Summary
During infection, Legionella pneumophila creates a replication vacuole within eukaryotic cells and this process requires a Type IVb secretion system (T4bSS). IcmQ is a critical component of this translocase and associates with IcmR. In this report, we show that the N-terminal domain of IcmQ (Qn) mediates IcmQ dimerization, while the C-terminal domain with a linker region promotes a stable membrane association of IcmQ. We then determined crystal structures of Qn with the interacting domain of IcmR. In this complex, each protein forms an α-helical hairpin within a parallel 4-helix bundle. We find that IcmR binding to IcmQ prevents dimerization of IcmQ and blocks membrane permeabilization. However, IcmR does not completely block membrane association of IcmQ. The amphipathic nature of Qn suggests two models for how IcmQ may permeabilize membranes. The Rm-Qn structure also suggests how hyper-variable IcmR-like proteins in other Legionellae may interact with their IcmQ partners to regulate IcmQ function.
Keywords: IcmQ, IcmR, Type IVb secretion system, Legionella pneumophila
Introduction
Legionella pneumophila is a Gram negative bacterium that grows within amoebae (Fields, 1996). This pathogen is passed to humans in contaminated aerosols (Doebbeling & Wenzel, 1987) and is engulfed by alveolar macrophages (Horwitz & Silverstein, 1980; Horwitz, 1983a; Horwitz & Maxfield, 1984; Coers et al., 1999). After uptake into host cells, vesicles surround the Legionella-containing vacuole and this compartment is transformed into a haven for bacterial replication (Roy & Tilney, 2002; Tilney et al., 2001; Swanson & Isberg, 1995; Horwitz, 1983b). Remodeling of the endocytic pathway depends upon protein effectors that are translocated into the host cell by a Type IVb secretion system (T4bSS; Christie & Vogel, 2000). Repeated cycles of infection, intracellular replication and cell lysis may result in a severe pneumonia, known as Legionnaires' disease. In addition, Legionella pneumophila (Lp) is emerging as a significant problem in the context of hospital-acquired diseases and is often fatal to patients.
The T4bSS is encoded by ∼27 genes and is required for growth within host cells (Berger & Isberg, 1993; Marra et al., 1992). These translocase genes have overlapping designations and are named dot, for defective in organelle traffic and icm for intracellular multiplication. The Dot/Icm translocase is related to plasmid-based, DNA conjugation systems such as R64 and Collb-P9 (Segal et al., 1998; Vogel et al., 1998; Pohlman et al., 1994). However, the primary function of the Dot/Icm translocase during infection is to deliver protein effectors to the host cell cytoplasm (Chen et al., 2004; Luo & Isberg, 2004; Conover et al., 2003; Nagai et al., 2002). Current estimates suggest that the Dot/Icm apparatus may translocate more than 100 different effector proteins into target cells (Altman and Segal, 2008; de Felipe et al., 2005; Kubori et al., 2008; Luo and Isberg, 2004; Shohdy et al., 2005; Zuman et al., 2007; RI, unpublished). Hence, secretion signals may be present in many of these translocated proteins (Ninio et al., 2005). An almost identical T4bSS is present in Coxiella burnetii (Zamboni et al., 2003), which causes Q-fever/endocarditis in humans, and related translocases are found in Rickettsiella, an arthropod pathogen (Leclerque & Kleespies, 2008) and in Bartonella (Saenz et al., 2007).
IcmR and IcmQ are not found in some plasmid-based conjugation systems (cf. Incl; Komano et al., 2000) and presumably have unique roles in the Dot/Icm translocase (Sexton & Vogel, 2002). Legionella with an icmQ knockout are not viable for intra-cellular growth (Segal et al., 1998; Vogel et al., 1998) and exhibit salt resistance in their growth on defined media (Katz and Hashemi, 1982). Since salt sensitivity requires an intact translocase, IcmQ and IcmR are thought to help assemble or maintain the Dot/Icm translocase. IcmR and IcmQ form a complex in bacterial cytoplasm (Coers et al., 2000) and aggregates of over-expressed IcmQ can be dispersed by IcmR, an activity which is similar to that observed in some chaperones (Duménil & Isberg, 2001). In these studies, IcmQ was shown to interact with membranes and release calcein molecules from dye-loaded vesicles. However, IcmQ did not release a larger dextran which suggested that discrete pores or limited membrane disruptions may be formed when IcmQ interacts with a membrane. The middle region of IcmR (Rm) binds to the N-terminal domain of IcmQ (Qn) and this interaction blocks dye release by IcmQ (Duménil et al., 2004).
To further our understanding, we have characterized the protein fold, oligomerization and membrane association of IcmQ and IcmR. In particular, we found that the C-terminal domain of IcmQ with a trypsin sensitive linker (Qcl) is well folded. This Qcl region is responsible for the stable association of Qn with membranes but is not able to permeabilize membranes. Membrane association of the Qn domain in the context of IcmQ could lead to membrane permeabilization or alternatively, other factors that may bind to the Qn domain and thus, would be targeted to the membrane.
Crystal structures were determined of the interacting domains of IcmR and IcmQ (the Rm-Qn complex). In these structures, the Rm and Qn domains form helical hairpins within a 4-helix bundle that is stabilized by hydrophobic packing. Mutation studies revealed that Qn is required for IcmQ function. Our structure suggests that hyper-variable homologs of IcmR in Legionellae and in Coxiella (Feldman and Segal, 2004; Feldman et al., 2005) may interact with their cognate IcmQs by forming a similar four-helix bundle that uses similar hydrophobic residues. This in turn, suggests how IcmR and its orthologs may regulate IcmQ within the Dot/Icm translocase. The amphipathic nature of the Qn helical hairpin also provides a rationale for how IcmQ may permeabilize or disrupt membranes in the absence of IcmR.
Results
IcmQ and IcmR
IcmQ is comprised of an N-terminal domain, denoted Qn (amino acids 1-57), and the C-terminal Qc domain (aa 72-191). A trypsin-sensitive linker is present between the two domains (Figure 1A; Duménil et al., 2004). IcmQ tends to aggregate in the absence of IcmR during its expression (Duménil & Isberg, 2001), so we devised a sequential purification that removed IcmQ aggregates (Experimental Procedures). In the last step, purified IcmQ was eluted in high salt from a Hi-Trap S column (Figure 1B, lane 1). Purified IcmQ runs as a dimer on a calibrated Superose-12 HR column (Figure 1E, S_Table 1) and crosslinking with DTSSP verified the existence of a dimer (Figure 1B, lane 2). However, aggregates of IcmQ reformed when the protein was concentrated to ∼2-5 mg/ml. We characterized the folded state of IcmQ and found that the circular dichroism (CD) spectrum in the far UV is typical of an α-helical protein (Figure 1B, right). This suggests that the Qn and Qc domains are folded and mostly α-helical within IcmQ.
Figure 1.
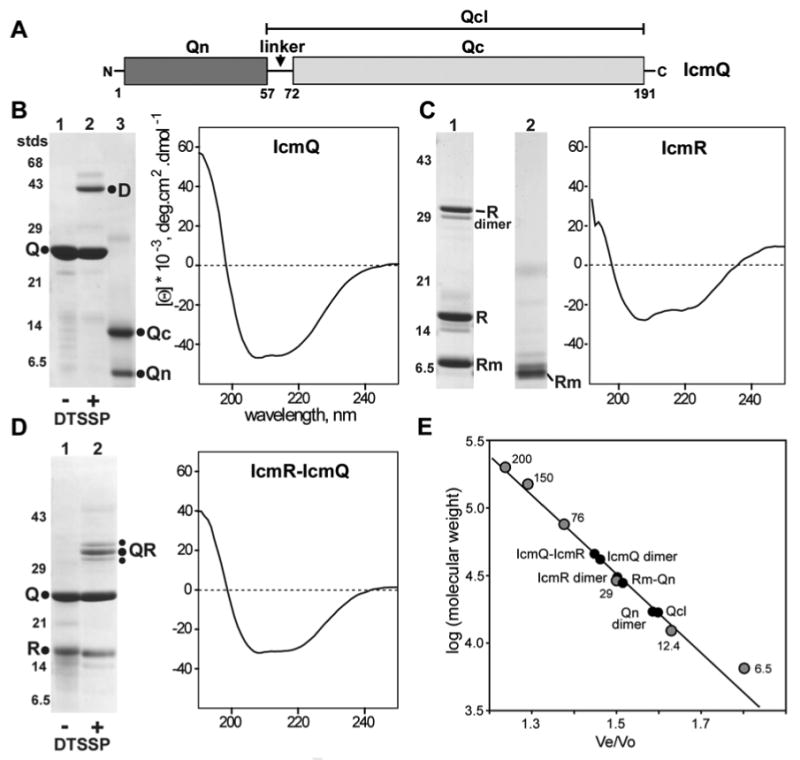
Purification and characterization of IcmQ, IcmR and IcmR-IcmQ.
A. A domain diagram is shown for IcmQ.
B. (left) Purified IcmQ is shown on an SDS gel (lane 1). Crosslinking with DTSSP reveals an IcmQ dimer (lane 2). Trypsinization of IcmQ overnight at a 1:1000 ratio (wt/wt) generates Qn and Qc domains (lane 3). (right) A CD spectrum of IcmQ shows that the protein is mostly α-helical.
C. (left) Purified IcmR is shown in lane 1 and the moderately trypsin resitant product is shown in lane 2. (right) IcmR is strongly α-helical based on its CD spectrum.
D. (left) Purified IcmR-IcmQ complex is shown on an SDS gel (lane 1) and DTSSP crosslinking reveals a simple heterodimer (lane 2). (right) IcmR-IcmQ retains the α-helicity of its components, based on a CD spectrum.
E. Molecular weights of IcmQ, IcmQ-domains, IcmR and their complexes were determined on a calibrated Superose-12 HR column. A plot of the log of molecular mass versus elution position is shown for standards and the purified proteins (also see S_Table 1).
Next, we over-expressed and purified IcmR. During purification, IcmR is slowly digested by an endogenous bacterial protease to form a smaller fragment that can account for ∼30-40% of the sample (Experimental Procedures, Figure 1C, lane 1). This purified IcmR mixture may contain roughly equal amounts of α-helix and unstructured regions, based on a CD spectrum in phosphate buffer (Figure 1C, right). The main protein peak migrated on a Superose 12-HR column as an apparent IcmR dimer (Figure 1E, S_Table 1), but DTSSP cross-linking was inconclusive, perhaps due to the low pI of IcmR which reflects a relative paucity of basic residues. However, an SDS resistant dimer of IcmR was present on 15% gels (Figure 1C, lane 1). The middle region of IcmR (Rm) forms a stable complex with the Qn domain (Duménil et al., 2004) and this prompted us to carry out a limited trypsinization of IcmR. This digestion produced a shorter Rm-like fragment (Figure 1C, lane 2). We speculate that the central Rm region (E23-R95; Duménil et al., 2004) is probably not stable unless paired with a second copy of itself or another protein, such as IcmQ. Thus, we suggest that IcmR may form a homodimer under some conditions using its Rm region.
We then purified the IcmR-IcmQ complex after co-expressing the two proteins (Figure 1D, lane 1). A CD spectrum of IcmR-IcmQ shows that the complex is partly α-helical (Figure 1D, right). When run on a molecular sizing column, the IcmR-IcmQ complex has an apparent molecular weight of ∼50 kDa (Duménil & Isberg, 2001; also see Figure 1E). This places the IcmR-IcmQ complex midway between a heterodimer (38 kDa) and a dimer of heterodimers (76 kDa, see S_Table 1). We used crosslinking with DTSSP to resolve this issue. Under mild conditions, we found a prominent band at ∼35 kDa after SDS PAGE, along with two weaker bands which closely flanked the major band. The position of these bands is consistent with a 1:1 heterodimer of IcmR and IcmQ. The observed banding pattern probably results from heterodimers stabilized with different crosslinks, which would affect their mobility (Figure 1D, lane 2). In addition, analytical ultracentrifugation (AUC) in TN buffer gave a 2S peak consistent with a simple IcmR-IcmQ heterodimer at neutral pH (not shown).
We conclude that IcmR and IcmQ form a complex with a 1:1 stoichiometry and this is due primarily to interactions between the Rm and Qn domains (Duménil et al., 2004; see next section). In addition, our data show that IcmQ and IcmR may each form homodimers in the absence of their binding partner.
The Qn and Qcl domains
To study the modular nature of IcmQ, we trypsinized the protein at a trypsin to IcmQ ratio of ∼1:1000 (wt/wt) and found that both the N-terminal and C-terminal domains of IcmQ remain intact after proteolysis of the putative linker (Figure 1B, lane 3). This prompted us to further characterize the Qn domain. First, we expressed Qn(1-66)-6xHis and used trypsin to generate a stable product without the tag (Qn 1-57); Figure 2A, lanes 1-3). At this stage, the Qn domain (∼6 kDa) formed a population of oligomers that ran at ∼150-200 kDa on a molecular sizing column (not shown). The Qn domain was purified further to remove aggregates, following our approach with IcmQ (Experimental Procedures). We found that purified Qn(1-57) forms a simple dimer based on crosslinking with DTSSP (Figure 2A, lanes 4, 5) and its migration on a calibrated Superose-12 HR column in 0.1 M NaCl (Figure 1E, S_Table 1). However, the Qn dimer aggregates when the protein is concentrated, a behavior that is similar to IcmQ (not shown). A CD spectrum shows that the Qn dimer is strongly α-helical (Figure 2A, right).
Figure 2.
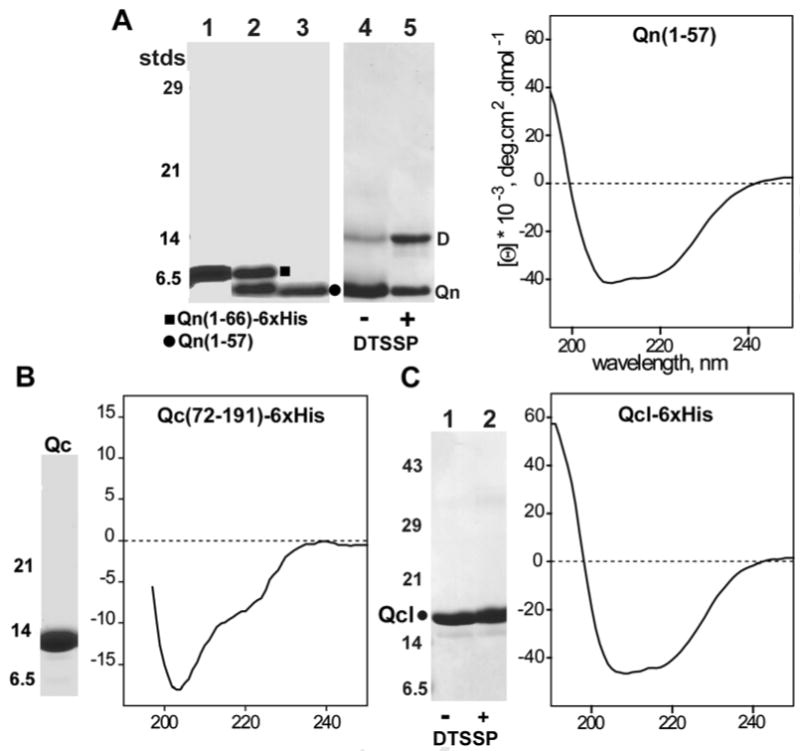
Characterization of the Qn, Qc and Qcl domains.
A. (left) A trypsinization time course is shown for Qn(1-66)-6xHis to form Qn(1-57) (lanes 1-3). Crosslinking with DTSSP shows that Qn is a dimer (lanes 4, 5). (right) Qn is mostly α-helical, based on a CD spectrum.
B. (left) Purified Qc is shown on an SDS gel. (right) The Qc domain is only partly-folded.
C. (left) Purified Qc(49/50-191) is shown on an SDS gel (lane 1) and cross-linking does not generate a visible product (lane 2). (right) A CD spectrum shows that Qcl is well-folded and strongly α-helical.
We next investigated the trypsin resistant Qc domain. After over-expression in E. coli, the purified Qc domain (Figure 2B, left) is stable to trypsinization but is only partly-folded, based on its CD spectrum (Figure 2B, right). In parallel studies, we recorded CD spectra of IcmQ during trypsinization and found a significant loss of α-helicity during proteolysis (not shown). Apparently, the destruction of the linker between Qn and Qc destabilizes the C-terminal Qc domain. We also over-expressed Qc with the trypsin-sensitive linker region, Qc (58-191). This produced an incompletely-folded protein (not shown). Hence, an intact Qn domain and linker region may be required to promote the proper folding of the Qc domain.
A helical wheel analysis of Qn suggested that this domain may form two amphipathic helices, which could promote dimerization of IcmQ. Thus, we created a double mutant (Leu17Asp, Leu39Asp) in the Qn domain to destabilize the predicted hydrophobic interface. Over-expression of this double mutant resulted in a degradation of the Qn domain, presumably because dimer interactions mediated by the predicted hydrophobic interface had been destabilized. Intriguingly, endogenous proteolysis of this double mutant created three longer versions of the Qc domain that retained the linker. These Qc proteins were well folded based on their CD spectrum (data not shown). Mass spectrometry provided information that allowed us to design an IcmQ mutant in which two additional lysine residues at possible protease clip sites were mutated. This quadruple mutant (Leu17Asp, Leu39Asp, Lys57Gln, Lys59Gln) was over-expressed and bacterial proteolysis produced a protein with its N-terminus at Lys48/Arg49 that contained the linker and Qc domain (Figure 2C, lane 1). We named this protein the Qcl domain, even though it retains a few C-terminal residues of Qn (Figure 1A). A normalized CD spectrum of purified Qcl shows that it retains significant α-helical character, similar to the Qn domain (Figure 2C, right panel). In addition, the Qcl domain migrated on a calibrated Superose-12 HR column as a monomer (Figure 1E, S_Table 1). This observation is consistent with the lack of inter-molecular crosslinking of this basic domain observed with DTSSP (Figure 2C, compare lanes 1 and lane 2).
In summary, IcmQ contains two domains connected by a trypsin sensitive linker (Duménil et al., 2004). The Qn domain forms a dimer and this domain (or the linker) may be required for the proper folding of Qc in vivo. The Qcl domain is a monomer and does not aggregate, as shown by a well dispersed, 2D HSQC spectrum (not shown). Hence, IcmQ dimerization is probably due to the α-helical Qn domain. As we show in a later section, Qn dimerization may involve the formation of a 4-helix bundle. In addition, IcmQ aggregation may be due to the Qn domain, as this region tends to aggregate strongly. This is supported by our observation that the Qcl domain remains monomeric at higher protein concentrations.
Qcl anchors IcmQ to membranes
In previous studies, IcmQ was shown to associate with membranes and release trapped calcein dye from vesicles (Duménil et al., 2004). The Qn domain by itself also released dye molecules; hence, it was proposed that Qn may form a pore when IcmQ is added to vesicles (Duménil et al., 2004). We repeated this experiment using freshly prepared Qn and Qcl domains to further dissect the processes of membrane association and dye release. In the membrane association experiments, we added proteins to preformed lipid vesicles made by extrusion with a PC-PG lipid mixture. Sucrose was then added to the vesicles (45% wt/wt) and the samples were over-laid with a step gradient. After spinning for 15 hrs at 55000 rpm, the pellet and floated vesicles were compared by SDS PAGE. We assayed “pore” formation using the calcein dye release assay (Duménil et al., 2004). In this approach, vesicles were preloaded with calcein and an increase in fluorescence was monitored due to the removal of inter-molecular quenching of released dye molecules.
First, we tested IcmQ and IcmR-IcmQ to provide positive and negative controls for calcein release, respectively (Table 1 and S_Figure 1; Duménil et al., 2004). Intriguingly, we also found that IcmQ caused vesicles to aggregate at higher protein concentrations, based on observed changes in sample turbidity and images of negatively-stained samples. This membrane aggregation may be due to extensive vesicle binding by the basic IcmQ molecules. We then tested freshly made Qn dimer, which was dialyzed into a low salt buffer, and verified that Qn at a low concentration (1 uM) does not associate with floated vesicles. However, it does release calcein dye from vesicles at 0.1 uM, which suggests that it may interact transiently with the membrane in the absence of Qcl (Table 1; Duménil et al. 2004). In addition, purified Qn did not associate with vesicles at higher protein concentration (25 uM), as seen previously (Duménil et al., 2004). This may be due to the fact that protein in the earlier studies was aggregated. In our experiments, we used freshly prepared Qn that was stored at ∼2 mg/ml and 4°C to prevent aggregation.
Table 1. Properties of IcmQ, Qn, Qcl and IcmR.
| Protein and salt concentration | Secondary structure and oligomeric state¶ | Vesicle association at 1 uM protein concentration | Vesicle association at 25 uM protein concentration | Calcein release 0.1 uM protein |
|---|---|---|---|---|
|
IcmQ (0.1 M salt) |
α-helical, dimer | Yes (>95%) | Yes (>95%) | Yes (64%) |
|
IcmQ (float with 0.5M salt) |
dimer | nd | No (< 2%) | nd |
|
Qn (aa 1-57) (0.1 M salt) |
α-helical, dimer | No | No | Yes (77%) |
|
Qcl (aa 48-191) (0.1 M salt) |
α-helical, monomer | Yes (>95%) | Yes (>95%) | No |
|
Qcl (float with 0.5 M salt) |
monomer | nd | No (< 5%) | nd |
|
IcmR-IcmQ† (0.1 M salt) |
α-helical with some unstructured regions; heterodimer |
+/- (~50/50) |
+/- (~50/50) |
No |
|
IcmR (0.1 M salt) |
α-helical (~50%), homodimer? | No | No | No |
| cytochrome c | ---- | Yes | Yes | No |
| GST | ---- | No | No | No |
nd- not determined.
The oligomer state was determined by comparing the elution position on a calibrated Superose-12 HR column (see Figure 1E, S_Table 1) and crosslinking with DTSSP.
Roughly half of the IcmR-IcmQ complex binds to membranes but will not form pores (see S_Figure 1).
1) The vesicle association and calcein release assays were done with an egg PC/PG mixture but similar results were obtained with E. coli lipids. See Experimental Procedures for details of the assays.
2) Protein association with vesicles is observed with Qcl. In contrast to Qc(72-191) studied previously, Qcl(48/49-191) is well folded as determined by CD spectroscopy.
3) Qn did not associate with membranes even at high protein concentrations (25 uM).
We then evaluated the role of Qcl and found that this basic domain binds to vesicles in the floatation assay at both 1 and 25 uM, but it did not release calcein dye molecules (Table 1). Additional controls showed that basic cytochrome c binds to vesicles, but does not release the calcein dye, while GST neither interacts with vesicles nor releases the dye. We then asked whether Qcl is located on the membrane surface. To test this, vesicles with IcmQ or Qcl were floated on a sucrose gradient in the presence of 0.5 M NaCl to disrupt electrostatic interactions between the basic proteins and the membrane. Under these conditions, very little IcmQ or Qcl floated with the vesicles (Table 1). Since the Qcl domain is released by high salt, this suggests that Qcl may remain on the surface of the vesicle during the targeting of IcmQ to the membrane.
Structure of the Rm-Qn complex
Interacting domains of IcmR and IcmQ correspond to the middle region of IcmR (Rm, E23-R95) and the Qn domain (Figure 3A; Duménil et al., 2004). To analyze this interaction, we expressed IcmR-6xHis and Qn(1-66)-6xHis separately, mixed the bacterial lysates and purified the IcmR-Qn(1-66) complex on a Nickel NTA column (Figure 3B, lane 1; Supplemental Methods). This complex was trypsinized to remove flexible residues from the two proteins. The resulting Rm-Qn complex was further purified with ammonium sulfate cuts (Figure 3B, lane 2 and blowup). The Rm-Qn complex is α-helical based on its CD spectrum (Figure 3C) and crystallization experiments produced crystals in space group P61. To solve the structure, we engineered an Rm(L84SeMet)-Qn complex, which gave crystals in space group P212121. A 3-wavelength MAD dataset allowed us to solve and refine the selenium derivative structure to 2.2Å resolution (S_Table 2). This model was then used to solve the native structure by molecular replacement and the final structure was refined to 2.1Å resolution (S_Table 2).
Figure 3.
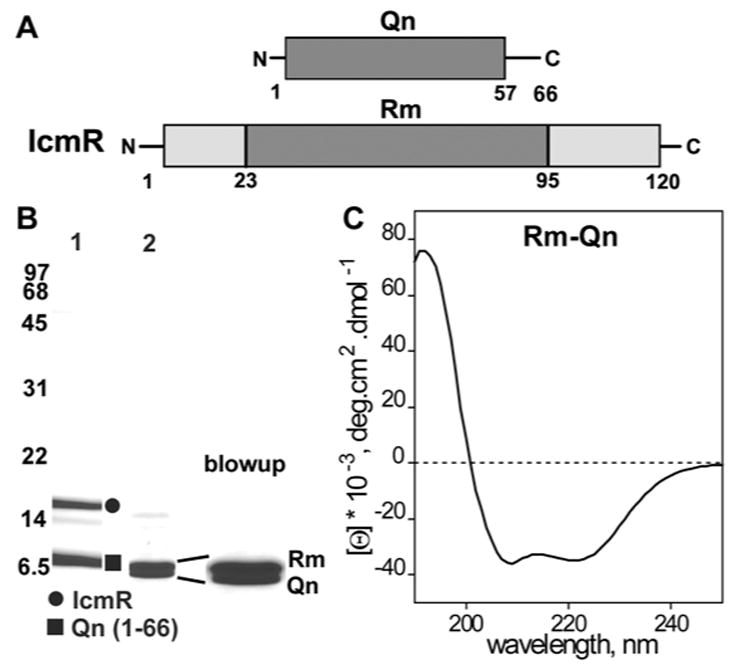
Purification and characterization of the Rm-Qn complex.
A. Domain diagrams are shown for IcmR and Qn and the interacting regions are shaded.
B. Purified IcmR-Qn(1-66)-6xHis complex is shown on an SDS gel (lane 1). Trypsinization generates the Rm-Qn complex (lane 2) and the similarly sized bands are shown in an expanded view on the right.
C. A CD spectrum indicates that the Rm-Qn complex is strongly α-helical.
A single Rm-Qn complex was present in the asymmetric unit of the derivative crystals with residues 4-53 for Qn and residues 29-86 of Rm in good density. The Rm-Qn complex forms a 4-helix bundle with two α-helices from Rm and two from Qn. These α-helices are aligned in parallel as two α-helical hairpins with a left-handed twist (Figure 4A). The positions of the α-helices in the Qn and Rm sequences are shown for these proteins in Figures 5A and 5B, respectively. In the 4-helix bundle, the α-helical hairpins of Rm and Qn are staggered (Figure 4A) and the Qn helices are also vertically offset with respect to each other. In addition, a proline is present in each of the inter-helical loops (Pro60 in Rm, Pro25 in Qn) and Pro44 causes a local break in the first Rm α-helix. In the native crystals, a dimer of Rm-Qn complexes was present in the asymmetric unit (S_Figure 2). Strikingly, the NCS dimer interface is formed by two Rm chains and creates a left-handed 4-helix bundle (S_Figure 2B). The overall structure of the Rm-Qn 4-helix bundle in the two crystal forms is nearly identical (S_Figure 2C), with the exception of a loop between the Rm α-helices. Differences in the Rm loop involve Phe58 which is flipped out to form a crystal contact with a neighboring basic residue through a pi-cation interaction in both crystal forms.
Figure 4.
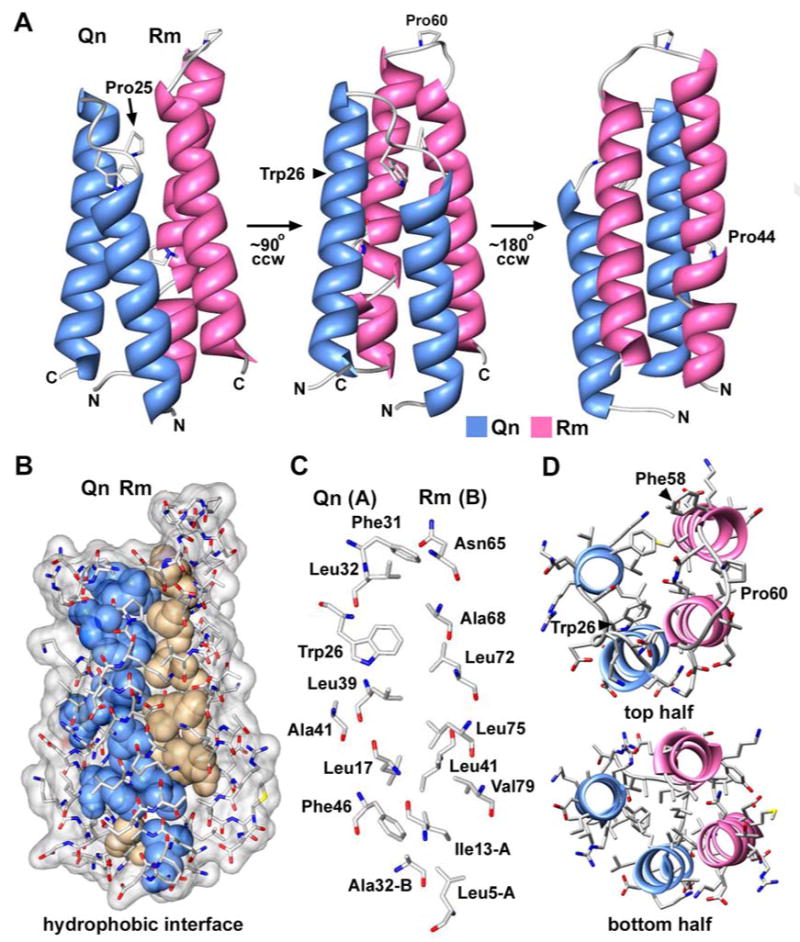
The structure of an Rm-Qn four helix bundle.
A. A rotation series of the Rm-Qn 4-helix bundle is presented with the molecules displayed as ribbons. This structure is from the derivative crystals.
B. Non-polar side chains in the Rm-Qn interface are shown as solid spheres and are color coded for those from Qn (blue) and from Rm (tan).
C. Side chains in the Rm-Qn interface are shown as stick models with CPK colors and are labeled.
D. A top view is shown of the Rm-Qn 4-helix bundle with α-helices displayed as ribbons and side chains as CPK stick models. The figure was made with Chimera.
Figure 5.
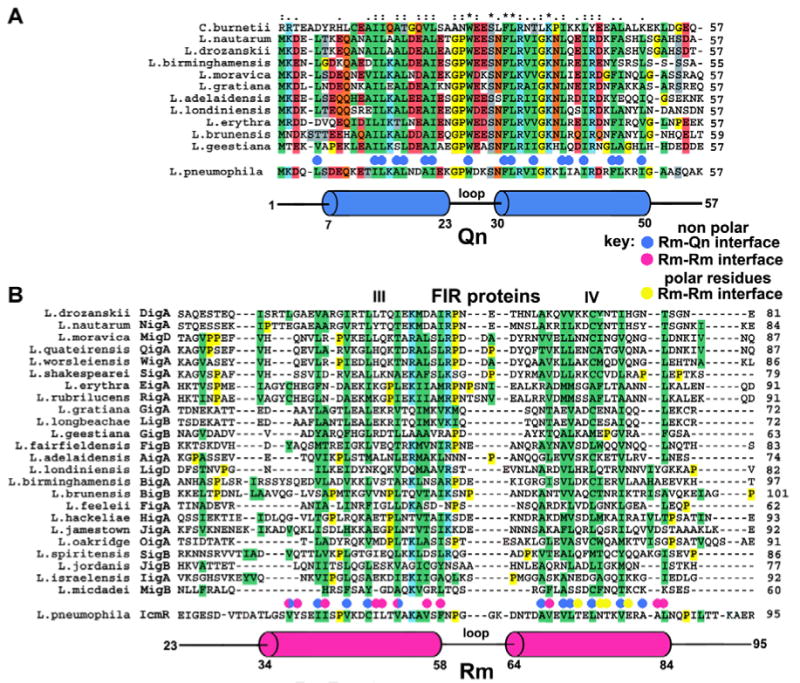
Structure-based sequence alignments of the interacting regions of IcmQ and IcmR.
A. A sequence alignment is shown for Qn domains from 11 representative Legionella species and from C. burnetti. Hydrophobic residues are highlighted in green. The Qn α-helices (in blue) are aligned below the sequences and hydrophobic residues in the Rm-Qn interface are marked with blue dots.
B. A sequence alignment is shown for the middle regions of 24 FIR proteins. The Rm α-helices (red) are aligned with IcmR and FIR sequences. Residues in the Rm-Qn interface are marked with blue dots. Hydrophobic residues in the Rm-Rm interface are marked with red dots, polar residues are marked with yellow dots.
The interface between the Rm and Qn α-helices in these crystals is formed by 27 hydrophobic residues and buries ∼2340 Å2 of surface area (Figures 4B-4D, S_Figure 2D, summarized in Figure 5). The properties of this interface are consistent with the estimated Kd of ∼20 nM for IcmR-IcmQ (Duménil & Isberg, 2001), although the actual Kd may be lower. Finally, salt bridges and two clusters of water-mediated interactions may stabilize the Rm-Qn 4-helix bundle (not shown). There is a salt bridge between Asp31 of Rm and Lys10 of Qn, while interactions between Ser36/Glu40 of Rm and Arg45/Arg49 of Qn are mediated by 4 waters. In a second cluster, interactions between Glu74/Lys78 of Rm and Lys23/Asp19 of Qn use 3 intervening waters.
We wondered about the relevance of the Rm dimer in native crystals grown at pH 4.7, since the IcmR-IcmQ complex is a simple heterodimer at neutral pH. In the NCS dimer, the Rm-Rm interface contains 16 non-polar residues, 8 polar residues and 4 buried water molecules (S_Figure 3). Low pH may favor formation of the Rm-Rm dimer in the crystal because one glutamate in the interface makes a pair of hydrogen bonds with two buried water molecules, while the second glutamate forms a hydrogen bond with a carbonyl group in the peptide backbone of the opposing polypeptide chain (S_Figure 3B). The second glutamate must be protonated for this interaction to occur. Thus, the Rm-Rm dimer may represent an unusual crystal contact that creates a 4-helix bundle at low pH which buries ∼2240 Å2 in the interface. In addition, only 5-6 hydrophobic residues in the Rm-Rm dimer interface are conserved in other members of the icmR/FIR gene family (Figure 5B). Hence, Rm dimerization at this interface may not be required for biological function. The precise nature of the putative IcmR dimer that forms in the absence of IcmQ remains to be determined, but it may utilize the extensive, apolar Rm interface that is normally used to associate with Qn, rather than the interface observed in the native crystals.
Together, these crystal structures provide insight into the roles of IcmR and IcmQ in the T4bSS. In particular, hydrophobic residues in the Rm-Qn interface are generally conserved in the IcmQ homolog from Coxiella burnetti (Figure 5A, blue dots). Overall, the formation of an extensive interface in the Rm-Qn 4-helix bundle suggests that IcmQ may be tightly bound by IcmR to prevent the Qn domain from destabilizing membranes or creating a pore.
The amphipathic Qn domain
Our data suggest that the Qn domain may form an α-helical bundle that mediates IcmQ dimerization and membrane permeabilization. When the Rm-Qn 4-helix bundle is pulled apart, the amphipathic nature of each Qn α-helix is evident in end-on views (Figure 6A). The N-terminal helix is acidic while the C-terminal helix is basic, as summarized in a helical wheel projection (Figure 6B). In the absence of IcmR, we theorize that Qn may form a stable helix bundle that promotes IcmQ dimerization. In this model, Qn α-helical hairpins could use their amphipathic interfaces to form a 4-helix bundle with either a parallel or an anti-parallel interface. However, the compact nature of the IcmQ dimer on a molecular sizing column (S_Table 1), would argue for a parallel dimer interface (Figure 6C), but this must be verified. In addition, the amphipathic nature of the Qn helices and their length suggests two possible mechanisms for calcein dye release. In one case, multiple Qn molecules would oligomerize and insert into the membrane, spanning the hydrophobic region of the bilayer to form a pore. In a second scenario, many Qn molecules could bind to the bilayer surface and destabilize the lipid packing thereby creating a membrane leak (see Shai, 2002).
Figure 6.
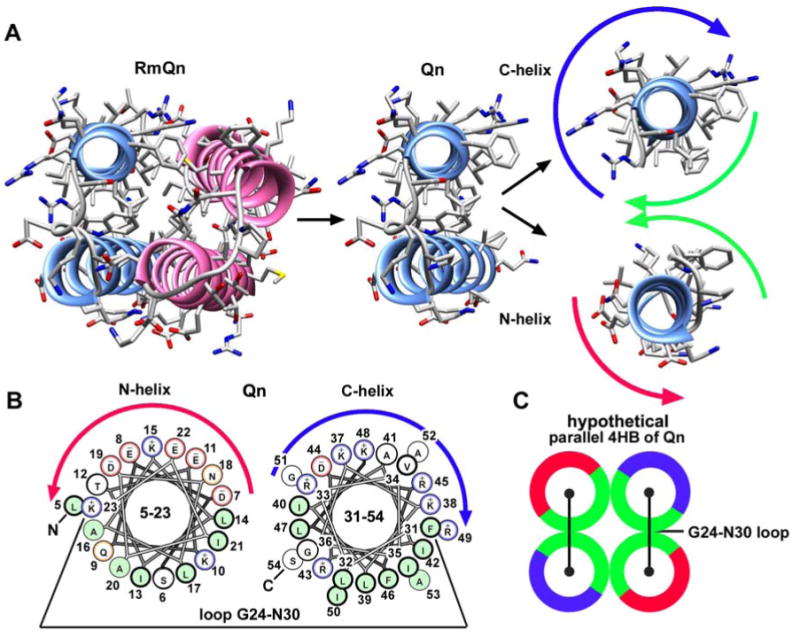
The Qn α-helices are amphipathic and show a strong charge segregation.
A. Qn α-helices are shown in the context of the Rm-Qn dimer (left) and are dissected out from the helix bundle (center, right). The Qn α-helices are amphipathic with a hydrophobic face (green arrows) and charged faces (blue arrow for basic and red arrow for acidic).
B. Helix wheel diagrams for the two Qn α-helices show the charge segregation between the two α-helices.
C. A possible model is shown for the hypothetical Qn-Qn 4-helix bundle that may mediate IcmQ dimerization.
To further define the role of Qn, a genetic screen was conducted to identify icmQ mutations that affected Dot/Icm-dependent functions. Because a functional Dot/Icm system interferes with L. pneumophila survival on medium containing 150 mM NaCl, icmQ alleles encoded on a mutagenized plasmid were identified based on their inability to restore salt sensitivity when transformed into a NaCl resistant, ΔicmQ strain of L. pneumophila (Supplemental Methods). In total, 4 mutant alleles were characterized including proteins with small deletions in Qn (Δ10-12, Δ40-42) or with amino acid substitutions, (Ileu3Asn and Pro25Leu/Ser29Phe), respectively. All of the IcmQ mutants were stably expressed in Legionella pneumophila (Figure 7A). IcmQ molecules containing the Δ40-42 and (Pro25Leu/Ser29Phe) mutations were unable to fully restore Dot/Icm-dependent activities, including L. pneumophila replication in macrophages (Figure 7B) and secretion of the DotA protein into the culture supernatant during growth in liquid medium (Figure 7C, Table 2). Amino acid substitutions in the inter-helical loop (Pro25Leu/Ser29Phe) or a deletion in the lower half of the interface (Δ40-42) did not affect IcmQ interactions with IcmR in vitro (Table 2), indicating that the defect in protein function caused by these icmQ mutations may not be related to a loss of IcmQ-IcmR complex formation per se. In addition, the (Pro25Leu/Ser29Phe) double mutant was fully active in calcein release. However, the Δ40-42 deletion mutant, which removed two isoleucines, was compromised in its ability to permeabilize the membrane (Table 2). We conclude that the Qn domain is critical to the function of IcmQ. However, we could not test the effects of a ΔQn mutant in Legionella because the resulting Qc domain is not completely folded when expressed in the absence of Qn, even though it is stable.
Figure 7.
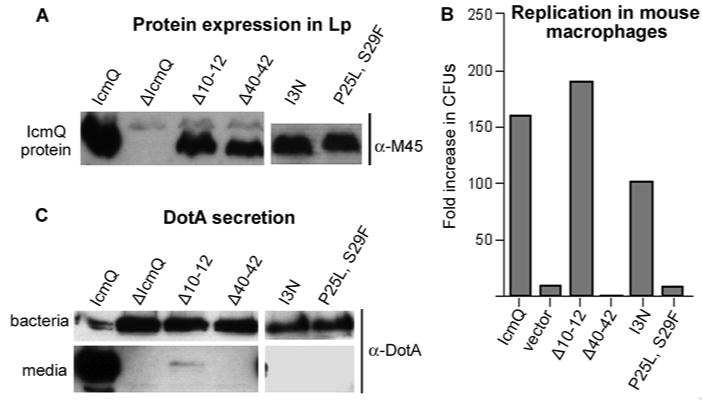
Mutations in Qn disrupt IcmQ function.
A. The stability of four IcmQ mutants expressed in L. pneumophila was ascertained by blotting with an antibody to the M45 epitope on the tagged IcmQ molecules. Cells infected with the vector without IcmQ (ΔIcmQ) did not express the protein, while the 4 mutants were made as full length proteins, albeit at lower levels than wild type IcmQ.
B. A plaque forming assay indicated that the Δ40-42 and [P25L, S29F] mutations in IcmQ inhibited growth of L. pneumophila in mouse macrophages.
C. In a DotA secretion assay, the protein was monitored in bacterial pellets and in the culture media by immuno-blotting. All 4 mutations in IcmQ prevented DotA secretion into the media, as this process is very sensitive to alterations in components of the T4bSS.
Table 2. Mutations in the Qn domain and their effects on IcmQ.
| Mutations | I13N | P25L, S29F | Δ10-12 | Δ40-42 |
|---|---|---|---|---|
| Stable IcmQ mutant protein in Legionella pneumophila | Yes | Yes | Yes | Yes |
| Intracellular growth in macrophages | +/- | No | Yes | No |
| DotA secretion | No | No | No | No |
| IcmR-IcmQ complexes† | nd | Yes | nd | Yes |
| Vesicle association of the IcmQ mutant protein | nd | Yes | nd | Yes |
| Calcein dye release by the IcmQ mutant protein* | nd | Yes (75%) |
nd |
+/- (23%) |
nd- not determined
Data for IcmQ stability in Legionella pneumophila, DotA secretion and intracellular growth in mouse macrophages are shown in Figure 7.
Complex formation was measured with recombinant proteins and is based on their mobility on a Superose-12 HR column and crosslinking with DTSSP.
Wild type IcmQ released ∼64% of the dye under these conditions.
Discussion
In this paper, we evaluated the functions of the N- and C-terminal domains of IcmQ. The Qn domain promotes IcmQ dimerization, while the Qcl region targets Qn to the membrane. IcmR binds to IcmQ and blocks membrane permeabilization, but does not completely block membrane association of IcmQ. Crystal structures of the interacting domains of IcmR and IcmQ revealed that the Rm and Qn domains form α-helical hairpins within a 4-helix bundle. This explains how a small and partly structured protein like IcmR can sequester the membrane active Qn domain. It also suggests how members of the hyper-variable family of IcmR homologs (the FIR proteins) may interact with cognate IcmQs in other Legionellae and Coxiella.
IcmR, IcmQ and domain functions
Studies of the Dot/Icm protein translocation machinery are at an early stage. However, some Dot/Icm proteins must form the translocase itself, while others regulate assembly or act as receptors and chaperones for cargo. Recent work has identified core constituents of the channel that span the inner and outer bacterial membranes (Vincent et al., 2006). Many of the ∼27 genes in the T4bSS share some sequence similarity with genes in plasmid-based, DNA conjugation systems (Segal et al., 1998; Vogel et al., 1998; Pohlman et al., 1994). However, translocation of proteins into target host cells presumably required the addition of 9 novel genes in the T4bSS that included icmR and icmQ (Christie and Vogel, 2000).
Functional properties of IcmQ and IcmR are summarized in Figure 8. In particular, IcmQ forms dimers and this process is mediated by the amphipathic and highly charged Qn domain. The geometry of the IcmQ dimer remains to be determined but may be formed by a parallel interaction between Qn domains (Figure 8 top left). IcmQ forms soluble aggregates at higher concentrations in the absence of IcmR (Figure 8, left; Duménil & Isberg, 2001) and this process may be due to the highly-charged Qn domain. In our studies, we confirmed that the amphipathic Qn domain interacts with IcmR (Figure 8, top right) and permeabilizes membranes (Figure 8, bottom left). In addition, IcmR may dimerize in the absence of IcmQ, while IcmR and IcmQ interact to form a simple heterodimer. We hypothesize that the central Rm region could mediate dimer formation by IcmR, due to an apolar surface which is conserved in all FIR proteins. This idea leads to a simple helix-pair exchange model, in which IcmR and IcmQ use their Rm and Qn domains respectively, to form self-dimers and to form the IcmR-IcmQ complex.
Figure 8.
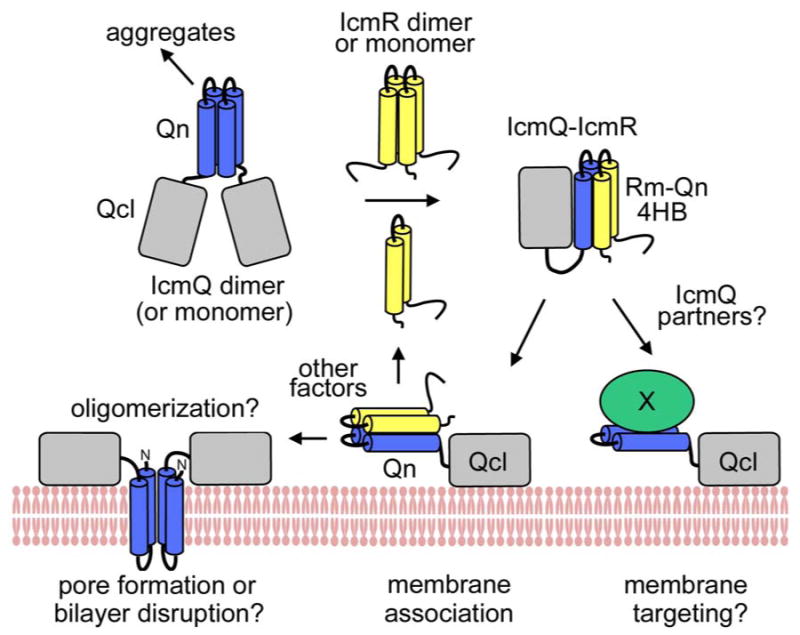
A model for IcmR and IcmQ function.
(top left) IcmQ dimers are formed by pairwise interactions between the Qn domains and the resulting interface may be either parallel or anti-parallel (not shown). IcmQ also aggregates in the absence of IcmR which may be mediated by Qn. (top middle) IcmR may form a dimer using interactions between Rm regions in the absence of IcmQ. (top right) IcmR interacts with the Qn domain to form a 4-helix bundle which “disrupts” the IcmQ dimer and also dis-aggregates IcmQ. (bottom right) Possible IcmQ binding partners could displace IcmR and would potentially be targeted to the cell membrane by the Qcl region. (bottom middle) IcmR does not completely block membrane association of IcmQ which is mediated by binding of Qcl to the bilayer surface. (bottom left) IcmR must be displaced from IcmQ to allow Qn to interact with the membrane.
Our studies also showed that the C-terminal Qc domain is not well-folded even when the linker is present during bacterial expression. Hence, the folding of Qc in vivo may require a folded Qn domain and/or an intact linker. In the next step, we made a well-folded Qcl domain using endogenous proteolysis of an IcmQ quadruple mutant. We found that the α-helical Qcl domain associates strongly with membranes using electrostatic interactions. While Qn can release calcein dye molecules from vesicles, it does not associate stably with membranes on its own. Thus, we suggest that the basic Qcl domain targets Qn to the membrane. This interaction is even capable of targeting IcmR-IcmQ complexes to the membrane (Figure 8, bottom middle). Other factors could release the Qn domain from IcmR, allowing Qn to form a pore and/or destabilize the membrane (Figure 8, bottom left). Alternatively, Qn may interact with other proteins that displace IcmR and target these proteins to the membrane surface via the Qcl domain (Figure 8, right).
Interactions between a chaperone-substrate pair
We determined crystal structures of the interacting domains of IcmR and IcmQ and found that each domain in the Rm-Qn complex is comprised of an α-helical hairpin. These helices have complimentary hydrophobic surfaces and interact to form a parallel 4-helix bundle. Thus, IcmR may keep IcmQ in an inactive state until another factor can disrupt this stable complex.
The site of IcmQ function and its precise role in the Dot/Icm translocase are not known. However, salt sensitivity of bacterial growth requires an intact T4bSS (Katz and Hashemi, 1982; Buscher et al., 2005) and growth becomes salt resistant when the icmQ gene is deleted. It is possible that IcmQ could be targeted to the inner bacterial membrane, as it readily interacts with membranes made from E. coli phospholipids (Duménil et al., 2004). Cell fractionation has shown that IcmR and IcmQ are both present in the cytoplasm before Legionella is exposed to a host cell (Duménil and Isberg, 2001). When IcmQ is over-expressed, it can be detected on the surface of L. pneumophila and this exposure is dependent upon bacterial contact with macrophages (Duménil et al., 2004). Unfortunately, this experiment is not definitive because protein localization after over-expression may not reflect the primary target site. Also, properties of the inner and outer membrane are quite different in Gram negative bacteria. Hence, the simplest model would invoke IcmQ association with the inner bacterial membrane, and additional work is needed to identify the site(s) of IcmQ function.
When taken together, the data allow us to propose a model in which IcmQ association with the membrane may help to assemble or stabilize the Dot/Icm translocase. Alternatively, IcmQ may target other proteins to the membrane which are required for a functional protein translocase. The role of IcmQ in this process is probably regulated by IcmR. The mechanism of membrane permeabilization or disruption by IcmQ and a possible function for this activity remains to be determined. However, the amphipathic nature of the two α-helices of Qn might allow the protein to insert into a membrane and form a pore by oligomerization or alternatively, IcmQ binding to the membrane surface might allow Qn to destabilize the lipid bilayer.
Structural conservation of FIR-IcmQ pairs
The icmQ and icmR genes are contiguous on the L. pneumophila chromosome (Segal et al., 1998). Although the icmQ gene is highly conserved in bacteria with a Dot/Icm apparatus, in species other than L. pneumophila the gene directly upstream from icmQ shows undetectable sequence homology to icmR (Feldman et al., 2005). However, each of these proteins forms a complex with its cognate IcmQ and thus, must have conserved structural features even though there are no obvious sequence similarities (Feldman et al., 2005; Feldman and Segal, 2004). These “functional homologs of IcmR” (FIR) exhibit a wide size range (71-135 residues) and vary greatly in their isoelectric points (4.7-9.5). Even so, cognate FIR-IcmQ pairs can rescue intracellular growth in an icmR/icmQ deficient strain of L. pneumophila. Therefore, the primary function of IcmR orthologs is to pair with their IcmQ partners (Feldman et al., 2005).
Our structure provides a basis for identifying common features in the FIR-IcmQ pairs that allow complex formation. In the Rm-Qn model, a parallel 4-helix bundle is formed with a hydrophobic interface. We suggest that conserved hydrophobic residues in the FIR proteins could participate in a similar interface. The idea that non-identical, hydrophobic side chains in FIR proteins may contribute to complex formation is supported by the observation that some of these proteins form complexes with the L. pneumophila IcmQ (Feldman et al., 2005).
To test this idea, we carried out a sequence alignment of 24 FIR proteins with Clustal X (Thompson et al., 1997), and the results are shown in Supplemental Figure 4 and Figure 5B. This analysis reveals that FIR sequences can be divided into 5 regions (labeled as blocks I through V; Supplemental Figure 4). Sequence similarity is limited to blocks III and IV, which correspond to the A and B helices in the Rm helical hairpin. Within these helices, a pattern of ∼9 conserved hydrophobic residues is present in the FIR proteins, to give an overall similarity of ∼8-10%. As these hydrophobic residues span the two amphipathic α-helices in Rm, we propose that regions III and IV in the FIR proteins could form similar α-helices. Moreover, this model can be extended to the CoxigA-IcmQ pair in the Coxiella burnetti T4bSS (Feldman et al., 2005; Zamboni et al., 2003).
The conservation of hydrophobic residues in the otherwise hyper-variable FIR proteins reinforces the idea that the helical and apolar interface of IcmR-IcmQ is critically important, and further suggests that the major role of the FIR proteins is to recognize their IcmQ partners. The function of non-conserved and presumably unstructured regions in the FIR proteins is not known. Although it is not clear why the fir locus has undergone such a dramatic sequence variation, this property may point to an unknown link between FIR-IcmQ complexes and the function of the Dot/Icm system (Feldman et al., 2005).
Experimental Procedures
Protein purification, crosslinking and CD studies
c-DNAs encoding IcmR, IcmQ, Qn(1-66) and Qc(72-191) were cloned into the pQE70 expression vector to create proteins with a C-terminal 6xHis-tag. For co-expression of IcmQ and IcmR, the IcmQ clone was moved to the pET-24a vector, which allowed for selection with kanamycin, and the C-terminal 6xHis-tag was removed in this clone. The expression vectors were transformed separately into XL-1 blue cells or into the BL21 (DE3) cell line for IcmQ (and mutant IcmQs), Qn, Qc(72-191) and IcmR. Cells were grown in 100 ug/ml ampicillin and expression was induced at 37°C with 0.5-1 mM IPTG for ∼3 hr. Frozen cell pellets were thawed in 50 mM Tris-HCl or Hepes (pH 8.0), 100 mM NaCl, 10 mM MgCl2 in the presence of a protease cocktail (1 mM EDTA, 1 mM PMSF, 1 mM benzamidine, 100 uM chymostatin and 10 uM E64). In addition, 1 uM leupeptin and 125 uM aprotinin were added in some cases. To induce lysis, resuspended cells were treated with lysozyme (0.3 mg/ml of lysate), deoxycholic acid (1.3 mg/ml) and DNase I (30 ug/ml).
To make IcmQ, the high speed lysate (100,000 × g) was bound to Ni-NTA resin and washed extensively with 20 mM Hepes (pH 7.2), 1.0 M NaCl and 20 mM imidazole. The protein was eluted in 20 mM Hepes (pH 7.2), 1.0 M NaCl and 350 mM imidazole and dialyzed extensively to remove the imidazole and excess NaCl against 20 mM Hepes (pH 7.2), 50 mM NaCl (HN buffer) at 4°C overnight. Next, IcmQ was run over a Hi-Trap Q column to remove impurities. IcmQ flows through this column and was then applied to a Hi-Trap S column, where it binds and elutes in two fractions. The first fraction was eluted in 400 mM NaCl and was comprised mostly of IcmQ aggregates, while the second fraction was eluted in 900 mM NaCl and contained IcmQ dimers. The second fraction was dialyzed extensively against HN buffer at 4 °C overnight and stored at -80°C after flash freezing.
Qn57 was purified in a manner similar to IcmQ with the addition of a trypsinization step. After elution from the Hi-Trap S column (900 mM NaCl), 6xHis-Qn66 (which was previously described as Qn (1-72) due to a sequencing error) was treated with trypsin (2000:1 wt/wt) for 2 hr at room temperature. Trypsin was removed with benzamidine beads and the resulting Qn57 dimer was dialyzed overnight against HN buffer at 4 °C. For studies with Qn, gels were fixed with 12% TCA for 30 min before staining with Coomassie blue. The IcmR dimer was purified in the same manner as IcmQ. However, the IcmR dimer did not bind to either the Hi-Trap Q or the Hi-Trap S columns. The flow-through from both columns was collected and dialyzed against HN buffer at 4°C overnight.
The Qcl domain (aa49/50-191) was made by surmounting the Qc folding problem in two steps. First, we engineered two mutations in IcmQ to destabilize the Qn domain (L17D, L39D). This led to selective degradation of Qn by an endogenous bacterial protease. In a second step, two additional point mutations were made in the linker (K57Q, K59Q). These mutations allowed proteolysis to occur at an upstream lysine-arginine pair (aa49/50) and created a larger Qc which retained the linker. This strategy allowed us to make a well-folded, α-helical Qcl domain when over-expressing the IcmQ quadruple mutant in E. coli. The purification of Qcl was similar to IcmQ, with the exception of the final Hi-Trap S column. At this step, Qcl binds the column and elutes in a single fraction in 900 mM NaCl and is not aggregated. The Qcl protein was flash frozen and stored at -80° C.
The co-expressed IcmR-IcmQ complex was made by transforming BL21 (DE3) cells with the pQE-70 vector encoding IcmR with a 6xHis-tag and the pET-24a vector with IcmQ. The cells were grown in the presence of 100 ug/ml ampicillin and 25 ug/ml kanamycin. Protein purification was carried out in a manner similar to IcmQ with the exception of the Hi-Trap S column. In this case, the IcmQ-IcmR complex binds to the column and elutes as a single fraction in 900 mM NaCl. The resulting complex was dialyzed against HN buffer at 4 °C overnight and flash frozen.
For crosslinking studies, proteins were diluted into HN buffer to ∼50 uM. DTSSP was used to crosslink IcmQ, IcmR-IcmQ, Qn-(1-66)-6xHis, Rm-Qn and IcmR. Freshly prepared crosslinker was added to the proteins at a final concentration of ∼250 uM (1:5 mol/mol ratio). The reaction was allowed to proceed at room temperature for 30 min and was stopped using 1 M Tris HCl, pH 7.5, which was added to a final concentration of 50 mM. The proteins were subjected to SDS-PAGE on 15% gels. For CD studies, AVIV 215 and 62DS spectrometers were used to record far-UV spectra from samples at 0.1 mg/ml in ∼5 mM phosphate buffer, pH 7.5 or in phosphate buffer with 100 mM NaCl in a 0.5 mm cell. The CD signal was expressed as molar residue ellipticity.
Membrane association and permeability assays
L-6-phosphatidylcholine (egg PC) was purchased in small ampules (Avanti Polar Lipids, Inc.) and dissolved in chloroform, while 1,2-dimyristoyl–sn-glycero-3-[phospho-rac-(1-glycerol)] (PG) was purchased as a dry powder. The PG was dissolved in chloroform to a concentration of 6 mg/ml. The PC and PG lipids were combined in the ratio of 3:1 (wt/wt), mixed thoroughly, and dried down under nitrogen in 1 mg aliquots. The aliquots were placed under a vacuum overnight to remove trace organics and then stored at −20°C. Vesicles for insertion experiments were made as follows. Lipid aliquots were re-suspended in HN buffer (total volume = 250 ul). Vesicles were made by extrusion through a 0.1 uM porous membrane (Duménil et al., 2004). Proteins were added to the preformed vesicles (3-4 mg/ml) at protein concentrations of ∼1 and 25 uM in HN buffer. For the membrane association assay, a sucrose step gradient was used to separate vesicles from free protein by flotation (Duménil et al., 2004). In this assay, the protein and vesicle solution was mixed with 60% sucrose (prepared in HN buffer) to a final concentration of 45%. A 30% layer and a 5% layer of sucrose in HN buffer were sequentially layered on top of the 45% layer. Samples were spun for 15 hr at 55000 rpm in an RP-55S swinging bucket rotor at 17°C and equivalent volumes were analyzed by SDS-PAGE with either Coomassie or silver staining, to ascertain the efficiency of protein association with the vesicles.
For the membrane permeation assay, 2 mg of lipids were re-suspended in 20 mM Hepes (pH 7.2), 50 mM NaCl and 10 mM EDTA (HNE buffer, total volume = 500 ul) with the addition of 80 mM calcein. The vesicles were made by extrusion and excess calcein was removed by filtration on a 5 ml G-75 column equilibrated in HNE buffer. To ensure complete removal of free calcein, the vesicles were dialyzed against HNE buffer at 4°C overnight (in the dark). The vesicles were then diluted to a concentration of ∼1.2 ug lipid/ml and placed in a cuvette in an ISA SPEX FluoroMax-2 fluorimeter and subjected to constant stirring. Proteins were mixed with vesicles to a concentration of 0.1 uM and the fluorescence emission was monitored at 515 nm as a function of time after excitation at 490 nm.
Crystallographic and mutation studies
Preparation of the Rm-Qn complex, crystallization and structure determination are described in the Supplement, and includes S_Table 2, which summarizes the crystallographic data collection and refinements. A description of the methods used to screen for mutants in Qn and in vivo tests for protein stability, DotA secretion and intra-cellular growth in mouse macrophages are also given in the Supplement. Figures were made with Chimera (Goddard et al., 2005) and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank Xinchao Yu for his help with Chimera, I.V. Akey for technical discussions and K. Hartman and Dr. T. Laue for analytical ultracentrifugation experiments. We also thank the staff of beamlines X8C and X12C at the NSLS for help during data collection. Structure files have been deposited in the PDB under accession numbers (xxxx, yyyy). R.I. is an Investigator of the Howard Hughes Medical Institute. J.D.F. and T.P.M. were supported by NIH T32 (training) grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman E, Segal G. The response regulator CpxR directly regulates the expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;190:1985–1896. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Buscher BA, Conover GM, Miller JL, Vogel SA, Meyers SN, Isberg RR, Vogel JP. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J Bacteriol. 2005;187:2927–2938. doi: 10.1128/JB.187.9.2927-2938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–61. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Monahan C, Roy CR. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nature Cell Biology. 1999;1:451–453. doi: 10.1038/15687. [DOI] [PubMed] [Google Scholar]
- Coers J, Kagan JC, Matthews M, Nagai H, Zuckman DM, Roy CR. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebbeling BN, Wenzel RP. The epidemiology of Legionella pneumophila infections. Semin Respir Infect. 1987;2:206–21. [PubMed] [Google Scholar]
- Duménil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- Duménil G, Montminy TP, Tang M, Isberg RR. IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. J Biol Chem. 2004;279:4686–4695. doi: 10.1074/jbc.M309908200. [DOI] [PubMed] [Google Scholar]
- Feldman M, Zusman T, Hagag S, Segal G. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc Natl Acad Sci U S A. 2005;102:12206–12211. doi: 10.1073/pnas.0501850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Segal G. A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect Immun. 2004;72:4503–4511. doi: 10.1128/IAI.72.8.4503-4511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS. The molecular ecology of Legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure (Camb) 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Horwitz MA, Silverstein SC. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J Clin Invest. 1980;66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MA. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome lysosome fusion in human monocytes. J Exp Med. 1983a;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MA. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983b;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MA, Maxfield FR. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol. 1984;99:1936–1943. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SM, Hashemi S. Electron microscopic examination of the inflammatory response to Legionella pneumophila in guinea pigs. Lab Invest. 1982;46:24–32. [PubMed] [Google Scholar]
- Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- Kubori T, Hyakutake A, Nagai H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol. 2008;67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- Leclerque A, Kleespies RG. Type IV secretion system components as phylogenetic markers of entomopathogenic bacteria of the genus Rickettsiella. FEMS Microbiol Lett. 2008;279:167–173. doi: 10.1111/j.1574-6968.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Cholon DM, Cambronne ED, Roy CR. The Legionella IcmS-IcmW protein complex is important for Dot/Icm mediated protein translocation. Mol Microbiol. 2005;55:912–926. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- Pohlman RF, Genetti HD, Winans SC. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- Roy CR, Tilney LG. The road less traveled: transport of Legionella to the endoplasmic reticulum. J Cell Biol. 2002;158:415–419. doi: 10.1083/jcb.200205011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC, Dehio C. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet. 2007;39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- Samuels AL, Lanka E, Davies E. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol. 2000;182:2709–2715. doi: 10.1128/jb.182.10.2709-2715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114:4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, Vogel JP. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2006;62:1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2003;49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


