These additional coordinate systems are required for determining the DNA bending and twisting at their points of attachments to the nucleosome cores.
We also define Euler angles αi, βi, and γi (where i = 1, N - 1) that transform the coordinate system of one linker DNA bead to that of the next along the oligonucleosome chain, i.e., {ai, bi, ci → {ai+1, bi+1, ci+1}. When the i + 1 component is a nucleosome, αi, βi, and γi transform {ai, bi, ci} to , and when i is a nucleosome, αi, βi, and γi transform to {ai, bi, ci}. An additional set of Euler angles , and are required to transform the coordinate system of the nucleosome core (i.e., when i ∈ IC) to that of the exiting linker DNA trajectory . The linker DNA twist at each bead location is then calculated as the sum of Euler angles αi and γi , and linker DNA bending angle is given by βi. Further details on Euler angles and their determination is provided elsewhere.26,30
Oligonucleosome energy
The total potential energy E of the oligonucleosome is given by the sum of seven different components:
| (1) |
The first three terms denote the stretching, bending, and torsional energy of linker DNA given by:
| (2) |
| (3) |
| (4) |
where h, g, and s denote the stretching, bending, and torsional modulus of DNA, li denotes the separation between the DNA beads, and l0 denotes the equilibrium separation distance between beads of relaxed DNA (=3 nm).
The fourth term represents the total stretching energy of the histone tails, which is composed of two terms: stretching of tail beads, and stretching of the histone tail bead from its assigned attachment site, as given by:
| (5) |
where Nbj is the number of beads in tail j, is the stretching constant of the bond between the k and k + 1 beads of the jth histone tail, and lijk and ljk0 represent the distance between tail beads k and k + 1, and their equilibrium separation distance, respectively. Also, htc is the stretching bond constant of the spring attaching the histone tail to the nucleosome core, tij is the position vector of the “attachment” tail bead in the coordinate system of its parent nucleosome, and tij0 is its ideal position vector in the crystal configuration.
The fifth term represents the intramolecular bending contribution to the histone tail energies, as given by:
| (6) |
where θijk and θjk0 represent the angle between three consecutive tail beads k, k + 1, and k + 2, and their equilibrium angle, respectively, and is the corresponding bending force constant.
The sixth term, EC, represents the total electrostatic interaction energy of the oligonucleo-some, which includes ten types of interactions: nucleosome/nucleosome, nucleosome/linker, nucleosome/tail, nucleosome/linker histone, linker/tail, linker/linker, linker/linker-histone, tail/tail, tail/linker-histone, and linker-histone/linker-histone. All these interactions are modeled using the Debye-Hückel potential that accounts for salt screening:
| (7) |
where qi and qj are the charges on two interacting charges separated by a distance rij in a medium with a dielectric constant of ∊ and an inverse Debye length of κ, ∊0 is the electric permittivity of vacuum. The parameter κ depends on salt concentration and is computed as 0.736 nm-1, where Cs is the monovalent salt concentration (Molal units) and T is the temperature (in Kelvins). It should be noted that the charges on the nucleosome and on the linker histone beads belonging to the same parent nucleosome do not interact electrostatically amongst/with each other as the nucleosome charges and linker histone beads are rigidly attached to the nucleosome making these interactions redundant. Neighboring linker DNA beads and histone tail beads belonging to the same chain (i.e., beads j and j ± 1) also do not interact electrostatically with each other as their interactions are already accounted through the intramolecular force field (harmonic spring). Finally, linker DNA beads and histone tail beads directly attached to the nucleosome also do not interact electrostatically with the nucleosomal pseudo charges. This is required to ensure that the attachment tail and linker DNA beads remain as close as possible to their equilibrium locations/trajectories prescribed by the nucleosome crystal structure.
The last energy term, EV, represents the total excluded volume (van der Waals) interaction energy of the oligonucleosome. It is composed of eight interactions: nucleosome/nucleosome, nucleosome/linker, nucleosome/tail, nucleosome/linker-histone, linker/tail, linker/linker-histone, tail/tail, and tail/linker-histone. Note that the linker DNA beads carry a large negative charge making them strongly repulsive. Thus, they do not require any additional linker/linker excluded volume interactions to prevent their mutual overlap. Likewise, we do not require any excluded volume interactions between separate linker histone beads as each linker histone bead carries a large positive charge. The excluded volume interactions are modeled using the Lennard-Jones potential and the total excluded colume energy is given by:
| (8) |
where σij is the effective diameter of the two interacting beads and kij is an energy parameter that controls the steepness of the excluded volume potential. For the same reasons cited in the case of electrostatic interactions (see above), the nucleosome pseudo charges and the linker histone beads belonging to the same parent nucleosome, neighboring tail beads belonging to the same histone chain, and linker DNA beads and histone tail beads directly attached to the nucleosome do not interact through excluded volume interactions. Table 1 lists the values of kij and σij used in the modeling.
2.2 Monte Carlo Simulations
Monte Carlo (MC) simulations are well suited for sampling large biomolecular systems that exhibit a wide range of thermally accessible states. Over the past few years we have developed a tailored MC methodology to efficiently sample the ensemble of oligonucleosome conformations under constant temperature conditions.31,32 The four MC moves consist of the following.
Global pivot rotation. For one randomly chosen linker bead or nucleosome core, and selected axis that passes through the chosen component, we rotate the shorter part of the oligonucleosome about this axis by an angle chosen from a uniform distribution within [0, 20°]. The attempted move is then accepted/rejected based on the standard Metropolis criterion.
Local translation. A chosen component (linker bead or core) is shifted along a randomly oriented axis passing through that element by a distance sampled from a uniform distribution in the range [0, 0.6 nm]. The move is then accepted/rejected based on the standard Metropolis criterion.
Local rotation. A chosen component (linker bead or nucleosome core) is rotated about a randomly selected axis by an angle uniformly sampled from the range [0, 36°]. The MC move is accepted/rejected based on the standard Metropolis criterion.
Tail regrowth. For efficient sampling of histone-tail conformations, based on the configurational bias MC method,60-62 we regrow a randomly selected histone tail bead-by-bead by using the Rosenbluth scheme,63 beginning with the bead attached to the nucleosome core. To prevent histone tail beads from penetrating the nucleosome core during tail regrowth, the volume enclosed within the nucleosomal surface is discretized, and any insertion attempts that place the tail beads within this volume are rejected automatically.
The four MC moves-pivot, translation, rotation, and tail regrowth-are attempted with relative frequencies of 0.2:0.1:0.1:0.6, respectively, in all our simulations of mesoscale oligonucleosomes, Typical ensemble sizes vary from 50-100 million MC steps. We employ four different oligonucleosome configurations (zigzag with parallel and perpendicular nucleosomes, and solenoid with parallel and perpendicular nucleosomes) as starting configurations in our MC simulations, as described in detail elsewhere.31 Because, inspection of representative oligonucleosomes reveals that fiber-like morphologies representative of chromatin fibers emerge more clearly for longer arrays (24 and 48 nucleosomes),31 we employ 24-unit or longer oligonucleosomes for our analyses, except for the computation of sedimentation coefficients where we use 12-unit oligonucleosomes to facilitate comparison with experimental data
To assess the role of histone tails in chromatin folding in this study, we have conducted Monte Carlo simulations of the enhanced mesoscale oligonucleosomes at both physiological and non-physiological conditions, i.e., with/without monovalent and divalent salt, and with/without linker histones at a temperature of 300 K. Specifically, we conduct simulations under four conditions: (a) 10 mM monovalent salt only (loMS); (b) 150 mM monovalent salt only (hiMS); (c) 150 mM monovalent salt + linker histone (MS-LH); and (d) 150 mM monovalent salt + linker histone + 1 mM divalent salt (MS-LH-Mg). We have also performed preliminary calculations for the case with 150 mM monovalent salt and 1 mM divalent salt (i.e., MS-LH-Mg without linker histone), and the results (not shown) do not provide any additional insights into the role of histone tails, linker histones, and magnesium ions.
2.3 Model Validation
Extensive validation studies are summarized below.
Salt-induced compaction of chromatin. Our mesoscale oligonucleosome model reproduces the experimental salt-dependent sedimentation coefficients of 12-unit oligonucleosomes representing chicken erythrocyte chromatin over a broad range of monovalent salt concentrations,64 as discussed in detail in our earlier study.30 Here, we show that our model also qualitatively reproduces the dramatic increase in the sedimentation coefficients of 12-unit oligonucleosomes upon the addition of linker histone and magnesium ions (Table 2, Figure 3). We have also computed the compaction of 12-unit oligonucleosomes in terms of the nucleosome packing ratio, resulting in excellent agreement with measurements from various experimental groups65-67 (see Table 2). Taken together, these results suggest that the mesoscale oligonucleosomes display similar global structure (size and compaction) as the experimental nucleosomal arrays under different conditions.
Dynamics of short oligonucleosomes. The diffusion coefficients of mesoscale mononucleosomes, dinucleosomes, and trinucleosomes computed via Brownian dynamics simulations match the experimental diffusivity values,68-70 as discussed in our earlier study.30 Our modeling/simulations also reproduce the salt dependent behavior of the diffusion coefficient observed experimentally.
Salt-dependent extension of histone tails. We have quantified the salt-dependent extension of histone tails for mononucleosomes over a broad range of monovalent salt concentrations through the quantities Dmax and Rg that denote the maximum diameter of the nucleosome and the radius of gyration of the nucleosome, respectively.30 We find that both Dmax and Rg match the values measured by Livolant and coworkers via small-angle X-ray scattering,71 and also reproduce the salt-dependent extension of histone tails from the nucleosome core.
Irregular zigzag topology of chromatin. Our mesoscale oligonucleosomes exhibit an irregular zigzag topology with straight or slightly-bent linkers under monovalent salt conditions in the absence of the linker histone,31 consistent with the models proposed by Bednar et al.67,70,72,73The structure comprises of a mixture of open and crossed linker DNA conformations, also in agreement with recent experiments.58 Our current work, discussed below, suggests that this irregular structure becomes more regular upon linker histone binding, very similar to the two-start zigzag structure proposed by Schalch et al.72 We can also capture using our model the mean entry/exit angle of the linker DNAs at the nucleosomes measured via electron microscopy, e.g., we obtain an entry/exit angle of 71° at 200 mM monovalent salt without linker histones that reduces to 39° upon addition of the linker histone, consistent with angles measured experimentally.10,13,67
Internucleosomal interaction pattern and energies. A detailed analyses of the internucleosomal interactions mediated by the histone tails from our latest mesoscale model suggests an interaction pattern dominated by i ± 2 interactions (i.e., interactions between alternate nucleosomes along the oligonucleosome chain dominate) and followed by i ± 3 interactions. The pattern becomes more sharply peaked at i ± 2 with the addition of the linker histone. The above interaction patterns are consistent with latest experimental measurements by the Grigoryev et al.57 and with the two-start zigzag structure of Schalch et al.72 Our simulations also allow a direct measurement of the strength of tail-mediated internucleosomal interactions. We obtain a value of -1.5 kcal/mol per nucleosome at 200 mM monovalent salt with linker histone, close to the experimentally measured value of -2.0 kcal/mol per nucleosome by Cui and Bustamante using optical tweezers.74
Table 2.
Chromatin compaction assessment by sedimentation coefficients of 12-unit oligonucleosomes at four different conditions with regular tails, selectively neutralized tails, and all tails neutralized (experimental values are provided in square brackets)
| Array type | S20,w, ΔS20,w (S) a | |||
|---|---|---|---|---|
| loMSb | hiMSc | MS-LHd | MS-LH-Mge | |
| regular | 30.0 ± 0.2 [29.8f] | 38.5 ± 1.2 [37.5f] | 48.8 ± 1.7 [55.6g] | 53.2 ± 2.5 [60g] |
| all tails neutral | 25.8 ± 0.2 (-4.2) [25h] | 31.2 ± 0.2 (-7.3) [32h] | 44.9 ± 1.3 (-3.9) | 48.7 ± 1.9 (-4.5) |
| H3 neutral | 25.8 ± 0.2 (-4.2) | 33.2 ± 0.3 (-5.3) | 45.6 ± 1.2 (-3.2) | 49.5 ± 2.0 (-3.7) |
| H4 neutral | 29.7 ± 0.2 (-0.3) | 34.9 ± 0.5 (-3.6) | 46.2 ± 1.5 (-2.6) | 49.4 ± 1.9 (-3.8) |
| H2A neutral | 29.6 ± 0.2 (-0.4) | 36.3 ± 0.8 (-2.2) | 48.5 ± 1.5 (-0.3) | 52.1 ± 2.2 (-1.1) |
| H2B neutral | 29.6 ± 0.2 (-0.4) | 35.1 ± 0.6 (-3.4) | 48.0 ± 1.4 (-0.8) | 52.3 ± 2.3 (-0.9) |
ΔS20,w shown in parenthesis is computed as the change in the sedimentation coefficient of tail neutralized oligonucleosomes relative to regular oligonucleosomes under same salt/linker histone conditions
loMS = 0.01 M Na+
hiMS = 0.15 M Na+
MS-LH = 0.15 M Na+, with linker histones
MS-LH-Mg = 0.15 M Na+, with linker histones and Mg2+
Experimental values from Grigoryev et al57
Experimental values from Fletcher et al79
Figure 3.
Sedimentation coefficients of 12-unit oligonucleosomes with regular tails and neutralized tails at the four conditions: loMS, hiMS, MS-LH, and MS-LH-Mg (see Table 2). The open circles represent experimental results.19,57,79
3 Results
We discuss the role of histone tails in chromatin compaction at the different external conditions (loMS, hiMS, MS-LH, MS-LH-Mg, where MS, LH, and Mg denote monovalent salt, linker histone, and magnesium ions, respectively) obtained from conducting various analyses on the ensemble of oligonucleosome configurations generated by Monte Carlo simulations. Specifically, we describe the impact of the addition of monovalent salt (low = 0.01 M, high = 0.15 M), linker histones, and magnesium ions on the global structure of chromatin. Our focus is on explaining the roles of the histone tails in compacting chromatin by determining the following: (a) contribution of each tail to chromatin compaction via selective neutralization of each tail; (b) positional distribution of each tail relative to its parent nucleosome; (c) frequency of internucleosomal and intranucleosomal interactions mediated by each tail; (d) positional distribution of the tails' interaction sites on nonparent nucleosomes; and (e) strength of internucleosomal interactions mediated by the histone tails. A separate work focuses on the internal structure of chromatin as influenced by linker histones and divalent cations.57 We conclude by discussing the relevance our results in the context of chromatin function and regulation.
3.1 Global oligonucleosome features
We assess the global extent of compaction of chromatin from sedimentation coefficients, S20,w, of 12-unit oligonucleosomes obtained from the ensemble average, using the Kirkwood-Bloomfield formulation,30,75,76 and the overall nucleosomal packing ratios. Analyses of our oligonucleosome ensembles reveal that the sequential addition of monovalent salt, linker histone, and divalent salt causes dramatic condensation of chromatin (Table 2 and Figs. 3 and 4). Specifically, an increase in monovalent salt from low (loMS) to physiological concentrations (hiMS) causes the sedimentation coefficient and nucleosome packing ratio to rise from 30 S (2.5 nucleosomes/11 nm) to 38.5 S (4.0 nucleosomes/11 nm). Addition of linker histones (MS-LH) followed by magnesium (MS-LH-Mg) cause further compaction to 48.8 S (6.7 nucleosomes/11 nm) and 53.2 S (7.8 nucleosomes/11 nm), respectively. The sedimentation coefficients are in excellent agreement with experimental values at low and physiological monovalent salt,64 but the slightly smaller computed values (by ~5 S) with the linker histone and magnesium ions77,78 possibly reflect sampling limitations and additional interactions (such as hydrogen bonding that are not in the model) that compact chromatin further53 (see Fig. 3).
Figure 4.
Representative oligonucleosomes obtained from our simulation ensemble highlighting the differences in the global morphology and internal structure of chromatin at the four conditions investigated in this study: (a) low monovalent salt (loMS); (b) high monovalent salt (hiMS); (c) high monovalent salt + linker histones (MS-LH); and (d) high monovalent salt + linker histone + magnesium cations (MS-LH-Mg). For clarity, a 12-unit oligonucleosome is presented in (a) and 24-unit oligonucleosomes are presented in (b-d). In (c,d), odd and even numbered nucleosome cores are colored white and blue, respectively, to highlight the predominant i ± 2 interactions; and severely bent linker DNAs are colored green, as characterized by an angle of bending greater than 90°. This bending angle is defined by the angle formed between the linker DNA exiting one nucleosome and entering the next nucleosome.
Figure 4 shows representative oliognucleosome configurations for each of the four conditions examined. As discussed earlier,14,30,31 the oligonucleosomes adopt an extended “beads-on-a-string” conformation at low monovalent salt concentrations due to the strong electrostatic repulsion between the linker DNA (Fig. 4a). At physiological salt, these repulsive interactions are considerably screened and balanced by electrostatic attraction between nucleosomes mediated by the histone tails. This results in compaction of chromatin to moderately folded state with an overall irregular zigzag topology with straight/gently-bent linker DNA (Fig. 4b), consistent with EM measurements.67,70 The oligonucleosomes also reveal a high degree of sharp bending resulting in significant long-ranged (fiber/fiber) interactions, bridged by histone tails (see below). Further addition of linker histones and magnesium ions results in more dramatic compaction of the oligonucleosomes as seen clearly in Figs. 4c and 4d.
As detailed earlier,57 the linker histone compacts chromatin by significantly reducing the entryexit angle of the linker DNA at the nucleosome (“triplet angle”), consistent with experiments.10,13,70 This effect leads to the formation of a rigid linker DNA “stem” that promotes strong internucleosomal interactions between alternate nucleosomes (i.e., between nucleosomes i and i ± 2) and brings them into closer proximity. Magnesium (divalent) cations enhance chromatin compaction in two ways. First, they help neutralize the linker DNA and significantly reduce mutual repulsion among them. Second, they promote bending of a fraction of linker DNAs that reduces the number of linker DNAs crossing at the chromatin fiber axis. The severely bent linker DNA are colored green in Fig. 4d, as characterized by bending angles greater than 90°. Both effects facilitate packing of linker DNAs along chromatin axis leading to greater compaction of chromatin. The addition of linker histones and magnesium also causes the oligonucleosomes to becomes stiffer, as determined by the reduced fluctuations in the ensemble of oligonucleosome configurations, consistent with the highly increased internucleosomal interactions of histone H3 tails in the presence of magnesium ions.21
3.2 Histone tail contributions to chromatin compaction
To dissect the contribution of each histone tail in chromatin compaction at different conditions, we have selectively neutralized each tail and studied its impact on the sedimentation coefficient (Table 2). The extent of the decrease in the sedimentation coefficient (ΔS20,w) helps interpret the importance of the modified interaction; a large decrease signifies an important tail contribution and a small change a lesser effect.
Table 2 suggests that at low monovalent salt, only the H3 tails seem to be important for chromatin compaction as their neutralization results in a 4.2 S decrease in S20,w, while neutralization of the other three tails results in little change. At physiological monovalent salt conditions, both the H3 and H4 tails become more important for compaction; their neutralization results in unfolding of chromatin by ΔS20,w = -5.3 S and -3.6 S, respectively. With the addition of the linker histone and divalent ions, the H3 and H4 histone tails remain most important for chromatin compaction. Less important are the H2A and H2B tails, under all conditions. Our analyses also suggest that the histone tails are collectively more important for chromatin compaction at physiological (high) monovalent salt conditions without linker histone (ΔS20,w = -7.3). They become less important at low monovalent salt conditions (S20,w = -4.2) or in the presence of linker histones (S20,w = -3.9) and divalent cations (S20,w = -4.5). This makes intuitive sense since the linker histones and the histone tails both have neutralizing effects, but their combined effects are not additive.
The above results at low and physiological monovalent salt agree well with experimental sedimentation coefficient measurements of reconstituted nucleosomal arrays with trypsinized histone tails.19,79 Our results with linker histones and magnesium ions cannot be directly compared to experiments. However, the results can be compared with two experiments conducted at slighly different conditions: one with tailfree linker-histone bound nucleosomal arrays at low magnesium concentration80 and another with acetylated H4 tails without linker histone and high magnesium concentration.23 Both these experiments observe greater unfolding of nucleosomal arrays upon the removal of tails and/or acetylation of H4 tail (from 55 S to 40-44 S) as compared to our predictions (49 S to 45 S). This discrepancy may arise from inherent sampling limitations as well as neglect of specific interactions between H4 tail and the nucleosomal acidic patch that may prevent our oligonucleosomes from compacting to their full extent. Our sedimentation coefficients of tailfree arrays, however, agree well with experimental measurements, suggesting that macroscopic properties are reasonable.
Further analyses (discussed in detail elsewhere57) suggest that linker-histone induced chromatin compaction is driven by the following features in decreasing order of importance: (a) geometrical constraints imposed on the linker DNA through stem formation; (b) internucleosomal interactions mediated by the histone tails; and (c) screening of electrostatic repulsion between linker DNAs belonging to different nucleosomes due to linker histones.
3.3 Histone tail positional distribution
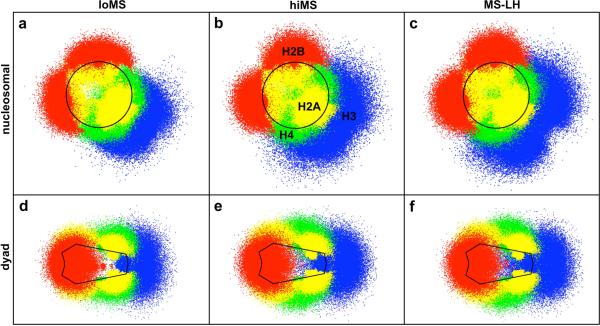
We can also compute the distribution in the positions of each histone tail as a function of salt concentration and the absence/presence of the linker histone from oligonucleosome ensembles. We analyze these distributions by determining the position vector of each tail bead tij in the frame of reference of the parental nucleosome core with center of mass position ri and orientation {aiI, bi, ci}. The resulting “projected” distribution is then denoted by , where , and . This three-dimensional distribution may be viewed in two dimensions along the “nucleosomal” plane given by or along the “dyad” plane given by .
Figure 5 shows the distribution of the histone tail beads for loMS chromatin (Fig. 5a,d), hiMS chromatin (Fig. 5b,e), and MS-LH chromatin (Fig. 5c,f). The individual dots in the figure represent the collection of projected tail bead positions sampled in our simulations. All tails except H3 exhibit narrow distributions, close to the parent nucleosome, at low salt, indicating that the tails remain near the nucleosome core (Fig. 5a,d). As the salt concentration is increased to physiological levels, the histone tail distributions become broader and extend further away from the core (see Fig. 5b,e). Thus, the electrostatic attraction between the tails and nucleosome core at low salt dominates their entropic freedom and they collapse onto the parent nucleosome. With the addition of salt, enhanced salt-screening reduces the attractive electrostatic interactions between the tails and the nucleosomal core, and tails extend further outwards from the core. These results are in close agreement with the observations from Livolant and coworkers,71 who noted an increase in the maximal diameter of the nucleosomes with monovalent salt concentration from their diffraction data. Further, the broad distributions of the tails underscores their highly flexible and dynamic nature, which is generally neglected in the models of chromatin with fixed tails.14,33
Figure 5.
Positional distribution of histone tails under different conditions: low monovalent salt (a,d); high monovalent salt (b,e); and high monovalent salt with bound linker histones (c,f). The upper panel (a-c) represents the histone tail positions projected onto the nucleosomal plane of the parent nucleosomes, and the lower panel (e-f) represents the positions projected onto the dyad plane of the parent nucleosomes. The tails are colored as: H2A, yellow; H2B, red; H3, blue; and H4, green. The nucleosome boundaries are indicated by the solid black lines.
We also note intriguing differences between the positional distribution of the four histone tails. Due to their origin from flat side of the nucleosome, the H4 and H2A tails spread in the direction normal to the nucleosomal plane while the H3 and H2B tails spread along the nucleosomal plane, primarily due to their point of origin in between the wound DNA gyres. Significantly, the H3 tail distribution centers around the mean position of the linker DNAs, suggesting that the H3 tails prefer to attach to the entering/exiting linker DNAs. Thus, the H3 tails help screen electrostatic repulsion between the linker DNAs, facilitating the compaction of chromatin. The H3 tails' tendency to attach to linker DNAs also explains why their neutralization among all tails results in unfolding of chromatin at low salt (Table 2). Since linker/linker repulsion dominates over nucleosome/nucleosome attraction at low salt, a reduction in nucleosome/nucleosome attraction due to tail neutralization results in little additional unfolding, while an increase in the linker/linker repulsion due to H3 tail neutralization causes greater unfolding. We also note that the H4 tails spread the most among all tails and are the most likely to mediate interactions with neighboring nucleosomes.
The addition of the linker histone affects slightly the distribution of the H3 tails (Fig. 5c,f). The H3 tails no longer attach as strongly/frequently to the linker DNA because the strongly positively charged linker histone takes over that role. This allows the H3 tails to spread more in the direction normal to the nucleosomal plane, increasing their tendency to mediate internucleosomal interactions. The other tails' distributions remain unaffected by the addition of the linker histone as they are located at distances farther than the Debye screening length (8 Å at physiological salt concetrations. Further addition of Mg2+ does not alter the distribution of any histone tail in our model, likely because we only treat the magnesium ions' impact on the screening of linker DNA repulsion and their bending but neglect interactions with the histone tails and/or nucleosome core. A finer resolution model is required to study such effects.
3.4 Histone tail interactions within chromatin
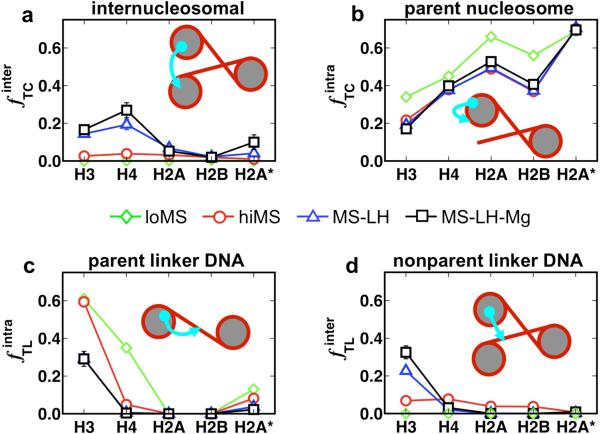
We also compute in Fig. 6 the frequencies f with which histone tails interact with the various components within chromatin: parent nucleosomes, neighboring nucleosomes (internucleosomal interactions), parental linker DNAs, and nonparental linker DNAs. The interaction frequencies for each histone tail are analyzed for the four conditions loMS, hiMS, MS-LH, and MS-LH-Mg, as calculated by the number of times a histone tail attaches to the chromatin component divided by the total number of sampled tail configurations.“Attachment” here refers to a distance approach within 0.8σ, where σ is the size parameter associated with the excluded volume interaction between the tails beads and the core charges. Such a strict distance criterion ensures that only tail beads that are strongly attracted to other components (nucleosome core charges or linker DNA beads) are counted and tails waving in the solvent that do not generally overlap are not counted.
Figure 6.
Histone tail interactions within chromatin captures in terms of the frequency with which they mediate: (a) internucleosomal interactions; (b) attach to parent nucleosomes; (c) attach to linker DNA associated with the parent nucleosome; and (d) linker DNA not associated with the parent nucleosome. The results are presented for the four conditions: chromatin at low monovalent salt (green down triangles); high monovalent salt (red circles); high monovalent salt with linker histones (blue up triangles); and high monovalent salt with linker histone and Mg2+ (black squares), respectively. The frequencies are calculated as the number of times a histone tail attaches to the chromatin component divided by the total number of sampled tail configurations.
Clearly, the histone tails mediate very little internucleosomal interactions at low monovalent salt (Fig. 6a; green diamonds, flat pattern). Because the zigzag fiber is so open, all tails interact with the parent nucleosomes or the linker DNAs of parent nucleosomes and not neighboring nucleosomes (Fig. 6b,d; green diamonds). An increase in monovalent salt to physiological concentrations increases the amount of internucleosomal interactions (Fig. 6a, red circles), but the overall tail interactions remains fairly low as indicated by the nearly flat red patterns in Fig. 6b and 6d (~4% and ~2% for H4 and H3 tails, respectively).
Upon the addition of linker histones and Mg2+ ions, the histone tails mediate significantly more internucleosomal interactions, a direct result of the closer proximity of nucleosomes to each other (Fig. 6a and 6d, blue triangles and black squares). This effect is especially striking for the H3 and H4 tails which spend as much as ~27% and ~18% of their time mediating internucleosomal interactions when both the linker histone and Mg2+ are present. Thus, we find that the H3 and H4 tails mediate the most while the H2A and H2B tails mediate the least number of internucleosomal interactions under all external conditions assessed here.
The above analyses also indicates that the H4 tails uniformly mediate the most number of inter-nucleosomal interactions within chromatin, especially in chromatin compacted by magnesium ions and linker histone. This property of H4 tails arises from their optimal location on the flat region of the nucleosome surface close to the linker DNA and the chromatin fiber axis (Fig. 5c), which makes them highly suited for interacting with either the wound DNA or the acidic patch53 of neighboring nucleosomes oriented almost parallel to the parent nucleosome. Our results agree with several experimental observations from different groups that capture significant unfolding of chromatin upon complete removal of the H4 tail or mutation/modifition of some of its residues.22,23
The H3 tails, on the other hand, display a more complex behavior. In the absence of linker histones and irrespective of monovalent salt concentration, the H3 tails attach almost 60% of the time to the linker DNAs, in agreement with the observed tail's positional distribution (Fig. 5a-c). This may be explained by their nucleosomal location close to the linker DNAs and their large extension length. However, the addition of linker histones at the nucleosome dyad decreases their propensity to attach to linker DNAs due to electrostatic repulsion. This allows the H3 tails to mediate more internucleosomal interactions (from ~2% for hiMS to ~18% for MS-LH-Mg) and attach to other linker DNAs at positions far from the linker histone (from ~7% for hiMS to ~32% for MS-LH-Mg). Still, the H3 tails attach frequently to the linker DNA (30%) and help reduce the electrostatic repulsion between linker DNA in the middle of the chromatin fiber. Thus, the H3 tails facilitate compaction via two mechanisms: through internucleosomal interactions, and by screening the electrostatic repulsion between the linker DNA.
The H2A and H2B tails mediate very few internucleosomal interactions because they originate from the curved face of the nucleosome cores (H2B), are located far from fiber axis (N-terminal H2A), or are too short to mediate interactions (C-terminal H2A). Only in the most compact form of chromatin does the C-terminal of H2A tail begin to mediate some internucleosomal interactions ~10% for MS-LH-Mg). Interestingly, we observe that interdigitation of fibers occurs at high monovalent salt (hiMS) (see, for example, the representative configuration in Fig. 4b) and these sharply bent configurations are almost always stabilized through the H2A and H2B tails, which mediate attractive interactions between distant portions of the fiber. In the presence of linker histones and magnesium ions, the oligonucleosomes become stiffer and these fiber/fiber interactions disappear. The above observations suggest that the H2A and H2B tails likely play important roles in mediating fiber/fiber interactions in higher-order structures of chromatin.
3.5 Positional distribution of H3 and H4 tails' attachment sites
To further investigate internucleosomal interactions mediated by the H3 and H4 tails, we compute the positional distribution of the sites on the nucleosomal surface at which the histone tails of neighboring nucleosomes attach and mediate internucleosomal interactions. A nucleosomal attachment site is defined as the position of the nucleosomal surface in close proximity to the histone tail beads of neighboring nucleosomes, where |tij - rc,kl| < σtc. Here, tij refers to the position of tail bead j of nucleosome i, rc,kl is the position of the l psuedo charge on the surface of nucleosome k (k ≠ i, and σtc is the effective excluded volume of tail bead/nucleosome charge interactions. We compute this distribution of nucleosomal attachment sites in the same spirit as the positional distribution of the tails outlined in Sec. 3.3, i.e., we project the absolute positions of these nucleosomal attachment sites from our ensemble of oligonucleosome configurations onto the nucleosome's coordinate frame.
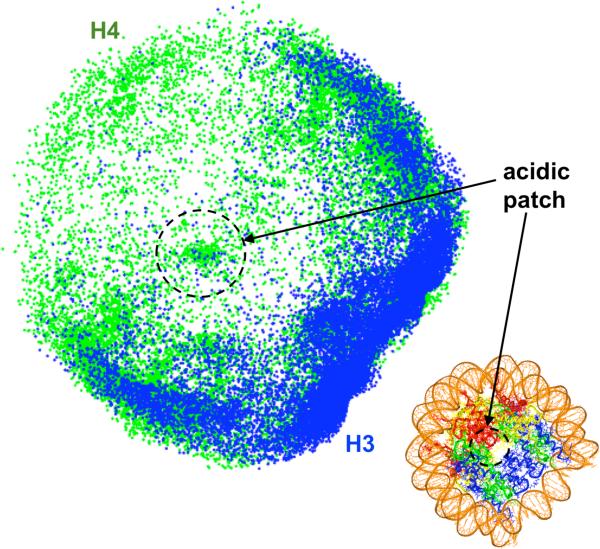
Figure 7 plots these distributions along the nucleosomal plane {a, b} for compact oligonucleosomes (MS-LH-Mg). Clearly, the H3 tail mediates internucleosomal interactions through the wound DNA of nucleosomes. Moreover, these attachment sites are concentrated on the portions of wound DNA nearest to the linker DNA. The H4 tails, on the other hand, mediate internucleosomal interactions through two regions: the entire wound DNA segment and a region located near the center of the nucleosome defined by seven acidic residues (glutamic acids) belonging to core H2A and H2B domains (circled in Fig. 7). This so called “acidic patch”53 has previously been implicated in mediating internucleosomal interactions through the H4 tail, and more specifically the 16-23 H4 tail residues.22,23
Figure 7.
Distribution of the histone tails' (H3, blue and H4, green) attachment sites on the surface of non-parental nucleosomes for MS-LH-Mg conditions. The distributions have been projected onto the plane of the nucleosome. The black circle denotes the acidic patch. The figure in the lower right corner shows the atomistic nucleosome with the same color coding as Fig. 1.
3.6 Internucleosomal interaction energy
The magnitude of internucleosomal interactions as mediated by the histone tails can also be estimated for the three conditions: hiMS, MS-LH, hMS-LH-Mg. Since the excluded volume energies in our model are weak compared to electrostatic energies, we only report the electrostatic component of the internucleosomal energies. We compute the average electrostatic component of the histone-tail/nucleosome-core interaction energy from the ensemble of oligonucleosomes configurations and divide it by the number of nucleosomes. These energies per nucleosome result in the following values: -0.75 kcal/mol (hiMS), -1.5 kcal/mol (MS-LH), and -1.9 kcal/mol (MS-LH-Mg). Thus, the absolute strength of internucleosomal interactions mediated by the histone tails in our oligonucleosomes increases with the degree of compaction. Our electrostatic internucleosomal interaction strength at MS-LH condition can also be compared with energy values obtained via optical tweezers by the Bustamante group (~2.0 kcal/mol).74 Note that the experimental values contain contributions to the internucleosomal energies from specific (hydrogen-bonding) interactions between the H4 tail and the acidic patch and solvation/desolvation energies that are not included in our model. Hence, our somewhat smaller values compared to the experimental values may be attributed to these missing specific and solvation energies in our model. No experimental data exist for comparing the computed internucleosomal energies at the other two conditions.
4 Discussion
The results presented here from mesoscale modeling/simulation have several implications in chromatin biology and raise several important questions regarding the role of histone tails and linker histones in chromatin compaction and regulation.
First, our analyses propose a detailed mechanism by which H4 tails make chromatin compact and why their chemical modification or removal triggers dramatic unfolding of nucleosomal arrays in in vitro reconstituted chromatin.22,23 Our results suggest that the H4 tails mediate a largest number of internucleosomal interactions (Fig. 6), likely due to their favorable point of origin from the flat face of the histone octamer (Figs. 5,7). Our model also predicts that most of H4 tails' internucleosomal interactions are mediated through the wound DNA on neighboring nucleosomes (Fig. 7), and a smaller fraction of them are mediated through the so-called acidic patch on nucleosomes. Experimental results, however, indicate a more critical role of the acidic patch in maximally compaction of chromatin.81,82 To capture these detailed effects and achieve the fully compact 60 S state, all-atom details might be needed. Nonetheless, it is encouraging that our mesoscale model correctly predicts the propensity of the H4 tails to occupy locations close to the acidic patch and also to mediate the largest number of internucleosomal interactions among all histone tails.
Our results also suggest how the H3 tails could promote chromatin compaction by both mediating internucleosomal interactions and attaching to linker DNAs and screening their electrostatic repulsion to allow them to better pack along the chromatin axes. This observation also supports suggestions by Kan et al.21 that the H3 tails likely serve multiple functions in chromatin compaction and regulation. This common function of the H3 tail and the linker histone in electrostatic screening of linker DNAs also explains the postulated redundancy between the H3 tails and the linker histone observed by Leuba et al.12
We have also confirmed previous experimental observations that the H2A/H2B tails are not important for chromatin compaction.22 Interestingly, we have uncovered a new possible role of the H2A and H2B tails: that of mediating fiber/fiber interactions across distant portions of the chromatin fibers, an effect with direct implications on the higher-order folding of chromatin. This new role of the H2A/H2B tails explains recent experimental observations by Gordon et al.5 that oligonucleosomes lacking the H2A and H2B tail regions fail to oligomerize into aggregates even at high magnesium concetrations. On the other hand, oligonucleosomes missing the H3 and H4 tails still oligomerize at higher magnesium concentrations.
Our tail neutralization analysis of Table 2 suggests that removal of individual tails and/or all tails does not result in complete unfolding of chromatin when the linker histones are present (both with and without magnesium ions). This implies that the linker histones possibly play the important role of stabilizing moderately folded fiber states (S20,w~ 45 S) during processes like chromatin remodeling to compensate for the loss of internucleosomal interactions through contacts with external proteins. Template-directed transcription of genes, on the other hand, requires a greater degree of chromatin unfolding for gene accessibility and might require dissociation of linker histones from their binding regions on the nucleosome. Recent evidence indicates that linker histone binding is indeed a very dynamic process involving rapid binding and dissociation of linker histones from the nucleosomes, where their residence time is modulated by post-transciptional modifications.83
It is also interesting to note that the correlation between the total charge carried by a histone tail and the degree to which chromatin unfolds (ΔS20,w) upon its neutralization is only moderate (see Table 2). This observation may be explained by realizing that the tails do not condense chromatin through the electrostatic screening effect alone (which would lead to a strong correlation between charge and compaction) but also by intermittently attaching to linker DNAs and nucleosome cores, a phenomenon which depends strongly on the location and length of the histone tail in addition to the charge by the tail. We also note that the individual contributions from each tail towards chromatin folding are not additive. For example, in hiMS conditions, the sum of ΔS20,w from each tail (=14.5) exceeds the actual compaction brough by the tails collectively (=7.3). This implies that there is a fair degree of redundancy in the role of each tail towards chromatin compaction. Furthermore, since ΔS20,w is a nonlinear function of the nucleosome-nucleosome distances, which themselves are complex functions of the nucleosome and tail charges, we do not anticipate the combined effect of histone tails to be additive. In sum, Nature has designed the tails to serve very specific purposes, but there is some redundancy involved in their function, which likely serves to make chromatin structure more robust towards perturbations.
Our study also postulates and expands a detailed mechanism by which linker histones and magnesium ions consort to produce highly compact chromatin.57 Briefly, the linker histones contrict the entry-exit angle of the linker DNA at the nucleosome and reduce the electrostatic repulsion between linker DNAs to promote a compact two-start zigzag arrangement of nucleosomes where the strongest internucleosomal interactions occur between alternate nucleosomes. Indeed our oligonucleosome configurations agree well with the various zigzag models proposed in the literature, e.g., by Dorigo et al,84 Wong and Mozziconacci,85 Bednar et al,67,70 Williams and Langmore,65 and Kepper et al.29 Magnesium ions further enhance compaction by strongly screening electrostatic repulsion among linker DNAs, promoting sharp bending of a small fraction, similar to linker DNA bending proposed in various solenoid models of chromatin by Thoma et al,86 McGhee et al,87 Widom and Klug,88 and Robinson et al.78 Both effects result in a denser packing of linker DNAs along the chromatin axis. Significantly, this proposed polymorphic structure of chromatin consisting of straight and bent linker DNAs, also verified by crosslinking experimental studies,57 reconciles the apparently divergent models proposed for chromatin, namely the solenoidal and zigzag.72,78 A detailed analyses of chromatin conformations in the presence of linker histone only and both the linker histone and magnesium ions is described separately.57
Finally, our results also pave the way to new experimentation for testing our additional predictions. For example, our model indicates a dramatic increase in the bending stiffness of chromatin fibers upon the addition of linker histones, suggesting a plausible mechanism by which the higher-order folded structure of chromatin could be modulated by linker histone binding and dissociation. Indeed, the cis entanglement between distant fiber portions that we capture without linker histones (Fig. 4b) become unfavored due to loss of order and accessibility of the genome template (Fig. 4c,d). This could be tested through measurement of the bending stiffness of chromatin associated with and deficient of linker histones. A systematic experimental study on the impact of histone tail removal on the sedimentation coefficient of nucleosomal arrays and higher-order folding under the different conditions investigated here could also be conducted and compared with our predictions of ΔS20,w. Our predictions on the frequencies of various interactions mediated by the histone tails (internucleosomal, intranucleosomal, parent linker DNA, neighboring linker DNA) could also be tested via experimentation. Further, details about the internucleosomal attachment sites of histone tails could also be verified experimentally.
5 Conclusions
Mesoscale modeling and simulation was used to systematically dissect the role of histone tails, linker histone, and magnesium ions in compacting physiological chromatin. Our model extends upon the prior mesoscale oligonucleosome model by incorporating both linker histones and magnesium ions and improving the nucleosome-DNA geometry according to the latest nucleosome geometry. The refined chromatin model was simulated using a tailored Monte Carlo methodology consisting of local and global moves to obtain a thermodynamically consistent ensemble of oligonucleosome configurations at different conditions in terms of low/physiological monovalent salt concentration and presence/absence of linker histone and magnesium ions. The resulting ensembles were examined using different analyses: positional distributions of histone tails relative to parent nucleosomes; attachment sites, frequencies, and energetics of internucleosomal interactions mediated by the histone tails; sedimentation coefficient calculation of regular oligonucleosomes and modified oligonucleosomes with selectively neutralized histone tails and linker histone domains; and global and internal oligonucleosome structure.
Our analyses uncover a detailed physical mechanism for chromatin compaction: linker histones alter the trajectory of linker DNAs emerging from the nucleosomes to promote greater interactions between alternate nucleosome; divalent ions allow sharp bending of a small portion of linker DNAs to allow greater compaction by reducing the number of linker DNA crossings at the chromatin axis; H2A and H2B tails mediate internucleosomal interactions across distant portions of the fiber (fiber/fiber interactions); and the H3 and H4 histone tails stabilize a compact state of chromatin by mediating attractive internucleosomal interactions (H3 and H4) and attaching to linker DNAs (H3). We also find that the H2A and H2B tails may be more important for mediating fiber/fiber interactions than internucleosomal interactions. Our results highlight the dynamical nature of histone tails and suggest how chemical modifications of their residues could alter chromatin morphology to regulate gene transcription. Such epigenetic factors are of great current interest.
Acknowledgments
G. Arya would like to thank the Hellman Fellowship for supporting part of this work. This work was also supported by NSF grant MCB-0316771 and NIH grant R01 GM55164 to T. Schlick. Acknowledgment is also made to the donors of the American Chemical Society Petroleum Research Fund for support (or partial support) of this research (Award PRF39225-AC4 to T. Schlick). Research described in this article was also supported in part by Philip Morris USA Inc. and Philip Morris International through an award to T. Schlick. We thank Dr. Nagasuma Chandra for providing the structure of the rat H1d linker histone, and Dr. Sergei Grigoryev for valuable discussions.
References
- 1.Felsenfeld G, Groudine M. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Horn PJ, Peterson CL. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 3.Tse C, Hansen JC. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 4.Hansen JC, Tse C, Wolffe AP. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 5.Gordon F, Luger K, Hansen JC. J. Biol. Chem. 2005;280:33701–3706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 6.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JJ, Pruss D, Wolffe AP. Proc. Natl. Acad. Sci. USA. 1994;91:7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown DT, Izard T, Misteli T. Nat. Struct. Mol. Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan L, Roberts VA. Proc. Natl. Acad. Sci. USA. 2006;103:8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Holde K, Zlatanova J. Proc. Natl. Acad. Sci. USA. 1996;93:10548–10555. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. J. Mol. Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 12.Leuba SH, Bustamante C, van Holde K, Zlatanova J. Biophys. J. 1998;74:2830–2839. doi: 10.1016/S0006-3495(98)77990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kepert JF, Tóth KF, Caudron M, Mucke N, Langowski J, Rippe K. Biophys. J. 2003;85:4012–4022. doi: 10.1016/S0006-3495(03)74815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Zhang Q, Schlick T. Proc. Natl. Acad. Sci. USA. 2005;102:8180–8185. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widom J. J. Mol. Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz PM, Hansen JC. J. Biol. Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 17.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouzina I, Bloomfield VA. Biophys. J. 1998;74:3152–3164. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore SC, Ausió J. Biochem. Bioph. Res. Co. 1997;230:136–139. doi: 10.1006/bbrc.1996.5903. [DOI] [PubMed] [Google Scholar]
- 20.Zheng C, Lu X, Hansen JC, Hayes JJ. J. Biol. Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 21.Kan P-Y, Hayes JJ. Methods. 2007;41:278–285. doi: 10.1016/j.ymeth.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Dorigo B, Schalch T, Bystricky K, Richmond TJ. J. Mol. Biol. 2003;372:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 23.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Science. 2006;10:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 24.Katritch V, Bustamante C, Olson WK. J. Mol. Biol. 2000;295:29–40. doi: 10.1006/jmbi.1999.3021. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Haim E, Lesne A, Victor J-M. Phys. Rev. E. 2001;64:051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 26.Beard DA, Schlick T. Structure. 2001;9:105–114. doi: 10.1016/s0969-2126(01)00572-x. [DOI] [PubMed] [Google Scholar]
- 27.Wedemann G, Langowski J. Biophys. J. 2002;82:2847–2859. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langowski J, Schiessel H. Theory and computational modeling of the 30 nm chromatin fiber. In: Zlatanova J, Leuba SH, editors. Chromatin structure and dynamics: State-of-the-art. Elsevier B. V.; Amsterdam, The Netherlands: 2004. [Google Scholar]
- 29.Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K. Biophys. J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arya G, Zhang Q, Schlick T. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arya G, Schlick T. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arya G, Schlick T. J. Chem. Phys. 2007;126:044107:1–044107:12. doi: 10.1063/1.2428305. [DOI] [PubMed] [Google Scholar]
- 33.Beard DA, Schlick T. Biophys. J. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, Beard DA, Schlick T. J. Comput. Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 35.Schlick T, Fogelson A. ACM Trans. Math. Softw. 1992;18:46–70. [Google Scholar]
- 36.Schlick T, Fogelson A. ACM Trans. Math. Softw. 1992;18:71–111. [Google Scholar]
- 37.Gilson MK, Sharp KA, Honig BH. J. Comput. Chem. 1988;9:327–335. [Google Scholar]
- 38.Sharp KA, Honig B. Ann. Rev. Biophys. Biophys. Chem. 1990;19:327–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 39.Sharp KA, Honig B. J. Phys. Chem. 1990;94:7684–7692. [Google Scholar]
- 40.Connolly ML. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 41.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 42.Allison SA, Austin R, Hogan M. J. Chem. Phys. 1989;90:3843–3854. [Google Scholar]
- 43.Stigter D. Biopolymers. 1977;16:1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 44.Bharath MMS, Chandra NR, Rao MRS. Nucl. Acids Res. 2003;31:4264–4274. doi: 10.1093/nar/gkg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bharath MMS, Chandra NR, Rao MRS. Proteins. 2002;49:71–81. doi: 10.1002/prot.10204. [DOI] [PubMed] [Google Scholar]
- 46.Vlachy V. Annu. Rev. Phys. Chem. 1999;50:145–165. doi: 10.1146/annurev.physchem.50.1.145. [DOI] [PubMed] [Google Scholar]
- 47.Bloomfield VA. Biopolymers. 1998;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 48.Korolev N, Lyubartsev AP, Nordenskiold L. Biophys. J. 2006;90:4305–4316. doi: 10.1529/biophysj.105.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai L, Mu Y, Nordenskiold L, van Maarel JRC. Phys. Rev. Lett. 2008;100:118301. doi: 10.1103/PhysRevLett.100.118301. [DOI] [PubMed] [Google Scholar]
- 50.Carnie SL, Torrie GM. Adv. Chem. Phys. 1984;56:141–263. [Google Scholar]
- 51.Pillardy J, Czaplewski C, Liwo A, Wedemeyer WJ, Lee J, Ripoll DR, Arlukowicz P, Oldziej S, Arnautova YA, Scheraga HA. J. Phys. Chem. B. 2001;105:7299–7311. [Google Scholar]
- 52.Scheraga HA, Pillardy J, Liwo A, Lee J, Czaplewski C, Ripoll DR, Wede-meyer WJ, Arnautova YA. J. Comput. Chem. 2002;23:28–34. doi: 10.1002/jcc.1154. [DOI] [PubMed] [Google Scholar]
- 53.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 54.Miyazawa S, Jernigan RL. J. Mol. Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 55.Sitkoff D, Sharp KA, Honig B. J. Phys. Chem. 1994;98:1978–1988. [Google Scholar]
- 56.Sheng S, Czjkowsky DM, Shao Z. Biophys. J. 2006;91:L35–L37. doi: 10.1529/biophysj.106.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grigoryev S, Arya G, Correll S, Woodcock C, Schlick T. submitted. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toth K, Brun N, Langowski J. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 59.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 60.Siepmann JI, Frenkel D. Mol. Phys. 1992;75:59–70. [Google Scholar]
- 61.de Pablo JJ, Laso M, Suter UW. J. Chem. Phys. 1992;96:2395–2403. [Google Scholar]
- 62.Frenkel D, Mooij GCAM, Smit B. J. Phys.: Cond. Matter. 1992;4:3053–3076. [Google Scholar]
- 63.Rosenbluth MN, Rosenbluth AW. J. Chem. Phys. 1955;23:356–359. [Google Scholar]
- 64.Hansen JC, Ausio J, Stanik VH, van Holde KE. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 65.Williams SP, Langmore JP. Biophys. J. 1991;59:606–618. doi: 10.1016/S0006-3495(91)82276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerchman SE, Ramakrishnan V. Proc. Natl. Acad. Sci. USA. 1987;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Kostner AJ, Woodcock CL. Proc. Nat. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao J, Lowary PT, Widom J. Proc. Natl. Acad. Sci. USA. 1990;87:7603–7607. doi: 10.1073/pnas.87.19.7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao J, Lowary PT, Widom J. Biochemistry. 1991;30:8408–8414. doi: 10.1021/bi00098a019. [DOI] [PubMed] [Google Scholar]
- 70.Bednar J, Horowitz RA, Dubochet J, Woodcock CL. J. Cell. Biol. 1995;131:1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bertin A, Leforestier A, Durand D, Livolant F. Biochemistry. 2004;43:4773–4780. doi: 10.1021/bi036210g. [DOI] [PubMed] [Google Scholar]
- 72.Schalch T, Duda S, Sargent DF, Richmond TJ. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 73.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C. Proc. Natl. Acad. Sci. USA. 1994;91:11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui Y, Bustamante C. Proc. Nat. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirkwood JG. J. Polym. Sci. 1954;12:1–14. [Google Scholar]
- 76.Bloomfield V, Dalton WO, van Holde KE. Biopolymers. 1967;5:135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- 77.Huynh VAT, Robinson PJJ, Rhodes D. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 78.Robinson PJJ, Fiarall L, Huynh VAT, Rhodes D. Proc. Natl. Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fletcher TM, Hansen JC. J. Biol. Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 80.Carruthers LM, Hansen JC. J. Biol. Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 81.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. Nat. Struct. Mol. Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 83.Bustin M, Catez F, Lim J-H. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 84.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 85.Wong H, Victor J-M, Mozziconacci J. PLoS ONE. 2007;2:e877:1–8. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thoma F, Koller T, Klug A. J. Cell. Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGhee JD, Nickol JM, Felsenfeld G, Rau DC. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- 88.Widom J, Klug A. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]